Abstract
A novel H10N8 influenza A virus has been detected in three humans in China since December 2013. Although this virus was hypothesized to be a novel reassortant among influenza viruses from wild birds and domestic poultry, its evolutionary path leading to human infection is unknown. Sporadic surveillance at the live poultry market (LPM) suspected to be the source of infection for the first H10N8 patient has shown a gradual increase in influenza virus prevalence culminating with a predominance of H10N8 viruses. Influenza viruses detected in the LPM up to 8 months prior to human infection contributed genetic components to the zoonotic virus. These H10N8 viruses have continued to evolve within this LPM subsequent to the human infection, and continuous assessments of these H10N8 viruses will be necessary. Serological surveillance showed that the virus appears to have been present throughout the LPM system in Nanchang, China. Reduction of the influenza virus burden in LPMs is essential in preventing future emergence of novel influenza viruses with zoonotic and pandemic potential.
Keywords: H10N8, avian influenza virus, human infection, live poultry market
In December 2013, a novel H10N8 influenza virus emerged, causing three human infections in Nanchang, China (Chen et al., 2014). Albeit it was apparent that the virus was a reassortant with the endemic H9N2 viruses circulating in poultry in the region, the source of the infection remained undetermined (Garcia-Sastre and Schmolke, 2014). The first index female patient had visited a live poultry market (LPM) four days before illness onset. LPMs have been well recognized as high-risk areas for zoonotic transmission of avian influenza viruses (Mounts et al., 1999; Wan et al., 2011; Zhou et al., 2009).
The particular LPM that the first index case visited was in the Donghu district of Nanchang City, a market where we have been conducting sporadic virologic surveillance since April 2013. The number of poultry retailers within this LPM ranged from 20 to 30, and a total of around 1,500 birds were sold per day; and the number of poultry retailers and birds being sold varied with season change and other factors. Typically about 90% of birds sold in this market were chickens and ducks with wild birds occasionally being sold as well. Here, we report that the epidemiological surveillance at this particular LPM is suspected to be the source of infection for the first H10N8 case as well as the presence of H10 subtype virus among four other LPMs in this region.
Between April 5, 2013, and January 1, 2014, we collected 361 samples from the market on ten separate sampling occasions (Supplementary Table S1). These samples included 192 paired oropharyngeal and cloacal swabs from chickens, pheasant, guinea fowl, pigeon, and turtle doves, as well as 122 cloacal swabs from ducks and geese. In addition, 47 environmental samples were also collected by swabbing feces in the cages and from the floor of this LPM, especially when the samples from live birds were inaccessible.
The proportion of influenza A virus positive samples among these samples as measured by M-gene real time PCR(WHO, 2011), increased from 4.3% on April 5, to 38.5% on August 24, and to 87.0% on December 12, 2013. The proportion of influenza A virus positive samples, albeit still high, dropped to 48.0% on January 1, 2014, sampling. Among these M-gene positive specimens, ten were subtyped as H7 by H7 specific real time PCR (Spackman et al., 2002) and sixteen as H10 by H10 specific hemagglutination inhibition assays (Supplementary Table S1).
To determine if the H10N8 viruses, had been and had continued to circulate in this market, we sequenced positive samples collected on the April, December, and January sampling occasions (the first index case was admitted to the hospital in late November). From the April sampling we sequenced all five positive samples and detected H7 and H9 hemagglutinin (HA) genes in conjunction with N2 and N9 neuraminidase (NA) genes. Although no H10 or N8 genes were detected, we found PB2, PA, NP, MP, and NS genes were genetically similar to the human H10N8 virus. From the December sampling, we successfully generated sequences from 44 of the 86 positive samples. Twelve samples contained H10 genes, all of which also contained N8 genes, though some did appear to be mixed infections containing gene segments from other viruses such as H9N2. From the January sampling results, six samples had both H10 and N8 genes (Supplementary Table S2).
The presence of mixed infections makes it difficult to determine the exact genotypes of the viruses circulating. For example, there were sixteen samples that contained H10 genes, twelve that contained H9 genes, two that contained H6 genes, and three that contained two genes (two H10/H9 and one H7/H9). Among the sixteen samples that contained H10 viruses, only two were of the N8 subtype; the remaining fourteen samples were mixed infections with both N2 and N8 NA genes present. The two mixed H10/H9 samples had both N2 and N8 NA genes as well. Besides the mixed HA and NA samples, 12 had at least two copies of the same gene segments, such as PB2, PB1, NP, MP, and NS, and these segments were genetically distinct. Of the 57 samples sequenced, 25 samples were determined to have at least two genetically distinct influenza viruses.
Phylogenetic analyses of the generated sequences showed that eighteen H10 and nineteen N8 genes were present with these genes forming a monophyletic clade with the corresponding genes of the human H10N8 virus (Figure 1). The H10 and N8 genes were genetically close to those of viruses isolated from migratory birds: H10 from the Eurasian lineage and N8 from the North American lineage. In contrast, the internal genes sequenced were closer to those genes derived from avian influenza viruses circulating in domestic poultry with some also being similar to those present in the human H10N8 virus (Figure 2). These data confirm that nucleotide sequences of the viruses more than 99% identical to those of the human H10N8 virus were circulating in this LPM, supporting the likelihood that the zoonotic transmission had occurred here.
Figure 1. Phylogenetic analysis of H10 (A) and N8 (B) genes recovered from the samples collected from the LPM which the index patient visited.
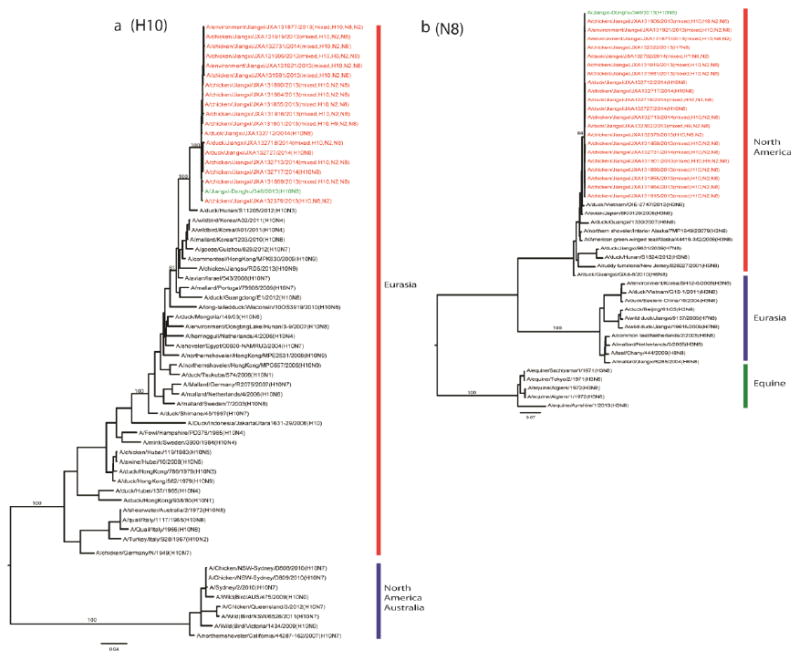
The phylogenetic analyses were performed using maximum likelihood by GARLI (Gutell and Jansen, 2006) and bootstrap resampling analyses using PAUP* 4.0 Beta (Wilgenbusch and Swofford, 2003) to apply a neighborhood joining method, as described earlier (Wan et al., 2008). The genes from this surveillance are marked in red, and those from the human H10N8 isolate in green. The bootstrap values for representative lineages are marked.
Figure 2. Phylogenetic analysis of PB2 (A), PB1 (B), PA (C), NP (D), MP (E), and NS (F) genes recovered from the samples collected from the LPM which the index patient visited.
