Supplemental Digital Content is available in the text.
Keywords: animal models, cardiomyopathy, Drosophila, genetics, proteomics
Abstract
Background—
The Drosophila heart is an important model for studying the genetics underpinning mammalian cardiac function. The system comprises contractile cardiomyocytes, adjacent to which are pairs of highly endocytic pericardial nephrocytes that modulate cardiac function by uncharacterized mechanisms. Identifying these mechanisms and the molecules involved is important because they may be relevant to human cardiac physiology.
Methods and Results—
This work aimed to identify circulating cardiomodulatory factors of potential relevance to humans using the Drosophila nephrocyte–cardiomyocyte system. A Kruppel-like factor 15 (dKlf15) loss-of-function strategy was used to ablate nephrocytes and then heart function and the hemolymph proteome were analyzed. Ablation of nephrocytes led to a severe cardiomyopathy characterized by a lengthening of diastolic interval. Rendering adult nephrocytes dysfunctional by disrupting their endocytic function or temporally conditional knockdown of dKlf15 led to a similar cardiomyopathy. Proteomics revealed that nephrocytes regulate the circulating levels of many secreted proteins, the most notable of which was the evolutionarily conserved matricellular protein Secreted Protein Acidic and Rich in Cysteine (SPARC), a protein involved in mammalian cardiac function. Finally, reducing SPARC gene dosage ameliorated the cardiomyopathy that developed in the absence of nephrocytes.
Conclusions—
The data implicate SPARC in the noncell autonomous control of cardiac function in Drosophila and suggest that modulation of SPARC gene expression may ameliorate cardiac dysfunction in humans.
Heart disease is a major cause of mortality worldwide, so identifying mechanisms that regulate cardiac physiology is a crucial step toward its treatment and prevention. Both vertebrate and invertebrate models contribute to our understanding of cardiovascular physiology and the related disease processes affecting humans. The Drosophila heart comprises contractile cardiomyocytes and neighboring pericardial nephrocytes that clear circulating colloids, macromolecules, and immune peptides from the hemolymph (blood).1–3 This organ system has proven to be a tractable model that permits genetic screens to identify novel pathways relevant to human cardiac performance.4 In addition, functional studies have contributed to our understanding of how alterations in structural proteins, including adhesion proteins such as fermitins/kindlins5 and contractile proteins, such as myosin and troponin-T, translate to cardiomyopathy.6 In addition, Drosophila has facilitated the elucidation of genetic pathways regulating cardiac aging7,8 and diet-induced cardiac and kidney podocyte dysfunction.9–11
Editorial see p 104
Clinical Perspective on p 129
There is evidence that cardiac phenotypes develop in the Drosophila model as a result of nephrocyte dysfunction; however, the mechanisms are not well characterized.12–15 It has also been reported that these cardiac phenotypes may depend on developmental changes to the nephrocyte–cardiomyocyte niche rather than a contribution by nephrocytes to cardiac homeostasis in adulthood.16 Characterizing the noncell autonomous regulation of heart function in flies is important because it may provide insights into molecules that regulate human cardiac physiology.
The Drosophila ortholog of the mammalian transcription factor Klf15 (also known as Kidney Kruppel-like factor) has recently been identified as a nephrocyte-restricted gene critical for the cells’ differentiation and function.17 Pericardial nephrocytes in flies homozygous for a dKlf15 loss of function allele develop normally during embryonic cardiogenesis but then fail to differentiate during larval development and undergo attrition before pupation, hence adult flies have no pericardial nephrocytes. This enables the nephrocytes’ impact on the circulating proteome in adults to be analyzed and for nephrocyte-dependent cardiomyocyte control mechanisms to be identified.
In this work, we took advantage of the nephrocyte–cardiomyocyte system in Drosophila to identify circulating cardiomodulatory factors of potential relevance to humans. Proteomics was used to establish the composition of the circulating proteome in flies with and without nephrocytes. It was found that either the loss of nephrocytes or their function during development led to cardiomyopathy, and contrary to previous reports, loss of nephrocyte function in adulthood also led to cardiomyopathy. Analysis of the hemolymph proteome established that nephrocytes had a broad impact on the circulating secretome. By coupling the proteomics data with genetic experiments, we showed that nephrocytes regulated circulating levels of the matricellular protein secreted protein acidic and rich in cysteine (SPARC) and prevent SPARC-dependent cardiomyopathy. SPARC plays multiple roles in mammals. SPARC levels increase in metabolic syndrome and aging, and it is well documented as contributing to pathological tissue fibrosis; however, reduced SPARC expression can lead to heart rupture in pressure overload models.18 The current findings suggest that SPARC’s role in the mammalian heart may be evolutionarily conserved and that its modulation may ameliorate cardiac dysfunction. The work also highlights the importance of Drosophila for the identification and study of cardiomodulatory signals of relevance to human cardiac physiology.
Materials and Methods
Strains Used in This Study
The w1118 (used as wild-type strain in this study), dKlf15NN (CG2932, FBgn0025679; previously known as Bteb2f06447 was described in the study by Ivy et al17), Dorothy-Gal4 (dot-Gal4, originally described in the study by Kimbrell et al19), UAS-mCherry (TRiP control line), dSparcMI00329 (with an MiMIC insertion in the 5-prime region of the dSparc locus; as described in the study by Venken et al20), and Tub-Gal80ts lines were all from the Bloomington Stock Center (Bloomington, IL). The HandC-Gal4 (Hand-Gal4) line was described in the study by Sellin et al21). The 2 RNAi lines for knocking-down dKlf15 were from the Vienna Drosophila RNAi stock Center (with a targeting hairpin inserted into the second chromosome, VDRC) and Bloomington (Klf15JF02420, a Harvard TRiP line with a dKlf15 targeting hairpin inserted into the third chromosome). All genetic combinations were generated by standard crosses. Generation of TARGET flies (Hand-Gal4; Tub-Gal80ts) was achieved by standard crosses.22
Husbandry and Propagation of Flies
Flies were propagated routinely on a cornmeal-molasses diet at 25°C under a 12:12 hour light:dark schedule. For TARGET experiments, flies were reared at 18°C and then transferred to 29°C within 1 to 5 days of eclosing. Flies remained at 29°C for 1 to 2 weeks. After analysis of heart function, flies were transferred to 25°C for 24 hours. To reduce the effect of genetic background, the dKlf15NN mutant was backcrossed onto the w1118 for >20 generations.
Quantitative PCR
See Methods section in the Data Supplement.
Imaging the Adult Heart
See Methods section for more detailed description of the method in the Data Supplement. Unless stated otherwise, 2- to 3-week-old adult female flies were anaesthetized with Flynap (Carolina Biological Supply Company, Burlington, NC), dissected and hearts stained as described previously.5,23
Analysis of Adult Heart Function
Two- to three-week-old adult flies were anesthetized with Flynap (Carolina Biological Supply Company), and the beating adult heart in semi-intact preparations was visualized with an Ionoptix Myocam S high frame rate video camera (Ionoptix Ltd, Dublin, Ireland) attached to a Zeiss AxioLab A1 with a water-dipping 10× objective. Approximately 15 s of video footage was collected using Micro-Manager open source microscopy software,24 with a frame rate capture of between 120 and 150 frames per second. Videos were converted to audio video interleave files using ImageJ and analyzed using semiautomated optical heart analysis software,25 www.sohasoftware.com; as previously described.5 Quantified data are presented as the mean (±SEM) of at least 20 different flies per genotype.
