Abstract
Somatic hypermutation (SHM) and class switch recombination (CSR) increase the affinity and diversify the effector functions of antibodies during immune responses. Although SHM and CSR are fundamentally different, their independent roles in regulating B cell fate have been difficult to uncouple because a single enzyme, activation-induced cytidine deaminase (encoded by Aicda), initiates both reactions. Here, we used a combination of Aicda and antibody mutant alleles that separate the effects of CSR and SHM on polyclonal immune responses. We found that class-switching to IgG1 biased the fate choice made by B cells, favoring the plasma cell over memory cell fate without significantly affecting clonal expansion in the germinal center (GC). In contrast, SHM reduced the longevity of memory B cells by creating polyreactive specificities that were selected against over time. Our data define the independent contributions of SHM and CSR to the generation and persistence of memory in the antibody system.
Introduction
During immune responses, B cells diversify their immunoglobulin genes in germinal centers (GCs) to produce the high affinity, class-switched antibodies that mediate humoral immunity (Allen et al., 2007a; Rajewsky, 1996; Victora and Nussenzweig, 2012). Antibody gene diversification is accomplished by somatic hypermutation (SHM) and class switch recombination (CSR). Whereas SHM generates a pool of antibody variants with differing affinities, CSR exchanges the antibody constant region to produce antibodies with a diverse set of effector functions (Bournazos et al., 2015; Stavnezer et al., 2008). A single enzyme, activation-induced cytidine deaminase (AID), which is expressed primarily in the GC, initiates both SHM and CSR (Muramatsu et al., 2000). Although mutant forms of AID bias the reaction to SHM or CSR, the two diversification reactions are never completely separated (Barreto et al., 2003; Shinkura et al., 2004; Ta et al., 2003; Wei et al., 2011). It has therefore been difficult to delineate the precise contributions of changes in affinity versus alterations in isotype to regulating the antibody response.
B cells expressing high affinity antibody variants are selectively expanded in the GC and preferentially seed the plasma cell compartment (Phan et al., 2006; Smith et al., 1997; Victora and Nussenzweig, 2012). As a result, serum antibody affinity increases over time, a phenomenon known as affinity maturation (Eisen and Siskind, 1964).
Although IgE expression is associated with limited bone marrow plasma cell and memory B cell formation (He et al., 2013; Yang et al., 2012) and IgA expression promotes plasma cell differentiation (Duchez et al., 2010), the independent roles of SHM and IgG antibody class switching in regulating B cell fate are not well defined. Experiments using a transgenic IgG1 antigen receptor specific for hen egg lysozyme indicates that this isotype enhances clonal expansion and might bias B cells to become plasmablasts (Horikawa et al., 2007; Martin and Goodnow, 2002). However, an IgG1 BCR specific for 4-hydroxy-3-nitrophenylacetyl (NP) within the endogenous antibody locus fails to show the same effect (Kometani et al., 2013). Moreover, clonal analysis of wild-type and Aicda-deficient B cells that are unable to class switch showed that they were equally capable of producing plasmablasts (Taylor et al., 2015). Thus, whether the observed differences in B cell fate are due to antibody isotype, transgene-induced alterations in B cell development (Pogue and Goodnow, 2000; Roth et al., 1993; Roth et al., 1995), or differences in the antigen and/or affinity remains to be determined.
In addition to effects on B cell fate, differences in antibody isotype have also been associated with differences in the longevity of memory B cells (Pape et al., 2011). IgG+ memory B cells appear to have a far shorter half-life than IgM+ memory B cells. However, the antibodies expressed by IgG+ memory cells are more mutated than their IgM+ counterparts (Dogan et al., 2009; Pape et al., 2011). Thus, the relative contribution of hypermutation versus isotype switching to memory cell persistence remains to be determined.
Here, we report on experiments designed to uncouple CSR and SHM in order to examine their independent contributions to antigen-specific polyclonal B cell responses.
Results
A polyclonal system that separates CSR and SHM
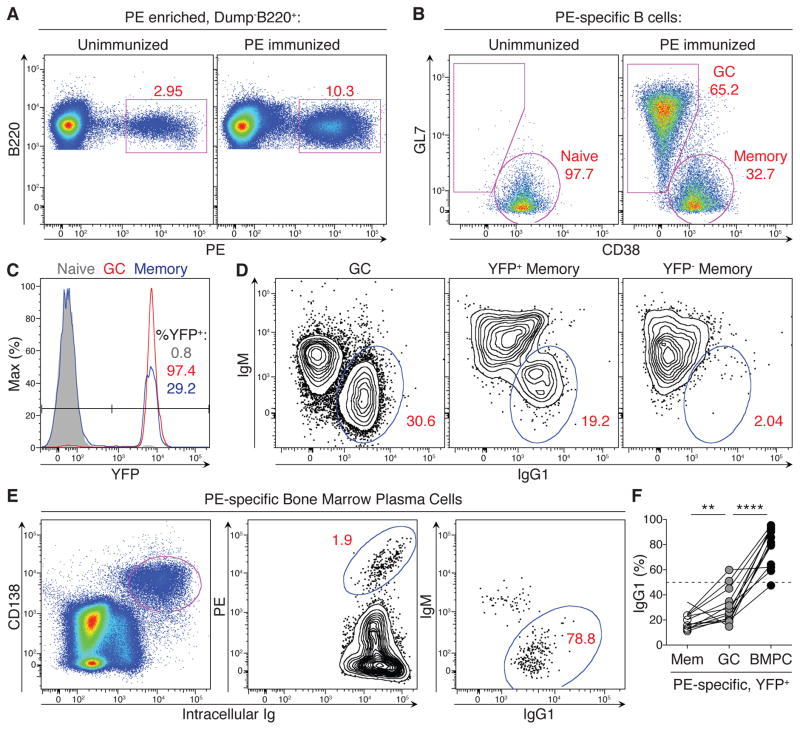
To examine the role of class-switching during a polyclonal immune response, we immunized mice with R-phycoerytherin (PE) in complete Freund’s adjuvant (CFA) (Pape et al., 2011). To track the fate of cells that had expressed Aicda, we combined a genetically targeted allele of Aicda, in which cre recombinase replaces the endogenous AID protein (Aicdacre/+), with the R26LSL-YFP allele, in which cre-mediated recombination leads to permanent yellow fluorescent protein (YFP) expression (Robbiani et al., 2008; Srinivas et al., 2001). Antigen-specific cells were enumerated by magnetic bead-based enrichment (Figure 2 and Figures S1A–C) (Pape et al., 2011). As expected, the majority of antigen-specific GC B cells and bone marrow plasma cells (BMPCs) had expressed Aicda as determined by the YFP marker (90.5% and 83% YFP+, respectively), and most of these cells were class-switched (95.6% and 95.7%, respectively) (Figure S1B). In contrast, only 28.5% of antigen-specific memory cells were YFP+, of which only 48% were class-switched (Figure S1B).
Figure 2. Antigen-specific B lineage cells and positive selection for the bone marrow plasma cell fate.
(A) PE-binding among Dump-B220+ cells in naïve and immunized Aicdacre/− Igh96K/+ R26LSL-YFP mice following column-based cell enrichment. (B) GC and memory cells among antigen-specific B cells gated in (A) and identified based on surface expression of CD38 and GL7. (C) Expression history of Aicda among antigen-specific naïve B cells and GC and memory B cells, gated as in (A–B). (D) IgM and IgG1 surface expression in indicated antigen-specific populations, gated as shown in (A–C). (E) Plasma cells were enriched in immunized mice from the bone marrow based on CD138 surface expression and assessed for intracellular antigen-binding and isotype expression. BMPCs were further gated as Dump−YFP+. (F) Fraction of IgG1+ cells among YFP+ cells in the antigen-specific memory, GC, and BMPC compartments of Aicdacre/− Igh96K/+ R26LSL-YFP mice 40–59 days after immunization with PE. Results are combined from 3 independent experiments; each line connects the indicated cell populations from the same animal. **** p <0.0001, ** p = 0.009. Two-tailed Mann-Whitney test was used to assess significance. See also Figure S1.
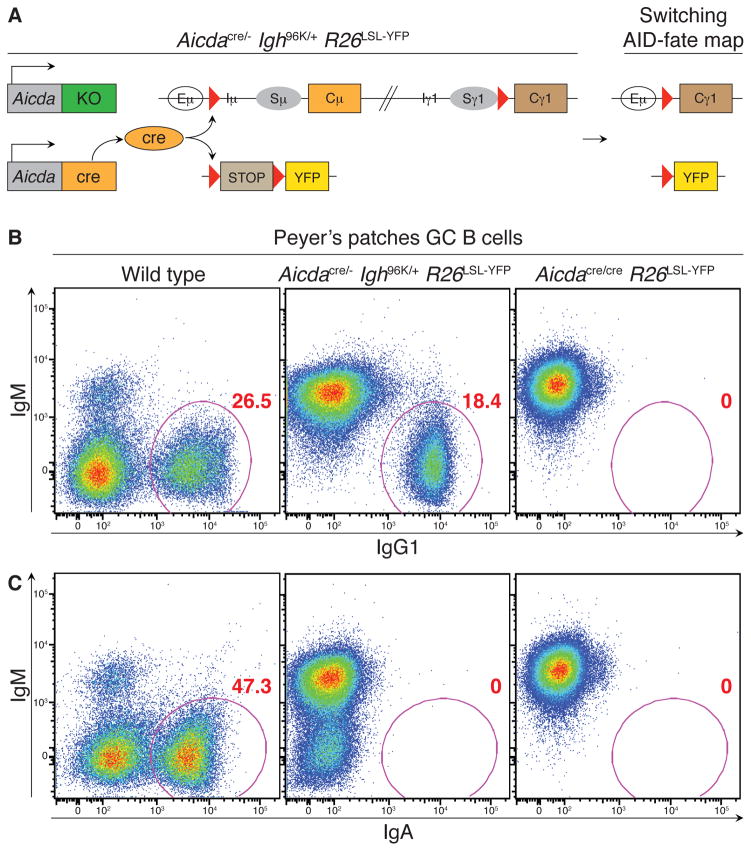
To examine the contribution of class switching to the B cell response, we generated mice in which class switching takes place in the absence of AID and SHM (Fig. 1A). To do so, we combined an Aicda-deficient allele (Aicda−) with the aforementioned Aicdacre allele (Aicdacre/−) (Muramatsu et al., 2000). These Aicda mutant alleles were further crossed to the R26LSL-YFP fate-mapping allele and to a genetically targeted Ig heavy chain (Igh) allele carrying loxP sites upstream of the μ and γ1 constant regions (Igh96K) (Bothmer et al., 2010). Activated B cells in the resulting Aicdacre/− Igh96K/+ R26LSL-YFP mice that transcribed the endogenous Aicda locus expressed cre in place of AID protein; cre expression recombines the loxP sites in the Igh96K and R26LSL-YFP loci thereby class switching the BCR to IgG1 while marking cells as YFP+ (Figure 1A). Because B cells in these mice do not express AID protein, they did not undergo SHM or suffer AID-induced DNA damage, thus allowing CSR to take place independently of SHM.
Figure 1. Uncoupling CSR and SHM in the polyclonal system.
(A) Diagram of allele combinations used to separate CSR and SHM. Aicdacre/− Igh96K/+ R26LSL-YFP mice express cre recombinase instead of AID protein, leading to cre-mediated class switching to IgG1 and fate-mapping based on Aicda expression in the absence of SHM. (B and C) Class switching to IgG1 (B) and IgA (C) among all GC B cells in Peyer’s patches of indicated mice. Results represent two independent experiments with at least two mice per genotype in each experiment.
Flow cytometric analysis of Peyer’s patches confirmed that GC B cells in Aicdacre/− Igh96K/+ R26LSL-YFP mice class switch robustly and exclusively to IgG1 (Figure 1B). The absence of class switching to IgA in Peyer’s patches of Aicdacre/− Igh96K/+ R26LSL-YFP mice confirmed the absence of endogenous AID activity (Figure 1C).
The IgG1 BCR mediates selection into the BMPC fate
Due to allelic exclusion, the vast majority of B cells express only one of their two Igh alleles (Cebra et al., 1966; Pernis et al., 1965). Thus, naïve B cells in Aicdacre/− Igh96K/+ R26LSL-YFP mice expressed either the Igh96K allele or the wild-type Igh allele (Figure S3A, see below). The ~50% of B cells with a productive V(D)J rearrangement in their Igh96K allele could class switch to IgG1 upon cre expression; the other half of the B cell population, expressing the wild-type Igh, could not class switch because of the absence of both AID protein expression and loxP sites in the constant region of their expressed Igh gene.
After immunization, nearly all antigen-specific GC B cells in Aicdacre/− Igh96K/+ R26LSL-YFP mice became YFP+ due to high amounts of transcription of the Aicdacre allele (Figure 2A–C). Among GC B cells, ~31% were IgG1+ (Figures 2D and 2F). Taking into consideration that only half of the B cells could class switch, this number of class-switched cells was similar to the frequency of class-switched GC cells in Aicdacre/+ R26LSL-YFP mice immunized with PE (Figure S1). The antigen-specific memory pool contained 28% YFP+ cells in a resting state (Figures S1D and S2B), of which only 19% were class-switched, and this too was similar to Aicdacre/+ R26LSL-YFP mice (Figures 2D and 2F and Figure S1). In contrast, antigen-specific BMPCs were 85% YFP+, of which nearly 79% of cells were class-switched (Figures 2E and 2F). Since only half of the responding B cells in Aicdacre/− Igh96K/+ R26LSL-YFP mice can class switch after activation (Igh96K-expressers), no antigen-specific compartment should exceed ~50% class-switched unless the IgG1 BCR itself promotes such a fate. Thus, the finding that 79% of antigen-specific BMPCs were class-switched reveals IgG1-mediated positive selection into this compartment, which is independent of SHM.
IgG1 alters GC LZ gene expression
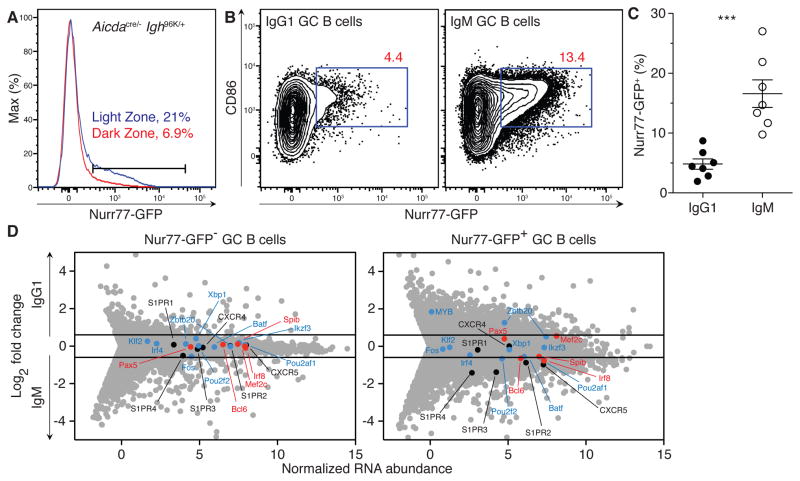
To investigate differences in signaling by IgG1 and IgM BCRs that may account for the differential fate of these cells, we incorporated a Nurr77-GFP transgene that acts as a reporter of BCR signaling (Zikherman et al., 2012). Consistent with recent reports (Mueller et al., 2015), Nurr77 reporter expression was higher among light zone (LZ) cells than among dark zone (DZ) cells (Figure 3A). Moreover, transcriptional profiles between IgG1+ and IgM+ dark zone (DZ) cells were nearly indistinguishable (r = 0.96; Figure S2A), and a short pulse with the nucleotide analog 5-ethynyl-2′-deoxyuridine (EdU) revealed similar proliferation rates among IgM+ and IgG1+ GC cells (Figure S2B). In contrast, transcriptional profiles of IgG1+ and IgM+ LZ cells differed to a greater extent (r = 0.86; Figure S2A). A greater fraction of IgM+ LZ B cells were Nurr77+ compared to IgG1+ LZ B cells in Aicdacre/− Igh96K/+ Nurr77-GFP mice (Figure 3B and 3C). We therefore purified and sequenced RNA from IgG1+ and IgM+ LZ cells that were Nurr77− and Nurr77+ (Figure 3D).
Figure 3. Isotype-specific signaling in the GC.
(A) Histogram showing Nurr77-GFP expression among light zone (CXCR4loCD86hi) and dark zone (CXCR4hiCD86lo) GC B cells 32–41 days after immunization with PE. (B) Representative flow cytometry plots showing Nurr77-GFP expression among IgM+ and IgG1+ GC B cells. (C) Fraction of Nurr77-GFP+ cells among IgM+ and IgG1+ GC B cells, gated as shown in (B). Line indicates mean. (D) RNA sequencing analysis showing selected chemokine receptor genes (black) and negative (red) and positive (blue) regulators of plasma cell differentiation (see Materials and Methods) among IgM+ and IgG1+ LZ cells that were Nurr77− (left plot) and Nurr77+ (right plot). Lines represent cut-offs for genes up- or down-regulated by a fold-change of at least 0.6 (log2). Results represent two independent experiments for A–D with n = 3–5 mice per genotype for each experiment. Error bars = SEM. *** p = 0.0006. Two-tailed Mann-Whitney test was used to assess significance. See also Figure S2 and Table S1.
Although we found little difference between Nur77− LZ cells of the two isotypes, Nurr77+ LZ cells differed in their transcriptional profiles to a far greater extent (Table S1 and Figure 3D), including differences in expression of chemokine receptors implicated in B cell migration in a manner that potentially presages the exit of IgG1+ cells from the GC and their differentiation into the BMPC fate (Figure 3D) (Cyster, 2003; Green and Cyster, 2012; Green et al., 2011; Hargreaves et al., 2001; Kabashima et al., 2006). For example, IgG1+ Nurr77+ cells had lower expression of CXCR5 and S1PR2 transcripts than their IgM+ counterparts (Figure 3D and Figures S2C and S2D), and expression of both of these receptors is essential to B cell retention within the follicle and the GC, respectively (Ansel et al., 2000; Forster et al., 1996; Green et al., 2011). In addition, expression of c-Myb, which has been implicated in CXCR4-mediated migration of plasma cells to the bone marrow (Good-Jacobson et al., 2015), was increased. We conclude that BCR signaling in the LZ differs in an isotype-specific manner, consistent with the biased fate choice made by IgG1+ B cells to the BMPC fate.
The IgG1 BCR disfavors the memory cell fate
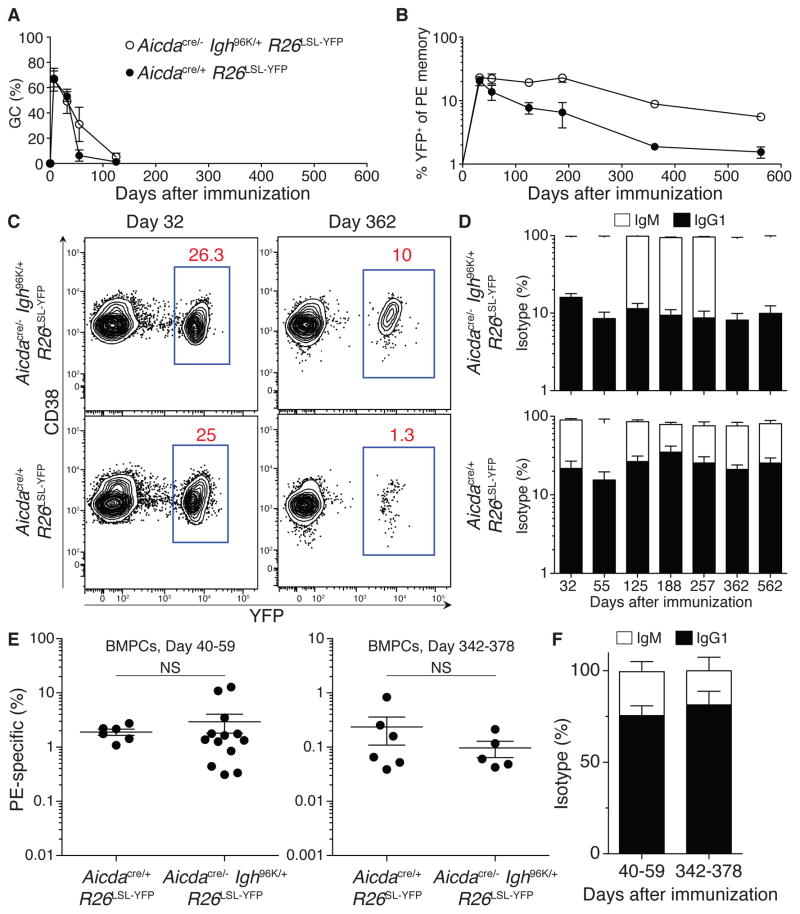
Whereas IgG1+ cells constitute a majority of GC B cells and BMPCs, they represent a minority of the memory pool (Figures 2D and 2F and S1) (Dogan et al., 2009; Pape et al., 2011; Slifka et al., 1995). This raises the possibility that class switching biases responding B cells away from entering the memory pool. Alternatively, IgG1+ memory cells may not persist as long as IgM+ memory cells; however, this possibility was ruled out because once the memory pool was formed, the fraction of memory cells that were IgM and IgG1 remained stable over ≥ 1.5 years (Figure 5D and see below).
Figure 5. Role of CSR and SHM in memory and plasma cell longevity.
(A) Mean fraction of GC cells among antigen-specific B cells in indicated mice after immunization with PE in CFA. (B) Mean fraction of YFP+ cells among antigen-specific memory B cells in indicated mice after immunization with PE in CFA. (C) Representative flow cytometry plots showing YFP expression among antigen-specific memory B cells in indicated mice at 32 and 362 days after immunization. (D) Mean fraction of IgG1+ and IgM+ cells within the PE-specific YFP+ memory cell compartment in the indicated mice after immunization. (E) Frequency of PE-specific cells in the BMPC compartment of the indicated mice at early (days 40–59) or late (days 342–378) time points after immunization. Lines represent means. (F) Mean fraction IgG1+ and IgM+ cells among PE-specific BMPCs at the indicated time points after immunization of Aicdacre/− Igh96K/+ R26LSL-YFP mice. Results are from 2–3 independent experiments for each genotype and at each time point. Error bars = SEM. Each dot represents one animal in (E). Significance was assessed using a two-tailed Mann-Whitney test. Error bars = SEM. See also Figure S4.
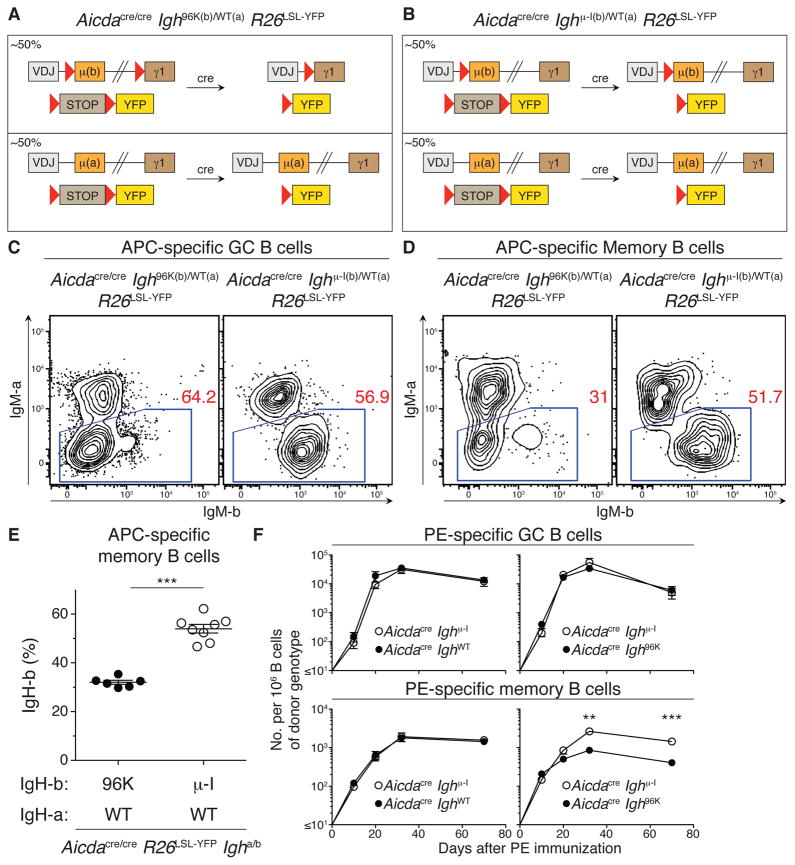
To test the notion that the IgG1 BCR counter-selects against the memory cell fate choice, we used Igh allotype-marked mice in which the Ighb allele was able to undergo isotype switching and the Igha allele was not (Aicdacre/cre Igh96k(b)/WT(a) R26LSL-YFP; Figure 4A). In the absence of counter-selection, Ihga- and Ighb-expressing B cells should each comprise ~50% of the memory compartment (Figure S3A). However, we found that B cells expressing the Ighb allele that can undergo class switching were under-represented in the memory compartment (Figure 4C–E and S3). In contrast, control B cells carrying only a single loxP site in the Ighb allele (Robbiani et al., 2008), and thus unable to class switch (Aicdacre/cre Ighμ-I(b)/WT(a) R26LSL-YFP; Figure 4B), were not counter-selected in the memory compartment (Figures 4C–E and S3).