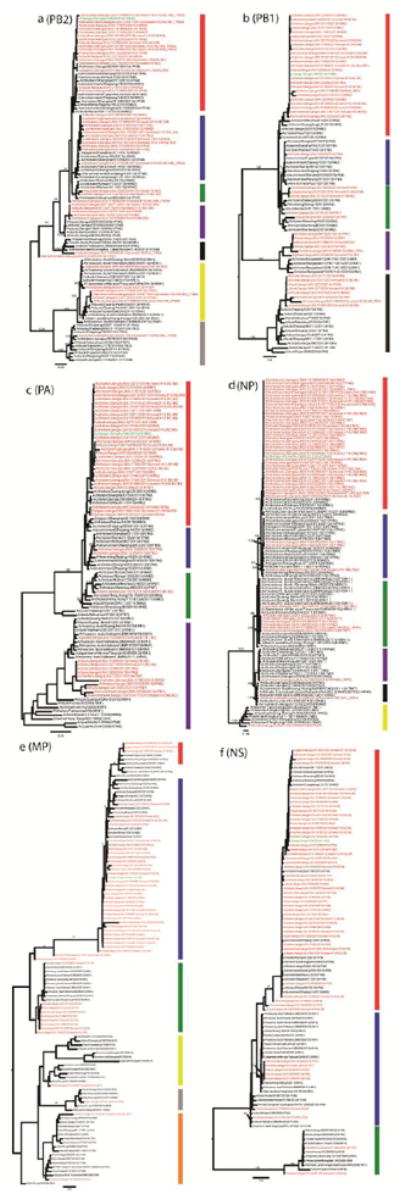
The details of these genes are shown in Supplementary Table S2. The genes from this surveillance are marked in red, and those from the human H10N8 isolate in green. The bootstrap values for representative lineages are marked.
The diversity of internal genes detected revealed that multiple distinct viruses were circulating in this individual LPM. The PB2 genes were clustered into seven distinct genetic lineages, PB1 into five, PA into four, NP into six, MP into six, and NS into three (Figure 2). Among these genes, PB2 from these eighteen samples alone belong to five distinct genetic lineages. In contrast, NP, MP, and NS of these samples with H10 genes were similar. None of the internal genes of these H10N8 samples were unique from those found in H9 and other subtypes of influenza A viruses from this LPM. The genetic lineages from the samples collected on April 5, 2013, were surprisingly active in these emerging H10 viruses in December 2013.
The residues at the receptor binding sites of the H10 proteins encoded by the identified genes from this market were identical to those in the human H10N8 isolate, and the majority of these sites except 128 (137, H3 position) and 181 (193, H3 position) were shown to be divergent from those H10 avian viruses from public databases: the emerging H10 viruses had R128 (100%) and I181(61%) while those in public databases were predominantly K (67.8%) and T(97.7%), respectively (Supplementary Tables S3 and S4). The human H10N8 isolate has R128 and T181. The impact of I181T on host or tissue tropism of the H10N8 virus is unknown, nevertheless, the receptor binding properties are predicted to be avian-like (Chen et al., 2014).
Analysis of NA gene sequences (Supplementary Table S4) suggests that these viruses are sensitive to oseltamivir and other neuraminidase inhibitors (Hurt et al., 2009). However, 94.1% of the M2 sequences have S31N(Hay et al., 1985) (Supplementary Table S4) indicating that they are resistant to amantadine, as is the human H10N8 isolate (Wan et al., 2013). All PB2 genes had E627 whereas the PB2 gene of the human H10N8 isolate had K627, a marker of mammalian adaptation(Shinya et al., 2004) (Supplementary Table S4).