Epifluorescence Microscopy of Adult Fly Tissues
See Methods section in the Data Supplement.
Collection of Hemolymph From Adult Flies
One-week-old adult female flies were rendered immobile at 4°C for 5 minutes. To remove contaminating food and feces flies were placed into a 1.5-mL centrifuge tube and 500-μL of 50% ice cold ethanol in water and the tube upturned several times. This step was repeated a further 2×, first with 50% ethanol and then with 50-mmol/L ammonium bicarbonate. Flies were then tipped onto an upturned 30-mm petri dish containing ice. The dorsal cuticle of the thorax of at least 100 flies was pricked with a 25G needle and then flies were collected into a centrifuge tube containing a 0.2-μm filter insert and 1 mL of 50-mmol/L ammonium bicarbonate. Flies were centrifuged at 4°C for 10 s at 2000 rpm (g value, 448g). The filtrate was removed and replaced into the upper filter cassette and centrifuged again. This step was repeated once more. The protein concentration of the filtrate was quantified by the Bradford assay using 50-mmol/L ammonium bicarbonate as a blank. Samples from flies that had not been pricked contained no protein. Samples contained 100 to 150 μg of total protein and were frozen at −20°C; volumes were adjusted with 50-mmol/L ammonium bicarbonate to normalize the total protein content between the samples. The mean spectral count of 3 independent biological replicates from the reference (w1118) and dKlf15NN mutant genotypes was used to infer protein abundance.
Proteomic and Bioinformatics Analysis of Hemolymph Proteome
Hemolymph samples were lyophilized and resuspended in 200 μL with 50-mmol/L ammonium bicarbonate. Protein reduction was done by adding 4 μL of Tris(2-carboxyethyl)phosphine to 200 μL of samples at 60°C for 30 minutes. Iodoacetamide was added (to 20 mmol/L), and proteins were alkylated at 30 minutes at room temperature in the dark. Mass spectrometry grade trypsin (Promega) was added (1:20 ratio) for overnight digestion at 37°C on thermomixer set on 700 rpm. After digestion, formic acid was added to the peptide solution (to 2%), followed by desalting by Microtrap (Michrom-Bruker) and then on-line analysis of peptides by high-resolution, high-accuracy LC–MS/MS, consisting of a Michrom HPLC, an Agilent Zorbax C18 peptide trap, a 15-cm Michrom Magic C18 column, a low-flow ADVANCED Michrom MS source, and an LTQ-Orbitrap XL (Thermo Fisher Scientific). A 120-minute gradient of 10% to 30%B (0.1% formic acid and 100% acetonitrile) was used to separate the peptides. The total LC time was 140 minutes. The LTQ-Orbitrap XL was set to scan precursors in the Orbitrap followed by data-dependent MS/MS of the top 10 precursors. The LC–MS/MS raw data were submitted to Sorcerer Enterprise v.3.5 release (Sage-N Research Inc) with SEQUEST algorithm as the search program for peptide/protein identification. SEQUEST was set up to search the target-decoy Swiss-Prot Drosophila melanogaster fasta protein database indexed with trypsin for enzyme with the allowance of ≤2 missed cleavages, Semi Tryptic search, and precursor mass tolerance of 50 ppm. The search results were viewed, sorted, filtered, and statically analyzed by using comprehensive proteomics data analysis software, Peptide/Protein prophet v.4.6.1 (Institute for Systems Biology, Seattle). The minimum transproteomic pipeline protein probability score was set to 0.8 to 0.90, to assure low error (much less than false discovery rate 2%) with reasonably good sensitivity. The differential spectral count analysis was done by QTools, an open source in-house developed tool for automated differential peptide/protein spectral count analysis and Gene Ontology.26 SignalP 4.1 was used to identify proteins having a signal peptide sequence in their N terminus; default settings with a D cutoff of 0.45 were used.27
Statistics
When >2 genotypes or treatments were used in an experiment, 1-way ANOVA was used to test the hypothesis that genotype may have affected heart function, and post hoc test (Tukey honest significant difference) was used to establish P values between control and the different genotypes. An unpaired 2-tailed student’s t test was used to compare 2 means. GeneProf28 was used to calculate the probability that hemolymph samples may be enriched with proteins predicted to have an N-terminal signal peptide versus a background data set (the proportion of all known Drosophila genes predicted to encode for an N-terminal signal peptide).
Results
Genetic Ablation of Nephrocytes Using dKlf15 Loss of Function
It has recently been demonstrated that dKlf15 is a nephrocyte-restricted transcription factor critical for the viability and differentiation of Drosophila’s 2 nephrocyte populations, the garland cells and pericardial nephrocytes.17 In flies homozygous for a dKlf15 loss of function allele (dKlf15NN), the nephrocyte populations undergo attrition during late embryogenesis (garland cells, compare Figure 1B′ and 1B″ and the L3 stage of larval development, so that adults are completely devoid of nephrocytes (compare Figure 1A′ and 1A″).
Figure 1.
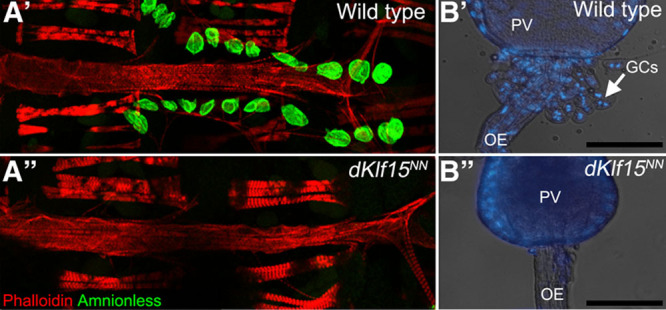
Loss of pericardial nephrocytes and garland cells in dKlf15NN mutants. A, Adult control (w1118; A′) and dKlf15NN mutant flies (A″) were dissected and the heart fixed and stained with phalloidin to visualize the heart’s actin cytoskeleton and antibodies to the pericardial nephrocyte marker Amnionless (CG11592). All pericardial nephrocytes fail to differentiate in the mutants, undergoing attrition during late larval development so that by adulthood there are none. B, Third instar larvae were dissected and the garland cells visualized after staining with Hoechst. In control flies (w1118; B′) garland cells (GCs) are binucleate and situated at the interface between the proventriculus (PV) and esophagus (OE). In contrast, the garland cells fail to develop normally and are lost in the dKlf15NN mutants (B″). Scale bar, 100 μm.
Loss of Nephrocyte dKlf15 Expression Leads to Cardiomyopathy
It is increasingly clear that Drosophila heart function is modulated by noncell autonomous mechanisms controlled by the neighboring pericardial nephrocytes.14,15 To confirm that nephrocytes modulate cardiac function in the Drosophila model, adult hearts in wild-type and dKlf15NN mutants were monitored by videomicroscopy. Homozygous dKlf15NN mutant females (and hemizygous dKlf15NN mutant males) had significantly longer heart periods (the time between the initiation of successive cardiac contractions) compared with controls, primarily because of a significant lengthening of the diastolic interval (Figure 2A and 2B for data from males see below). The mutants also had a modest increase in the arrhythmicity index (Figure 2B), a measure of the heart’s beat-to-beat variability. In addition, end-diastolic and end-systolic diameters (EDD and ESD) were greater in mutants than in controls; however, this was not associated with a change in fractional shortening (the ratio of EDD to ESD—the relative distance that the heart wall travels during a contraction). To establish if the heart phenotype was because of the specific loss of nephrocyte dKlf15 expression, dKlf15 was silenced specifically in nephrocytes using dorothy-Gal4. Knockdown of dKlf15 in nephrocytes led to a heart phenotype that was almost identical to that of the dKlf15NN mutants; however, the arrhythmicity index, although trending toward being increased, was not statistically different from that of the controls (Figure 2B).
Figure 2.
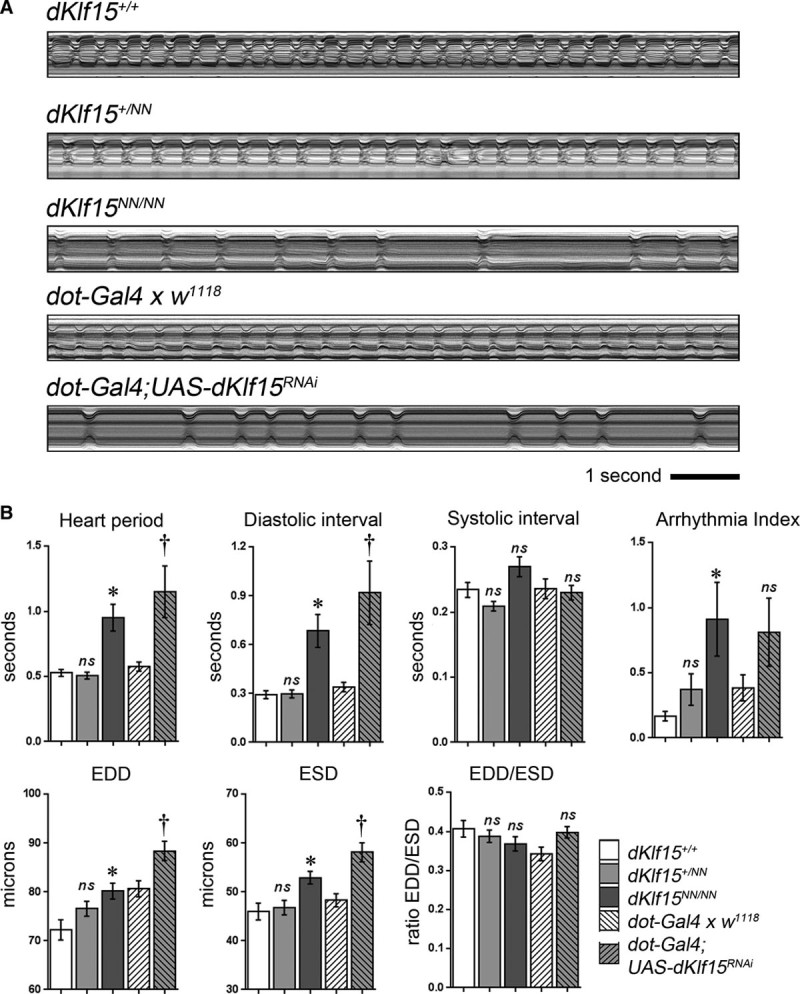
Loss of nephrocytes leads to cardiomyopathy. A, M-mode records of adult hearts. Regular contractions can be seen in wild-type (dKlf15+/+), dKlf15+/NN heterozygote, and dot-Gal4 outcrossed once to the w1118 control line (dot-Gal4 x w1118); whereas flies homozygous for the dKlf15NN allele, or in which dKlf15 has been silenced in nephrocytes (dot-Gal4; UAS-dKlf15RNAi), there is an abnormally long diastolic interval and periods of arrhythmia. B, Adult heart function. EDD indicates end-diastolic diameter; ESD, end-systolic diameter; and ns, not statistically different from dKlf15+/+ or dot > w1118 control; *P<0.01 compared with dKlf15+/+; †P<0.01 compared with dot >w1118); n=41 to 45 flies per genotype.
Nephrocytes Mediate Normal Cardiac Function in Adults
There is a doubt as to whether normal heart function in adult flies is dependent or not on sustained interactions between cardiomyocytes and nephrocytes. Sustained dKlf15 expression is required for adult nephrocyte function,17 so to establish if nephrocytes were required for normal cardiac function in adult flies, the TARGET system22 was used to silence dKlf15 in the nephrocytes of adult flies (Figure 3). Using this system, it was possible to allow functional nephrocytes to develop normally and then silence dKlf15 in adults to cause nephrocyte dysfunction. Accordingly, nephrocytes dedifferentiated (showed reduced Amnionless protein expression) and lost their ability to accumulate dextran. In association with this, the flies developed a cardiomyopathy, which recapitulated that seen in the dKlf15NN mutants as well as dorothy-Gal4–driven dKlf15 silencing experiments (Figure 3B).
Figure 3.
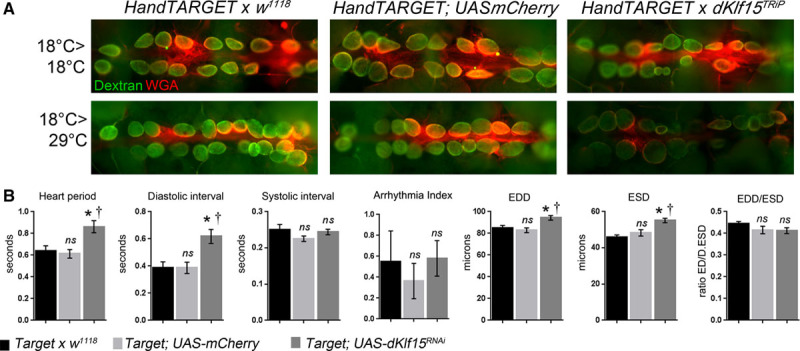
Conditional loss of nephrocyte function in adults leads to cardiomyopathy. dKlf15 was conditionally silenced in the adult fly heart using the temperature-sensitive TARGET system driven by Hand-Gal4. Gene silencing is prevented at 18°C but permitted at higher temperatures (29°C). Flies were reared at 18°C until they eclosed and then maintained at this temperature to prevent gene silencing or moved to the higher temperature to allow dKlf15 silencing. The Hand-TARGET parent line crossed to w1118 line or UAS-mCherry was used as controls. A, The endocytic function (ability to take-up fluorescently labeled dextran) was used to assess nephrocyte function. At the nonpermissive temperature, nephrocytes in all genotypes were able to accumulate dextran. When shifted to the permissive temperature the nephrocytes in control flies were still able to accumulate dextran, but flies in which dKlf15 had been silenced could not. B, Quantification of heart function in adult flies reared at 18°C until eclosion and then transferred to 29°C for 2 weeks. The beating heart was imaged in semi-intact preparations using high frame rate video microscopy. n=20 for each genotype. EDD indicates end-diastolic interval; ESD, end-systolic diameter; and ns, not significantly different from Target or w1118; * and †P<0.01 compared with Target, w1118 or Target; UAS mCherry controls, respectively.
Reduction of Nephrocyte Amnionless Expression Is Associated With Cardiomyopathy
Amnionless is crucial for nephrocyte function,29 so it was hypothesized that loss of Amnionless may be sufficient to cause cardiomyopathy. Silencing Amnionless in nephrocytes did not cause nephrocyte death but did impair nephrocyte endocytic function (Figure 4A and 4B). Importantly, silencing Amnionless affected cardiac function by increasing the heart period because of a lengthening of the diastolic interval (Figure 4C), similar to the phenotype in dKlf15 loss-of-function experiments.
Figure 4.
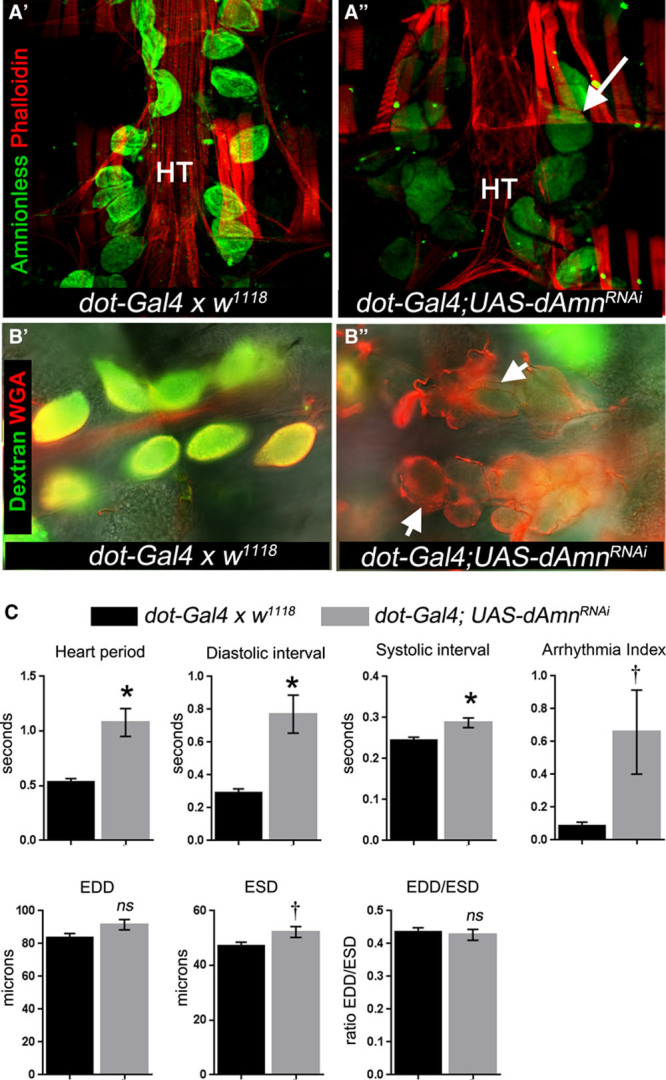
Loss of Amnionless function in nephrocytes leads to cardiomyopathy. A, Amnionless was silenced in nephrocytes using dot-Gal4. As a negative control, the parent driver line was outcrossed once to the w1118 line (dot-Gal4 x w1118) and offspring analyzed. Micrographs show adult hearts stained with anti-Amnionless antibodies (green) and phalloidin (red). Amnionless protein was localized to nephrocytes in controls (A′), whereas silencing led to reduction in its detection but not the loss of pericardial nephrocytes (A″; arrow indicates a pericardial nephrocyte); HT indicates heart tube. B, Semi-intact heart preparations were incubated with fluorescently tagged 10-kDa dextran (green) and wheat germ agglutinin (red) for 30 minutes, washed and imaged. Arrows indicate nephrocytes. Dextran accumulated in controls (B′) but not in Amnionless-silenced nephrocytes (B″). WGA indicates wheat germ agglutinin. C, Quantification of heart function in flies with Amnionless silenced nephrocytes. Hearts were analyzed by high frame rate video microscopy of semi-intact adult heart preparations. EDD indicates end-diastolic interval; ESD, end-systolic diameter; EDD/ESD, fractional shortening of the heart contraction; and ns, not statistically different from control genotype (dot-Gal4 x w1118); *P<0.01, †*<0.05; n=18 to 20 per genotype.
Disruption of the Hemolymph Proteome in dKlf15 Loss of Function Flies
Given that disruption of nephrocyte endocytosis was associated with the development of cardiomyopathy, it was hypothesized that nephrocytes may regulate levels of circulating, cardiomodulatory signals. We therefore examined the hemolymph proteome of control and dKlf15NN mutants using a method similar to that used by others to identify >700 larval hemolymph peptides.30 Signals corresponding to 495 different proteins were identified. Of these, 209 were identified in the hemolymph of both genotypes, 192 were identified only in the control strain, and 94 were found only in dKlf15NN mutants (Figure 5A).
Figure 5.
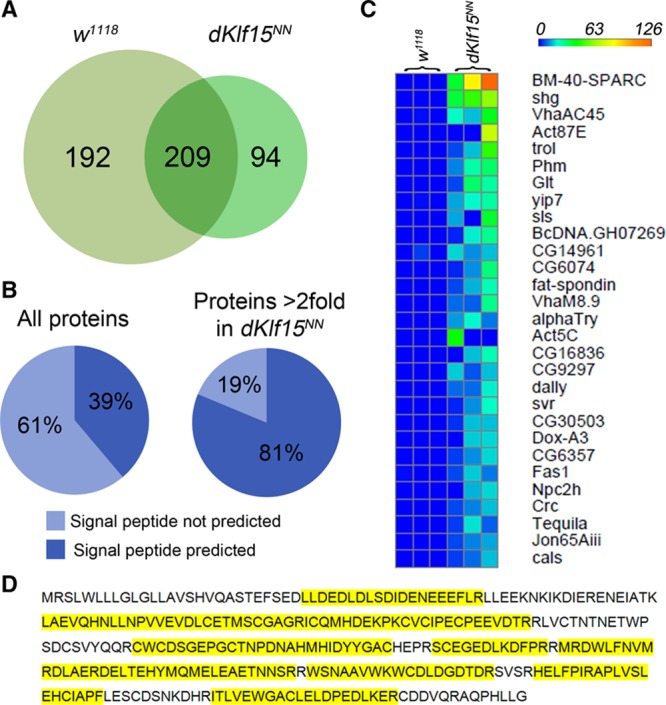
Proteomic analysis of adult Drosophila hemolymph. Hemolymph was collected from adult flies and the proteome analyzed. A, Number of different proteins identified in the hemolymph of adult control (w1118) and mutant dKlf15NN flies. B, The number of proteins predicted to have a signal peptide by SignalP 4.1. C, Heat map showing a truncated list of proteins identified only in the hemolymph of dKlf15NN mutants. Proteins are rank ordered according to spectral count; each row represents 3 independent samples from each genotype (Table I in the Data Supplement). D, Coverage of secreted protein acidic and rich in cysteine (SPARC) protein sequence by detected peptides (yellow corresponds to regions detected in proteomics).
Proteins were allocated to 5 nonoverlapping groups (Table I in the Data Supplement); (group 1) unique to dKlf15NN mutants; (group 2) increased at least 2-fold in dKlf15NN mutants; (group 3) present in both genotypes and within 0.8- to 2-fold different in mutants relative to controls; (group 4) reduced by >0.8-fold in the mutants relative to controls; and (group 5) unique to controls.
It was predicted that ablation of the nephrocytes would lead to the accumulation of secreted proteins in circulation. To address this possibility, the SignalP 4.1 informatics tool was used to identify proteins predicted to have a signal peptide in their N-terminal region, a sequence associated with transport to the extracellular space.27 Of the 448 proteins identified in the circulation of the reference strain, 39% were predicted to contain an N-terminal signal peptide (Figure 5B). In contrast, the hemolymph of the nephrocyte-free mutants was enriched for proteins predicted to contain a signal peptide (81%). In comparison with the total number of Drosophila genes predicted to encode signal peptides (3173 of 17 559 known genes, 18%), it was established that hemolymph proteome is enriched for proteins with signal peptides (P=3.827e-30), and that in the absence of nephrocytes, there is further enrichment for proteins with signal peptides compared with wild-type hemolymph (P=7.408e-35). These data are consistent with nephrocytes having a broad impact on the circulating proteome and suggest that loss of nephrocyte function causes the accumulation in the circulation of a large subset of secreted proteins.
Of the proteins found only in the dKlf15NN mutants (group 1), the most abundant signals were for the matricellular protein BM-40-SPARC (an ortholog of mammalian SPARC (Figures 5C, 5D, and 6A)31) and the cell adhesion protein DE-cadherin (encoded by shotgun, shg). Analysis of the SPARC peptide peak areas confirmed the absence of SPARC in the wild-type hemolymph (Figure I in the Data Supplement). Proteins significantly upregulated in the hemolymph of dKlf15NN mutants compared with wild-type flies (group 2; Figure 6B) included 3 genes with unknown functions (CG18067, CG15293, and CG14961; upregulated 27-, 9-, and 8.5-fold, respectively; P<0.05) as well as several proteins involved in immunity and clotting (gelsolin, immune-induced peptides 10 and 23 and the defence protein l(2)34F; P<0.05). The immune modulatory serpin necrotic1 trended toward a 3-fold accumulation in the mutants’ hemolymph.
Figure 6.
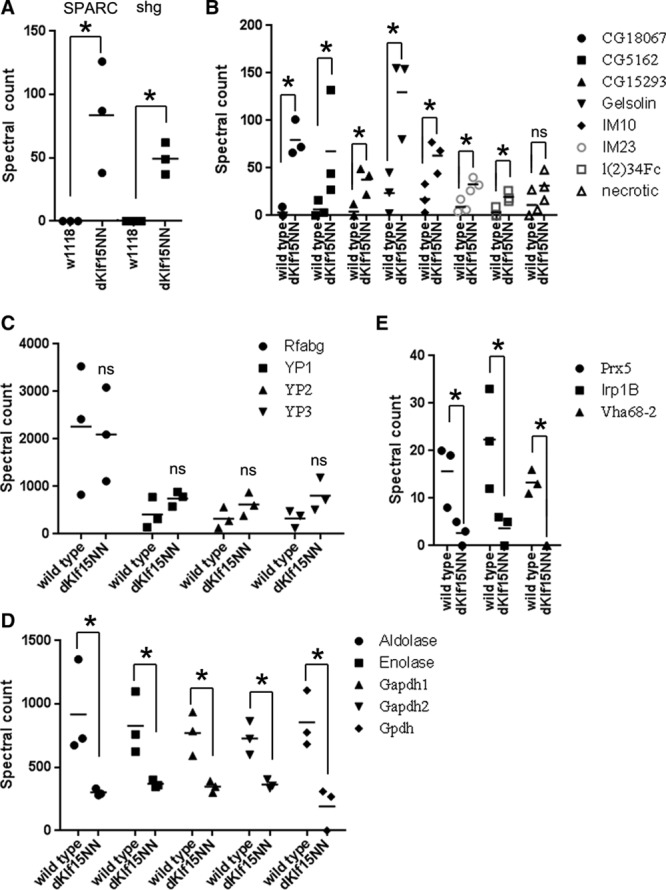
Mean spectral counts of hemolymph proteins. A, Counts for 2 most abundant proteins found only in the dKlf15NN mutants hemolymph. B, Proteins showing an increase in the mutant’s hemolymph. C, The most abundant proteins in the hemolymph of wild-type and dKlf15NN mutants. D, Proteins significantly reduced in the hemolymph of the dKlf15NN mutants. E, Proteins showing the largest, statistically significant, decrease in the mutant’s hemolymph; n=3 independent hemolymph samples from ≈100 flies of each genotype; *P<0.05, ns indicates no significant difference. SPARC indicates secreted protein acidic and rich in cysteine.
Of the proteins common to both genotypes and at similar levels (group 3, Figure 6C), the largest spectral counts corresponded to the lipophorin, retinoid-binding and fatty acid–binding glycoprotein (Rfabg; spectral count of 2260±785 and 2097±570 for wild-type and mutant hemolymph; P=0.88) and the yolk proteins/vitellogenins (VIT 1, 2, and 3). Proteins with large spectral counts in the wild-types that were significantly downregulated in the mutants hemolymph (group 4; Figure 6D) included several intracellular cytoskeletal and metabolic proteins (aldolase, enolase, and subunits of glyceraldehyde phosphate dehydrogenase enzyme). Finally, proteins unique to wild-type hemolymph (group 5, Figure 6E) included peroxiredoxin 5 (Prdx5; spectral count of 16±4 in wild-type and 2±1 in the mutant, P<0.05), iron regulatory protein 1B (Irp1B; spectral count of 22±6 in wild-type and 4±1 in mutant, P<0.05), and vacuolar H+ ATPase 68-kDa subunit 2A (Vha68-2; spectral count of 13±2 in wild-type, undetected in the mutant, P<0.05).
A dSparcMI00329 Mutation Corrects the Cardiomyopathy in dKlf15NN Mutants
Of the proteins unique to the dKlf15NN mutants’ hemolymph, SPARC was notable because of its role in collagen deposition and several growth factor signaling pathways thought to affect cardiac function.32,33 We therefore tested whether SPARC contributed to the observed cardiomyopathy. We obtained a recessive lethal Drosophila SPARC allele (dSparcMI00329) caused by the insertion of a 7.3-kb MiMIC transposon in the 5-prime untranscribed region of the dSparc open reading frame.20 Homozygous adults do not develop, however, dSparcMI00329 heterozygotes are viable, fertile, and develop to adulthood, albeit with dSparc gene expression reduced by 60% (Figure 7A). Homozygous dKlf15NN mutant females were crossed with males carrying the dSparcMI00329 allele, and then the heart function of male progeny (ie, those hemizygous for dKlf15NN and heterozygous for the dSparcMI00329 allele) was analyzed. It was found that the dKlf15NN mutant males had no nephrocytes and exhibited cardiomyopathy characterized by long diastolic intervals, similar to the phenotype of homozygous dKlf15NN mutant females (cf Figures 2 and 6). However, heart function parameters in dSparcMI00329 heterozygotes (w1118; dSparcM100329) were not different from those of control w1118 flies, and specifically there was no increase in heart period/diastolic intervals as observed for hearts from hemizyogous dKlf15NN mutants (Figure 7B). In contrast, when Sparc was silenced in wild-type cardiomyocytes, there was a significant impact on heart function (Figure II in the Data Supplement). Importantly, reducing dSparc expression rescued the abnormal heart phenotype in hemizygous dKlf15NN mutants ((dKlf15NN; dSparcM100329+/−; Figure 7B), despite these flies having no nephrocytes. The findings demonstrate that heterozygosity for SPARC leading to reduced gene expression has no direct impact on cardiac function in the wild-type flies, whereas it ameliorates the cardiomyopathy caused by Klf15-induced loss of nephrocytes.
