Abstract
Inter-alpha Inhibitor Proteins (IAIPs) are a family of related serine protease inhibitors. IAIPs are important components of the systemic innate immune system. We have identified endogenous IAIPs in the central nervous system (CNS) of sheep during development and shown that treatment with IAIPs reduces neuronal cell death and improves behavioral outcomes in neonatal rats after hypoxic-ischemic brain injury. The presence of IAIPs in CNS along with their exogenous neuroprotective properties suggests that endogenous IAIPs could be part of the innate immune system in CNS. The purpose of this study was to characterize expression and localization of IAIPs in CNS. We examined cellular expressions of IAIPs in vitro in cultured cortical mouse neurons, in cultured rat neurons, microglia, and astrocytes, and in vivo on brain sections by immunohistochemistry from embryonic (E) day-18 mice and postnatal (P) day-10 rats. Cultured cortical mouse neurons expressed the light chain gene Ambp and heavy chain genes Itih-1, 2, 3, 4, and 5 mRNA transcripts and IAIPs proteins. IAIP proteins were detected by immunohistochemistry in cultured cells as well as brain sections from E18 mice and P10 rats. Immunoreactivity was found in neurons, microglia, astrocytes and oligodendroglia in multiple brain regions including cortex and hippocampus, as well as within both the ependyma and choroid plexus. Our findings suggest that IAIPs are endogenous proteins expressed in a wide variety of cell types and regions both in vitro and in vivo in rodent CNS. We speculate that endogenous IAIPs may represent endogenous neuroprotective immunomodulatory proteins within the CNS.
Keywords: Brain, cell culture, fetal, Inter-alpha Inhibitors, neonate, neurons, rodent
INTRODUCTION
Endogenous proteases are found in all organisms from microbes to mammals and participate in various physiological and pathological processes including coagulation, immune responses, and cancer metastasis (Ranasinghe and McManus, 2013). In the central nervous system (CNS), a subfamily of proteases, serine proteases, and their inhibitors have been identified in both neurons and glia (Choi et al., 1990, Weinstein et al., 1995, Vivien and Buisson, 2000). Accumulating evidence suggests that serine proteases play important roles in neuronal development, plasticity, and pathology (Turgeon and Houenou, 1997) and that their inhibitors modulate neuronal cell death and exert neuroprotective properties in brain injury (Vivien and Buisson, 2000, Reuther et al., 2014).
Inter-alpha Inhibitor Proteins (IAIPs) are a group of serine protease inhibitors that have been detected in many tissues including liver, intestine, kidney, stomach, placenta and brain signifying their diverse biological functions (Itoh et al., 1996, Daveau et al., 1998, Takano et al., 1999, Spasova et al., 2014). Although liver is the primary site of IAIPs synthesis and the major source of the IAIPs found in blood, our recent findings suggest that IAIPs are also present in brain and other organs during development (Spasova et al., 2014).
In hepatocytes, IAIP molecules are encoded by an α1-microglobulin/bikunin precursor (Ambp) light chain gene and at least 5 structurally associated inter alpha-trypsin inhibitor heavy chain genes (Itih 1–5) (Salier et al., 1996, Zhuo et al., 2004). The genes of Ambp and Itih 1–5 code for the polypeptide precursors of the light chain (AMBP) and heavy chains (H1P, H2P, H3P, H4P, and H5P), which subsequently undergo post-translational processing including removal of signal peptides and trimming of C-terminal ends (Kaumeyer et al., 1986, Salier et al., 1987, Bratt et al., 1993, Salier et al., 1996, Zhuo and Kimata, 2008). The core mature polypeptide of the light chain (LC) is called bikunin because it contains two Kunitz type domains making it a serine protease inhibitor (Potempa et al., 1989). Bikunin is covalently linked to one or two mature polypeptide(s) of the heavy chains (HCs) by a glycosaminoglycan forming a bikunin-HC complex before secretion by liver into blood (Jessen et al., 1988, Zhuo and Kimata, 2008). There are two major bikunin-HC complexes found in blood: Inter-alpha Inhibitor (IaI, 250 kDa), in which bikunin is linked to HC-1 and HC-2 and Pre-alpha Inhibitor (PaI, 120 kDa), in which bikunin is linked to HC-3 (Enghild et al., 1989, Odum et al., 1989). In these complexes, bikunin remains inactive until it is cleaved by partial proteolytic degradation (Fries and Blom, 2000).
Currently, IAIPs are gaining increasing attention because of their potential therapeutic beneficial effects. Administration of exogenous bikunin and IAIPs has been shown to attenuate systemic inflammation (Lim et al., 2003, Singh et al., 2010) and inhibit tumor metastasis (Kobayashi et al., 2003, Suzuki et al., 2004). IAIPs exert beneficial effects in sepsis and systemic inflammatory disorders both in neonates and in adults (Lim et al., 2003, Singh et al., 2010). Very little information is available regarding IAIPs in CNS. We recently have identified endogenous IaI and PaI proteins in cerebral cortex, choroid plexus and cerebral spinal fluid of sheep during development and have shown that exogenous treatment with IAIPs reduce neuronal cell death and improve behavioral outcomes in neonatal rats exposed to hypoxic-ischemic brain injury (Spasova et al., 2014, Threlkeld et al., 2014, Gaudet et al., 2016). Others have shown beneficial effects of the light chain, bikunin, in cerebral ischemia-reperfusion injury and experimental autoimmune encephalomyelitis (Yano et al., 2003, Koga et al., 2010, Shu et al., 2011). Although IAIPs in blood are mainly of hepatic in origin, our recent findings have suggested that IAIPs in brain and other tissues may be endogenously produced (Spasova et al., 2014). Even though it is plausible that IAIPs might cross the blood-brain barrier via a yet to be demonstrated specific transport mechanism (Banks, 2015), their ability to readily traverse the blood-brain barrier seems unlikely given the large size of the IAIPs and the early development of blood-brain barrier integrity in the fetus (Stonestreet et al., 1996). It is far more likely that endogenous IAIPs (Spasova et al., 2014). are produced within specific brain cells. Hence, the major goal of this current study was to determine the presence of IAIPs mRNA and protein within specific cell types of the immature rodent brain.