That we were able to detect the H10N8 virus in December and early January following human infection raised the question of how widely this virus might be spread within LPM in the region. To estimate this, we conducted serologic surveillance within four additional LPMs in Nanchang city. We collected a total of 800 sera from chickens and ducks from five LPMs, from February 25, 2014, to March 27, 2014 (Figure 3). These five LPMs cover a geographic area of about 160 square kilometers and the major metropolitan area of Nanchang. Using hemagglutination inhibition assays with a H10N8 virus isolated from the surveillance described above and a cutoff of 1:20, we found that 9.4% of the 800 sera were positive. The H10 positive sera were distributed across all five sampled LPMs and from both ducks and chickens. The highest H10 positive percentile on a single market and a single sampling period (34.0%) was detected on March 9, at LPM A, which the index patient visited. Of note, LPM B located across the Gan River from LPM A, had close to 50% H10 positivity by the end of March. These data showed that H10 viruses were widespread in the region’s LPMs. Although we are unable to determine from these serologic studies the exact nature of the H10 virus, two further human cases with the H10N8 virus in Nanchang (in January and February 2014) are consistent with it being the zoonotic virus. Of these two additional human cases, the second human case was documented with a visit to a local LPM prior to illness onset whereas the exposure history of the third human case was unclear.
Figure 3. Distribution of H10 seropositive samples collected from February 25 to March 27, 2014 and across five LPMs in Nanchang city.
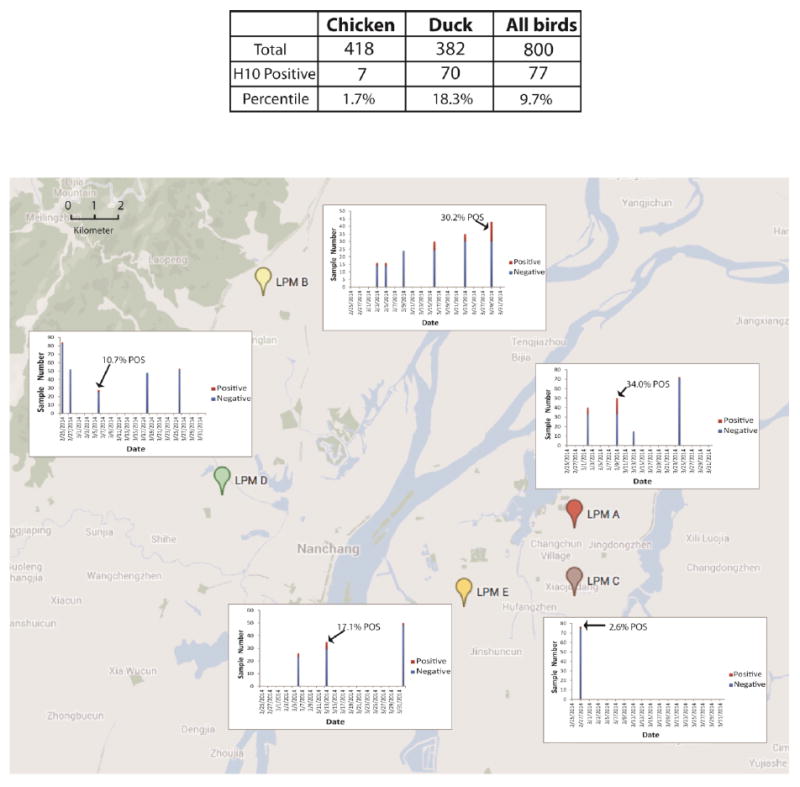
The seropositive samples were determined by hemagglutination inhibition assays with a H10N8 virus isolated from the surveillance described above and a cutoff of 1:20. LPM A was the one that the first index H10N8 patient visited in November 2013. The highest H10 positive percentile in a single sampling period was marked for each LPM.
In summary, we show here that the LPM visited by the index H10N8 female patient was very likely the source of her infection and that the virus appears to have been present throughout the LPM system in Nanchang, China. However, the findings in this study were limited by the small number of samples we collected and by the fact that the H10N8 virus prior to the emergence of the first human case was lack. It is still unclear whether these H10N8 viruses were generated in the LPM or prior to being introduced into the LPM system. Continuous influenza surveillance is needed to monitor the epidemiology of this novel H10N8 virus in Nanchang as well as those areas that share poultry movements with Nanchang. In addition to those minor poultry species, there are a variety of chicken and duck species in the LPM; further studies will be needed to identify the reservoir for the H10N8 virus among these bird species, and such information will be useful for developing effective strategies for prevention and control of the H10N8 virus at the LPM.