Figure 7.
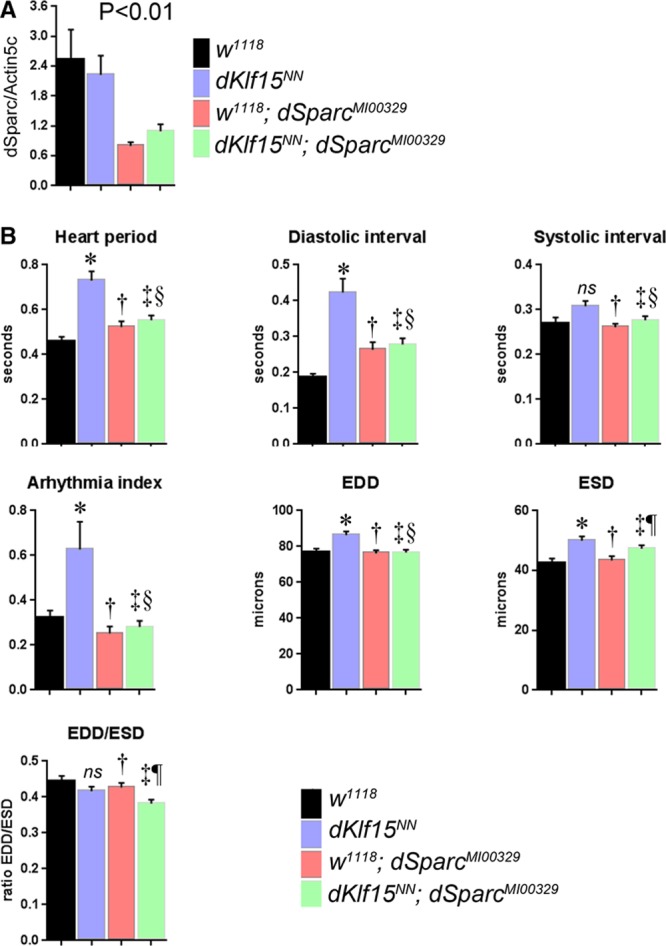
Secreted protein acidic and rich in cysteine (SPARC) mediates cardiomyopathy in dKlf15NN mutants. The heart function of 2-week-old male flies of different genotypes was analyzed in semi-intact fly preparations using high frame rate videomicroscopy. Quantified data for several parameters are presented. EDD indicates end-diastolic diameter; and ESD, end-systolic diameter. *Different from w1118 (P<0.01); †Not different from w1118 (P>0.05); ‡Different from dKlf15NN (P<0.01); §Not different from w1118; dSparcMI00329 (P>0.05); ¶Not different from dKlf15NN (P>0.05); ns, not different from w1118 (P>0.05); n=40 to 69 flies per genotype.
Discussion
The Drosophila heart model represents a highly tractable genetic system with which to study mammalian cardiac physiology. Although at an anatomic level the links between fly nephrocytes and cardiomyocytes may not be evolutionarily conserved, the high degree of gene conservation supports the use of this model for the identification of genetic pathways underlying human heart function. We used both proteomics and genetics to identify SPARC as an important component of cardiac function in Drosophila, highlighting the possibility that SPARC’s role in the heart may be evolutionarily conserved from flies to humans. By using a cell-specific and temporally conditional nephrocyte loss of function paradigm, we establish that pericardial nephrocytes sustain normal heart function in adult Drosophila. Importantly, it was shown that nephrocytes prevent the development of a SPARC-dependent cardiomyopathy, a finding of considerable importance because SPARC is emerging as a clinically important target for the control of tissue fibrosis in humans.33,34 Collectively, these findings highlight the importance of the Drosophila heart model as a means of identifying and studying cardiomodulatory signals of relevance to human cardiac function and suggest that changes to SPARC in humans may contribute to cardiac dysfunction in disease and aging.
Our data reaffirm that pericardial nephrocytes mediate noncell autonomous mechanisms controlling Drosophila heart function. The findings also suggest that the changes in heart morphology and the increased arrhythmias seen in dKlf15NN mutants were not only because of interactions between the cardiomyocytes and nephrocytes during preadult stages but also that adult heart rate is mediated by an on-going interaction between the cardiomyocytes and nephrocytes. Thus, it can be concluded that loss of nephrocytes or nephrocyte function both developmentally or acutely in adults, leads to cardiac dysfunction. Our data also suggest that the cardiomyopathy caused by loss of nephrocytes or nephrocyte function is linked to the nephrocytes’ role in peripheral clearance.
To our knowledge, our data set represents the first examination of the adult Drosophila hemolymph proteome. The most abundant protein in the hemolymph of both wild-type and dKlf15NN mutants was Rfabg. Rfabg is a lipid transporter found in insect hemolymph and known to be required for Hedgehog and Wingless signaling.35 There were also large signals for several important intracellular metabolic proteins (eg, aldolase, enolase, and subunits of the glyceraldehyde phosphate dehydrogenase enzyme). The presence of intracellular proteins is a feature of the human plasma proteome, suggesting that intracellular proteins are a constituent of circulating fluids in animals. We also recorded an expected absence or near-absence from the adult hemolymph of the larval serum proteins LSP1α, LSP1β, LSP1γ, and LSP2, all of which are among the most highly represented proteins in larval hemolymph.30 The LSPs are metabolized during the nonfeeding third instar and pupal stages, hence our data indicate that by the first week of adulthood, they are difficult to detect in the hemolymph.
In addition, there were proteins identified in both genotypes, which were significantly upregulated or downregulated in the hemolymph of dKlf15NN mutants (group 2). The most highly upregulated signals were ascribed to genes with unknown functions (CG18067, CG15293, and CG14961) and proteins involved in immunity and clotting (gelsolin, immune-induced peptides 10 and 23, and the Defence protein l(2)34F). Although not reaching statistical significance, necrotic, an immune modulatory serpin removed from circulation by nephrocytes,1 trended toward accumulating in the mutants’ hemolymph of the mutants. In addition, there was a significant reduction in the mutants’ hemolymph of peroxiredoxin 5, an antioxidant that also negatively regulates the immune response.36 Collectively, these findings suggest that the mutant flies may have modified immune responses, and this is currently under investigation.
The most abundant proteins identified only in the hemolymph of the dKlf15NN mutants were DE-cadherin and SPARC. DE-cadherin mediates cell adhesion and is critical for embryonic development and is present in the medulla of the lymph gland,37 whereas SPARC stabilizes basal lamina by interacting with collagen IV, an interaction critical for normal development.38,39 In contrast, the role of SPARC in postembryonic and adult Drosophila remains unclear. Mammalian SPARC directly binds to collagen as well as growth factors40 and is associated with a diverse range of pathologies, including the maintenance of cardiac integrity41 and metabolic syndrome.42 Our data indicate that a SPARC-dependent cardiomyopathy is prevented in the Drosophila model via a nephrocyte-mediated clearance mechanism. Peripheral clearance of macromolecules is fundamentally important to tissue homeostasis but difficult to study in mammals. Although few studies exist, it is interesting to note that disruption of peripheral clearance by liver sinusoidal cells in stabilin-1 and stabilin-2 knockout mice led to local and systemic tissue fibrosis, albeit without an increase in circulating SPARC levels being detected.43 It remains to be verified whether the increased SPARC levels directly cause cardiomyopathy or whether it is because of other hemolymph factors that are increased in dKlf15NN mutants that act via a SPARC-dependent pathway.
Although abnormal cardiomyocyte function in dKlf15NN mutants could be rescued by reducing SPARC gene dosage, we could not confirm whether this was an effect of reduced SPARC expression in the cardiomyocytes, because SPARC knockdown in wild-type cardiomyocytes led to a severe cardiomyopathy, characterized by reduced fractional shortening (Figure II in the Data Supplement). Hence, rescue of the cardiomyopathy by reducing SPARC gene dosage in the nephrocyte-free dKlf15NN mutants may have been because of either a less severe reduction in SPARC expression in the cardiomyocytes or reduced SPARC expression in cells other than cardiomyocytes. Interestingly, moderate reductions in ILK expression in whole flies can extend lifespan and retard cardiac ageing, yet strong knockdown in cardiomyocytes has a profound negative impact on cardiac function.8 Thus, different phenotypes can develop in the heart as a consequence of differing levels of gene silencing/gene dosage.
In summary, the Drosophila heart can develop a SPARC-dependent cardiomyopathy as a result of nephrocyte loss. These findings identify Drosophila as a highly tractable model system with which to study the important relationship between tissue homeostasis and peripheral clearance, especially as it relates to human cardiac physiology. The next step will, therefore, be to establish how SPARC contributes to cardiac function in Drosophila and explore whether these mechanisms are conserved and relevant to the human heart.
Acknowledgments
Dr Hartley acknowledges support of the British Heart Foundation Centre of Research Excellence award (RE/08/001). Dr Hartley is especially grateful to Dr Georg Vogler and Ms. Hanah Catan (Sanford Burnham Medical Research Institute); Dr Richard Marley and Ms. Ashleigh Cheyne (University of Edinburgh) for comments and assistance with experiments. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks used in this study.
Sources of Funding
Dr Hartley is supported by a British Heart Foundation Intermediate Basic Science Fellowship (FS/13/17/29905). This work was also supported by a British Heart Foundation Centre of Research Excellence award to the University of Edinburgh and by a subsistence grant from the Scottish Universities Life Science Alliance to Dr Hartley. This work was also supported by grants from the National Institutes of Health to R. Bodmer. K. Ocorr is supported by American Heart Association award 0835243N.
Disclosures
None.
Supplementary Material
Footnotes
The Data Supplement is available at http://circgenetics.ahajournals.org/lookup/suppl/doi:10.1161/CIRCGENETICS.115.001254/-/DC1.
CLINICAL PERSPECTIVE
In this work, we identified a genetic pathway in Drosophila linking cardiac function with the matricellular protein secreted protein acidic and rich in cysteine (SPARC). Changes to SPARC gene expression or protein levels in humans are associated with cardiac aging, chronic inflammatory disease, and metabolic syndrome. SPARC controls collagen deposition as well as mitogenic signaling and, as such, it is a potential clinical target with which to control scarring and organ dysfunction. It has been noted that Drosophila heart function is regulated by pericardial nephrocytes, highly endocytic kidney-like cells that flank the heart and filter the fly’s blood. Our research aims to understand this relationship because the mechanisms involved may be relevant to the regulation of the human heart. To address this, we genetically ablated the nephrocytes and analyzed heart function as well as the blood proteome, in the hope of discovering cardiomodulatory peptides. We found that nephrocyte loss led to a severe cardiomyopathy, characterized by tissue remodeling and a slow, irregular heartbeat. Proteomics revealed an accumulation of many circulating proteins, most notably SPARC. We then conducted genetic loss of function experiments and established that the cardiomyopathy was dependent on SPARC. It remains to be seen whether this link involves SPARC’s role in collagen deposition or a novel, uncharacterized mechanism. Nevertheless, by establishing a link between SPARC and cardiac function in Drosophila we now have a new, powerful tool in the research armoury with which to ascertain how SPARC contributes to cardiac function in humans.
References
- 1.Soukup SF, Culi J, Gubb D. Uptake of the necrotic serpin in Drosophila melanogaster via the lipophorin receptor-1. PLoS Genet. 2009;5:e1000532. doi: 10.1371/journal.pgen.1000532. doi: 10.1371/journal.pgen.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das D, Aradhya R, Ashoka D, Inamdar M. Macromolecular uptake in Drosophila pericardial cells requires rudhira function. Exp Cell Res. 2008;314:1804–1810. doi: 10.1016/j.yexcr.2008.02.009. doi: 10.1016/j.yexcr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Mills RP, King RC. The pericardial cells of Drosophila melanogaster. Q J Microsc Sci. 1965;106:261–268. [PubMed] [Google Scholar]
- 4.Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. doi: 10.1016/j.cell.2010.02.023. doi: 10.1016/j.cell.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catterson JH, Heck MM, Hartley PS. Fermitins, the orthologs of mammalian Kindlins, regulate the development of a functional cardiac syncytium in Drosophila melanogaster. PLoS One. 2013;8:e62958. doi: 10.1371/journal.pone.0062958. doi: 10.1371/journal.pone.0062958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viswanathan MC, Kaushik G, Engler AJ, Lehman W, Cammarato A. A Drosophila melanogaster model of diastolic dysfunction and cardiomyopathy based on impaired troponin-T function. Circ Res. 2014;114:e6–17. doi: 10.1161/CIRCRESAHA.114.302028. doi: 10.1161/CIRCRESAHA.114.302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura M, Kumsta C, Kaushik G, Diop SB, Ding Y, Bisharat-Kernizan J, et al. A dual role for integrin-linked kinase and β1-integrin in modulating cardiac aging. Aging Cell. 2014;13:431–440. doi: 10.1111/acel.12193. doi: 10.1111/acel.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birse RT, Choi J, Reardon K, Rodriguez J, Graham S, Diop S, et al. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010;12:533–544. doi: 10.1016/j.cmet.2010.09.014. doi: 10.1016/j.cmet.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Na J, Musselman LP, Pendse J, Baranski TJ, Bodmer R, Ocorr K, et al. A Drosophila model of high sugar diet-induced cardiomyopathy. PLoS Genet. 2013;9:e1003175. doi: 10.1371/journal.pgen.1003175. doi: 10.1371/journal.pgen.1003175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Na J, Sweetwyne MT, Park AS, Susztak K, Cagan RL. Diet-induced podocyte dysfunction in Drosophila and mammals. Cell Rep. 2015;12:636–647. doi: 10.1016/j.celrep.2015.06.056. doi: 10.1016/j.celrep.2015.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buechling T, Akasaka T, Vogler G, Ruiz-Lozano P, Ocorr K, Bodmer R. Non-autonomous modulation of heart rhythm, contractility and morphology in adult fruit flies. Dev Biol. 2009;328:483–492. doi: 10.1016/j.ydbio.2009.02.013. doi: 10.1016/j.ydbio.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujioka M, Wessells RJ, Han Z, Liu J, Fitzgerald K, Yusibova GL, et al. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ Res. 2005;97:1108–1114. doi: 10.1161/01.RES.0000191546.08532.B2. doi: 10.1161/01.RES.0000191546.08532.B2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson AN, Burnett LA, Sellin J, Paululat A, Newfeld SJ. Defective decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila. Genetics. 2007;176:1609–1624. doi: 10.1534/genetics.107.073569. doi: 10.1534/genetics.107.073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim HY, Wang W, Chen J, Ocorr K, Bodmer R. ROS regulate cardiac function via a distinct paracrine mechanism. Cell Rep. 2014;7:35–44. doi: 10.1016/j.celrep.2014.02.029. doi: 10.1016/j.celrep.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das D, Aradhya R, Ashoka D, Inamdar M. Post-embryonic pericardial cells of Drosophila are required for overcoming toxic stress but not for cardiac function or adult development. Cell Tissue Res. 2008;331:565–570. doi: 10.1007/s00441-007-0518-z. doi: 10.1007/s00441-007-0518-z. [DOI] [PubMed] [Google Scholar]
- 17.Ivy JR, Drechsler M, Catterson JH, Bodmer R, Ocorr K, Paululat A, et al. Klf15 is critical for the development and differentiation of Drosophila nephrocytes. PLoS One. 2015;10:e0134620. doi: 10.1371/journal.pone.0134620. doi: 10.1371/journal.pone.0134620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frangogiannis NG. Matricellular proteins in cardiac adaptation and disease. Physiol Rev. 2012;92:635–688. doi: 10.1152/physrev.00008.2011. doi: 10.1152/physrev.00008.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimbrell DA, Hice C, Bolduc C, Kleinhesselink K, Beckingham K. The Dorothy enhancer has Tinman binding sites and drives hopscotch-induced tumor formation. Genesis. 2002;34:23–28. doi: 10.1002/gene.10134. doi: 10.1002/gene.10134. [DOI] [PubMed] [Google Scholar]
- 20.Venken KJ, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sellin J, Albrecht S, Kölsch V, Paululat A. Dynamics of heart differentiation, visualized utilizing heart enhancer elements of the Drosophila melanogaster bHLH transcription factor hand. Gene Expr Patterns. 2006;6:360–375. doi: 10.1016/j.modgep.2005.09.012. doi: 10.1016/j.modgep.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 22.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 23.Park SK, Venable JD, Xu T, Yates JR., III. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010;Chapter 14:Unit14.20. doi: 10.1002/0471142727.mb1420s92. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007;104:3943–3948. doi: 10.1073/pnas.0609278104. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brill LM, Motamedchaboki K, Wu S, Wolf DA. Comprehensive proteomic analysis of Schizosaccharomyces pombe by two-dimensional HPLC-tandem mass spectrometry. Methods. 2009;48:311–319. doi: 10.1016/j.ymeth.2009.02.023. doi: 10.1016/j.ymeth.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 28.Halbritter F, Vaidya HJ, Tomlinson SR. GeneProf: analysis of high-throughput sequencing experiments. Nat Methods. 2012;9:7–8. doi: 10.1038/nmeth.1809. doi: 10.1038/nmeth.1809. [DOI] [PubMed] [Google Scholar]
- 29.Zhang F, Zhao Y, Chao Y, Muir K, Han Z. Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J Am Soc Nephrol. 2013;24:209–216. doi: 10.1681/ASN.2012080795. doi: 10.1681/ASN.2012080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handke B, Poernbacher I, Goetze S, Ahrens CH, Omasits U, Marty F, et al. The hemolymph proteome of fed and starved Drosophila larvae. PLoS One. 2013;8:e67208. doi: 10.1371/journal.pone.0067208. doi: 10.1371/journal.pone.0067208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer P, Hohenadl C, Hohenester E, Göhring W, Timpl R, Engel J. The C-terminal portion of BM-40 (SPARC/osteonectin) is an autonomously folding and crystallisable domain that binds calcium and collagen IV. J Mol Biol. 1995;253:347–357. doi: 10.1006/jmbi.1995.0557. doi: 10.1006/jmbi.1995.0557. [DOI] [PubMed] [Google Scholar]
- 32.de Castro Brás LE, Toba H, Baicu CF, Zile MR, Weintraub ST, Lindsey ML, et al. Age and SPARC change the extracellular matrix composition of the left ventricle. Biomed Res Int. 2014;2014:810562. doi: 10.1155/2014/810562. doi: 10.1155/2014/810562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kos K, Wilding JP. SPARC: a key player in the pathologies associated with obesity and diabetes. Nat Rev Endocrinol. 2010;6:225–235. doi: 10.1038/nrendo.2010.18. doi: 10.1038/nrendo.2010.18. [DOI] [PubMed] [Google Scholar]
- 34.Trombetta-Esilva J, Bradshaw AD. The function of SPARC as a mediator of fibrosis. Open Rheumatol J. 2012;6:146–155. doi: 10.2174/1874312901206010146. doi: 10.2174/1874312901206010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 36.Radyuk SN, Michalak K, Klichko VI, Benes J, Orr WC. Peroxiredoxin 5 modulates immune response in Drosophila. Biochim Biophys Acta. 2010;1800:1153–1163. doi: 10.1016/j.bbagen.2010.06.010. doi: 10.1016/j.bbagen.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- 38.Martinek N, Shahab J, Saathoff M, Ringuette M. Haemocyte-derived SPARC is required for collagen-IV-dependent stability of basal laminae in Drosophila embryos. J Cell Sci. 2008;121(pt 10):1671–1680. doi: 10.1242/jcs.021931. doi: 10.1242/jcs.021931. [DOI] [PubMed] [Google Scholar]
- 39.Pastor-Pareja JC, Xu T. Shaping cells and organs in Drosophila by opposing roles of fat body-secreted collagen IV and perlecan. Dev Cell. 2011;21:245–256. doi: 10.1016/j.devcel.2011.06.026. doi: 10.1016/j.devcel.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44:480–488. doi: 10.1016/j.biocel.2011.12.021. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schellings MW, Vanhoutte D, Swinnen M, Cleutjens JP, Debets J, van Leeuwen RE, et al. Absence of SPARC results in increased cardiac rupture and dysfunction after acute myocardial infarction. J Exp Med. 2009;206:113–123. doi: 10.1084/jem.20081244. doi: 10.1084/jem.20081244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tartare-Deckert S, Chavey C, Monthouel MN, Gautier N, Van Obberghen E. The matricellular protein SPARC/osteonectin as a newly identified factor up-regulated in obesity. J Biol Chem. 2001;276:22231–22237. doi: 10.1074/jbc.M010634200. doi: 10.1074/jbc.M010634200. [DOI] [PubMed] [Google Scholar]
- 43.Schledzewski K, Géraud C, Arnold B, Wang S, Gröne HJ, Kempf T, et al. Deficiency of liver sinusoidal scavenger receptors stabilin-1 and -2 in mice causes glomerulofibrotic nephropathy via impaired hepatic clearance of noxious blood factors. J Clin Invest. 2011;121:703–714. doi: 10.1172/JCI44740. doi: 10.1172/JCI44740. [DOI] [PMC free article] [PubMed] [Google Scholar]


