Summary
Intracellular membrane fusion is mediated by dynamic assembly and disassembly of soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (SNAP) receptors (SNAREs). α-SNAP guides NSF to disassemble SNARE complexes after membrane fusion. Recent experiments showed that α-SNAP also dramatically enhances SNARE assembly and membrane fusion. How α-SNAP plays roles in these opposing activities is not known. Here, we examined the effect of α-SNAP on the step-wise assembly of the synaptic SNARE complex using optical tweezers. We found that α-SNAP destabilized the linker domain (LD) of the SNARE complex but stabilized its C-terminal domain (CTD) through a conformational selection mechanism. In contrast, α-SNAP minimally affected assembly of the SNARE N-terminal domain (NTD), indicating that α-SNAP barely bound the partially assembled trans-SNARE complex. Thus, α-SNAP recognizes the folded CTD for SNARE disassembly with NSF and subtly modulates membrane fusion by altering the stabilities of the SNARE CTD and LD.
Graphical abstract
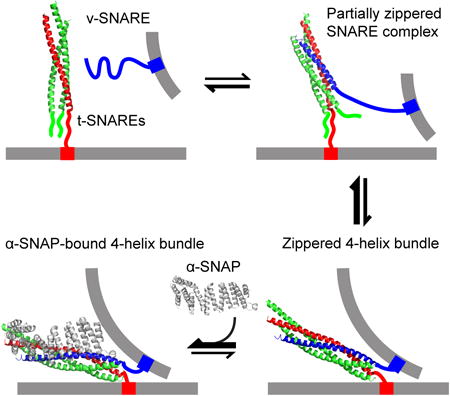
Introduction
Alpha SNAP is a ubiquitous protein essential for membrane fusion in eukaryotes (Clary et al., 1990). Deletion of α-SNAP is lethal in mice, and mutation of α-SNAP causes hydrocephaly (Chae et al., 2004). Changes in the expression level of α-SNAP have been associated with neurological disorders, cancer, and diabetes (Andreeva et al., 2006). α-SNAP mainly serves as an adaptor protein for the hexameric AAA+ ATPase, NSF, to disassemble the fully assembled cis-SNARE complex after membrane fusion has occurred, thereby recycling individual SNARE proteins for the next round of fusion (Sollner et al., 1993). NSF and α-SNAP disassemble nearly all fully assembled cis-SNARE complexes in the cell (Zhao et al., 2015) as well as some partially- and mis-assembled SNARE complexes in vitro (Vivona et al., 2013). However, NSF and α-SNAP do not disassemble the trans-SNARE complex that bridges two opposing membranes (Lobingier et al., 2014; Weber et al., 2000), an essential intermediate for calcium-triggered synaptic exocytosis (Sudhof and Rothman, 2009). Because NSF does not directly interact with SNARE proteins (Sollner et al., 1993; Zhao et al., 2015), these observations reveal that α-SNAP, either alone or together with other proteins, recognizes certain global and common features unique to cis-SNARE complexes. Whereas all cis-SNARE complexes exhibit a shared four-helix bundle structure (Stein et al., 2009; Sutton et al., 1998), evidence suggests that trans-SNARE complexes are partially assembled (Gao et al., 2012; Zorman et al., 2014). Accordingly, Zhao et al. proposed that the partially assembled SNARE complexes may not be a favored substrate for α-SNAP and NSF (Zhao et al., 2015). Alternatively, Sec1/Munc18 and other proteins are suggested to protect trans-SNARE complexes from NSF- and α-SNAP-mediated disassembly (Lobingier et al., 2014; Sollner et al., 1993). In particular, the yeast homotypic fusion and vacuole protein sorting (HOPS) complex prevents disassembly of vacuolar trans-SNARE complexes (Xu et al., 2010). α-SNAP alone primarily binds cytoplasmic SNARE complexes in a 1:1 stoichiometry. However, in the presence of membranes or NSF, α-SNAP is found to associate with SNARE complexes in different stoichiometries, ranging from one to four α-SNAP molecules per SNARE complex (Marz et al., 2003; Shah et al., 2015; Wimmer et al., 2001; Zhao et al., 2015; Zhou et al., 2015). How α-SNAP binds SNARE complexes with different stoichiometries remains mysterious.
Recently, α-SNAP was reported to regulate SNARE assembly and membrane fusion. Park et al. found that α-SNAP arrests the trans-SNARE complex in a half-zippered state, thereby preventing fusion in a membrane curvature-dependent manner (Park et al., 2014). Ryu et al. observed that α-SNAP destabilizes the C-terminus of the SNARE complex but not enough to form the half-zippered state (Ryu et al., 2015). In contrast, Zick et al. found that the yeast α-SNAP homologue, Sec17, strongly promotes trans-SNARE folding and triggers membrane fusion (Zick et al., 2015). However, the mechanism by which α-SNAP directly attenuates or enhances SNARE zippering is not fully understood.
Studying regulated SNARE assembly and disassembly is difficult using traditional ensemble-based experimental approaches (Gao et al., 2012). Therefore, we developed a single-molecule approach using high-resolution optical tweezers to characterize the energetics and kinetics of SNARE assembly (Gao et al., 2012; Ma et al., 2016; Zorman et al., 2014). We found that synaptic SNARE complexes zip up in at least three stages, each with potentially distinct functions: while NTD association initiates SNARE assembly and is responsible for vesicle docking, rapid CTD zippering directly drives membrane fusion. In addition, the SNARE complexes that are responsible for different membrane trafficking pathways in multiple different species share the same assembly pathway and kinetics (Zorman et al., 2014). In this work, we extended the single-molecule approach to pinpoint the role of α-SNAP during synaptic SNARE complex assembly. We find that α-SNAP attenuates LD zippering, but enhances CTD zippering, via conformational selection.