Avian influenza viruses were identified in LPMs in China many decades ago. The first H9N2 low pathogenic avian influenza virus was initially isolated from domestic poultry in 1994 (Guo et al., 2003), and has since been found to be endemic in domestic poultry in China (Li et al., 2005). Besides infecting poultry, this H9N2 virus has caused sporadic infections in humans (Cheung et al., 2007; Yoon et al., 2005). The H9N2 virus has undergone rapid evolution, and contemporary H9N2 viruses are both genetically and antigenically diverse (Choi et al., 2004; Chu et al., 2011; Cong et al., 2007; Liu et al., 2003). The internal genes of H9N2 viruses contributed largely to the genomic diversity of H5N1 highly pathogenic avian influenza viruses in China (Guan et al., 2000; Lin et al., 2000). The recent emergence of H7N9 low pathogenic avian influenza viruses in Yangze Delta has six genes derived from H9N2 viruses, and mixed infections with H7N9 and H9N2 were very common (Gao et al., 2013; Liu et al., 2013).
This study suggested further that novel H10N8 influenza A virus were frequently co-infected with H9N2 viruses, and their internal gene constellations were similar to each other. It seems that this diverse genetic pool is potentially more dangerous than any other single virus. With any chance to interact with other subtypes of HA and NA genes, novel reassortants could emerge, including some antigenically distinct from those contemporary human influenza viruses. These emerging viruses, including H5N1, H7N9, H9N2, and H10N8, have posed and will continue to pose potential threats to public health.
A large influenza vaccine campaign against highly pathogenic H5N1 viruses has been conducted in China since 2004. H9N2 vaccines have also been distributed to domestic poultry farms but their implementation has been comparatively less effective. With the substantial subtype and genetic diversity of viruses within LPMs, vaccination is not a realistic strategy to reduce the levels of virus circulation and subsequent zoonotic infections. Other intervention strategies must, therefore, be used to control viral flow into the LPMs. As it seems impossible to close all LPMs simultaneously, more practical policies and approaches such as routine surveillance and regular market disinfection should be urgently implemented.
Supplementary Material
Highlights.
Our surveillance showed that the live poultry markets were likely the infection source of H10N8 influenza virus;
H10N8 virus appears to present throughout the live poultry market system in Nanchang, China.
Acknowledgments
This study was partially supported by P20GM103646 from the U.S. National Institutes of Health. RW and SSW are supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, USA (contract no. HH-SN266200700005C).
We are thankful to Scott Olsen and Dana Roeber from the Genome Sequencing Facility at St. Jude’s Children’s Research Hospital for their technical supports and to Karen Templeton and Dena Pruett for their proofreading this manuscript. We would like to thank Dr. Lusheng Huang for his assistance in influenza surveillance implementation and Dr. Jun Ren from Jiangxi Agricultural University for his technical assistance in quantitative PCR and genomic sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. The Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Cheung CL, Vijaykrishna D, Smith GJ, Fan XH, Zhang JX, Bahl J, Duan L, Huang K, Tai H, Wang J, Poon LL, Peiris JS, Chen H, Guan Y. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol. 2007;81:10402–10412. doi: 10.1128/JVI.01157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YK, Ozaki H, Webby RJ, Webster RG, Peiris JS, Poon L, Butt C, Leung YH, Guan Y. Continuing evolution of H9N2 influenza viruses in Southeastern China. J Virol. 2004;78:8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YC, Cheung CL, Hung Leung CY, Man Poon LL, Chen H, Peiris JS, Guan Y. Continuing evolution of H9N2 influenza viruses endemic in poultry in southern China. Influenza Other Respi Viruses. 2011;5(Suppl 1):68–71. [PubMed] [Google Scholar]