In contrast to the increasing number of studies examining the exogenous effects of IAIPs, the endogenous localization, specific cellular expression, and regional distribution of IAIPs in CNS have not been previously examined, particularly in fetal and neonatal subjects. In addition, the information that is available regarding localization of IAIPs in CNS remains controversial. Some studies report the presence of the bikunin gene in neurons but not in astrocytes (Takano et al., 1999), whereas, others report bikunin immunoreactivity in astrocytes, but not in neurons (Yoshida et al., 1991a, Yoshida et al., 1994). Although we identified both the IaI and PaI proteins in the cerebral cortex of sheep (Spasova et al., 2014), more detailed information regarding the expression and localization of IAIPs in different cell types in the fetal and neonatal brain is lacking. Expression of endogenous IAIPs in cortical and hippocampal neurons may be of great importance given our previous observation that exogenous treatment with IAIPs attenuated cortical neuronal loss, improved behavioral outcomes, and decreased the number of basal dendrites per CA1 pyramidal neuron in the hippocampus of neonatal rats exposed to hypoxic ischemic brain injury (Threlkeld et al., 2014, Gaudet et al., 2016). Therefore, we reasoned that it would also be important to determine whether endogenous IAIPs were expressed by neurons in these critical brain regions.
The objective of the current study was to establish the presence and expression of IAIPs in fetal and neonatal CNS in an effort to begin to elucidate the cellular and regional distribution of endogenous IAIPs in the immature CNS. The current study also begins to explore the concept that endogenous IAIPs could be locally produced in diverse cell types and brain regions and potentially have paracrine/autocrine modes of action. Therefore, we examined the cellular expression of IAIPs in vitro in cultured cortical mouse neurons, in cultured rat neurons, microglia, and astrocytes, and in vivo in brain sections by immunohistochemistry from embryonic day 18 (E18) mice and postnatal day 10 rats (P10).
EXPERIMENTAL PROCEDURES
Experimental Animals
Wild type C57BL/6 mice and Wistar rats were obtained from The Jackson Laboratory (Bar Harbor, Maine, USA) and Charles River Laboratories (Wilmington, MA, USA), respectively. The present study was performed with approval by the Institutional Animal Care and Use Committees of the Alpert Medical School of Brown University and Women & Infants Hospital of Rhode Island and in accordance with the National Institutes of Health Guidelines for the use of experimental animals.
Primary cortical neuron, microglia, and astrocyte cultures
Mice on embryonic day 18 (E18) and rats on postnatal day 1–2 (P1–2) were used for the primary cortical neuronal cultures. The mouse brain samples used in this study were available from terminal mouse studies that did not use the CNS. The techniques for the neuronal cultures were performed as previously described in detail (Beaudoin et al., 2012). Briefly, the mice or rats were decapitated and the brains immediately transferred into a dissection medium [97.5% Hank’s balanced salt solution (Invitrogen, Frederick, MD, USA, 0.11 mg/ml sodium pyruvate (Invitrogen), 0.1% glucose, 10 mM HEPES (pH 7.3, Sigma-Aldrich), and 1000 U/ml penicillin/streptomycin (P/S, Invitrogen)]. The meninges were removed and the cerebral cortices dissected. Cerebral cortical tissues were dissociated with 0.25% trypsin (Worthington Biochemical Corp. Lakewood, NJ, USA) for 20 minutes at 37°C followed by 0.1% DNase (Sigma-Aldrich Inc., St. Louis, MD, USA) treatment for 5 minutes. Then, tissues were carefully triturated and the resulting cells were plated in minimum essential medium (MEM) with Earle’s Balanced Salt Solution (Invitrogen), 10% fetal bovine serum (FBS, Invitrogen), 0.45% glucose, 1 mM sodium pyruvate, 2 mM glutamine, and 1000 U/mL P/S) onto poly-D-lysine (Sigma-Aldrich)-coated 18-mm glass coverslips at a density of 260 cells per mm2. On the second day of plating, the culture medium was changed to maintenance media (neurobasal media, Invitrogen) containing 2% B27 supplement (Invitrogen), 2 mM glutamine, and 1000 U/mL P/S) without added serum or mitosis inhibitors. Two days after incubation at 37°C with 5% CO2, 5-μM cytosine arabinoside (AraC, Sigma-Aldrich) was added. The experiments for total RNA and protein extraction, and immunohistochemical detection of IAIPs were performed between 10–14 days after cell plating. The purity of the neuronal cultures was verified as 94.8 ± 1.4% (mean ± SD) by immunostaining with the neuronal marker microtubule-associated protein 2 (MAP-2), astrocytic marker glial fibrillary acidic protein (GFAP), and microglial marker Isolectin IB4 (data not shown).
Primary microglia and astrocytes were cultured as follows: P 1–2 rats were used and prepared as previously described (Faustino et al., 2011). Briefly, the brains were gently placed into a petri dish containing cold Dulbecco’s modified eagle medium (DMEM, Lonza, Allendale, NJ, USA). Cerebral cortical tissues treated as described above. The cell mixture was centrifuged for 10 minutes at 168 x g. The pellet was re-suspended in 10 ml of warmed DMEM 10:10:1 [(DMEM, 10% of FBS, 10% of horse serum (Invitrogen), 1% P/S]. The cell suspension was seeded on poly-D-Lysine covered 75 cm2 flasks (Corning Inc., Corning, NY, USA). The cells were incubated in a humid incubator at 37°C with 5% CO2 for 7 to 10 days without changing the culture medium. After incubation, the flask was shaken at 230 rpm for three hours. The microglial cell pellet was obtained by centrifuging the cell suspension for 10 minutes at 168 x g, and then re-suspending the suspension with warmed DMEM 10:10:1 and plating the cells onto glass coverslips at a density of 280 cells per mm2.
Astrocytes were isolated as follows. The flask containing the cell suspension was immediately filled with 10 ml of warmed DMEM 10:10:1, closed and then shaken at 260 rpm overnight. After washing the flask twice with phosphate-buffered saline (PBS), the cells were treated with five ml of trypsin (0.05%) plus EDTA (0.02%) for 5–10 minutes in the incubator. The cell suspension was collected by adding five ml of DMEM 5:5:1 (DMEM, 5% of FBS, 5% of horse serum, 1% P/S) into the flask. After centrifugation for 10 minutes at 168 x g, the pellet was re-suspended with five ml of DMEM 5:5:1 and seeded onto glass coverslips at a density of 340 cells per mm2. The purity and ratios of microglia and astrocytes to neurons were verified by immunostaining with specific cell markers. The purity was 99.1 ± 0.6% and 98.7 ± 1.2%, respectively (data not shown).
Primary antibodies
Polyclonal antibodies (pR-21) were raised against rat IAIPs generated by immunizing rabbits with highly purified IAIPs derived from rat plasma. The pR-21 cross-reacts with IAIPs including human, mouse, rat, and sheep, and detects both 250 kDa IαI and 125 kDa PαI molecules in Western immunoblot (data not shown). The primary antibodies used in this study are as follows: pR-21 (1:500, ProThera Biologics, Inc., Providence, RI, USA), mouse anti-Fox3/neuronal nuclei (NeuN) monoclonal antibody (mAb, 1:500, Abcam, Cambridge, MA, USA), mouse anti-microtubule-associated protein-2 (MAP-2) mAb (1:500, Abcam), chicken anti-glial fibrillary acidic protein (GFAP) polyclonal antibody (pAb, 1:1000, Abcam), mouse anti-ionized calcium binding adaptor molecule 1 (Iba-1) mAb (1:200, Abcam), griffonia simplicifolia anti-isolectin B4 (IB4) antibody (10 μg/ml, Invitrogen), mouse anti-A2B5 mAb (1:1000, Abcam), mouse anti-2′, 3′-cyclic nucleotide 3′-phosphodiesterase (CNPase) mAb (1:500, Abcam).
Total protein and RNA extraction
Total protein and RNA were extracted from the primary cultured mouse neurons. Cells grown on coverslips were rinsed briefly with PBS three times. One ml of ice-cold RIPA buffer (150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate, 50 mM Tris (pH 8.0) with 1% complete protease inhibitor cocktail (Roche, Nutley, NJ, USA) was added to each 22 mm diameter well of the culture plates and incubated with the cells for five minutes on ice. The cells were centrifuged at 10,000-x g for 20 minutes at 4°C. The supernatant was collected and the protein concentration determined (Bicinchoninic acid protein assay, BCA, Pierce, Rockford, IL, USA).
RNA was extracted from neurons as follows. One ml of TRIZOL (Invitrogen) reagent was added to the 35 mm diameter culture dish after briefly rinsing the neuronal monolayer with PBS three times. Adult mouse liver was used as a control. One ml of TRIZOL reagent per 50 mg of liver sample was added. The liver tissues were then homogenized using a glass-Teflon® homogenizer and centrifuged at 12,000-x g for 15 minutes at 4°C. The homogenized liver tissue samples were incubated for five minutes at room temperature. After the addition of 0.2 ml of chloroform (Sigma-Adrich), samples containing the neurons and liver were vortexed thoroughly for 15 seconds, incubated at room temperature for three minutes, and then centrifuged at 12,000-x g for 15 minutes at 4°C. The RNA pellet was precipitated with isopropyl alcohol (Sigma-Aldrich), washed with 75% ethanol twice, and air-dried at room temperature. The concentration of resultant RNA was determined by Spectrophotometer (Nanodrop 2000, Thermo Scientific, Wilmington, DE, USA).
RNA and protein quantitation
Ten and 20 μg protein of total protein extracted from neurons were fractionated by SDS-PAGE under reducing conditions and transferred onto a Polyvinylidene fluoride (PVDF) membrane (0.2 mm, Bio-Rad Laboratories, Hercules, CA) using a semi-dry technique. Membranes were incubated with pR-21 at a dilution of 1:5000 overnight at 4°C. After three washes with Tris-Buffered Saline-0.1% Tween-20 (TBS-T), membranes were incubated with HRP-conjugated goat anti-rabbit antibody (1:10,000) for one hour. Binding of the secondary antibody was detected with enhanced chemiluminescence (ECL plus, Amersham Pharmacia Biotech, Inc., Piscataway, NJ, USA) before exposure to autoradiography film (Daigger, Vernon Hills, IL, USA). β-actin (Santa Cruz Biotechnology, Inc. Dallas, Texas, USA) was used as a loading control.
One μg of total RNA was treated with one μl of deoxyribonuclease-I (DNase, Amp Grade, 1 U/μl, Invitrogen) for 15 minutes before First-Strand cDNA synthesis. First-strand cDNA was synthesized by SuperScript® III First-Strand Synthesis SuperMix (Invitrogen) according to the manufacturer’s protocol, in which Random hexamer was used and recombinant RNase is included to protect from target RNA degradation because of RNase contamination. OneTag Hot Start DNA polymerase (New England Biolabs Inc, Ipswich, MA, USA) was used to amplify cDNA sequences according to the manufacturer’s protocol.
Polymerase chain reaction (PCR) amplification was performed in a 25-μl reaction mixture using a Veriti® 96-Well Thermal Cycler (Grand Island, NY, USA). The mixture contains one μl of cDNA, 0.625 units OneTag Hot Start DNA polymerase (New England Biolabs), 1 x OneTag buffer (New England Biolabs), 200 μM dNTPs (New England Biolabs), 0.2 μM of forward and reverse specific primers (IDT Inc., Skokie, IL, USA). The touch-down PCR was programed as follows: 1) step one: initial denaturation at 94°C for 30 seconds; 2) step two: denaturation at 94°C for 30 seconds, annealing for 45 seconds at 65°C for Ambp, Itih-2 and Itih-4, 68°C for Itih-1 and Itih-3, and 67°C for Itih-5, elongation at 68°C for 45 seconds for Ambp, Itih-1 and Itih-3, for 60 sec for Itih-2, Itih-4 and Itih-5; (10 cycles were performed, and the annealing temperature was decreased by 1°C per cycle); 3) step three: denaturation at 94°C for 30 seconds, annealing for 45 sec at 56°C for Ambp, Itih-2 and Itih-4, 59°C for Itih-1 and Itih-3, and 58°C for Itih-5, elongation at 68°C for 45 seconds for Ambp, Itih-1 and Itih-3, for 60 sec for Itih-2, Itih-4 and Itih-5 (40 cycles were performed for Ambp, Itih-1 and Itih-3, 35 cycles were performed for Itih-2, Itih-4 and Itih-5); 4) step four: elongation at 68°C for five minutes and end of reaction at 4°C for long-term storage. The primer sequences and predicted PCR fragment sizes are listed in Table 1. PCR products were separated and visualized on a 2% agarose gel containing 10 ng/ml of ethidium bromide, ChemiDoc XRS System (Bio-Rad).
Table 1.
Primers used for mouse IAIP nucleotide sequence amplification.
| Gene | Accession No. | Primer sequences (5′–3′) | Size (bps) |
|---|---|---|---|
| Bikunin (Ambp) | NM_007443 | F: GCCTTTGCAGCCCCCGTAGTG | 208 bp |
| R: CATGGCCTGCGAGACCTTTCAATA | |||
| Itih-1 | NM_008406 | F: AAGGCGTTCATTGGGGACATAAAG | 397 bp |
| R: TCACAGTGGGGCGGAAGAGC | |||
| Itih-2 | NM_010582 | F: CGCCGATAAGCTGACCGTTGACT | 371 bp |
| R: TCCGCTTACTGCCATGGTGATAGA | |||
| Itih-3 | NM_008407 | F: CCATCGGGGGCAAGTTCC | 514 bp |
| R: AGGGCCTCCGCTGTGATGT | |||
| Itih-4 | NM_018746 | F: GCCGAAGCCCAGAAACAATACAGT | 295 bp |
| R: CCGGGGTCATGAAGGTGCTCT | |||
| Itih-5 | NM_172471 | F: ACAACGGGCAAGCACAGGTAGA | 247 bp |
| R: AGCGCATAGCAAGAGCCAAGGTAG |
Immunohistochemistry of primary cultured cells
Primary cultured cells were rinsed with PBS three times for five minutes each, fixed in 4% paraformaldehyde/4% sucrose in PBS for 15 minutes, and permeabilized in PBS with 0.3% triton X-100 (PBS-T) for 10 minutes. Non-specific binding of antibodies was blocked by incubating cells with 1% bovine serum albumin (BSA, Sigma-Aldrich) and 5% normal goat serum (Vector Laboratories Inc. Burlingame, CA, USA) in PBS-T for one hour. Cells were stained with primary antibodies overnight at 4°C. After washing three times in PBS five minutes each, cells were incubated with appropriate secondary antibodies (Alexa Fluor® 488 goat anti-rabbit IgG, 1:500, Invitrogen; Alexa Fluor® 555 goat anti-mouse IgG, 1:500, Invitrogen; Alexa Fluor® 488 goat anti-rabbit IgG, 1:500, Invitrogen; Alexa Fluor® 555 goat anti-chicken IgG, 1:1000, Invitrogen) for one hour in the dark. Coverslips were then mounted with DAPI containing media (Vector Laboratories).
Immunohistochemistry of paraffin-embedded brain sections
Coronal brain sections (10 μm thick) obtained from E18 and P10 rats were treated as follows. Sections were deparaffinized and rehydrated. Brain sections were immersed in sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) and heated in an autoclave at 120°C for 10 minutes. After heat-induced epitope retrieval, the slides were rinsed with Tris-buffered saline (TBS, 10 mM) containing 0.025% triton-X100 (TBS-T, Sigma-Aldrich) twice for five minutes each. After blocking in buffer (1% BSA and 5% normal goat serum in TBS-T) for two hours, the brain sections were incubated with the primary antibodies at 4°C overnight. On the second day, the slides were rinsed with TBS-T twice for five minutes and incubated with fluorophore-conjugated secondary antibodies for one hour in the dark. Counterstaining and mounting were performed as described above. Slides stained with 3,3′-diaminobenzidine (DAB) and were treated with 0.3% H2O2 (Sigma-Aldrich) in TBS for 10 minutes to block the endogenous peroxidase before incubating with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies (Alpha Diagnostic, San Antonio, TX, USA) for one hour. Slides were rinsed with TBS three times for two minutes each, and incubated with DAB solution (R&D Systems, Minneapolis, MN, USA). After DAB staining, the slides were rinsed in distilled water, dehydrated with 70%, 95%, and 100% ethanol two times for 3 minutes each, cleared in xylene two times for five minutes each, and mounted with cytoseal (Thermo Scientific, Waltham, MA, USA). The DAB staining was used in addition to the immunofluorescent methodology to confirm similar distributions of the reaction product using a different chromogen with greater long-term stability. No differences in staining patterns were observed between the two methods. Pre-immune serum (PIS, 1:500, ProThera Biologics) obtained from a rabbit before immunization with rat IAIPs was used as a negative control in each staining procedure. The images were digitized and visualized using a Zeiss AxioImager M2 microscope (Carl Zeiss Meditec, Peabody, MA, USA) and digital analysis software Stereo Investigator (MBF Bioscience, Williston, VT, USA). Anatomical descriptions of rodent brain regions as described in the results for Figs. 3–5 were determined by E.G.S with the use of standard mouse and rat neuroanatomical references(Paxinos and Watson, 1997, Paxinos and Franklin, 2001).
Fig. 3.
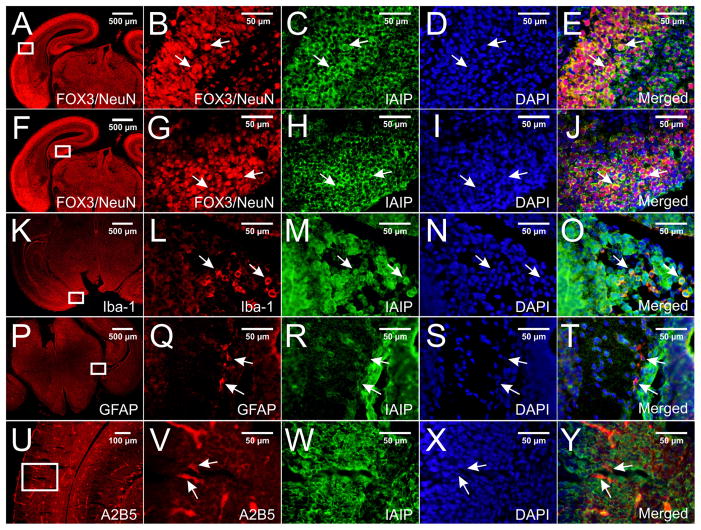
Double immune-staining with pR-21 and with specific brain cell markers in paraffin-embedded E18 mouse brain sections. IAIPs immunoreactive signals were detected in different brain regions including cortex and hippocampus. Figs. 3E and 3J show co-localization of IAIPs with FOX3/NeuN (Figs. 3B and G) in neurons in the entorhinal cortex and hippocampus, respectively. Figs. 3O and 3T show co-localization of IAIPs with Iba-1 (L) and GFAP (Q) in the microglial cells and astrocytes in the area of medial hippocampus, respectively. A2B5-positive (U and V) glial progenitors were not found to co-localize with IAIP (W and Y). Specific IAIPs staining was mainly localized to cytoplasm and perinuclear of cells (E, J, O, T arrows). DAPI was used as a counterstaining (D, I, N, S, X).
Fig. 5.
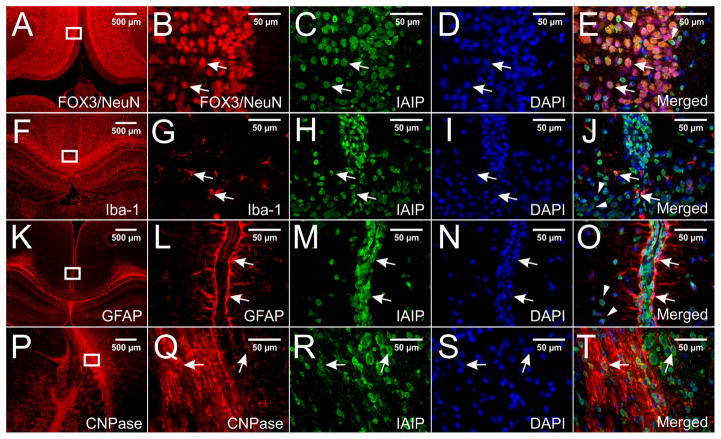
Double immune-staining with pR-21 and with specific brain cell markers in paraffin-embedded P10 rat brain sections. Immunoreactivity of IAIPs were detected in neurons, microglial cells, astrocytes, and oligodendrocytes in different brain regions. Figs 5E, J, and O show co-localization of IAIPs with FOX3/NeuN (B), Iba-1, and GFAP in neurons, microglial cells, and astrocytes in the retrosplenial granular cortex, respectively. Fig. 5T shows co-localization of IAIPs with CNPase (Q) in the oligodendrocytes in the medial hippocampus adjacent to the geniculate body. Specific IAIPs staining was mainly localized to perinuclear of cells (E, J, O, T, arrows). DAPI was used to stain nuclei (D, I, N, S).
RESULTS
Cultured mouse neurons: IAIPs gene and protein expression
Cultured neurons from E18 mouse cerebral cortex were used to identify the gene and protein expression of IAIPs in CNS. We used neurons at this embryonic stage because they are more robust and less susceptible to damage during the culture procedures (Banker and Cowan, 1977, 1979, Beaudoin et al., 2012), and embryonic cultures produce a higher yield of healthy viable cells with a lower proportion of non-neuronal cells compared with postnatal cultures (Beaudoin et al., 2012).
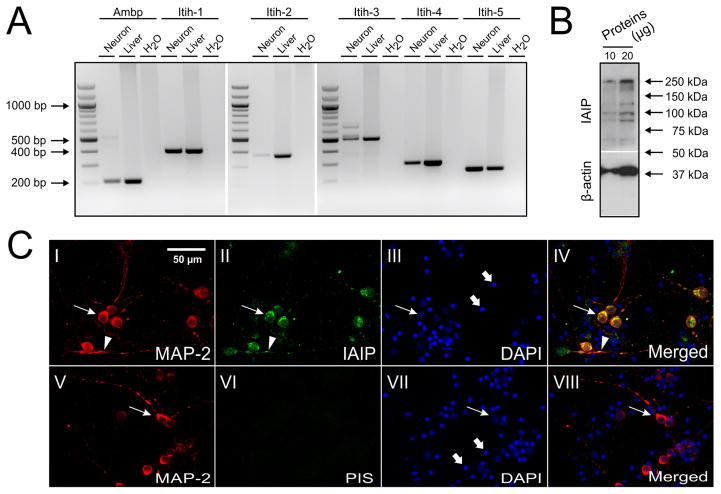
The mRNA transcripts of Ambp and Itih-1, 2, 3, 4, 5 were determined by reverse transcript (RT)-PCR, which was performed on cDNAs prepared from mouse neuronal cultures. As shown in Fig. 1A, mouse neurons growing in culture express Ambp and Itih-1, 2, 3, 4, and 5 with the expected product base pair size (bp, Table 1). The RT-PCR bands for Ambp and Itih 1, 2, 3, 4, and 5 from mouse neurons and liver were of same bp size, indicating that similar mRNA transcripts of IAIPs are expressed in both neurons and liver cells.
Fig. 1.
Gene and protein expressions of IAIPs in mouse cultured neurons. (A) RT-PCR shows that mRNAs of Ambp and Itih-1, 2, 3, 4, 5 are expressed in the neurons with expected bp size. Water and mRNA of mouse liver were used as negative and positive controls, respectively. (B) Bands of IaI (250 kDa) and PaI (120 kDa) were detected by Western-immunoblot. (C) Neurons were double-stained with antibodies against MAP-2 (I: red) and IAIPs (II: green). IAIPs are co-localized with MAP-2 and enriched in the cytoplasm (II and IV, arrows) and dendrites (II and IV, arrowheads). PIS and DAPI were used as negative (VI and VIII), and counter-stains, respectively (III and VII). In addition, neuronal nuclei (III and VII, arrows), fragmented cellular debris caused by the preparation procedure, and non-neuronal dividing cells undergoing cell death caused by AraC were stained by DAPI (III and VII, thick arrows).
IAIP proteins were also expressed in cultured mouse neurons (Fig. 1 B). IAIPs were detected by Western immunoblot using the polyclonal antibody (pR-21) against the rat IAIPs. The pR-21 recognizes the IAIP proteins including IaI at 250 kDa and PaI 125 kDa (Fig. 1B). The polyclonal antibody pR-21 binds to the heavy chains as well as the bikunin light chain of IAIPs. The additional bands at 90, 100, 175 kDa in the Western immunoblot (Fig. 1B) were most likely protein breakdown products of the IAIP complex including free heavy chains H1, H2 or H3 and a combination of a single heavy chains and the light chain or 2 heavy chains. Protein bands were not detected in the PIS treated Western immunoblots (data not shown).
In order to establish that the IAIPs mRNA detected by RT-PCR and protein expression by Western-immunoblot did not result from minor glia cell contamination within the neuronal cultures, the cultured neurons were also double-stained with the dendritic marker, anti-MAP-2, and the pR-21 antibodies (Fig. 1C). MAP-2 and IAIPs were colocalized in neurons with the greatest expression detected in the neuronal cytoplasm (Fig. 1C-II, IV, arrows) and dendrites (Fig. 1C-II, IV, arrowheads). Specific immunoreactive signals were not detected by double staining of MAP-2 and PIS (Fig. 1C-VI, VIII, arrows). DAPI also stained some structures found in the neuronal cultures (Fig. 1C-III, VII, arrows and Fig. 1C-III, VII, thick arrows), which are considerably smaller than neuronal nuclei and probably represented fragmented cell debris caused by the preparation procedure, and/or non-neuronal dividing cells undergoing cell death after AraC exposure (Beaudoin et al., 2012).
Cultured rat neurons, microglia, and astrocytes: IAIPs immunoreactivity
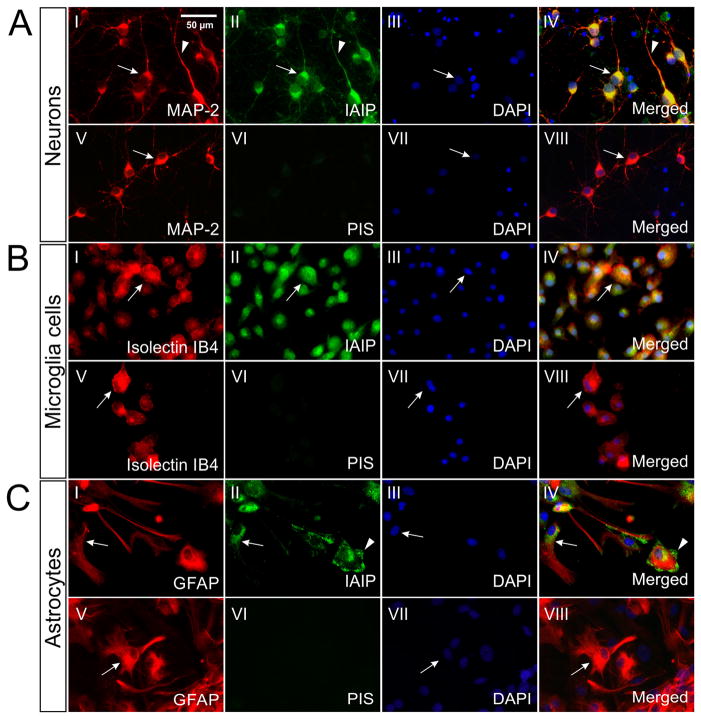
IAIPs expression was further confirmed in rat neuronal cultures by co-localization of IAIPs with MAP-2 (Fig. 2 A). The confirmation of IAIPs in both mouse and rat neuronal cultures further validates the presence of IAIPs in neurons because substantial differences exist between CNS tissues in mice and rats (Giussani et al., 1998, Rosenzweig and Luu, 2010). Consistent with the results from cultured mouse neurons, immunoreactive signals of IAIPs are detected in rat neurons and primarily expressed in cytoplasm (arrow) and dendrites (arrow head, Fig. 2A-I, II, IV). Our findings strongly suggest that IAIPs proteins are expressed in cultured neurons of two different rodent species.
Fig. 2.
IAIPs immunoreactivity in cultured rat cortical neurons, microglial cells and astrocytes. (A): Double immune-staining for MAP-2 (AI) and IAIPs (AII) shows that IAIPs were confined to neurons [cell body (arrow)] and dendrites (arrowhead), AIV). (B): Double immune-staining for Isolectin IB4 (BI, arrow) and IAIPs (BII, arrow) demonstrated the presence of IAIPs in the cytoplasm of microglial cells (BIV, arrow). (C): Double staining for GFAP (CI, arrow) and IAIPs (CII, arrow) revealed a perinuclear localization of IAIPs in astrocytes (CIV, arrow). Localization of IAIPs in the end-feet area of astrocytes was observed (CII and CIV, arrowhead). PIS negative staining and DAPI counter-staining were performed for each experiments. Bars: 50 μm.
Cultured microglial cells and astrocytes were also double-stained with pR-21 and cell specific markers in order to define further IAIPs expression in CNS. IAIPs co-localized with isolectin IB4 in microglial cells (Fig. 2B-I, II, IV, arrows). IAIPs immunoreactivity was mainly detected in cytoplasm. IAIPs immunoreactivity was also detected in GFAP-stained astrocytes, mainly in perinuclear and cytoplasmic distributions (Fig. 2C-I, II, IV, arrows). Interestingly, immunofluorescence for IAIPs was also identified at astrocytic end-feet (Fig. 2C-II, IV, arrowheads). Therefore, our findings of IAIPs in the in vitro cultured neurons, microglia and astrocytes firmly establish that these cell types endogenously express IAIPs.
E18 mouse CNS: IAIPs immunoreactivity
Embryonic and postnatal stages are the most important periods of mammalian brain development (Han et al., 2009). Neuronal development is established by E18 in mouse brain (Han et al., 2009, Watkins et al., 2012). Therefore, we investigated the expression of IAIPs in neurons with immunohistochemical staining using Fox3/NeuN as a neuronal marker in the cerebral cortex and in the hippocampus of the E18 mouse brain (Figs. 3A–B and 3F–G). IAIPs expression is shown in the neurons of the cerebral cortex such as the entorhinal cortex (Figs. 3C and E), and hippocampus (Figs. 3H and J) in the E18 mouse brain. Although for the purposes of illustration, Figure 3 contains select regions of the cerebral cortex and hippocampus, IAIPs expression was widely evident in neurons in most brain regions.
Mature astrocytes and oligodendrocytes can be first detected at about E16 and birth, respectively (Qian et al., 2000), and microglia derived from myeloid progenitors have already started to infiltrate into mouse brain around E9.5 (Alliot et al., 1999, Ueno et al., 2013). Consequently, we also examined E18 mouse brain sections for co-localization of IAIPs with Iba-1 (microglia marker, Figs. 3K–O), GFAP (mature astrocyte marker, Figs. 3P–T), and CNPase (mature oligodendrocyte marker, data not shown).
IAIPs co-localization was detected in neurons in E18 mouse cortex such as entorhinal cortex (Figs. 3 C–E, arrows) and hippocampus (Figs. 3 H–J, arrows). The Fox3/NeuN antibody staining identifies both neuronal nuclei and distal cytoplasmic processes (Kim et al., 2009). Therefore, we used a DAPI counterstain along with the Fox3/NeuN stain to demonstrate that IAIPs co-localization was more abundant in the perinuclear and cytoplasmic areas than within the nucleus (Figs. 3C–E, arrows). We also found that IAIPs are co-localized with Iba-1 in the medial hippocampus of the mouse brain (Figs. 3L–O, arrows) and with GFAP in the medial hippocampus (Figs. 3Q–T, arrows) primarily in the perinuclear and cytoplasmic areas of astrocytes. However, the number of Iba-1- and GFAP-positive cells at this stage of brain development was relatively low. On the other hand, CNPase-positive cells were not detected in the E18 mouse brain sections by immunohistochemical staining (data not shown). Therefore, we examined whether IAIPs were expressed in glial cells at E18 using an A2B5 antibody. A2B5 is a cell surface ganglioside epitope that is expressed on glia cell progenitors during the early stages of glia differentiation (Bottenstein et al., 1988, Salmaso et al., 2014). Although cells could be visualized with A2B5 positive expression by fluorescence microscopy in brain regions such as white matter of the occipital cortex at E18 mouse (Figs. 3U–V, arrows), IAIP co-localization was not detected in the A2B5 positive cells (Figs. 3W–Y, arrows).
P10 Rat CNS: IAIPs immunoreactivity
IAIPs expression was also identified in neonatal brain tissue sections from P10 rats in addition to the primary cell cultures. P10 rats were selected because after primary cells have been in culture for 10 days, they exhibit some important developmental features such as synapse maturation (Grabrucker et al., 2009). In addition, oligodendrocyte differentiation peaks at P10–14 (Richardson, 2009) and the brain development of P10 neonatal rats is similar to that of full-term human infants (Dobbing and Sands, 1979, Towfighi et al., 1991, Jansen and Low, 1996).
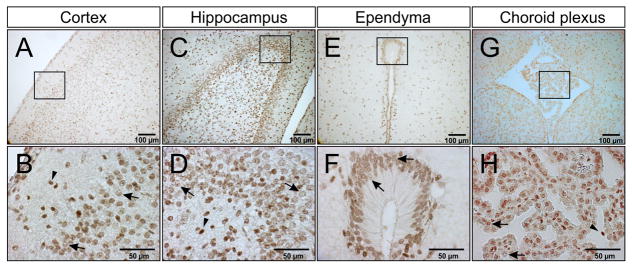
Figure 4 illustrates that IAIPs were detected by the pR-21 antibody on immersion fixed paraffin-embedded sections of P10 rat CNS and by DAB staining. Although for the purpose of illustration, Figure 4 contains select regions of the P10 rat brain, IAIPs were diffusely expressed in the cerebral cortex and in most other brain regions. DAB positive cells are widely distributed in P10 rat brain including cerebral cortical layers two and three shown in Figs. 4A–B and the hippocampus (dentate gyrus shown in Figs. 4C–D), striatum (data not shown), ependymal lining of the CSF-filled ventricles such as the subcommissural organ and cerebral aqueduct (Figs. 4E–F), choroid plexus (Figs. 4G–H), and vascular structures (data not shown). Specific staining was mainly observed in the cytoplasm (Figs. 4B, D, F, H, arrows), and occasionally in the nucleus (Figs. 4B, D, F, H, arrowheads). IAIPs were detected in ependymal cells (Figs. 4E–F, and Figs. 4G–H) and endothelial cells (data not shown) by DAB staining. In addition, IAIPs were also found to be widely expressed in neurons, microglia, astrocytes, and oligodendrocytes in different rat P10 brain regions by double immunofluorescence staining as described in the methods. For example, IAIPs were found in neurons (Figs. 5B–E arrows), and microglial cells (Figs. 5G–J arrows), of the retrosplenial granular cortex, astrocytes of the retrosplenial granular cortex above the corpus callosum (Figs. 5 L–O arrows) and oligodendrocytes of the medial hippocampus adjacent to the geniculate body (Figs. 5 Q–T arrows). IAIPs were observed to be more abundant in the perinuclear areas than in the nucleus of neurons, microglial cells, astrocytes, and oligodendrocytes (Figs. 5 E, J, O, T arrowheads). Although IAIPs were found in the oligodendrocyte soma (Figs. 5 Q, R, T arrowheads), co-localization of IAIPs with CNPase in the myelin sheaths was not detected.
Fig. 4.
IAIPs DAB staining in P10 rat brain tissues. Immunohistochemistry (peroxidase, DAB) for IAIPs is positive in different P10 brain regions including cerebral cortex (A), hippocampus (C), ependyma (E), and choroid plexus (G). Representative images of IAIPs DAB staining were shown in the molecular layers two and three of cortex (B), dental gyrus of hippocampus (D), ependyma of subcommissural organ (F), choroid plexus of third ventricle (H).
Overall, positive IAIP cells were widely observed in different brain regions including cerebral cortex, hippocampus, striatum (data not shown) and corpus callosum (data not shown). IAIPs immunoreactivity was specific, as it was not detected by DAB or immunofluorescent staining in PIS control samples (data not shown).
DISCUSSION
The purpose of the current study was to investigate the expression and localization of IAIPs in the immature CNS as an initial step to understand further the implications of the widespread abundance of IAIPs in brain (Spasova et al., 2014). In this study, we used primary cortical cell cultures in vitro, and E18 and P10 brain sections in vivo to demonstrate the wide range of endogenous expression and localization patterns of IAIPs in CNS. There were four main findings of our study. First, genes and proteins of IAIPs were expressed in cultured mouse neurons. Second, IAIPs immunoreactivity was also found in neurons, microglia cells, and astrocytes in isolated cultured rat cerebral cortical cells. Third, IAIPs immunoreactivity was detected in discrete brain regions including cerebral cortex, hippocampus, ependyma, choroid plexus, and microvessels in neonatal rats. Fourth, IAIPs were detected specifically in neurons, microglia cells, and astrocytes in paraffin embedded sections from the fetal and neonatal rodent brain. Consequently, there is extensive expression of endogenous IAIPs genes and proteins in a variety of brain cells and regions in vitro and in vivo in both mouse and rat CNS. These findings support our contention (Spasova et al., 2014) that endogenous IAIPs represent a previously unrecognized important component of normal brain composition and most likely function. The widespread expression of IAIPs both in vitro cultured cells and in vivo strongly suggests that these proteins are locally produced in brain cells.
Tissue-specific expression of one or more IAIPs family-related Ambp and Itih mRNA transcripts has been reported in a variety of animal models (Milland et al., 1990, Chan et al., 1995, Salier et al., 1996, Daveau et al., 1998). The brain has been suggested to be a predominant extrahepatic site of IAIPs expression (Daveau et al., 1998). However, only Itih-2 and -3 transcripts have been previously reported in both adult mouse and rat brains at low levels (Chan and Salier, 1993, Chan et al., 1995, Daveau et al., 1998). We have detected Ambp, and Itih-1, -4, and -5 transcripts for the first time in cultured mouse neurons in addition to the Itih-2 and -3 transcripts. Our observation of the presence of Ambp in mouse neurons is consistent with previous work by Takano, et. al (Takano et al., 1999). However, the presence of Ambp and Itih-1 and -4 in mouse neurons is in contrast to the work of Chan, et. al and Daveau, et. al, in which the Ambp transcript was not detected by RT-PCR in mouse brain (Chan et al., 1995) and Itih-1 and -4 transcripts were restricted to the liver (Daveau et al., 1998). These discrepancies could be attributed to improved sensitivity and accuracy of the RT-PCR methodologies. Transcriptions of Ambp and Itih-1, -2, and -3 imply that the mature IAIPs protein complexes of IαI and PαI are present in brain, which we confirmed by Western immunoblot and by double immunostaining of cultured mouse neurons. Our findings in rodent cultured neurons are consistent with the same assembly of IAIPs as found in the liver. Gene expressions of Ambp/Itih-2, -3, -4 and Itih-5 were apparently also regulated in turpentine-induced systemic inflammation and breast ductal carcinoma (Daveau et al., 1998, Himmelfarb et al., 2004) suggesting that endogenous gene expression of IAIPs in neurons could potentially have an important role in neuroinflammation and brain tumor metastasis.
The presence of IAIPs immunoreactivity in cultured rat neurons is consistent with our findings of IAIPs in cultured mouse neurons suggesting that IAIPs genes and proteins are present in neurons of both rodent species (Daveau et al., 1998). In addition, our finding that IAIP genes are present within the cultured neurons strongly suggests the potential for local synthesis of IAIPs with in the neurons. Immunoreactivity of IAIPs was also detected in cultured rat microglial cells. To the best of our knowledge, our findings are the first demonstration of immunoreactivity of IAIPs in microglial cells in vitro. In addition to neurons and microglial cells, cultured astrocytes also were stained with the pR-21 antibody. Interestingly, IAIPs expression in cultured neuronal and microglial cells appeared to be restricted to intracellular compartments, whereas expression was mainly localized to the perinuclear areas and occasionally on the end-feet area of astrocytes, suggesting that astrocytes could be a major origin of cellular IAIPs secretion in brain. Yoshida, et. al also reported the presence of immunoreactivity for IAIPs in astrocytes (Yoshida et al., 1991b, Yoshida et al., 1994). However, the presence of immunoreactivity for IAIPs was detected in astrocytes under pathological conditions such as human brain with tumors (Yoshida et al., 1994) and Alzheimer’s disease (Yoshida et al., 1991b).
Although the presence of Itih-1 has not been formerly reported in brain, we have previously demonstrated that the IaI along with PaI complex was present in newborn sheep cerebral cortex and choroid plexus (Spasova et al., 2014). However, in our earlier work the cell types expressing IAIPs were not determined in fetal or neonatal brain. In the current study, IAIP protein expression was detected in numerous brain regions including cerebral cortex, hippocampus, striatum, corpus callosum, ependyma, and microvessels in P10 rats. Furthermore, neurons and glial cells including microglial cells, astrocytes, and oligodendrocytes also exhibited IAIPs expression by immunohistochemistry consistent with our results in vitro.
The timing of neurogenesis and gliogenesis in the developing mammalian CNS are distinct. In the mouse brain, neurogenesis begins on E10–E11 and is completed around the time of birth (Bayer and Altman, 1991, Jacobson, 1991), whereas the macroglia including astrocytes and oligodendrocytes begin to appear in the cortex around mid-gestation at very low levels (Cameron and Rakic, 1991, Mission et al., 1991, Qian et al., 2000). The first immature oligodendrocytes that express the O4 marker and GFAP-positive cells can both be detected at around E17 but remain at low levels throughout the embryonic period (Abney et al., 1981, Qian et al., 2000). The myeloid progenitor-derived microglia infiltrate into mouse brain around E9.5 and accumulate along the areas of subcerebral and callosal projection axons starting from P1 to P3 (Alliot et al., 1999, Ueno et al., 2013). Consistent with our findings in P10 rats, we detected IAIP protein expression in neurons, microglia, and mature astrocytes in E18 mice. The presence of IAIPs in GFAP labeled isolated cultured rat astrocytes (Fig. 2C), and by immunohistochemical detection in E18 mouse brain (Figs. 3P–T) and in P10 rats (Figs. 5K–O) brain strongly suggest that IAIPs are expressed in astrocytes (Levitt and Rakic, 1980, Malatesta et al., 2008) in the developing brain and that they could have important functions in these supporting cells.
The ubiquitous nature of the expression of IAIPs in a variety of cell types in the brain of two different rodent species and in sheep brain during ovine development (Spasova et al., 2014), strongly suggest that IAIPs most likely have important endogenous roles in diverse cell types during normal brain development. Interactions among neurons, microglial cells, and astrocytes are important during development because of many critical physiological and pathophysiological changes occur during development. