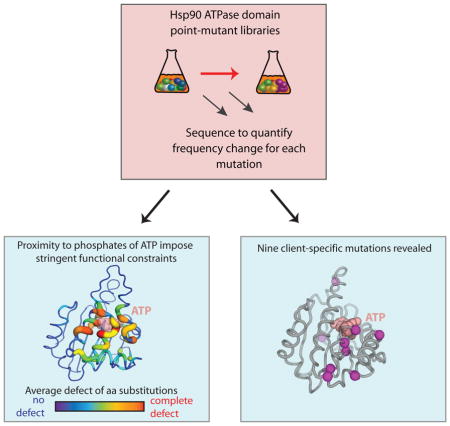
Summary
To probe the mechanism of the Hsp90 chaperone that is required for the maturation of many signaling proteins in eukaryotes, we analyzed the effects of all individual amino acid changes in the ATPase domain on yeast growth rate. The sensitivity of a position to mutation was strongly influenced by proximity to the phosphates of ATP, indicating that ATPase driven conformational changes impose stringent physical constraints on Hsp90. To investigate how these constraints may vary for different clients, we performed biochemical analyses on a panel of Hsp90 mutants spanning the full range of observed fitness effects. We observed distinct effects of nine Hsp90 mutations on activation of v-src and glucocorticoid receptor (GR), indicating that different chaperone mechanisms can be utilized for these clients. These results provide a detailed guide for understanding Hsp90 mechanism and highlight the potential for inhibitors of Hsp90 that target a subset of clients.
Graphical Abstract
Introduction
The Hsp90 chaperone continues to be an intense focus of research because of its central roles in protein homeostasis in normal (Wayne et al., 2011) and disease states (Neckers and Trepel, 2014) and in the accumulation and propagation of mutations in evolution (Rohner et al., 2013). The chaperone function of Hsp90 can rescue mutations in clients that would otherwise lead to functional defects. This ability of Hsp90 to rescue mutations in clients enables the accumulation of genetic diversity that can contribute to the evolution of new adaptive traits (Rohner et al., 2013). Hsp90 plays a central role in many physiological processes through its substrates or client proteins. The majority of Hsp90 clients are associated with signal transduction, and include nuclear steroid hormone receptors (Guy et al., 2015) and roughly half of all kinases (Taipale et al., 2012). The upregulation of signaling clients in many cancers leads to an increased dependence on Hsp90. Inhibitors of Hsp90 can revert the transforming phenotypes of oncogenes in cell culture (Whitesell et al., 1994) and have shown promise as therapeutic agents to treat cancer (Neckers and Trepel, 2014). Of note, it has been possible to develop inhibitors that are specific for the ATPase site of Hsp90 because Hsp90 harbors an unusual Gyr-B like active site that is distinct from the Walker motifs in most ATPases (Prodromou et al., 1997). Because Hsp90 is essential in all cells, broad inhibition of Hsp90 function is toxic in healthy cells and therapeutic benefits occur with treatment regimens that partially inhibit Hsp90 (Neckers and Trepel, 2014). These partial inhibition strategies are sufficient to selectively inhibit many oncogenic clients, though the detailed mechanism for this selectivity is unclear (Kamal et al., 2003).
Tremendous progress has been made in understanding the mechanism of Hsp90, but many important questions remain unanswered. Structural and biochemical analyses have shown that Hsp90 contains three domains: an N-terminal ATPase domain (Prodromou et al., 1997), a middle domain implicated in binding to clients (Hawle et al., 2006; Meyer et al., 2003), and a C-terminal domain that forms a constitutive homodimer (Harris et al., 2004; Richter et al., 2001). Hinge regions between each domain of Hsp90 impart flexibility on the full-length protein (Ali et al., 2006; Shiau et al., 2006). The “lid” region of the ATPase domain adopts distinct nucleotide-bound conformations (Ali et al., 2006; Prodromou et al., 1997) including transient dimerization when ATP and the co-chaperone p23 are bound that in turn mediate dramatic conformational changes of the full-length Hsp90 dimer (Southworth and Agard, 2008). Purified Hsp90 can act as an anti-aggregation chaperone (Pursell et al., 2012), but activation of signaling clients including kinases and nuclear steroid receptors in vitro requires multiple co-chaperones including p23, Hop, Cdc37, and Hsp70 (Arlander et al., 2006; Boczek et al., 2015; Guy et al., 2015). Exciting recent views of Hsp90 bound to clients and co-chaperones (Karagoz et al., 2014; Kirschke et al., 2014; Vaughan et al., 2006) indicate that different clients can bind to distinct surfaces of Hsp90. Despite these advances, we do not currently have a clear understanding of how ATPase driven conformational changes in Hsp90 activates most clients. For example, while nucleotide binding causes dramatic changes to the conformation of purified Hsp90, experiments with engineered conformation-restricted variants of Hsp90 indicate that fully open conformations are not required for chaperone function (Pullen and Bolon, 2011). In addition, analyses of client binding affinities to Hsp90 indicate that the rules governing client binding are complex (Taipale et al., 2012), consistent with the emerging structural views of clients binding to different surfaces on Hsp90 (Karagoz et al., 2014; Kirschke et al., 2014; Vaughan et al., 2006). It remains unclear to what extent Hsp90 utilizes ATPase-driven mechanisms that are common to all clients compared to mechanisms that are client specific.
To address these questions, we probed the physical requirements for function at each position in the ATPase domain of Hsp90. Using a mutational scanning approach (Boucher et al., 2014; Fowler and Fields, 2014) that we refer to as EMPIRIC (Hietpas et al., 2011), we analyzed the effects of each amino acid change in an otherwise wild-type Hsp90 sequence background on the growth rate of engineered budding yeast. This screen probes the impacts of Hsp90 mutants integrated over multiple clients that contribute to yeast growth. Hsp90 function is strongly conserved in the eukaryotic lineage, where it is essential in all species that have been tested including budding yeast (Borkovich et al., 1989), and worms (Birnby et al., 2000). In addition, human Hsp90 can complement yeast knockouts, and vertebrate clients including the v-src oncogene as well as the glucocorticoid receptor (GR) exhibit Hsp90 dependent activation in yeast, further indicating the functional conservation of Hsp90 in eukaryotes (Picard et al., 1990; Xu and Lindquist, 1993). Because of the strong conservation of Hsp90 function across the eukaryotic lineage, studies of yeast Hsp90 and model clients including v-src and GR provide valuable guides for understanding Hsp90 mechanism.