- Cong YL, Pu J, Liu QF, Wang S, Zhang GZ, Zhang XL, Fan WX, Brown EG, Liu JH. Antigenic and genetic characterization of H9N2 swine influenza viruses in China. J Gen Virol. 2007;88:2035–2041. doi: 10.1099/vir.0.82783-0. [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A, Schmolke M. Avian influenza A H10N8--a virus on the verge? Lancet. 2014;383:676–677. doi: 10.1016/S0140-6736(14)60163-X. [DOI] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Chin PS, Dyrting KC, Ellis TM, Webster RG, Peiris M. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J Virol. 2000;74:9372–9380. doi: 10.1128/jvi.74.20.9372-9380.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Liao M, Xin C. Sequence of HA gene of avian influenza A/Chicken/Guangdong/SS/1994 (H9N2) virus. Avian Dis. 2003;47:1118–1121. doi: 10.1637/0005-2086-47.s3.1118. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Jansen RK. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion 2006 [Google Scholar]
- Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt AC, Holien JK, Parker MW, Barr IG. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs. 2009;69:2523–2531. doi: 10.2165/11531450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N, Hay A. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, Bi Y, Wu Y, Li X, Yan J. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. The Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- Liu J, Okazaki K, Ozaki H, Sakoda Y, Wu Q, Chen F, Kida H. H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathol. 2003;32:551–560. doi: 10.1080/0307-9450310001596728. [DOI] [PubMed] [Google Scholar]
- Mounts AW, Kwong H, Izurieta HS, Ho Y, Au T, Lee M, Buxton Bridges C, Williams SW, Mak KH, Katz JM, Thompson WW, Cox NJ, Fukuda K. Case-control study of risk factors for avian influenza A (H5N1) disease, Hong Kong, 1997. J Infect Dis. 1999;180:505–508. doi: 10.1086/314903. [DOI] [PubMed] [Google Scholar]
- Shinya K, Hamm S, Hatta M, Ito H, Ito T, Kawaoka Y. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–266. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT, Suarez DL. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X-F, Nguyen T, Davis CT, Smith CB, Zhao Z-M, Carrel M, Inui K, Do HT, Mai DT, Jadhao S. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One. 2008;3:e3462. doi: 10.1371/journal.pone.0003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan XF, Carrel M, Long LP, Alker AP, Emch M. Perspective on emergence and re-emergence of amantadine resistant influenza A viruses in domestic animals in China. Infect Genet Evol. 2013;20C:298–303. doi: 10.1016/j.meegid.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Wan XF, Dong L, Lan Y, Long LP, Xu C, Zou S, Li Z, Wen L, Cai Z, Wang W, Li X, Yuan F, Sui H, Zhang Y, Dong J, Sun S, Gao Y, Wang M, Bai T, Yang L, Li D, Yang W, Yu H, Wang S, Feng Z, Wang Y, Guo Y, Webby RJ, Shu Y. Indications that live poultry markets are a major source of human H5N1 influenza virus infection in China. J Virol. 2011;85:13432–13438. doi: 10.1128/JVI.05266-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. WHO information for molecular diagnosis of influenza virus in humans 2011 [Google Scholar]
- Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Current protocols in bioinformatics. 2003:6.4. 1–6.4. 28. doi: 10.1002/0471250953.bi0604s00. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Cooper VL, Schwartz KJ, Harmon KM, Kim WI, Janke BH, Strohbehn J, Butts D, Troutman J. Influenza virus infection in racing greyhounds. Emerg Infect Dis. 2005;11:1974–1976. doi: 10.3201/eid1112.050810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liao Q, Dong L, Huai Y, Bai T, Xiang N, Shu Y, Liu W, Wang S, Qin P, Wang M, Xing X, Lv J, Chen RY, Feng Z, Yang W, Uyeki TM, Yu H. Risk factors for human illness with avian influenza A (H5N1) virus infection in China. J Infect Dis. 2009;199:1726–1734. doi: 10.1086/599206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