Figure 4. Counter-selection by IgG1 BCR for memory cell fate.
(A and B) Diagrams of allele combinations used to assess IgG1-mediated counter-selection in the memory cell fate in C–E. (C and D) IgMa and IgMb were examined by flow cytometry 21 days after immunization with APC in CFA in the antigen-specific GC (C) and memory (D) compartments of indicated mice. (E) Fraction of antigen-specific YFP+ memory B cells expressing the Ighb allotype in indicated mice 40 days after immunization with APC in CFA. Lines represent means. (F) Mean number of antigen-specific GC (upper panels) and YFP+ memory (lower panels) B cells of indicated genotype per 106 total B cells of the donor genotype in bone marrow chimeras immunized with PE in CFA. Results represent 3 independent experiments with a total of 8–10 mice of each genotype in (C) and (D); two independent experiments with a total of 6–8 mice total of each genotype in (E); and two independent experiments with a total of 6–7 mice per group at each time point in (F). Error bars = SEM. ** p = 0.0022; *** p ≤ 0.0007. Two-tailed Mann-Whitney test was used to assess significance. See also Figure S3.
To corroborate these findings in an independent system, and to examine the kinetics of counter-selection of IgG1 memory cells, we generated competitive bone marrow chimeras. In control chimeras, neither of the two Igh alleles were able to class switch (50% Aicdacre R26LSL-YFP; 50% Aicdacre Ighμ-I R26LSL-YFP); neither of these B cell subsets expressed AID protein, and Ighμ-I could not undergo cre-mediated class switching. In experimental chimeras, 50% of the cells were able to class switch to IgG1 (50% Aicdacre Igh96K R26LSL-YFP; 50% Aicdacre Ighμ-I R26LSL-YFP), and counter-selection against IgG1+ B cells would result in a decrease in the frequency of Aicdacre Igh96K R26LSL-YFP B cells (CD45.2+) in the memory pool.
Chimeras were immunized with PE in CFA and antigen-specific B cells were enumerated over time. On day 10 after immunization, cells that were and were not able to undergo CSR were equally represented in the GC and YFP+ memory compartment (Figure 4F). In contrast, by day 32, when memory and GC populations had undergone significant expansion, B cells able to class switch became under-represented in the memory pool but not in the GC, an effect that persisted even 70 days after immunization (Figure 4F). Notably, counter-selection became most prominent between 20 and 32 days after immunization, excluding a major contribution by the extrafollicular B cell response, which resolves earlier (Jacob et al., 1991; Liu et al., 1991; Taylor et al., 2012b). We conclude that, independent of SHM or AID expression, the class-switched BCR biases the fate choice made by B cells entering the long-lived compartment of B lineage cells.
AID expression reduces the longevity of memory B cells
The persistence of memory cells is essential for humoral immunity (McHeyzer-Williams et al., 2012; Tarlinton and Good-Jacobson, 2013). Given that IgM+ memory cells that have relatively few mutations persist longer than IgG+ memory cells that have a higher load of hypermutation (Pape et al., 2011), we asked whether SHM or CSR have a role in memory cell persistence.
To address this question, we tracked long-term memory B cell responses in two groups of mice: (1) Aicdacre/+ R26LSL-YFP mice, in which activated B cells that express AID protein are marked by YFP expression; (2) Aicdacre/− Igh96K/+ R26LSL-YFP mice, in which B cells could not express AID protein but could nevertheless undergo CSR to IgG1 and were marked by YFP expression when they expressed cre from the Aicda locus.
GC responses in both sets of mice showed similar kinetics, peaking between 2–5 weeks after immunization and subsiding by 125 days (Figure 5A). In addition, nearly all antigen-specific IgG1+ memory cells in both groups of mice were YFP+. In contrast, only 16% and 22% of IgM+ memory cells were YFP+ in Aicdacre/+ R26LSL-YFP and Aicdacre/− Igh96K/+ R26LSL-YFP, respectively (Figure S4A). The majority of IgM+ YFP−memory B cells were bona fide memory cells because they were significantly expanded in number compared with naïve animals (Figure S4B). However, it remains possible that a minor subset of this population may not have responded to the immunization.
Consistent with previous observations (Pape et al., 2011), IgM+ memory cells that had not expressed AID (YFP−), which represent most of the IgM+ memory population, did not appreciably decline for 562 days after immunization in both sets of mice (Figure S4C). In contrast, AID-experienced (YFP+) memory cells in Aicdacre/+ R26LSL-YFP mice underwent a long-term decline (Figures 5B and 5C). As noted earlier, the fraction of IgM+ and IgG1+ cells within the surviving AID-experienced memory population did not change over time, indicating that the negative effect of AID on memory cell persistence was independent of BCR isotype (Figure 5D). Furthermore, a history of AID protein expression per se contributed to the decline of memory cells because YFP-marked IgM+ and IgG1+ memory cells generated in Aicdacre/− Igh96K/+ R26LSL-YFP mice, which transcribed the Aicda locus but did not express AID protein, persisted at far higher frequencies throughout the period of observation (Figures 5B and 5C and Figure S4D). Even after GC responses had dissipated in both sets of mice (day 125; Figure 5A), YFP+ memory cells continued to decline at a faster rate in Aicdacre/+ R26LSL-YFP mice than in Aicdacre/− Igh96K/+ R26LSL-YFP mice (Figure S4E). Thus, differences in GC kinetics could not account for the differences in memory cell longevity observed. In contrast to the memory cells, antigen-specific BMPCs of both isotypes persisted similarly in both genotypes of mice (Figures 5E and 5F). We conclude that AID expression negatively affects the persistence of memory B cells in a manner that is independent of class switching, suggesting that SHM may alter memory B cell longevity.
Memory B cells with polyreactive BCRs have diminished longevity
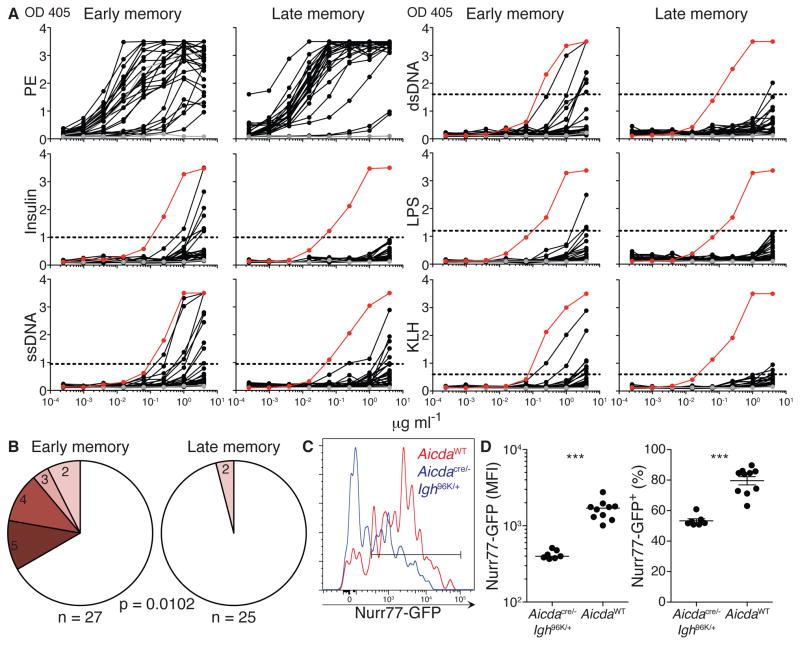
A large proportion of nascent B cells carry polyreactive antibodies, but this reactivity is removed from the repertoire at 2 distinct checkpoints during development (Wardemann et al., 2003). During the GC response, up to 40% of B cells reacquire polyreactivity by SHM (Koelsch et al., 2007; Mouquet et al., 2010; Tiller et al., 2007). To determine whether the reacquisition of polyreactivity accounts for the long-term decline of AID-experienced memory B cells, we cloned antibodies from single antigen-specific IgG1+ memory B cells at early (day 31) and late (day 347+) time points after immunization. Early and late memory antibodies that bound to the cognate antigen (PE; Figure 6A) were tested for polyreactivity. In healthy humans, a substantial fraction of non-specific IgG+ memory B cells are polyreactive (Tiller et al., 2007). Consistent with these findings, ~33% of antigen-specific early memory antibodies were highly polyreactivity (Figure 6A and 6B). In contrast, late memory B cells carried antibodies that were devoid of such reactivity (Figure 6A and 6B; p = 0.0102). As might be expected for cells that acquired polyreactivity, Nurr77-GFP expression was significantly higher among IgG1+ memory B cells that had expressed AID than among those generated in the absence of AID (Figure 6C and 6D). We conclude that AID expression is associated with increased polyreactivity, and that this reactivity is removed from the memory compartment over time.
Figure 6. Selection against memory cell polyreactivity.
(A) ELISAs measuring binding of early and late memory antibodies to PE, insulin, ssDNA, dsDNA, LPS, and KLH. Horizontal lines show cut-off values for positive reactivity. Grey lines represent negative control antibody mGO53; red lines represent positive control antibody ED38 (Meffre et al., 2004). (B) Pie charts of early and late memory antibodies showing fraction of antibodies that react with 2, 3, 4, or all 5 antigens tested in polyreactivity ELISAs. P value compares fraction of antibodies that bind to ≥ 3 structurally distinct antigens. (C) Nurr77-GFP expression among PE-specific IgG1 memory B cells of indicated genotypes at 32–42 days after immunization. (D) Median fluorescence intensity and fraction of Nurr77-GFP+ cells among PE-specific IgG1 memory B cells. Results are from two independent experiments. 3–5 mice per genotype were used in each experiment in (C) and (D). Each dot represents one animal and line indicates median (left graph) or mean (right graph) in (D). Error bars = SEM. *** p ≤ 0.0007. Fisher’s exact test and two-tailed Mann-Whitney test were used to assess statistical significance in (B) and (D), respectively. See also Figure S5.
Discussion
Humoral immune memory is essential for vaccines and protection against reinfection. Memory depends on B cell clonal expansion, differentiation, and long-term persistence of antigen-specific cells (McHeyzer-Williams et al., 2012; Rajewsky, 1996; Tarlinton and Good-Jacobson, 2013; Taylor et al., 2012a). Despite their importance, the roles of SHM and CSR in governing the development and longevity of memory in the antibody system are not well defined. Here we have addressed these issues by genetically separating SHM and CSR, which normally occur in parallel, thereby revealing their independent contributions to humoral immune memory.
The antibody response includes two waves of B cell differentiation. The extrafollicular B cell response generates an early wave of plasmablasts that are short-lived, and memory B cells that produce germline-encoded or sparsely-mutated antibodies. Concomitantly, the GC response forms and persists for up to several weeks after the extrafollicular response subsides (Victora et al., 2012). As a result, the GC supplies an essential source of somatically hypermutated and class-switched B lineage cells that enter the memory and plasma cell pools.
Within the GC, somatic antibody mutants are iteratively selected in an affinity-dependent manner. GC selection is mediated by BCR-dependent uptake of antigen for presentation as peptide-MHCII to CD4+ T follicular helper cells (Allen et al., 2007b; Shulman et al., 2014; Victora et al., 2010). High affinity clones thereby receive greater T cell help and consequently reside for a longer period of time in the DZ where they undergo accelerated cell cycles, allowing such clones to expand within the population (Gitlin et al., 2015; Gitlin et al., 2014). Although the BCR plays a key role in this process, by capturing and internalizing antigen, the contribution of BCR isotype and signaling to GC dynamics and output remain to be defined.
A multitude of signals can induce isotype-switching, including those derived from cytokines, toll-like receptor ligands, CD40 activation and BCR cross-linking (Stavnezer et al., 2008). Moreover, high affinity B cells may preferentially elicit many of these signals. Distinguishing the effects of these signal(s) and affinity-enhancing mutations, both of which may precede and lead to class switching, from the effect of the class-switched BCR itself has been difficult. For example, the BMPC compartment is enriched in both affinity-enhancing mutations and class-switched cells (Slifka et al., 1995; Smith et al., 1997). Thus, whether class-switching simply occurred en passant with affinity-enhancing mutations and other signal(s) that mediated selection into the BMPC pool, or whether the class-switched BCR per se played a deterministic role in BMPC formation, remained unclear.
Using a polyclonal system in which class-switching is mediated by cre recombinase in the absence of SHM, we found that class switching to IgG1 is sufficient to bias B cell fate choice into the long-lived compartment of effector cells without affecting expansion in the GC. It is possible that differences in germline variable gene usage could emerge among Igh96K-expressing and IghWT-expressing B cells in Aicdacre/− Igh96K/+ R26LSL-YFP mice. However, any such differences would necessarily arise due to the altered isotype of Igh96K-expressing B cells, since no other differences exist between these two types of allelically excluded B cells within the same mouse. Moreover, Igh96K-expressing B cells are unlikely to selectively utilize higher affinity germline variable genes, since these B cells had similar profiles of cognate antigen-binding in the GC and did not outcompete B cells that did not class switch.
Whereas class-switched B cells were positively selected into the BMPC pool, they were counter-selected from the memory compartment. Once the memory and BMPC pools had formed, the fractions of IgM and IgG1 cells within these compartments were stable for ≥ 1 year, suggesting that the class-switched BCR affects entry into, rather than persistence within, these compartments. The role of isotype in fate choice is likely to have physiologic relevance since the antibodies encoded by isotype-switched B cells possess unique Fc-mediated effector functions that are particularly well-suited for the antibody-secreting compartment of BMPCs. In contrast, IgM+ B cells may be better suited as a source of memory cells that can re-diversify their antibodies and subsequently class switch during secondary responses to antigenically related pathogens (Dogan et al., 2009; Pape et al., 2011; Purtha et al., 2011).
In addition to altering the membrane external constant region, isotype switching alters the cytoplasmic tail of the BCR in an evolutionarily conserved and isotype-specific manner (Achatz et al., 1997; Kaisho et al., 1997; Reth, 1992). In vitro experiments indicate that the IgG cytoplasmic tail alters the signal transducing effector molecules that associate with the BCR (Engels et al., 2009; Engels et al., 2014; Liu et al., 2012; Wakabayashi et al., 2002), increases BCR clustering (Liu et al., 2010), and crosslinking-induced Ca2+ responses (Horikawa et al., 2007; Waisman et al., 2007). We found that the IgG1 BCR differentially regulated signaling in the GC LZ as revealed by altered Nur77-GFP expression and chemokine receptor expression, suggesting a mechanism for isotype-specific B cell exit from the GC and preferential entry into the BMPC fate. Consistent with the idea that isotype switching per se can alter B cell fate, IgE expression has been associated with shortened residence in the GC and rapid differentiation into short-lived plasma cells, without prominent contribution to the memory or BMPC pools (He et al., 2013; Yang et al., 2012). Moreover, transgenic IgA expression has also been described to promote plasma cell differentiation (Duchez et al., 2010). Thus, isotype-specific BCR signaling appears to generally contribute to GC B cell fate decisions, in a manner unique to each isotype.
The maintenance of serum antibody varies considerably among different vaccines (Amanna et al., 2007). However, characterizing memory B cells has been challenging, especially in human studies that frequently rely on in vitro techniques to enumerate such cells by eliciting antibody secretion (Amanna et al., 2007; Crotty et al., 2004; Crotty et al., 2003). Nevertheless, the available data indicates that ~10% of the memory cell population survives 10 years after immunization and is then stably maintained (Crotty et al., 2003; Taylor et al., 2012a; Yu et al., 2008).
In mice, the persistence of memory B cells is independent of both helper T cells (Vieira and Rajewsky, 1990) and the persistence of cognate antigen (Anderson et al., 2006; Maruyama et al., 2000). However, longevity varies among memory B cells expressing different isotypes: IgM+ memory B cells, which as a group have a lesser amount of SHM, were found to be longer-lived than IgG+ memory B cells, which are more mutated, raising the possibility that memory cell persistence may be controlled by BCR isotype and/or SHM (Pape et al., 2011). We found that the rate of memory B cell decline was dependent on AID expression but not on CSR. Superficially, our findings may appear to be at odds with those of Jenkins and colleagues, but in fact they are entirely in keeping with their report of a difference in longevity between IgM+ and IgG+ memory B cells because IgG+ cells are uniformly AID-experienced while only a fraction of IgM+ memory B cells have expressed high amounts of AID (Dogan et al., 2009; Pape et al., 2011). More importantly, our experiments provide a mechanistic explanation for the observation that memory B cells that have expressed high amounts of AID have a shorter half-life, thereby revealing a previously undocumented tolerance checkpoint in the antibody system.
The nascent B cell repertoire contains a substantial fraction of antibodies with poly- and self-reactivity, both of which are removed at two distinct checkpoints during B cell maturation (Wardemann et al., 2003). However, up to 40% of antibodies regain these features by SHM in the GC (Tiller et al., 2007). Although the most highly self-reactive of these antibodies are eliminated in the GC upon encountering autoantigen (Chan et al., 2012), polyreactive antibody mutants frequently enter the memory pool (Koelsch et al., 2007; Mouquet et al., 2010; Tiller et al., 2007). Moreover, SHM is a frequent source of autoantibodies in diseases such as systemic lupus erythematosus (Mietzner et al., 2008). Therefore, while a checkpoint against autoreactivity exists in the GC, it appears to eliminate only the most autoreactive antibodies and/or GC-expressed autoantigens (Chan et al., 2012; Pulendran et al., 1995; Shokat and Goodnow, 1995). Notably, mature B cells that are transgenically engineered to encounter a high concentration of autoantigen in the periphery are rapidly eliminated by clonal deletion (Lam and Rajewsky, 1998; Ota et al., 2011). Whether autoreactive specificities that enter the memory pool are culled by a similar mechanism remains to be determined. We conclude that differential persistence of autoreactive antibody mutants in the memory compartment represents a tolerance checkpoint in the antibody system.
Experimental Procedures
Mice
Aicdacre, Aicda−, R26LSL-YFP, Ighμ-I, and Igh96K were described previously (Bothmer et al., 2010; Muramatsu et al., 2000; Robbiani et al., 2008; Srinivas et al., 2001). Nurr77-GFP mice were obtained from A. Weiss (Zikherman et al., 2012). B6.SJL mice and B6.Cg-Gpi1a Thy1a Igha/J were obtained from Jackson Laboratories. Bone marrow chimeras were made for experiments in Figure 4F and for experiments involving Nurr77-GFP transgenic cells in Figure 3B–D and Figure 6C and 6D. To generate chimeras, C57BL/6 or B6.SJL males were irradiated with 2 doses of 525 rad 3 hours apart. 5–10 × 106 bone marrow cells from donor genotypes were injected into recipients, which were analyzed at least 8 weeks later. All experiments were performed with authorization from the Institutional Review Board and the IACUC at The Rockefeller University.
Cell enrichments
Spleen and lymph node cells were purified by forcing tissue through 40 μm mesh into complete RPMI media (Gibco) containing 6% serum. PE-specific B cells were enriched as previously described (Pape et al., 2011; Taylor et al., 2012b). To enrich plasma cells, cell suspensions were made from bone marrow in PBS containing 1 mM EDTA and 2% serum. Anti-CD138-APC (BD Biosciences) and anti-APC microbeads (Militenyi Biotec) were used for enrichment with magnetized LS columns. In figure 3, APC-specific cells were enriched by negative selection using anti-CD4-biotin, anti-CD8-biotin, anti-Gr1-biotin, and anti-Ter119-biotin (eBioscience), streptavidin microbeads (Militenyi Biotec), and magnetized LS columns. Following column-based enrichment, samples were processed for flow cytometry. Cell counts from column-bound and flow-through samples were assessed using AccuCheck counting beads (Invitrogen) and cell numbers from both fractions were evaluated.
Immunizations and treatments
Mice were immunized with 15 μg of PE or cross-linked APC (Prozyme) in a 50 μl emulsion with CFA (Sigma) at a single subcutaneous site at the base of the tail. To examine proliferating cells, mice were injected with 1 mg EdU (Life Technologies) in PBS 2 hours before analysis. Cells were then stained for flow cytometry and processed using a Click-iT EdU-Pacific Blue kit (Life Technologies) according to manufacturers’ instructions.
Flow cytometry and cell sorting
Single cell suspensions were processed in PBS that was maintained at 4°C and contained 2% serum and 1 mM EDTA. Samples were treated at 4°C for 10 minutes with 1 μg/ml of anti-CD16/32 (2.4G2, Bio-X-Cell) and then stained for 25 minutes at 4°C. Antibodies used for flow cytometry were from eBioscience, BD Biosciences, Biolegend, and Life Technologies. Streptavidin-Qdot800 was from Life Technologies and Streptavidin-PerCP-Cy5.5 was from eBioscience. Cell fixation and permeabilization were performed using the Cytofix/Cytoperm kit (BD Biosciences). Samples were analyzed on a BD Fortessa. GC B cells were gated as live, single, and Dump(CD4, CD8, Gr-1, F4/80) − B220+CD38−GL7+ and PE–binding. Memory B cells were gated as live, single, and Dump−B220+CD38+GL7− and PE-binding (Pape et al., 2011; Ridderstad and Tarlinton, 1998; Taylor et al., 2012b). DZ and LZ surface phenotype was assessed using CXCR4 and CD86, as described previously (Victora et al., 2010). BMPCs were gated as live, single, and Dump−CD138+ and R26YFP+ and/or intracellular Ighigh. Antigen-specific BMPCs were identified by surface and intracellular staining with either PE or APC. CD45.1 and CD45.2 allotypic markers were were used to trace donor genotypes in bone marrow chimera experiments. Aicdacre R26LSL-YFP and Aicdacre Igh96K R26LSL-YFP cells were CD45.2+, and Aicdacre Ighμ-I R26LSL-YFP cells were CD45.1+. For RNA sequencing, cells were sorted directly into Trizol LS reagent (Invitrogen) that was maintained at 4°C using a FACS Aria II (Becton Dickinson). For antibody cloning, memory B cells were single-cell sorted into 96 well plates using a FACSAria II (Becton Dickinson). Antibodies from sorted cells were cloned and produced as described previously (Dosenovic et al., 2015).
Gene expression analysis
Purified RNA from sorted samples was processed using the SMARTer Ultra Low Input RNA for Illumina Sequencing kit (Clontech Laboratories). High throughput sequencing was performed using a HiSeq 2500 (Illumina). In Figure 3D, genes referred to as positive and negative regulators of plasma cell differentiation were curated based on the literature (Nutt et al., 2015). Positive regulators included Irf4, Zbtb20, Pou2af1, Pou2f2, Ikzf3, Batf, Fos, Klf2, and Xbp1. Negative regulators included Irf8, Bcl6, Pax5, Spib, and Mef2c. Chemokine receptors included CXCR4, CXCR5, S1PR1, S1PR2, S1PR3 and S1PR4. A SYBR Green based PCR mix (ThermoFisher Scientific) was used for quantitative PCR analysis. RNA-seq reads were aligned with STAR version 2.3.0 allowing unique alignments and using Mouse Ensembl genes as reference. Differential expression was calculated using Cufflinks with default settings.
ELISA
ELISA for PE binding was performed by overnight coating of high binding 96-well plates (Corning Incorporated) with 50 μl of 4 μg/ml PE in PBS. Plates were then washed with PBS containing 0.05% TWEEN 20 (Sigma) and then blocked with PBS containing 1 % PBS. Monoclonal antibodies were incubated for 1 hour at indicated dilutions in PBS. Plates were then washed and incubated with HRP conjugated anti-mouse IgG (Jackson Immunoresearch). Polyreactivity was assessed using LPS from E. coli (Sigma), human insulin (Sigma), dsDNA (Sigma) and ssDNA (prepared from dsDNA by heating at 95°C for 30 minutes), and KLH (Sigma) (Mouquet et al., 2010). High binding plates were incubated overnight with 50 μl of ssDNA, dsDNA, LPS, and KLH at 10 μg/ml each and insulin at 5 μg/ml. Plates were then washed with PBS containing 0.001% TWEEN 20. Plates were blocked for 2 hours at room temperature in PBS containing 1 mM EDTA, 0.05% TWEEN 20 and 2.5% bovine serum albumin (Sigma). After washing, monoclonal antibodies were incubated for 2 hours at indicated dilutions in PBS. Plates were then washed and incubated with HRP conjugated anti-mouse IgG (Jackson Immunoresearch) for 1 hour. After washing, plates were developed by adding HRP substrate (Life Technologies). Absorbance was measured at 405 nm.
Statistical analyses
Statistical significance was assessed using the tests indicated in the figures using Prism software v. 5.0 (Graphpad).
Supplementary Material
Acknowledgments
We thank T. Eisenreich and S. Hinklein for help with mouse colony management; S. Bournazos for reagents; M.K. Jenkins, J.J. Taylor, and K.A. Pape for generous advice; E.S. Gitlin, H. Mouquet, D. Mucida, G.D. Victora, and C.T. Mayer for discussion and advice; D.F. Robbiani, P.C. Rommel, A. Weiss, and J. Zikherman for mice; J. Golijanin, S. Ackerman, and K. Yao for technical help; B. Zhang and C. Zhao and the Rockefeller University Genomics Resource Center for assistance with high-throughput sequencing; K. Velinzon and N. Thomas for assistance with cell sorting; and all members of the Nussenzweig laboratory for discussion. Supported by NIH Medical Scientist Training Program grant T32GM07739 and National Institute of Allergy and Infectious Diseases, NIH, grant 1F30AI109903-01 (A.D.G.); grant UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award program (L.v.B.); NIH grants AI037526-19 and AI072529-06 (M.C.N.); and the NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-01 (M.C.N). Z.S. is a Human Frontiers of Science Program fellow (reference LT000340/2011-L). M.C.N. is an HHMI investigator.
Footnotes
Accession Numbers
The gene expression data have been deposited in the GEO database under accession number GSE76866.
Author Contributions
A.D.G. and M.C.N. conceived the study, interpreted the data, and wrote the manuscript. A.D.G. and Z.S. performed experiments. L.v.B. and A.G. performed antibody cloning and production. T.Y.O. performed bioinformatic analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achatz G, Nitschke L, Lamers MC. Effect of transmembrane and cytoplasmic domains of IgE on the IgE response. Science. 1997;276:409–411. doi: 10.1126/science.276.5311.409. [DOI] [PubMed] [Google Scholar]
- Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007a;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007b;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- Anderson SM, Hannum LG, Shlomchik MJ. Memory B cell survival and function in the absence of secreted antibody and immune complexes on follicular dendritic cells. J Immunol. 2006;176:4515–4519. doi: 10.4049/jimmunol.176.8.4515. [DOI] [PubMed] [Google Scholar]
- Ansel KM, Ngo VN, Hyman PL, Luther SA, Forster R, Sedgwick JD, Browning JL, Lipp M, Cyster JG. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, DiLillo DJ, Ravetch JV. The role of Fc-FcgammaR interactions in IgG-mediated microbial neutralization. J Exp Med. 2015;212:1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebra JJ, Colberg JE, Dray S. Rabbit lymphoid cells differentiated with respect to alpha-, gamma-, and mu-heavy polypeptide chains and to allotypic markers Aa1 and Aa2. J Exp Med. 1966;123:547–558. doi: 10.1084/jem.123.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TD, Wood K, Hermes JR, Butt D, Jolly CJ, Basten A, Brink R. Elimination of germinal-center-derived self-reactive B cells is governed by the location and concentration of self-antigen. Immunity. 2012;37:893–904. doi: 10.1016/j.immuni.2012.07.017. [DOI] [PubMed] [Google Scholar]
- Crotty S, Aubert RD, Glidewell J, Ahmed R. Tracking human antigen-specific memory B cells: a sensitive and generalized ELISPOT system. J Immunol Methods. 2004;286:111–122. doi: 10.1016/j.jim.2003.12.015. [DOI] [PubMed] [Google Scholar]
- Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- Cyster JG. Homing of antibody secreting cells. Immunol Rev. 2003;194:48–60. doi: 10.1034/j.1600-065x.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, Reynaud CA, Weill JC. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchez S, Amin R, Cogne N, Delpy L, Sirac C, Pascal V, Corthesy B, Cogne M. Premature replacement of mu with alpha immunoglobulin chains impairs lymphopoiesis and mucosal homing but promotes plasma cell maturation. Proc Natl Acad Sci U S A. 2010;107:3064–3069. doi: 10.1073/pnas.0912393107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen HN, Siskind GW. Variations in Affinities of Antibodies during the Immune Response. Biochemistry. 1964;3:996–1008. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- Engels N, Konig LM, Heemann C, Lutz J, Tsubata T, Griep S, Schrader V, Wienands J. Recruitment of the cytoplasmic adaptor Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells. Nat Immunol. 2009;10:1018–1025. doi: 10.1038/ni.1764. [DOI] [PubMed] [Google Scholar]
- Engels N, Konig LM, Schulze W, Radtke D, Vanshylla K, Lutz J, Winkler TH, Nitschke L, Wienands J. The immunoglobulin tail tyrosine motif upgrades memory-type BCRs by incorporating a Grb2-Btk signalling module. Nat Commun. 2014;5:5456. doi: 10.1038/ncomms6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–1047. doi: 10.1016/s0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- Gitlin AD, Mayer CT, Oliveira TY, Shulman Z, Jones MJ, Koren A, Nussenzweig MC. HUMORAL IMMUNITY. T cell help controls the speed of the cell cycle in germinal center B cells. Science. 2015;349:643–646. doi: 10.1126/science.aac4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good-Jacobson KL, O’Donnell K, Belz GT, Nutt SL, Tarlinton DM. c-Myb is required for plasma cell migration to bone marrow after immunization or infection. J Exp Med. 2015;212:1001–1009. doi: 10.1084/jem.20150191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JA, Cyster JG. S1PR2 links germinal center confinement and growth regulation. Immunol Rev. 2012;247:36–51. doi: 10.1111/j.1600-065X.2012.01114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JA, Suzuki K, Cho B, Willison LD, Palmer D, Allen CD, Schmidt TH, Xu Y, Proia RL, Coughlin SR, et al. The sphingosine 1-phosphate receptor S1P(2) maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat Immunol. 2011;12:672–680. doi: 10.1038/ni.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Hyman PL, Lu TT, Ngo VN, Bidgol A, Suzuki G, Zou YR, Littman DR, Cyster JG. A coordinated change in chemokine responsiveness guides plasma cell movements. J Exp Med. 2001;194:45–56. doi: 10.1084/jem.194.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JS, Meyer-Hermann M, Xiangying D, Zuan LY, Jones LA, Ramakrishna L, de Vries VC, Dolpady J, Aina H, Joseph S, et al. The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response. J Exp Med. 2013;210:2755–2771. doi: 10.1084/jem.20131539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K, Martin SW, Pogue SL, Silver K, Peng K, Takatsu K, Goodnow CC. Enhancement and suppression of signaling by the conserved tail of IgG memory-type B cell antigen receptors. J Exp Med. 2007;204:759–769. doi: 10.1084/jem.20061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Kassir R, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. I. The architecture and dynamics of responding cell populations. J Exp Med. 1991;173:1165–1175. doi: 10.1084/jem.173.5.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Haynes NM, Xu Y, Nutt SL, Allende ML, Proia RL, Cyster JG. Plasma cell S1P1 expression determines secondary lymphoid organ retention versus bone marrow tropism. J Exp Med. 2006;203:2683–2690. doi: 10.1084/jem.20061289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaisho T, Schwenk F, Rajewsky K. The roles of gamma 1 heavy chain membrane expression and cytoplasmic tail in IgG1 responses. Science. 1997;276:412–415. doi: 10.1126/science.276.5311.412. [DOI] [PubMed] [Google Scholar]
- Koelsch K, Zheng NY, Zhang Q, Duty A, Helms C, Mathias MD, Jared M, Smith K, Capra JD, Wilson PC. Mature B cells class switched to IgD are autoreactive in healthy individuals. J Clin Invest. 2007;117:1558–1565. doi: 10.1172/JCI27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kometani K, Nakagawa R, Shinnakasu R, Kaji T, Rybouchkin A, Moriyama S, Furukawa K, Koseki H, Takemori T, Kurosaki T. Repression of the transcription factor Bach2 contributes to predisposition of IgG1 memory B cells toward plasma cell differentiation. Immunity. 2013;39:136–147. doi: 10.1016/j.immuni.2013.06.011. [DOI] [PubMed] [Google Scholar]
- Lam KP, Rajewsky K. Rapid elimination of mature autoreactive B cells demonstrated by Cre-induced change in B cell antigen receptor specificity in vivo. Proc Natl Acad Sci U S A. 1998;95:13171–13175. doi: 10.1073/pnas.95.22.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Chen E, Zhao XW, Wan ZP, Gao YR, Davey A, Huang E, Zhang L, Crocetti J, Sandoval G, et al. The scaffolding protein synapse-associated protein 97 is required for enhanced signaling through isotype-switched IgG memory B cell receptors. Sci Signal. 2012;5:ra54. doi: 10.1126/scisignal.2002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Meckel T, Tolar P, Sohn HW, Pierce SK. Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity. 2010;32:778–789. doi: 10.1016/j.immuni.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat Immunol. 2002;3:182–188. doi: 10.1038/ni752. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Lam KP, Rajewsky K. Memory B-cell persistence is independent of persisting immunizing antigen. Nature. 2000;407:636–642. doi: 10.1038/35036600. [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E, Schaefer A, Wardemann H, Wilson P, Davis E, Nussenzweig MC. Surrogate light chain expressing human peripheral B cells produce self-reactive antibodies. J Exp Med. 2004;199:145–150. doi: 10.1084/jem.20031550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K, Gonzalez JB, Pascual V, Stichweh D, Wardemann H, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci U S A. 2008;105:9727–9732. doi: 10.1073/pnas.0803644105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller J, Matloubian M, Zikherman J. Cutting edge: An in vivo reporter reveals active B cell receptor signaling in the germinal center. J Immunol. 2015;194:2993–2997. doi: 10.4049/jimmunol.1403086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- Ota T, Ota M, Duong BH, Gavin AL, Nemazee D. Liver-expressed Igkappa superantigen induces tolerance of polyclonal B cells by clonal deletion not kappa to lambda receptor editing. J Exp Med. 2011;208:617–629. doi: 10.1084/jem.20102265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape KA, Taylor JJ, Maul RW, Gearhart PJ, Jenkins MK. Different B cell populations mediate early and late memory during an endogenous immune response. Science. 2011;331:1203–1207. doi: 10.1126/science.1201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernis B, Chiappino G, Kelus AS, Gell PG. Cellular localization of immunoglobulins with different allotypic specificities in rabbit lymphoid tissues. J Exp Med. 1965;122:853–876. doi: 10.1084/jem.122.5.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Paus D, Chan TD, Turner ML, Nutt SL, Basten A, Brink R. High affinity germinal center B cells are actively selected into the plasma cell compartment. J Exp Med. 2006;203:2419–2424. doi: 10.1084/jem.20061254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue SL, Goodnow CC. Gene dose-dependent maturation and receptor editing of B cells expressing immunoglobulin (Ig)G1 or IgM/IgG1 tail antigen receptors. J Exp Med. 2000;191:1031–1044. doi: 10.1084/jem.191.6.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Kannourakis G, Nouri S, Smith KG, Nossal GJ. Soluble antigen can cause enhanced apoptosis of germinal-centre B cells. Nature. 1995;375:331–334. doi: 10.1038/375331a0. [DOI] [PubMed] [Google Scholar]
- Purtha WE, Tedder TF, Johnson S, Bhattacharya D, Diamond MS. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J Exp Med. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Reth M. Antigen receptors on B lymphocytes. Annu Rev Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- Ridderstad A, Tarlinton DM. Kinetics of establishing the memory B cell population as revealed by CD38 expression. J Immunol. 1998;160:4688–4695. [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth PE, Doglio L, Manz JT, Kim JY, Lo D, Storb U. Immunoglobulin gamma 2b transgenes inhibit heavy chain gene rearrangement, but cannot promote B cell development. J Exp Med. 1993;178:2007–2021. doi: 10.1084/jem.178.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth PE, Kurtz B, Lo D, Storb U. lambda 5, but not mu, is required for B cell maturation in a unique gamma 2b transgenic mouse line. J Exp Med. 1995;181:1059–1070. doi: 10.1084/jem.181.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Shokat KM, Goodnow CC. Antigen-induced B-cell death and elimination during germinal-centre immune responses. Nature. 1995;375:334–338. doi: 10.1038/375334a0. [DOI] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, Nussenzweig MC. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka MK, Matloubian M, Ahmed R. Bone marrow is a major site of long-term antibody production after acute viral infection. J Virol. 1995;69:1895–1902. doi: 10.1128/jvi.69.3.1895-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KG, Light A, Nossal GJ, Tarlinton DM. The extent of affinity maturation differs between the memory and antibody-forming cell compartments in the primary immune response. EMBO J. 1997;16:2996–3006. doi: 10.1093/emboj/16.11.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- Tarlinton D, Good-Jacobson K. Diversity among memory B cells: origin, consequences, and utility. Science. 2013;341:1205–1211. doi: 10.1126/science.1241146. [DOI] [PubMed] [Google Scholar]
- Taylor JJ, Jenkins MK, Pape KA. Heterogeneity in the differentiation and function of memory B cells. Trends Immunol. 2012a;33:590–597. doi: 10.1016/j.it.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Pape KA, Jenkins MK. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J Exp Med. 2012b;209:597–606. doi: 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JJ, Pape KA, Steach HR, Jenkins MK. Humoral immunity. Apoptosis and antigen affinity limit effector cell differentiation of a single naive B cell. Science. 2015;347:784–787. doi: 10.1126/science.aaa1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P, Rajewsky K. Persistence of memory B cells in mice deprived of T cell help. Int Immunol. 1990;2:487–494. doi: 10.1093/intimm/2.6.487. [DOI] [PubMed] [Google Scholar]
- Waisman A, Kraus M, Seagal J, Ghosh S, Melamed D, Song J, Sasaki Y, Classen S, Lutz C, Brombacher F, et al. IgG1 B cell receptor signaling is inhibited by CD22 and promotes the development of B cells whose survival is less dependent on Ig alpha/beta. J Exp Med. 2007;204:747–758. doi: 10.1084/jem.20062024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi C, Adachi T, Wienands J, Tsubata T. A distinct signaling pathway used by the IgG-containing B cell antigen receptor. Science. 2002;298:2392–2395. doi: 10.1126/science.1076963. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Yang Z, Sullivan BM, Allen CD. Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity. 2012;36:857–872. doi: 10.1016/j.immuni.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature. 2008;455:532–536. doi: 10.1038/nature07231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zikherman J, Parameswaran R, Weiss A. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature. 2012;489:160–164. doi: 10.1038/nature11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.