Results
α-SNAP Destabilizes the LD
Synaptic SNARE complexes consist of syntaxin 1A and SNAP-25B, which are located on plasma membranes (t-SNAREs), and VAMP2, which is anchored in synaptic vesicles (v-SNARE) (Sollner et al., 1993) (Figure 1A). The fully assembled SNARE complex contains a parallel four-helix bundle (4HB), a two-stranded coiled coil that spans the LD and the transmembrane domain, and an N-terminal regulatory domain (NRD) in syntaxin (Stein et al., 2009; Sutton et al., 1998). The NRD consists of two functionally distinct domains: an N-terminal peptide of ∼15 amino acids and an autonomously folded three-helix bundle called the Habc domain (Shen et al., 2007; Zhou et al., 2013). We pulled a single cytoplasmic SNARE complex from the C-termini of syntaxin and VAMP2 via a 2,260-bp DNA handle (Cecconi et al., 2005; Gao et al., 2012) while their N-termini were crosslinked through a disulfide bridge (Figure 1A). The folding intermediates, energies, and kinetics of the same SNARE construct have recently been determined (Ma et al., 2016).
Figure 1. Experimental Setup and Derived States of SNARE Zippering and α-SNAP Binding.
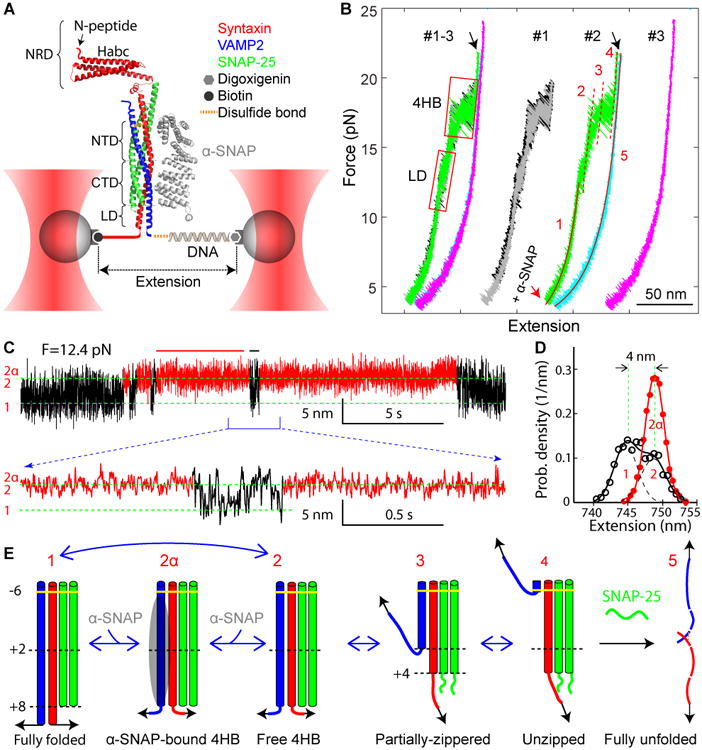
(A) A single SNARE complex was attached to two beads and pulled by moving one optical trap at a speed of 10 nm/s or by fixing the separation between two optical traps. The extension and the tension of the protein-DNA tether were detected at 10 kHz and used to derive conformations, energies, and transition kinetics of SNARE folding intermediates with the help of the crystal structure of the SNARE complex (Rebane et al., 2016). The crystal structure of the yeast α-SNAP homolog Sec17 (Rice and Brunger, 1999) was modeled to bind to the SNARE complex.
(B) Force-extension curves (FECs) of the SNARE-DNA tether obtained by pulling (black, green, and magenta) and relaxing (gray and cyan) a single SNARE complex in three consecutive rounds (#1-3) in the absence of α-SNAP (#1) or in the presence of 5 μM α-SNAP (#2 and #3). The FECs corresponding to individual rounds of pulling and relaxation (#1, #2, and #3) are shifted along the x-axis for comparison and clarity. Regions of the LD and the four-helix bundle (4HB) transitions are marked by red rectangles. The black arrow indicates t-SNARE unfolding. Continuous regions in the FECs #2 were fit by worm-like chain models for the DNA handle and the unfolded polypeptide (red solid lines), revealing conformations of the corresponding SNARE folding states (red numbers, see E). The red dashed lines mark the extension positions of transient intermediate states.
(C) Extension-time trajectories under a constant mean force of 12.4 pN that demonstrate LD transitions in the presence of α-SNAP. A close-up view of the indicated region is shown at the bottom. The α-SNAP-unbound and -bound regions are shown in black and red, respectively. Positions of different SNARE zippering states with their conformations shown in E are marked by green dashed lines. Data were plotted in 333 Hz.
(D) Probability density distributions of the extensions of the α-SNAP-unbound (black) and -bound (red) states corresponding to the regions marked by black and red bars in C. The distributions were fit by double- and single-Gaussian functions (solid lines) with individual Gaussian functions for the former plotted in black dashed lines. States corresponding to the distribution peaks are marked by the associated state numbers as in E.
(E) Conformations of different folding states and their transitions derived from this work and previous work (Gao et al., 2012; Ma et al., 2016). The states are numbered the same throughout the work: the free 4HB state 2, the α-SNAP-bound 4HB state 2α, the partially-zippered state 3, and the unzipped state 4. Individual SNAREs are colored as in A, and α-SNAP is depicted as a gray oval.
We first pulled and then relaxed a single SNARE complex for three consecutive rounds (Figure 1B, #1-3), with α-SNAP added at the end of the first round (Figure 1B). The resultant force-extension curves (FECs) exhibited discrete extension that flickered in two force ranges, one at 11-14 pN and the other at 16-20 pN (Figure 1B, regions marked by red rectangles), which was caused by reversible folding and unfolding of the LD and the 4HB, respectively. Closer inspection showed that the 4HB transition contained two overlapping transitions mediated by a partially zippered state (Figure 1B, state 3 in the green FEC). SNARE assembly and disassembly was fully reversible below a force of ∼22 pN, as is indicated by the overlapping FECs obtained by pulling and relaxing the SNARE complex (black and gray FECs). All of these observations are consistent with our recent report (Ma et al., 2016).
After adding 5 μM α-SNAP to the solution, we found that α-SNAP generally did not alter the FECs or the folding and unfolding pathways of the SNARE complex under these experimental conditions (Figure 2B, compare the green and grey FECs). Further pulling on the SNARE complex to ∼22 pN caused a small rip (Figure 1B, black arrow), indicating irreversible unfolding of the t-SNARE complex and dissociation of the SNAP-25 molecule (Gao et al., 2012; Ma et al., 2016). As a result, the SNARE complex failed to re-assemble when the tension was reduced to a low force, as is shown by the large hysteresis between the FECs (the cyan FEC) and confirmed by subsequent pulling (FEC #3). Thus, we identified at least five different SNARE folding states (Figure 1E).
Figure 2. α-SNAP Stabilizes the Folded SNARE Four-Helix Bundle (4HB).
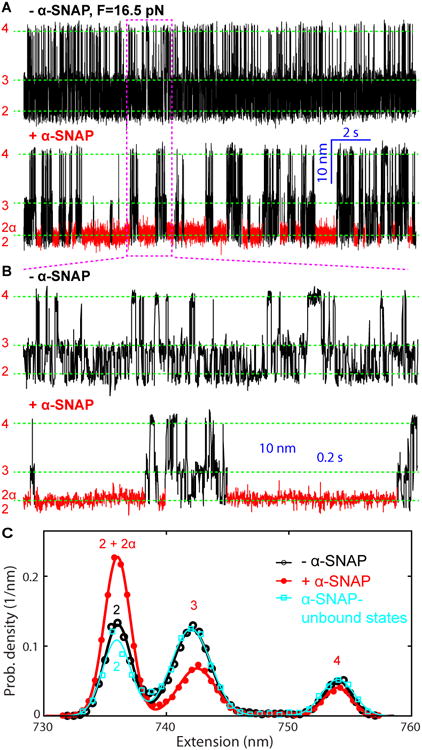
(A) Extension-time trajectories of a single SNARE complex at 16.5 pN mean force in the absence (- α-SNAP) and presence (+ α-SNAP) of 5 μM α-SNAP. The α-SNAP-bound state (2α) is highlighted in red, which was identified by hidden Markov modeling (HMM) shown in Figure 3. Dashed green lines indicate the positions of the different SNARE folding states depicted in Figure 1E.
(B) Close-up views of the region in A marked by the magenta dashed rectangle. The extension trajectories were mean-filtered to 1 kHz and plotted in A and B. More trajectories plotted at 5 kHz are shown in Figure S2.
(C) Probability density distributions of extensions shown in A (symbols) and their best fits with a sum of three Gaussian functions (lines).
We further examined the possible effect of α-SNAP on SNARE folding at higher spatiotemporal resolution by holding a single SNARE complex at a fixed trap separation. We first held the complex in the force region of the LD transition. In the absence of α-SNAP, the LD rapidly unfolded and refolded in a two-state manner with an extension change of 3-5 nm and a lifetime of 1-5 ms for both states, as seen in the extension-time trajectories (Figure S1) (Gao et al., 2012; Ma et al., 2016). In the presence of 5 μM α-SNAP, we found that the fast extension flickering frequently paused in a high extension state (Figure 1C, regions colored in red). The flickering states (regions in black) were indistinguishable from those seen in the absence of α-SNAP, indicating an intact LD transition. The flickering-paused state typically lasted for 0.1-10 seconds in an extension position similar to that of the unfolded LD state (Figures 1D and S1), revealing a new α-SNAP-bound state (state 2α). In addition, the new state occurred less frequently at a lower α-SNAP concentration (Figure S1). We analyzed the extension trajectories using three-state hidden-Markov modeling (HMM) (Ma et al., 2016; Rebane et al., 2016) and obtained apparent α-SNAP-SNARE association rates of 9 s-1 and 2 s-1 at 5 μM and 1 μM α-SNAP concentrations, respectively. These observations indicate that α-SNAP bound to the SNARE complex and stabilized its LD-unfolded state. This finding is consistent with a recent report (Ryu et al., 2015) and further reveals information about the kinetics of α-SNAP binding to the SNARE complex.
α-SNAP Stabilizes the SNARE CTD, but not the NTD
When held at a higher constant force in the range of 16-18 pN, the SNARE complex sequentially and rapidly transitioned between three states: the free 4HB state 2, the partially-zippered state 3, and the unzipped state 4 (Figure 2A, B, - α-SNAP, and Figure 1E), as was previously observed (Ma et al., 2016). In the partially-zippered state, the t-SNARE is largely structured as in the fully folded SNARE complex, but its C-terminus is frayed to approximately the +4 hydrophobic layer, and the v-SNARE is unzipped to approximately the +1 hydrophobic layer (Figure 1E).
We then added α-SNAP into the solution and repeated the experiment under otherwise identical conditions. In the presence of 5 μM α-SNAP, distinct long dwelling frequently appeared in the SNARE transitions (Figure 2A, B, + α-SNAP, red regions), revealing a fourth state in the 4HB transition. This state had an average extension equal to that of the free 4HB state 2 (Figure 2C), suggesting that α-SNAP bound to the 4HB structure. As expected, the α-SNAP-bound state occurred more frequently as α-SNAP concentration increased (Figure S2). We analyzed the extension trajectories in the absence or presence of α-SNAP based on three- or four-state HMM, respectively, and obtained good fits to the corresponding extension trajectories (Figure 3A and Figure 3B, Schemes 1 and 2). We determined the average extensions, lifetimes, populations, and transition rates associated with all states. Particularly, the HMM successfully separated the α-SNAP-bound state from the other three states based on their distinct lifetimes (Figures 2A, 3A, and 3C). The latter states (black regions in Figure 2A, - α-SNAP) exhibited approximately the same extension probability density distribution as the SNARE states in the absence of α-SNAP (Figure 2A, + α-SNAP, and Figure 2C), which suggests that these states with short lifetimes were α-SNAP-unbound states. In addition, the dwell time distributions of both the α-SNAP-bound and -unbound 4HB states were single exponentials (Figures 3C,D), as is expected for distinct Markovian states. The average lifetimes of the two states derived by fitting the distributions with exponential functions (212 ms and 3.0 ms) were close to the corresponding lifetimes calculated based on the best-fit HMM parameters, which were 223 ms for the α-SNAP-bound state and 4.6 ms for the unbound 4HB state (Figure 3B, Scheme 2). The long lifetime of the α-SNAP-bound state shows that α-SNAP significantly stabilized the 4HB state. In contrast, α-SNAP had minimal effects on the partially-zippered state 3 and the unzipped state 4 with respect to their extensions (Figure 2C), lifetimes, and transition rates (Figure 3B, Schemes 1 and 2). These observations suggest that α-SNAP specifically bound to the 4HB state (Weber et al., 2000).
Figure 3. α-SNAP Binds the 4HB by Conformational Selection.
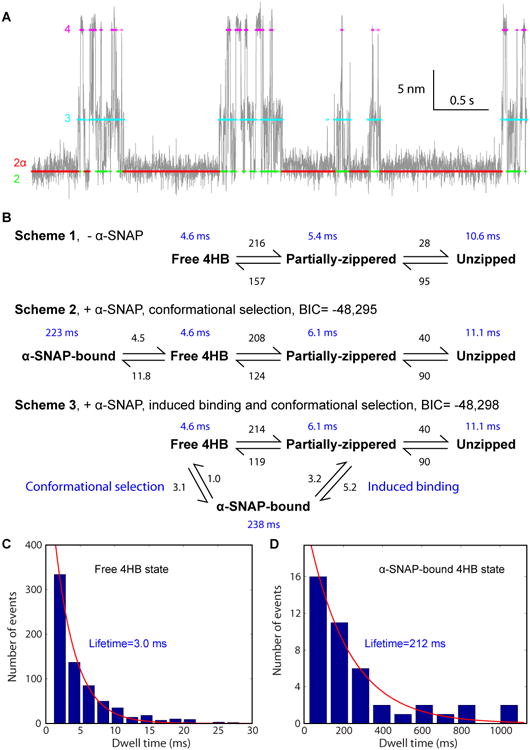
(A) Comparison of the measured (gray) and idealized (colored) extension trajectories in the presence of α-SNAP. The idealized trajectory was derived from the HMM based on reaction Scheme 2 in B. Different idealized states (Figure 1E) are color-coded. Data were analyzed and shown at 1 kHz bandwidth.
(B) Comparison of transition schemes, rates, and lifetimes of the SNARE complex derived from the extension trajectories shown in Figure 2A. Three- or four-state HMM was used to fit the extension trajectories in the absence (Scheme 1) or presence (Schemes 2 and 3) of α-SNAP, respectively, yielding the best-fit transition rates shown by numbers in black in units 1/s and state lifetimes shown by numbers in blue. Scheme 2 led to greater Bayesian information criterion (BIC) (Schwarz, 1978), indicating its better fit to the experimental data than Scheme 3. The BIC is defined as BIC = log (L) − 0.5 × N log (T), where L is the maximum likelihood observing the measured extension trajectory based on the HMM, N the number of fitting parameters (13 for Scheme 2 and 14 for Scheme 3), and T = 25,280 the number of data points in the trajectory.
(C-D) Histogram distributions of the dwell times of the free 4HB state (C) and the α-SNAP-bound 4HB state (D).
The two α-SNAP binding activities observed in the force regions of the LD and 4HB transitions could result from either the same or two distinct α-SNAP-SNARE binding modes. We found that α-SNAP showed close SNARE association rates and that the α-SNAP-bound 4HB state and the LD-unfolded state had similar lifetimes. Thus, we contended that the two activities were caused by a single α-SNAP-SNARE binding mode 2α. Moreover, due to step-wise SNARE folding, α-SNAP had to bind the unfolded LD state to attenuate its folding. In conclusion, α-SNAP directly bound to the LD and the CTD to simultaneously destabilize the LD and stabilize the CTD.
α-SNAP Binds the SNARE 4HB by Conformational Selection
In principle, α-SNAP could bind the SNARE 4HB by targeting either the pre-assembled 4HB (Figure 3B, Scheme 2, conformational selection), the partially-zippered SNARE complex (induced binding in Scheme 3), or both. In the induced binding model, α-SNAP first transiently bound the partially zippered SNARE complex in state 3 and then induced the SNARE complex to fold from state 3 to state 2α. An optimal model can be determined by HMM based on these potential models, as we have recently demonstrated (Jiao et al., 2015). We compared results from HMM containing only the conformational selection pathway (Scheme 2) or both pathways (Scheme 3). We found that the conformational selection pathway alone fit the extension trajectory better, as was judged by its higher likelihood in terms of the Bayesian information criterion (BIC) (Schwarz, 1978), a standard criterion for model selection. Extensive analyses confirmed that the conformational selection pathway generally fit our data better and yielded more consistent transition rates than the induced binding pathway (Figures 4 and S2). The fitting revealed an association rate of 5-12 s-1 and a dissociation rate of 4-8 s-1, which led to an association rate constant of 1-2×106 M-1s-1 and a dissociation constant of 2-6 μM. The dissociation constant was close to the recently reported measurement of 1.5 μM (Vivona et al., 2013). However, both association and dissociation rate constants we measured were greater than corresponding previous values, i.e., 1.7×105 M-1s-1 and 0.26 s-1, respectively, indicating that our SNARE zippering assay demonstrated different α-SNAP binding kinetics compared to those derived from previously used direct binding assays.
Figure 4. Force-Dependent SNARE Zippering and α-SNAP Binding.
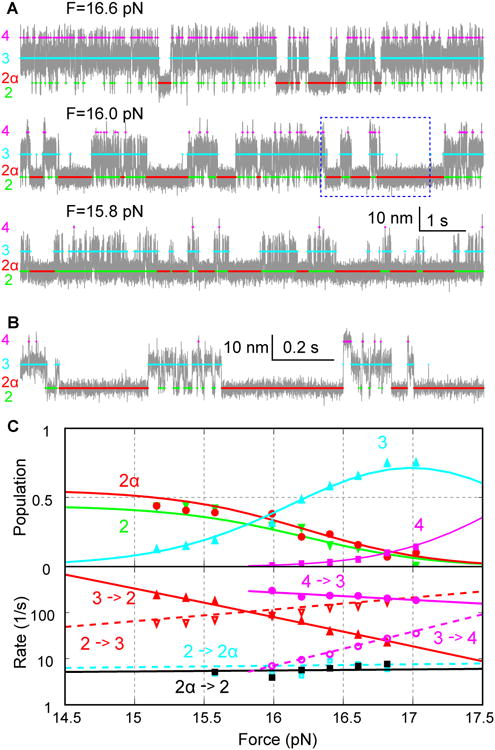
(A) Extension-time trajectories (gray) of a single SNARE complex under different mean forces F. The colored traces indicate the average extensions of different states derived from HMM (Figure 1E). The extensions were mean-filtered to 5 kHz and plotted.
(B) Close-up view of the region in A shown in the dashed box.
(C) Force-dependent state populations and transition rates determined by HMM (symbols). These data were fitted with a theoretical model (curves) to extrapolate folding energetics and kinetics to zero force (‘Experimental Procedures’). The SNARE complex without the NRD was used in this experiment.
The NRD is targeted by SNARE regulatory proteins, such as Munc18-1, to regulate SNARE assembly and membrane fusion (Ma et al., 2016; Shen et al., 2007; Sudhof and Rothman, 2009; Zhou et al., 2013). It is not clear whether the NRD plays a role in the α-SNAP-dependent SNARE assembly. To test the potential role of the NRD in SNARE zippering, we repeated our above experiments using a SNARE construct lacking the NRD. We found that the NRD did not significantly affect zippering of the SNARE complex (Ma et al., 2016) or binding of α-SNAP to the complex, as the SNARE complex without the NRD exhibited similar kinetics of SNARE zippering and α-SNAP binding as the SNARE complex with the NRD (compare extension trajectories in Figures 4 and 2). The similarity became more pronounced when the extension trajectories with and without the NRD were plotted at the same bandwidth, especially with respect to α-SNAP binding (Figure S2). Increasing the force applied to the SNARE complex proportionally reduced the populations of both the free and α-SNAP-bound 4HB states (Figure 4C, states 2 and 2α), corroborating the conformational selection mechanism of α-SNAP binding. Accordingly, force did not significantly change the α-SNAP association and dissociation rates. However, the lifetime of the α-SNAP-bound 4HB state changed more dramatically than that of the free 4HB state (Figure S3). Using a theoretical model to describe protein folding in optical traps (‘Experimental Procedures’) (Rebane et al., 2016), we fit the model to the measured state populations and forces, transition rates, and relative state extensions, and determined the model parameters at zero force. The fitting yielded the folding energies and lifetimes of different states in the absence and presence of 5 μM α-SNAP. We obtained the average free energy of the α-SNAP-bound 4HB state relative to the free 4HB state, ΔG=-0.2 (±0.6, standard deviation) kBT, which was measured from a total of 32 SNARE complexes. Accordingly, we calculated the dissociation constant as KD=P2×[α-SNAP]/P2α=[α-SNAP]×exp(ΔG/kbT)=4 (±3) μM, where P2 and P2α are the populations of the free 4HB state and the α-SNAP-bound 4HB state, respectively, with the α-SNAP concentration [α-SNAP]=5 μM. The dissociation constant is consistent with that derived in the presence of force. Taken together, α-SNAP was able to bind to the folded SNARE 4HB via a conformational selection mechanism, and the force applied to the SNARE complex affected the population of the free 4HB state to modulate α-SNAP binding yield.
Heterogeneity in α-SNAP Binding Kinetics
The α-SNAP binding kinetics were generally homogeneous, which indicated that α-SNAP primarily binds a single site on the folded 4HB in solution with a 1:1 stoichiometry, consistent with previous results (Vivona et al., 2013). Yet, we occasionally observed heterogeneity in α-SNAP binding kinetics. This was demonstrated by sudden switching in the binding kinetics (Figures 5A and S4), unusually long dwelling times in the folded state (Figure 5B), and distinct dwelling durations in the partially folded state (Figure 5C). These events occurred at a rate of less than 0.003 s-1, whereas the canonical α-SNAP binding event described before appeared at a rate of 1-6 s-1. The kinetic heterogeneity suggests that α-SNAP could occasionally bind to the folded 4HB state on alternative sites or with variable stoichiometry, or associate with the partially zippered SNARE complex. Particularly, when a SNARE complex is partially or completely unzipped, α-SNAP might specifically or nonspecifically bind to the SNARE complex with a low probability, leading to overestimation of the association and dissociation rates of α-SNAP detected in our assay.
Figure 5. Kinetic Heterogeneity in α-SNAP-SNARE Interactions.
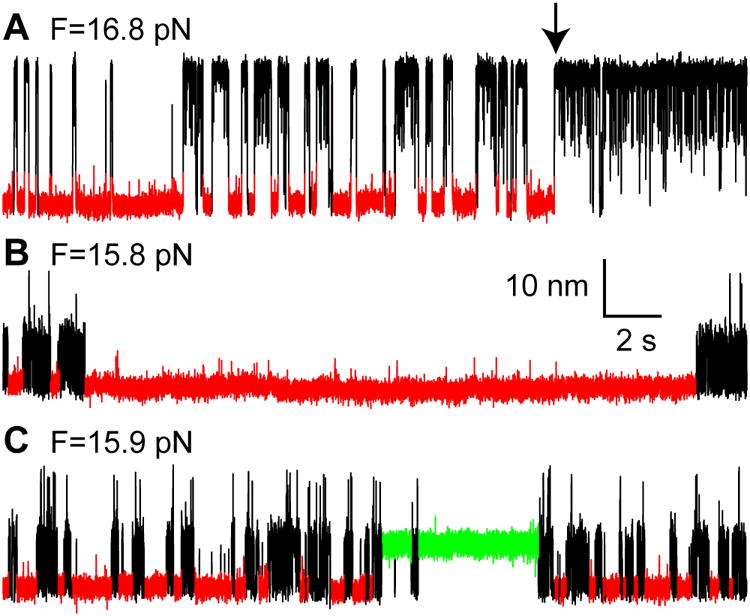
Extension-time trajectories at different mean forces (F) showing three kinds of kinetic heterogeneity: kinetic switching (A), unusually long dwelling in the folded 4HB state (B), and long dwelling in the partially zippered state (C). These events occurred at a rate of 1.1×10-3 s-1, 8.3×10-4 s-1, and 2.9×10-3 s-1, respectively, which were measured on 26 single SNARE complexes in 5 μM α-SNAP for an accumulated time of 7,219 seconds in the force range of 15-19 pN. The folded 4HB state and the partially zippered state stabilized by α-SNAP are highlighted in red and green, respectively. The black arrow in A indicates the moment of kinetic switching upon α-SNAP binding.
Discussion
Our results show that α-SNAP destabilized the LD, stabilized the CTD, and barely affected the NTD, which may help elucidate the functions of α-SNAP in SNARE assembly and disassembly. First, our findings suggest that α-SNAP preferentially binds the folded 4HB state that corresponds to the cis-SNARE complex, but not the partially-zippered state characteristic of the synaptic trans-SNARE complex. Intriguingly, yeast Sec17 is able to bind vacuolar trans-SNARE complexes and disassemble them in vitro with Sec18, an NSF homolog (Ungermann et al., 1998). The apparent discrepancy may be explained by the observation that trans-SNARE complexes alone are in dynamic equilibrium between a partially-zippered state and a 4HB state (Shin et al., 2014). Like α-SNAP, Sec17 may specifically bind to trans-SNARE complexes in the 4HB state to disassemble SNAREs. However, in the cell, the HOPS complex likely protects the 4HB state from disassembly (Xu et al., 2010). Thus, our observations corroborate the notion that α-SNAP specifically disassembles cis-SNARE complexes, but not trans-SNARE complexes, in a physiological context. Second, α-SNAP increased the SNARE CTD zippering energy by ∼12.4 kBT and decreased the LD zippering energy by ∼8 kBT under standard conditions (calculated from ∼4 μM dissociation constant), leading to a net energy gain of ∼4.4 kBT (Gao et al., 2012). This energy increase is expected to significantly enhance membrane fusion (Ma et al., 2016), but it does not likely impede SNARE disassembly because the energy gain is negligible compared to the ∼ 70 kBT total SNARE zippering energy (Gao et al., 2012; Ma et al., 2016) and the ∼200 kBT total energy necessary for NSF to disassemble a single SNARE complex (Shah et al., 2015). Third, the dual role of α-SNAP in SNARE zippering may cause the observed positive and negative effects of α-SNAP on membrane fusion (Park et al., 2014; Stein et al., 2009; Zick et al., 2015). CTD zippering serves as the major power stroke for membrane fusion (Ma et al., 2016). We suggest that Sec 17 enhances CTD zippering by conformational selection in a manner similar to that of α-SNAP, which accounts for the positive role of Sec17 in membrane fusion (Zick et al., 2015). The role of LD in membrane fusion is not well understood. Continued SNARE zippering to the LD is shown to be required for membrane fusion in some experiments in vitro (Stein et al., 2009) but not in other experiments (Kesavan et al., 2007). Additionally, both the syntaxin and VAMP2 LDs bind to membranes (Liang et al., 2013). Particularly, the syntaxin LD strongly interacts with phosphatidylinositol 4,5-biphosphate and mediates the formation of syntaxin clusters (van den Bogaart et al., 2011). Consequently, membranes may attenuate or inhibit association of α-SNAP to the LD, which reduces the negative role of α-SNAP in LD zippering and leads to a membrane curvature-dependent role of α-SNAP in membrane fusion (Park et al., 2014; Zick et al., 2015). Finally, our data support the finding that α-SNAP mainly binds the cytoplasmic SNARE 4HB in a 1:1 stoichiometry at up to a 10-μM α-SNAP concentration (Shah et al., 2015; Vivona et al., 2013). However, NSF and/or membranes promote up to four α-SNAP molecules to bind each SNARE 4HB (Zhao et al., 2015). In this case, binding of the first α-SNAP molecule to the primary binding site on the SNARE 4HB observed may nucleate binding of additional α-SNAP molecules to weaker binding sites. Future experiments will pinpoint how membranes and NSF influence α-SNAP-SNARE binding and its role in SNARE zippering (Winter et al., 2009; Zhao et al., 2015; Zhou et al., 2015)
Experimental Procedures
Proteins and sample preparation
Bovine α-SNAP, rat syntaxin 1A (amino acids 1-265, L205C), VAMP2 (1-96, Q36C), and SNAP-25B were expressed in BL21 E. Coli cells and purified using His-tags. Except for SNAP-25B, His-tags were cleaved for all proteins after purification. Four natural cysteine residues in SNAP-25B and one cysteine residue in syntaxin 1A were substituted with serine. An Avi-tag was added to the C-terminus of syntaxin 1A and biotinylated using biotin ligase (Gao et al., 2012). The SNARE complex was formed by mixing syntaxin 1A, VAMP2, and SNAP-25B at a molar ratio 0.8:1:1 in the presence of 1 mM tris(2-carboxyethyl)phosphine (TCEP) and purified using nickel-nitrilotriacetic acid (NTA) columns. After removing TCEP, the purified complex was immediately mixed with the thiol-containing DNA handle at a molar ratio of 50:1 in 100 mM phosphate buffer, pH 8.5, with 0.5 M NaCl, oxidized in air overnight, and stored at -20 °C before use.
Dual-trap optical tweezers
The optical tweezers were home-built and calibrated as previously described (Gao et al., 2012; Ma et al., 2016). The stiffness of the optical traps was measured based on the Brownian motion of a trapped bead and was typically 0.1-0.2 pN/nm for polystyrene beads that were ∼2 μm in diameter (Spherotech, IL, USA). A microfluidic chamber with four channels was used to inject buffers, α-SNAP solutions, and streptavidin-coated and anti-digoxigenin antibody-coated polystyrene beads. The optical tweezers had relative resolutions of ∼0.3 nm for extension and ∼0.05 pN for force measured on the same molecule, but they had systematic errors of >30 nm for extension and ∼10% for force measured on different molecules (Gao et al., 2012; Ma et al., 2016).
Single-molecule experiments
An aliquot of the crosslinked SNARE complex-DNA handle was first bound to 20 μL DIG beads, diluted in phosphate-buffered saline (PBS) to 1 mL, and injected to a microfluidic channel. The DIG bead was trapped and brought close to a streptavidin-coated bead in the other trap to form a single SNARE-DNA tether between the two beads. The experiment was carried out at room temperature (23 °C ± 1 °C).
Data analysis
Our methods of data analysis have been described in detail elsewhere (Rebane et al., 2016) and were used to derive the conformations shown in Figure 1E in the absence of α-SNAP (Ma et al., 2016). Briefly, extension-time trajectories were mean-filtered to 5 kHz or 1 kHz and analyzed by the histogram analysis (Figures 1D and 2C) and HMM (Rebane et al., 2016). The probability density distribution of the extension ρ(X) was calculated as the number of extension data points Ni falling into the i-th bin with an average extension of Xi, i.e., ρi = Ni/(NTdX), where NT was the total number of data points in all bins and dX was the extension size for each bin. Given a hidden Markov model, model parameters included average state positions, fluctuations, and transition rates. The likelihood of observing a specific extension trajectory was calculated using the forward-backward algorithm and maximized by optimizing the model parameters using either a gradient-search method or the expectation-maximization algorithm (Gao et al., 2012; Rebane et al., 2016). The state populations were calculated based on the optimized transition rates. The lifetime of a state was computed as the inverse of the sum of the rates leaving the state. Folding energies at zero force were calculated by non-linear least-squares fitting of a theoretical model of protein transitions in optical traps to the measured state populations, forces, extensions, and transition rates, as previously described (Gao et al., 2012; Ma et al., 2016; Rebane et al., 2016).
Supplementary Material
Acknowledgments
We thank James Rothman for discussion and Haijia Yu and Jingshi Shen for help. This work was supported by the NIH grants RO1GM093341 (to Y. Z.) and T32GM007223 and the Raymond and Beverly Sackler Institute at Yale.
Footnotes
Supporting Information: Supplemental Information includes four figures and can be found with this article online.
Author Contributions: L. M., J. J., A. R., and Y. Z. designed the experiments, L. M., Y. K., J. J., H. C., Z. X., and H. Q. performed the experiments, L. M., A. R., and Y. Z. analyzed the data, and L. M., J. J., A. R., and Y. Z. wrote the paper.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreeva AV, Kutuzov MA, Voyno-Yasenetskaya TA. A ubiquitous membrane fusion protein alpha SNAP: a potential therapeutic target for cancer, diabetes and neurological disorders? Expert Opin Ther Tar. 2006;10:723–733. doi: 10.1517/14728222.10.5.723. [DOI] [PubMed] [Google Scholar]
- Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- Chae TH, Kim S, Marz KE, Hanson PI, Walsh CA. The hyh mutation uncovers roles for alpha SNAP in apical protein localization and control of neural cell fate. Nat Genet. 2004;36:264–270. doi: 10.1038/ng1302. [DOI] [PubMed] [Google Scholar]
- Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane-fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zorman S, Gundersen G, Xi ZQ, Ma L, Sirinakis G, Rothman JE, Zhang YL. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337:1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao JY, Rebane AA, Ma L, Gao Y, Zhang YL. Kinetically coupled folding of a single HIV-1 glycoprotein 41 complex in viral membrane fusion and inhibition. Proc Natl Acad Sci USA. 2015;112:E2855–E2864. doi: 10.1073/pnas.1424995112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesavan J, Borisovska M, Bruns D. v-SNARE actions during Ca2+-triggered exocytosis. Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- Liang BY, Kiessling V, Tamm LK. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proc Natl Acad Sci USA. 2013;110:19384–19389. doi: 10.1073/pnas.1314699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobingier BT, Nickerson DP, Lo SY, Merz AJ. SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE 5 complexes to oppose SNARE disassembly by Sec18. Elife. 2014;3:e02272. doi: 10.7554/eLife.02272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Rebane AA, Yang G, Xi Z, Kang Y, Gao Y, Zhang YL. Munc18-1-regulated stage-wise SNARE assembly underlying synaptic exocytosis. eLIFE. 2016;4:e09580. doi: 10.7554/eLife.09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz KE, Lauer JM, Hanson PI. Defining the SNARE complex binding surface of alpha-SNAP - Implications for SNARE complex disassembly. J Biol Chem. 2003;278:27000–27008. doi: 10.1074/jbc.M302003200. [DOI] [PubMed] [Google Scholar]
- Park Y, Vennekate W, Yavuz H, Preobraschenski J, Hernandez JM, Riedel D, Walla PJ, Jahn R. alpha-SNAP interferes with the zippering of the snare protein membrane fusion machinery. J Biol Chem. 2014;289:16326–16335. doi: 10.1074/jbc.M114.556803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebane AA, Ma L, Zhang YL. Structure-based derivation of protein folding intermediates and energies from optical tweezers. Biophys J. 2016;110:441–454. doi: 10.1016/j.bpj.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice LM, Brunger AT. Crystal structure of the vesicular transport protein Sec17: Implications for SNAP function in SNARE complex disassembly. Mol Cell. 1999;4:85–95. doi: 10.1016/s1097-2765(00)80190-2. [DOI] [PubMed] [Google Scholar]
- Ryu JK, Min D, Rah SH, Kim SJ, Park Y, Kim H, Hyeon C, Kim HM, Jahn R, Yoon TY. Spring-loaded unraveling of a single SNARE complex by NSF in one round of ATP turnover. Science. 2015;347:1485–1489. doi: 10.1126/science.aaa5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. Estimating dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- Shah N, Colbert KN, Enos MD, Herschlag D, Weis WI. Three alpha SNAP and 10 ATP molecules are used in SNARE complex disassembly by N-ethylmaleimide-sensitive Factor (NSF) J Biol Chem. 2015;290:2175–2188. doi: 10.1074/jbc.M114.620849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JS, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Shin J, Lou XC, Kweon DH, Shin YK. Multiple conformations of a single SNAREpin between two nanodisc membranes reveal diverse pre-fusion states. Biochem J. 2014;459:95–102. doi: 10.1042/BJ20131668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Ungermann C, Sato K, Wickner W. Defining the functions of trans-SNARE pairs. Nature. 1998;396:543–548. doi: 10.1038/25069. [DOI] [PubMed] [Google Scholar]
- van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, Hubrich BE, Dier M, Hell SW, Grubmuller H, Diederichsen U, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivona S, Cipriano DJ, O'Leary S, Li YH, Fenn TD, Brunger AT. Disassembly of all SNARE complexes by N-ethylmaleimide-sensitive factor (NSF) is initiated by a conserved 1:1 interaction between alpha-soluble NSF attachment protein (SNAP) and SNARE complex. J Biol Chem. 2013;288:24984–24991. doi: 10.1074/jbc.M113.489807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Parlati F, McNew JA, Johnston RJ, Westermann B, Sollner TH, Rothman JE. SNAREpins are functionally resistant to disruption by NSF and alpha SNAP. J. Cell Biol. 2000;149:1063–1072. doi: 10.1083/jcb.149.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer C, Hohl TM, Hughes CA, Muller SA, Sollner TH, Engel A, Rothman JE. Molecular mass, stoichiometry, and assembly of 20 S particles. J Biol Chem. 2001;276:29091–29097. doi: 10.1074/jbc.M011292200. [DOI] [PubMed] [Google Scholar]
- Winter U, Chen X, Fasshauer D. A conserved membrane attachment site in alpha-SNAP facilitates N-ethylmaleimide-sensitive factor (NSF)-driven SNARE complex disassembly. J Biol Chem. 2009;284:31817–31826. doi: 10.1074/jbc.M109.045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jun Y, Thompson J, Yates J, Wickner W. HOPS prevents the disassembly of trans-SNARE complexes by Sec17p/Sec18p during membrane fusion. EMBO J. 2010;29:1948–1960. doi: 10.1038/emboj.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Pang ZPP, Yang XF, Zhang YS, Rosenmund C, Bacaj T, Sudhof TC. Syntaxin-1 N-peptide and H-abc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2013;32:159–171. doi: 10.1038/emboj.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Huang X, Sun S, Li XM, Wang HW, Sui SF. Cryo-EM structure of SNAP-SNARE assembly in 20S particle. Cell Res. 2015;25:551–560. doi: 10.1038/cr.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Orr A, Schwartz ML, Merz AJ, Wickner WT. Sec17 can trigger fusion of trans-SNARE paired membranes without Sec18. Proc Natl Acad Sci USA. 2015;112:E2290–E2297. doi: 10.1073/pnas.1506409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorman S, Rebane AA, Ma L, Yang GC, Molski MA, Coleman J, Pincet F, Rothman JE, Zhang YL. Common intermediates and kinetics, but different energetics, in the assembly of SNARE proteins. Elife. 2014;3:e03348. doi: 10.7554/eLife.03348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