Our results demonstrate that the experimental sensitivity to mutation varies dramatically for different positions in Hsp90. We identified eight critical positions where any amino acid change caused severe functional defects. All eight of these positions were either in direct contact with the phosphates of ATP or in second shell positions structurally poised to transmit ATPase dependent conformational changes. Across the entire ATPase domain, proximity to the phosphates of ATP was a strong determinant of sensitivity to mutation, indicating that local conformational changes propagated by ATP hydrolysis impose stringent functional constraints. Many positions in Hsp90 contribute to less stringent aspects of function. Based on the results of our EMPIRIC scan, we sampled a panel of mutations at these less stringent sites for potential client-specific effects. Our results demonstrate that mutations spread across the structure of the ATPase domain can differentially impact the maturation of GR, v-src and the yeast kinase Ste11, whose kinase activity is Hsp90-dependent (Louvion et al., 1998). Together, these observations indicate that conformational changes mediated by ATP hydrolysis are likely central to the maturation of most Hsp90 clients, but that client-specific features of the ATPase domain also contribute to chaperone function. These client-specific features suggest that it may be feasible to develop inhibitors of Hsp90 with client selectivity.
Results and Discussion
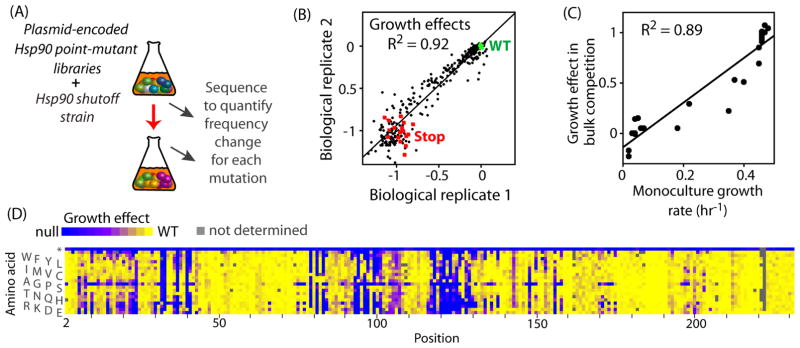
We analyzed the experimental fitness effects of all point mutations in the ATPase domain of Hsp90 using bulk competitions of engineered budding yeast (Fig. 1A). The effects of Hsp90 mutations on yeast growth rate were quantified based on changes in the frequency of each mutation during competitions where yeast growth rates are tightly coupled to the function of Hsp90 mutant variants by limiting the promoter strength driving their expression (Jiang et al., 2013). As in our previous work (Hietpas et al., 2012; Hietpas, 2013; Hietpas et al., 2011), focused deep sequencing was used to assess the frequency of each Hsp90 mutation in bulk competition samples. To assess the reliability of the scanning protocol, we performed full biological replicate analyses of mutations at 20 amino acid positions (~10% of the N-domain). These biological replicates indicated that our fitness measurements could clearly distinguish wild-type synonyms from stop codons (Fig. 1B). The correlation coefficient from these biological replicates (R2=0.92) is consistent with expected variation based on sequencing depth (Boucher et al., 2014) (average confidence interval of 9%, Table S1). Mutations with strongly deleterious effects exhibited greater variation between replicates consistent with expected uncertainty in estimating the frequency of mutations that deplete rapidly. To examine how growth rates may be influenced by cultures containing competing variants, we compared these to growth rates determined in monoculture for a panel of 21 individually cloned Hsp90 variants. Monoculture growth rates correlated with measurements from bulk competitions (Fig. 1C), indicating that growth rates were not dramatically impacted by the complexity of variants in the culture. Mutations that cause partial growth defects are poised on the edge of viability and are therefore sensitive to small changes in selection pressures. These mutations tended to have slightly more severe defects in the bulk competitions (Fig. 1C), hinting that selection strength in the bulk competitions may be slightly increased relative to monoculture. Of note, differences in genetic background and environmental conditions can both impact the fitness effects of mutations and lead to distinctions between studies where these parameters differ (Hietpas, 2013; Jiang et al., 2013). For example, a previous study observed that the E33D mutation in Hsp90 could support yeast viability (Obermann et al., 1998), but in our mutation scan we observed that the E33D mutation was severely deleterious, potentially due to distinctions in these experiments including genetic backgrounds and environmental conditions. While future studies will be required to address the dependencies of N-domain mutations on genetic background and environmental conditions, our results indicate that the estimates of fitness effects from bulk competitions appear robust for the genetic background and environmental conditions studied.
Figure 1. Effects of individual amino acid changes in the N-domain of Hsp90 on yeast growth rate.
(A) Outline of the bulk competition strategy utilized to analyze the effects of Hsp90 mutants on yeast growth rate. (B) Full biological replicates of Hsp90 mutations spanning amino acids 12-21 and 92-101 are strongly correlated. Growth effects are plotted as selection coefficients (0=no effect compared to wildtype, and −1=null). Silent mutations that do not change the protein sequence are plotted in green, stop codons are shown in red and amino acid changes in black. (C) Estimates of growth effects from bulk competitions correlate strongly with measurements of mono-culture growth rate for a panel of Hsp90 mutants. The following mutations were analyzed in monoculture: L18F, Y24L, R32A, N37S, D40E, K54E, E57K, R65L, Q72D, G83T, I96A, S99A, I117F, F120A, G121A, F124A, Y125W, L129M, E165K, I172D, and K191F. (D) Heatmap representation of the fitness landscape observed for individual amino acid changes across amino acids 2-231 of Hsp90. See also Figure S1 and tableS1.
Because the natural amino acids sample a variety of different physical properties, our data provides a broad description of the underlying physical requirements at each position in the ATPase domain for function. To provide an overview of these physical requirements, we generated a heatmap representation (Fig. 1D) of the fitness effects of each possible amino acid change (Table S1). Patterns for an amino acid across different positions indicate global impacts of particular amino acids. For example, the consistent blue horizontal stripe at the top of the heatmap indicates universal defects for stop codons at all positions, consistent with known requirements for downstream regions of Hsp90 for dimerization and binding to clients (Alvira et al., 2014; Karagoz et al., 2014; Vaughan et al., 2006; Wayne and Bolon, 2007). Other than stop codons, proline mutations caused the strongest average defect (Fig. S1A), consistent with the propensity of proline mutations to disrupt secondary structure (Strehlow et al., 1991). Across all mutations in our dataset, we observed a weak correlation between amino acid similarity and fitness effects (Fig. S1B), indicating that factors other than amino acid similarity (e.g., the structural context of each mutation) play important roles in determining impacts on function and fitness.
Comparison of experimental fitness effects with conservation in evolution
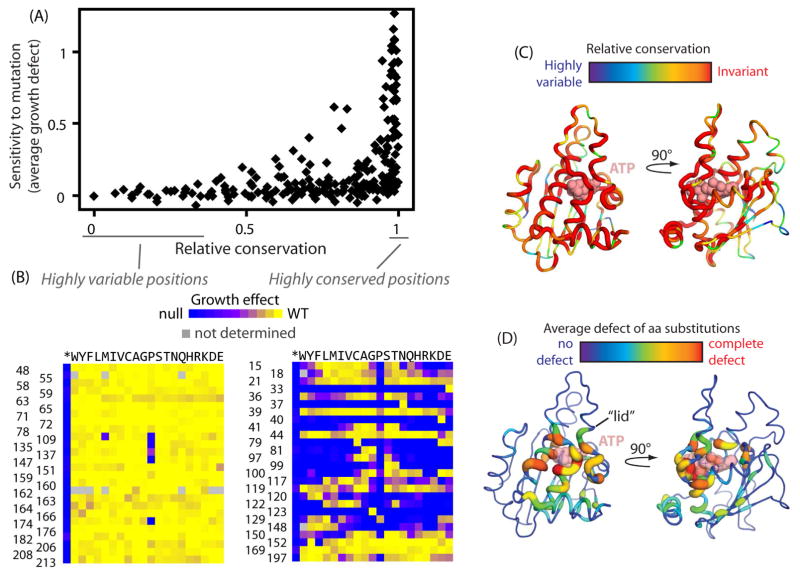
Vertical patterns in the heatmap indicate the tolerance of a position to mutations. Dramatic variations in these vertical patterns indicate large differences in the tolerance of positions to mutations (Fig. 1D). To explore how experimental fitness measurements relate to evolutionary selection pressures, we compared the experimental sensitivity of positions in yeast Hsp90 with conservation in an alignment of homologs from diverse species (Fig. 2A). Our biochemical expectations were that positions performing critical functions (e.g., catalytic positions) would exhibit strong conservation. A common and intuitive extension of this logic is that positions that don’t perform critical functions will vary across large evolutionary distances. According to these ideas, evolutionarily conserved positions should be consistently sensitive to mutations. Instead, we observe that strongly conserved positions exhibit extremely varied experimental sensitivity to mutation (Fig. 2B).
Figure 2. Relationship between evolutionary conservation and experimental fitness effects.
(A) Comparison of the experimental fitness sensitivity of each position in the N-domain to conservation observed in Hsp90 sequences from diverse eukaryotes. Positions exhibiting the strongest evolutionary conservation exhibit a broad range of experimental sensitivity to mutation while the most evolutionary variable positions are almost universally experimentally tolerant to mutations (B) Experimental fitness effects at amino acid positions that exhibited the greatest and least evolutionary variation. (C&D) Structural representations of the Hsp90 N-domain from 2CG9.PDB (Ali et al., 2006) where the width of the backbone trace and the color indicate the evolutionary variation (C) or experimental sensitivity to mutation (D) of each position.
Multiple factors contribute to distinctions between conservation patterns in evolution and experimental fitness effects. These explanations include distinctions between conditions in natural and laboratory selections (e.g., environmental variation, and timescales). Environmental conditions can have a dramatic impact on the fitness effects of Hsp90 mutations (Hietpas, 2013), and the conditions experienced in evolution likely impose pressures that cannot be fully captured by laboratory experiments. In addition, the strength of purifying selection in large natural populations over evolutionary timescales is far more stringent than can be distinguished by experimental evolution studies (Boucher et al., 2014; Ohta, 1973). The nearly-neutral window in natural evolution is inversely related to effective population size, and for yeast it has been estimated that a fitness defects of 0.0001% would be subject to purifying natural selection (Tsai et al., 2008). Thus, an amino acid position would be conserved in evolution if mutations caused even minor reductions to function that are beyond the resolution of laboratory experiments. From this perspective, evolutionary conservation in the face of purifying selection is not sufficient to distinguish a critical catalytic position from a position that makes more nuanced contributions to function. In contrast, positions that vary or diverge during evolution under purifying selection must maintain function. Of note, the least evolutionarily conserved positions in Hsp90 universally tolerate the vast majority of possible amino acid changes in our experiments (Fig. 2B). For Hsp90, and likely other proteins whose functions are conserved across evolution, divergent positions are predominantly experimentally tolerant to mutations while evolutionarily conserved positions have varied experimental sensitivity to mutation.
Given the distinction between evolutionary conservation and experimental sensitivity to mutation, we examined how each of these metrics related to structure (Fig. 2C&D). The most conserved positions in evolution are located broadly over roughly half of the ATPase domain (Fig. 2C). They surround the site of ATP binding, but also occur at sites spread sparsely throughout the structure. Based on evolutionary conservation, these sites likely make contributions to function that vary from critical catalytic sites to minor improvements in the efficiency of client maturation. Mapping the pattern of experimental sensitivity to structure provides a distinct profile (Fig. 2D). Few positions were experimentally ultra-sensitive to mutation and these were all clustered around the site of ATP binding.
Proximity to phosphates of ATP imposes stringent functional constraints
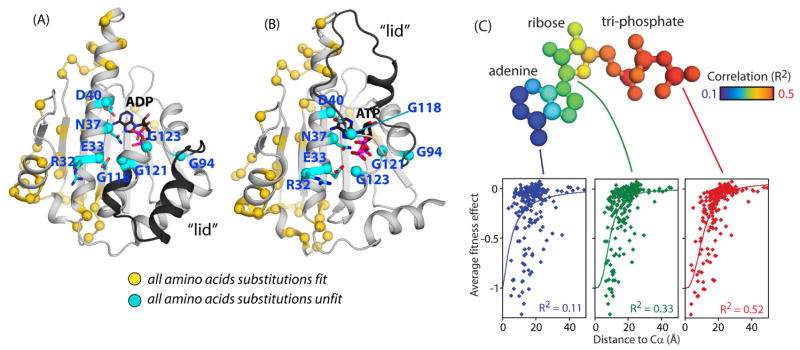
Experimental sensitivity distinguishes biochemically critical and non-critical sites. All possible amino acids were compatible with efficient function at 55 positions (Fig. 3A&B). At the other end of the spectrum, eight positions (R32, E33, N37, D40, G94, G118, G121, and G123) were experimentally ultra-sensitive such that even the most physically conservative amino acid changes caused a severe fitness defect. All eight of these critical positions are in proximity to the phosphates of ATP (Fig. 3B). Six of these critical positions (E33, N37, D40, G118, G121, and G123) directly contact the phosphates of ATP. E33 is a known catalytic amino acid that positions a water molecule for hydrolysis of the γ-phosphate (Panaretou et al., 1998). Our results indicate that all six positions that directly contact ATP are similarly critical for function. Structural inferences suggest that the critical amino acids that do not directly contact ATP (R32 and G94) may mediate conformational changes sparked by ATP hydrolysis. R32 forms a hydrogen bond to E33 as well as the main chain of a residue in the middle domain of Hsp90, and is poised to enable inter-domain conformational changes. G94 is located at a region that forms a hinge for the “lid” region (highlighted in black in Fig 3A&B), a loop whose conformation changes to close over ATP, but not ADP (Ali et al., 2006; Prodromou et al., 1997).
Figure 3. Structural distribution of experimentally sensitive and tolerant positions.
(A&B) Structural representations of the Hsp90 ATPase domain illustrating the location of the eight amino acids that are experimentally sensitive to any amino acid change (indicated with cyan spheres), and the 55 positions that experimentally tolerate any amino acid change (indicated with yellow spheres). Structural representations are shown for the ADP bound state from 1AM1.PDB (Prodromou et al., 1997) (A) and the ATP bound state from 2CG9.PDB (Ali et al., 2006) (B).(C) Sensitivity to mutations correlates with proximity to atoms in ATP. The atoms of ATP are colored based on the observed correlation between experimental fitness effects and distance to the Cα atoms of each amino acid residue. Distance to the γ-phosphate of ATP exhibits the strongest correlation and explains more than half of the observed variance in mutational sensitivity. See also Figure S2, S3, and S4.
We explored how proximity of a position to different atoms in ATP correlates with sensitivity to mutation (Fig. 3C and Fig. S2). Consistent with the structural location of critical amino acids surrounding the γ–phosphate of ATP, we observed the strongest correlation (R2=0.52) between the mutational sensitivity of a residue and the distances of Cα atoms to the γ-phosphate. To investigate influences of the directional orientation of side chains, we examined the angle of the Cα-Cβ bond of each side chain towards each atom in ATP, but parsing the positions using this metric did not have a dramatic impact on the observed distance relationships (Fig. S3). Of note, a weaker correlation (R2 = 0.27) was observed between distance to the γ–phosphate of ATP and evolutionary conservation (Fig. S4). The observation that half of the variation in experimental sensitivity to mutation can be explained by distance to the γ-phosphate was surprising given the number of different co-chaperones (Aha1, p23, Cdc37, Hsp70) and client proteins (GR, Tau, Cdk4) structurally observed to bind to different surfaces of the ATPase domain (Ali et al., 2006; Alvira et al., 2014; Karagoz et al., 2014; Kirschke et al., 2014; Retzlaff et al., 2010; Vaughan et al., 2006). While these structurally observed contacts with co-chaperones and clients clearly contribute to function, they do not appear to impose dominant constraints on amino acid mutations compared to proximity to the phosphates of ATP.
Positions that contact the adenine ring can tolerate mutations
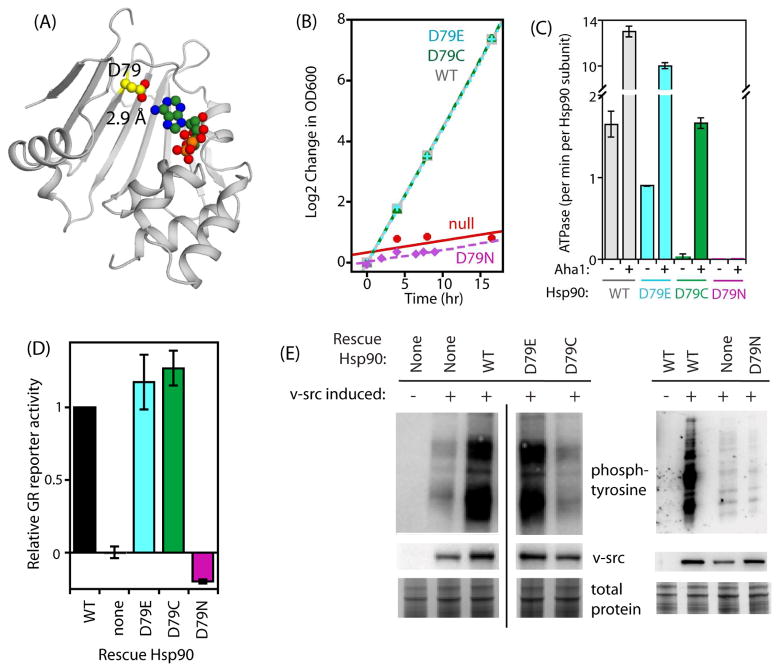
Distances from Hsp90 positions to different atoms in ATP exhibited distinct experimental fitness profiles. For example, correlations of mutational sensitivity drop off dramatically with the distance of positions from the phosphates of ATP compared with distance to atoms in the adenine ring (Fig. 3C). Consistent with this analysis, amino acids that directly contact the adenine ring could tolerate mutations. Because all contacts contribute to binding, these observations indicate that binding to the adenine ring is not as critical as the contacts between Hsp90 and the γ-phosphate that mediate hydrolysis-dependent conformational changes. Even D79, which forms a direct hydrogen bond with the adenine ring (Fig. 4A), could tolerate amino acid changes. Based on the results from bulk competitions (Fig. 1 and Table S1), both D79E and D79C were able to support yeast growth rates similar to wild-type. Previous studies had demonstrated that the D79N mutation prevents binding to ATP and is inviable in yeast (Obermann et al., 1998; Panaretou et al., 1998). Because of the similarities between aspartate and asparagine, including nearly identical steric footprints, the observation that D79N was inviable in yeast provided strong support for the hypothesis that ATP binding and hydrolysis are required for Hsp90 function despite the slow hydrolysis rates that had been observed in vitro. While a larger body of evidence now supports the conclusion that ATP hydrolysis by Hsp90 is essential, including observations in this work, it was perhaps fortuitous for the field that the D79N mutation was analyzed initially instead of D79E.
Figure 4. Contacts between D79 and adenine can be altered without compromising Hsp90 function.
(A) Structural representation of the N-domain of Hsp90 highlighting the near ideal geometry of the hydrogen bond formed between N6 of adenine and the side chain of D79. (B) Growth rate of budding yeast in monoculture for D79E, D79C, and D79N Hsp90 variants. (C) ATPase rates observed for purified Hsp90 proteins (2.5 μM) in the presence and absence of the co-chaperone Aha1 (10 μM). (D) Ability of D79E, D79C and D79N Hsp90 to mature the GR client in yeast. (E) Efficiency of v-src maturation supported by D79E, D79C, and D79N Hsp90 variants. The black vertical line indicates where lanes were removed for figure clarity. D79N along with appropriate controls were analyzed on a separate blot that is shown on the right side of this panel. See also Figure S5.
Given the historical relevance of engineered mutations at position 79 as well as the apparent tolerance to mutations at this location in our bulk analyses, we investigated the function of D79E and D79C in further detail. We generated these variants as individually isolated clones. Both D79E and D79C supported yeast growth rates in monoculture that were similar to wild-type (Fig. 4B), consistent with the observations from bulk competitions. As purified proteins, both D79E and D79C Hsp90 were capable of hydrolyzing ATP (Fig. 4C), though D79C activity was strongly dependent on the Aha1 co-chaperone, and even then was impaired relative to wild-type. This is in contrast to the D79N mutant that is unable to hydrolyze ATP in the presence or absence of Aha1 (Fig. 4C).
Importantly, both D79E and D79C were resistant to trypsin digestions (Fig. S5C), indicating that these mutations do not compromise the ability of Hsp90 to fold to a native state. In the absence of nucleotide, we note that the N and M domain bands are shorter lived, suggesting that D79C exhibits increased trypsin accessibility in these regions. The ability to resist trypsin digestion was enhanced in the presence of an ATP analogue further supporting their ability to bind nucleotide. In the presence of nucleotide, D79C is similar to WT in susceptibility to trypsin digestion. In addition, D79C is capable of hydrolyzing ATP in the presence of Aha1. Our interpretation of these results is that the D79C is partially destabilized but is capable of accessing the conformations required for ATP hydrolysis. The apparent binding affinity of Aha1 for D79C and D79E were within two-fold to that measured for wildtype Hsp90 (Fig. S5A). Because Aha1 stimulates the ATPase activity of Hsp90 by positioning the R380 side chain from the middle domain to stabilize the leaving phosphate (Li et al., 2013; Meyer et al., 2004b), our observations suggest that the D79C mutation may have difficulty positioning this catalytic arginine without the aid of Aha1. In the presence of Aha1, the Km for ATP was reduced for both D79C (30-fold) and D79E (8-fold) relative to wildtype Hsp90 (Fig. S5B). These observations suggest that D79C and D79E impact the interaction with nucleotide to a greater extent than binding to Aha1, and that Aha1 binding can partially rescue these nucleotide binding defects.
Previous studies (Prodromou et al., 2000) as well as this work demonstrate that Hsp90 mutations with a wide impact on ATPase are able to support yeast viability (Table 1 and Fig. S6), indicating that the relationship between ATPase rates in purified form and in vivo function of Hsp90 is complex, and highlighting the importance of studying Hsp90 function in vivo. We analyzed the ability of D79E and D79C variants to mature two Hsp90-dependent clients (GR and v-src) in yeast (Fig. 4D&E). As seen in previous studies, D79N Hsp90 was null for the activation of GR and v-src (Wayne et al., 2010). D79E Hsp90 could mature both clients efficiently. In contrast, D79C Hsp90 was compatible with efficient activation of GR, but caused a severe defect in the activation of v-src. These results indicate that different clients may place distinct demands on ATPase-dependent conformational changes in the N-domain.
Table 1.
Comparison of ATPase rates of purified Hsp90 protein with fitness effects.
| Mutation | Relative ATPase ratea | Fitness effectb | |
|---|---|---|---|
| − Aha1 | + Aha1 | ||
| None (WT) c | 1.0 | 7.9 | 0.0 |
| T22Id | 4.5 | nd | −0.64 |
| A41Vd | <0.1 | nd | −0.42 |
| D79Cc | <0.1 | 1.0 | −0.03 |
| D79Ec | 0.5 | 6.1 | −0.06 |
| D79Nc,d | <0.1 | <0.1 | −1.10 |
| T101Id | 0.1 | nd | −0.74 |
ATPase rate measured at 37 °C with or without the co-chaperone Aha1 normalized to the rate of hydrolysis for wildtype without Aha1.
Fitness effect observed with limiting expression level of Hsp90 from bulk competitions in this work.
ATPase measurements of Hsp90 variants generated in this work.
ATPase measurements from previous work (Prodromou et al., 2002).
Hsp90 mutations with client-specific effects
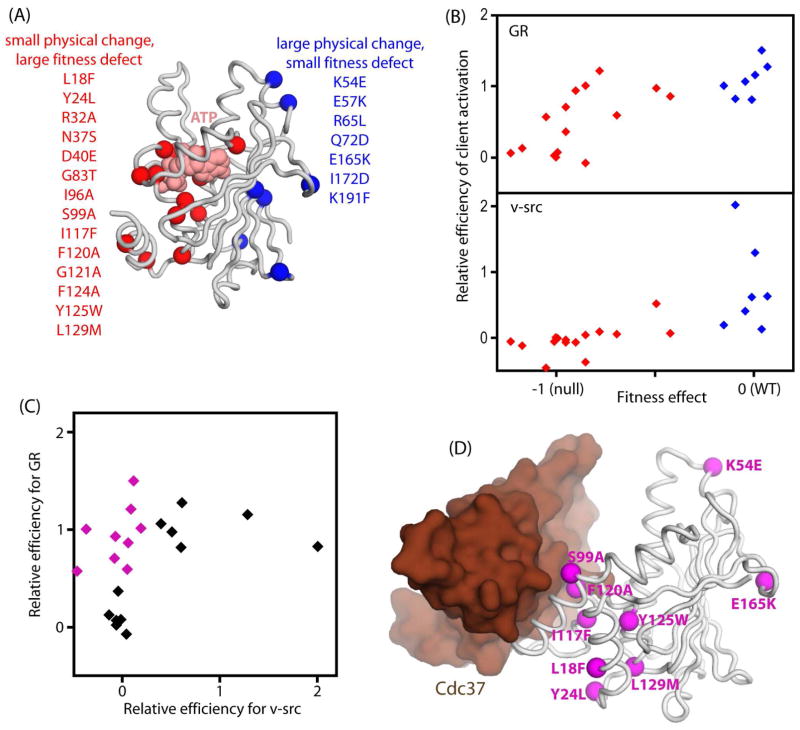
To further test the hypothesis that different clients impose distinct biochemical constraints on the Hsp90 ATPase domain, we used the results of our mutational scan as a guide to search for additional mutations that might have strong client-specific impacts. We focused our analyses on positions located near the surface of Hsp90 that could distinctly impact binding to different clients and/or co-chaperones. We reasoned that mutations with large physical changes in the nature of the amino acid side chain and small fitness effects akin to D79C would have the potential to disturb physical properties required for a subset of clients. These types of mutations were predominantly distal from ATP, and we manually selected a set for further analyses (Fig. 5A). To select these mutants, we first analyzed the fitness heatmap to identify tolerant positions spread across the surface of the ATPase domain. For each chosen tolerant position, we then manually identified a mutation with minor to no fitness effect (s>−0.1) that caused a large physical change. We also reasoned that severe growth defects caused by small physical changes could be due to specific impacts on critical clients. Motivated by this logic, we used an analogous procedure to manually select a set of mutations at sensitive positions that caused large fitness defects (s<−0.5) with small physical changes. We generated individual clones for the combined set of mutations and quantified the impacts of each Hsp90 variant on activation of GR, v-src, and Ste11 in yeast (Fig. 5B&C and Fig. 6A).
Figure 5. Identification of Hsp90 mutants that differentially impact the maturation of GR and v-src.
(A) Structural representation of the N-domain of Hsp90 indicating mutations identified as having a strong potential for separation of function. (B) The effects of individually cloned Hsp90 variants on activation of GR and v-src compared to effects on yeast growth rate. (C) Comparison of the effects of Hsp90 mutants on activation efficiency for GR versus v-src identifies eight mutations with severe deficiency for v-src that are capable of mediating the efficient activation of GR. (D) Structural representation of the N-domain of Hsp90 and the Cdc37 co-chaperone based on 1US7.PDB (Roe et al., 2004) indicating the location of the sites of mutations that caused client-specific effects. See also Figure S6.
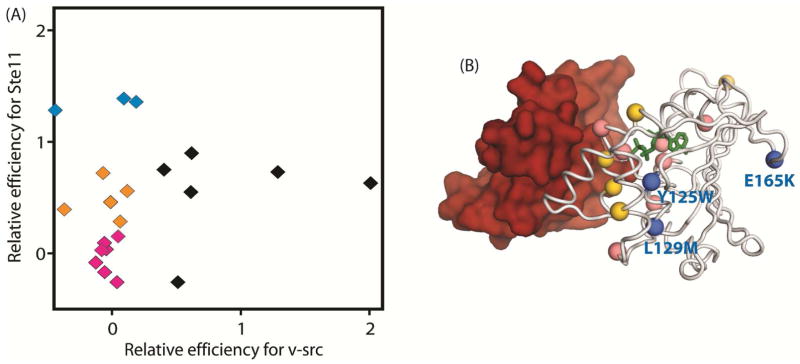
Figure 6. Distinct impact of Hsp90 mutations on maturation of v-src and Ste11 kinases.
(A) Comparison of the effects of Hsp90 mutants on activation efficiency for Ste11 versus v-src. Hsp90 mutations with severe defects for v-src activation exhibited impacts on Ste11 activation that ranged from severe defects (colored pink) to intermediate defects (colored orange) to no observable defect (colored blue). (B) The locations of mutations with severe defects for v-src activation are indicated on the structure of Hsp90 bound to Cdc37. The impacts of these mutations on Ste11 activation are indicated by color as in panel A. The location where nucleotide binds is indicated in green.
Mutations that caused little to no fitness effects (blue diamonds in Fig. 5B) universally exhibited efficient GR activation, but many were deficient in activating v-src. This observation indicates that maturation of v-src imposes greater constraints on the ATPase domain of Hsp90 than GR and the endogenous clients required to support rapid yeast growth. Conversely, mutations that caused strong fitness defects (red diamonds in Fig. 5B) were universally deficient in activating v-src, but many were able to efficiently mature GR. While the detailed mechanistic reasons for these observations remain to be elucidated, it is tempting to speculate that maturation of v-src may require more efficient utilization of ATP hydrolysis by Hsp90 than GR (e.g., efficient transmission of the energy of ATP hydrolysis to conformational changes that activate v-src) and thus v-src may be a more difficult client than GR to activate. Investigating this possibility in vitro is complicated because the hydrolysis of ATP by Hsp90 is strongly influenced by co-chaperones (Panaretou et al., 2002; Ratzke et al., 2014) and likely by clients, resulting in a complex relationship between in vitro and in vivo ATPase activity. The complexities of this relationship are exemplified by the in vitro ATPase activity of the nine Hsp90 variants that are capable of maturation of GR but not v-src (Fig. S6). Variants with severe fitness defects that retain their ability to mature GR exhibit a wide range of ATPase rates from extremely defective (Y125W, S99A and Y24L) to faster than WT (I117F and F120A). Future efforts will be needed to elucidate the molecular details of how ATP hydrolysis contributes to the maturation of different clients such as GR and v-src in the presence of the full complement of co-chaperones.
Previous analyses of a handful of mutations in the middle domain of Hsp90 identified three mutations (E431K, S485Y and T525I) that caused severe defects for GR maturation, but that were compatible with efficient v-src maturation in yeast (Hawle et al., 2006). Recent structural studies have observed that different clients bind to distinct surfaces of the middle domain (Karagoz et al., 2014; Kirschke et al., 2014; Vaughan et al., 2006), which provides a potential explanation for distinct impacts of amino acid changes in the middle domain on different clients.
Almost half of the Hsp90 mutations in our panel (9 of 21) exhibited strong client-specific effects (purple diamonds in Fig 5C). For comparison, a previous study identified five amino acid changes in the ATPase domain of yeast Hsp90 that conferred temperature sensitive growth (Meyer et al., 2004b). Four of the five Hsp90 temperature sensitive alleles (T22I, A41V, G81S and T101I) exhibited functional defects at permissive conditions, which meet part of the criteria that we used in identifying client-specific mutations. However, these criteria alone may not be sufficient to effectively identify client-specific mutations. All four of the temperature sensitive variants exhibited strong defects for both GR and v-src maturation (Meyer et al., 2004b). The other temperature sensitive mutation (G170D) efficiently matured both GR and v-src under permissive conditions, but exhibited defects for both of these clients at non-permissive temperatures. Thus, temperature sensitive mutations in the ATPase domain do not generally appear to have client-specific impacts. None of the temperature sensitive mutations met the full criteria that we developed to search for mutations with client-specific effects. In particular, the previously identified temperature sensitive mutations were all located in the solvent-inaccessible core, and most caused both large physical changes and large fitness defects in our experiments (Table S1). Our results indicate that subtle physical changes at sensitive surface positions and dramatic physical changes at insensitive surface positions provided an efficient approach to identify mutations with client-specific effects.
To investigate the potential mechanism of the client-dependent Hsp90 mutations, we mapped their location onto the structure of the Hsp90 ATPase domain determined in complex with Cdc37 (Roe et al., 2004) (Fig. 5D). The Cdc37 co-chaperone is specific for kinases, such that disruption of the Cdc37-Hsp90 interface may selectively disrupt maturation of kinases including v-src compared to non-kinase clients such as GR. Cdc37 contacts the “lid” region of nucleotide free Hsp90 and is in direct contact with two of the positions where we identified client-specific mutations (S99A and F120A). However, most of the client-specific mutations that we identified are not in direct contact with Cdc37, and two client-specific mutations (K54E and E165K) are located on a face of the Hsp90 ATPase domain opposite from the contact site with Cdc37. These structural observations suggest that multiple mechanisms including alteration to Cdc37 binding likely contribute to the observed client-specific impacts of mutations in Hsp90.
To further investigate the potential role of Cdc37 in mediating the distinct Hsp90 ATPase domain constraints that we observed for GR and v-src, we examined the effects of the same panel of Hsp90 mutations on activation of the Ste11 kinase (Fig. 6). The Ste11 kinase is a yeast homolog of the Raf serine/threonine kinase and was one of the first endogenous yeast Hsp90 clients identified. Its kinase activity is dependent on the presence of both Hsp90 and Cdc37 (Abbas-Terki et al., 2000; Louvion et al., 1998). Because Ste11 and v-src both require Cdc37 for maturation, the effects of Hsp90 ATPase domain mutants on Cdc37 binding affinity should have similar impacts on both of these kinase clients. Comparing the effects of our panel of Hsp90 mutations on v-src and Ste11 (Fig. 6A) indicates that most mutations that are compatible with efficient v-src maturation are also able to efficiently mature Ste11. Hsp90 mutations that are severely defective for v-src maturation had variable impacts on Ste11 maturation efficiency. This observation indicates that Ste11 imposes fewer constraints on the Hsp90 ATPase domain than v-src. While further studies will be required to investigate the molecular mechanism responsible for these client-specific constraints, the structural location of these mutations may provide valuable mechanistic hints. The seven Hsp90 mutations that were severely defective for both Ste11 and v-src maturation (colored pink in Fig 6A&B) are primarily in close proximity to either the Cdc37 interface or the nucleotide binding cavity (Fig. 6B), suggesting that direct contacts of Hsp90 with Cdc37 and nucleotide are critical for both of these kinase clients. In contrast, the three Hsp90 mutations that are defective for v-src but efficiently mature Ste11 (colored blue in Fig. 6A&B) are not in direct contact with either Cdc37 or nucleotide. This observation suggests that auxiliary Hsp90 contacts or conformational changes are critical for v-src maturation compared to Ste11.
Our observations indicate that distinct structural features of the Hsp90 ATPase domain contribute to the maturation of different clients. This observation suggests that it will be valuable to investigate the molecular mechanism of Hsp90 activation of multiple clients. Understanding client-specific mechanisms will likely contribute to our understanding of protein homeostasis in normal as well as cancerous cells, which often exhibit a heightened dependence on Hsp90 (Miyata et al., 2013; Whitesell et al., 1994). While the biophysical properties that determine the binding of clients to Hsp90 remain to be fully resolved, stability of the native state appears to be a contributing factor (Taipale et al., 2013). Consistent with the influence of folding stability on client interactions with Hsp90, subtle changes to the client sequence can influence Hsp90 dependence. For example, the c-src kinase that is nearly identical in sequence to v-src is only weakly dependent on Hsp90 for activation (Xu et al., 1999). Buchner and colleagues (Boczek et al., 2015) recently showed that three amino-acid differences between c-src and v-src mediate the majority of the differences in Hsp90 dependence. Analyses of the affinities of kinases for Hsp90 in mammalian cells demonstrated that v-src had one of the strongest observed levels of associations with Hsp90 (Taipale et al., 2012). It is possible that the heightened dependence of many cancer cells to Hsp90 may be due to an increased load of difficult to chaperone oncogenic clients similar to v-src, and that therapeutic interventions may be developed to target these clients with increased specificity.
Conclusions
Our results indicate that conformational changes mediated by ATP hydrolysis provide the main functional constraints in the ATPase domain of Hsp90. These constraints include strict physical requirements for the amino acids immediately contacting the phosphates of ATP and those that are structurally poised to propagate hydrolysis dependent conformational changes. Positions throughout the ATPase domain clearly contribute to function, though in ways less severely sensitive to amino acid changes than those mediating ATP hydrolysis. The critical functional steps mediated by ATP hydrolysis can differentially impact clients. The v-src kinase imposes more stringent constraints on hydrolysis driven steps than GR. Variation in the challenges that clients impose on hydrolysis driven steps indicate the potential for inhibitors that mediate these steps to provide increased client specificity.
Experimental Procedures
Bulk fitness competitions
The effects of point mutants on yeast growth were determined as previously described (Hietpas et al., 2012; Hietpas et al., 2011; Jiang et al., 2013). Libraries of Hsp82 (yeast Hsp90) point mutants were generated in p414ADHΔTER, which expresses Hsp82 at a reduced level relative to wild-type strains and provides a sensitive readout of the functional impacts of mutations (Jiang et al., 2013). Confidence intervals (95%) were calculated for the growth effects of each amino acid change (Table S1) based on the standard error (Sp) from sampling depth in sequencing (Boucher et al., 2014) using the equation below.
Where Rtotal is the total sequencing reads of all variants, Ei is the estimated frequency of a variant calculated based on sequencing reads (Ri/Rtotal). 95% confidence intervals for Ei were calculated as 2×Sp and propagated to estimate the impacts of sampling on selection coefficients. Reproducibility of fitness effects was assessed by performing a full biological replicate including separate yeast transformations for mutants at 20 amino acid positions. The reliability of the bulk competitions was further assessed by comparison to the growth effects of a panel of Hsp90 mutations determined in monoculture.
Assays for GR, v-src, and Ste11 activity
To facilitate the analyses of Hsp90 mutants with a full range of fitness effects, client assays were performed using a temperature sensitive strain (iG170D) where Hsp90 function can be rapidly inactivated prior to client analyses (Nathan et al., 1997). For analyses of clients at elevated temperature, we cloned Hsp90 mutations into p414GPD with an N-terminal His6 tag which produces near-endogenous levels of Hsp90 protein that is required for function at elevated temperatures (Borkovich et al., 1989). Glucocorticoid receptor (GR) function in yeast was analyzed using the P2A/GRGZ plasmid as described previously (Meyer et al., 2004b; Wayne and Bolon, 2007). To quantify v-src activity, we utilized a galactose inducible v-src plasmid (p316GALv-srcV5) described previously (Meyer et al., 2004b; Wayne and Bolon, 2007). Ste11 function in yeast was quantified using the FUS1-LacZ reporter plasmid pSB234 as described (Trueheart et al., 1987). See Supplemental Information for additional details.
ATPase activity of purified Hsp90 proteins
Hydrolysis of ATP by Hsp90 was determined using an enzymatic assay as previously described (Norby, 1988). ATPase assays were performed at 37°C using a Bio50 Spectrophotometer equipped with a Peltier temperature control unit (Cary) and a 1 cm pathlength cuvette. See Supplemental Information for additional details.
Statistical Analysis
The data presented are the representative or examples of three biological replicates unless it is specified. The error bars represent the SD for three independent experiments, unless it is indicated.
Additional Experimental Procedures can be found in Supplemental Information.
Supplementary Material
Highlights.
Functional requirement at each site of N-Hsp90 revealed by systematic mutagenesis
Conformational constraints depend on amino acid proximity to γ-phosphate of ATP
The v-src kinase imposes more stringent constraints on hydrolysis compared to GR
ATPase domain of Hsp90 uses client-specific features to mature different clients eTOC Blurb
Acknowledgments
We are thankful to members of the Bolon lab for conceptual discussions and editorial comments on the manuscript. This work was supported in part by a grant from the National Institutes of Health (R01-GM112844 to DNAB).
Footnotes
Author contributions
Conceptualization, P.M., J.M.F, D.N.B.; Methodology, P.M., J.M.F., T.N.S., D.N.B.; Investigation, P.M., J.M.F., T.N.S.; Writing – Original Draft, D.N.B., Writing – Review & Editing, P.M., J.M.F., T.N.S., D.N.B.; Visualization, P.M., J.M.F., T.N.S., D.N.B.; Project Administration, P.M., J.M.F., D.N.B.; Funding Acquisition, D.N.B.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbas-Terki T, Donze O, Picard D. The molecular chaperone Cdc37 is required for Ste11 function and pheromone-induced cell cycle arrest. FEBS letters. 2000;467:111–116. doi: 10.1016/s0014-5793(00)01134-0. [DOI] [PubMed] [Google Scholar]
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvira S, Cuellar J, Rohl A, Yamamoto S, Itoh H, Alfonso C, Rivas G, Buchner J, Valpuesta JM. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nature communications. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- Arlander SJ, Felts SJ, Wagner JM, Stensgard B, Toft DO, Karnitz LM. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J Biol Chem. 2006;281:2989–2998. doi: 10.1074/jbc.M508687200. [DOI] [PubMed] [Google Scholar]
- Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, Thomas JH. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in caenorhabditis elegans. Genetics. 2000;155:85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boczek EE, Reefschlager LG, Dehling M, Struller TJ, Hausler E, Seidl A, Kaila VR, Buchner J. Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1424342112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9:3919–3930. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher JI, Cote P, Flynn J, Jiang L, Laban A, Mishra P, Roscoe BP, Bolon DN. Viewing protein fitness landscapes through a next-gen lens. Genetics. 2014;198:461–471. doi: 10.1534/genetics.114.168351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy NC, Garcia YA, Sivils JC, Galigniana MD, Cox MB. Functions of the Hsp90-binding FKBP immunophilins. Sub-cellular biochemistry. 2015;78:35–68. doi: 10.1007/978-3-319-11731-7_2. [DOI] [PubMed] [Google Scholar]
- Harris SF, Shiau AK, Agard DA. The crystal structure of the carboxy-terminal dimerization domain of htpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure (Camb) 2004;12:1087–1097. doi: 10.1016/j.str.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Hawle P, Siepmann M, Harst A, Siderius M, Reusch HP, Obermann WM. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol Cell Biol. 2006;26:8385–8395. doi: 10.1128/MCB.02188-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas R, Roscoe B, Jiang L, Bolon DN. Fitness analyses of all possible point mutations for regions of genes in yeast. Nat Protoc. 2012;7:1382–1396. doi: 10.1038/nprot.2012.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Bank C, Jensen JD, Bolon DN. Shifting Fitness Landscapes In Response To Altered Enviornments. Evolution; international journal of organic evolution. 2013 doi: 10.1111/evo.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Jensen JD, Bolon DN. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Mishra P, Hietpas RT, Zeldovich KB, Bolon DN. Latent effects of hsp90 mutants revealed at reduced expression levels. PLoS Genet. 2013;9:e1003600. doi: 10.1371/journal.pgen.1003600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, Burrows FJ. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- Karagoz GE, Duarte AM, Akoury E, Ippel H, Biernat J, Moran Luengo T, Radli M, Didenko T, Nordhues BA, Veprintsev DB, et al. Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell. 2014;156:963–974. doi: 10.1016/j.cell.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157:1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Richter K, Reinstein J, Buchner J. Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat Struct Mol Biol. 2013;20:326–331. doi: 10.1038/nsmb.2502. [DOI] [PubMed] [Google Scholar]
- Louvion JF, Abbas-Terki T, Picard D. Hsp90 is required for pheromone signaling in yeast. Molecular biology of the cell. 1998;9:3071–3083. doi: 10.1091/mbc.9.11.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Liao C, Hu B, Mark Roe S, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. Embo J. 2004a;23:511–519. doi: 10.1038/sj.emboj.7600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Liao C, Hu B, Roe SM, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. Embo J. 2004b;23:1402–1410. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Nakamoto H, Neckers L. The therapeutic target Hsp90 and cancer hallmarks. Current pharmaceutical design. 2013;19:347–365. doi: 10.2174/138161213804143725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci U S A. 1997;94:12949–12956. doi: 10.1073/pnas.94.24.12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckers L, Trepel JB. Stressing the development of small molecules targeting HSP90. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:275–277. doi: 10.1158/1078-0432.CCR-13-2571. [DOI] [PubMed] [Google Scholar]
- Norby JG. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Slightly deleterious mutant substitutions in evolution. Nature. 1973;246:96–98. doi: 10.1038/246096a0. [DOI] [PubMed] [Google Scholar]
- Palmer CA, Watts RA, Houck LD, Picard AL, Arnold SJ. Evolutionary replacement of components in a salamander pheromone signaling complex: more evidence for phenotypic-molecular decoupling. Evolution; international journal of organic evolution. 2007;61:202–215. doi: 10.1111/j.1558-5646.2007.00017.x. [DOI] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. Embo J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Picard D, Khursheed B, Garabedian MJ, Fortin MG, Lindquist S, Yamamoto KR. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Prodromou C, Panaretou B, Chohan S, Siligardi G, O’Brien R, Ladbury JE, Roe SM, Piper PW, Pearl LH. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. Embo J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- Pullen L, Bolon DN. Enforced N-domain proximity stimulates Hsp90 ATPase activity and is compatible with function in vivo. J Biol Chem. 2011;286:11091–11098. doi: 10.1074/jbc.M111.223131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell NW, Mishra P, Bolon DN. Solubility-promoting function of hsp90 contributes to client maturation and robust cell growth. Eukaryot Cell. 2012;11:1033–1041. doi: 10.1128/EC.00099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzke C, Hellenkamp B, Hugel T. Four-colour FRET reveals directionality in the Hsp90 multicomponent machinery. Nature communications. 2014;5:4192. doi: 10.1038/ncomms5192. [DOI] [PubMed] [Google Scholar]
- Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, Richter K, Buchner J. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- Roe SM, Ali MM, Meyer P, Vaughan CK, Panaretou B, Piper PW, Prodromou C, Pearl LH. The Mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50(cdc37) Cell. 2004;116:87–98. doi: 10.1016/s0092-8674(03)01027-4. [DOI] [PubMed] [Google Scholar]
- Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342:1372–1375. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Southworth DR, Agard DA. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 2008;32:631–640. doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehlow KG, Robertson AD, Baldwin RL. Proline for alanine substitutions in the C-peptide helix of ribonuclease A. Biochemistry. 1991;30:5810–5814. doi: 10.1021/bi00237a026. [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Whitesell L, Santagata S, Zhang J, Liu Q, Gray NS, Lindquist S. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nature biotechnology. 2013;31:630–637. doi: 10.1038/nbt.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci U S A. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne N, Bolon DN. Dimerization of Hsp90 is required for in vivo function. Design and analysis of monomers and dimers. J Biol Chem. 2007;282:35386–35395. doi: 10.1074/jbc.M703844200. [DOI] [PubMed] [Google Scholar]
- Wayne N, Lai Y, Pullen L, Bolon DN. Modular control of cross-oligomerization: analysis of superstabilized Hsp90 homodimers in vivo. J Biol Chem. 2010;285:234–241. doi: 10.1074/jbc.M109.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne N, Mishra P, Bolon DN. Hsp90 and client protein maturation. Methods in molecular biology. 2011;787:33–44. doi: 10.1007/978-1-61779-295-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci U S A. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Singer MA, Lindquist S. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc Natl Acad Sci U S A. 1999;96:109–114. doi: 10.1073/pnas.96.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.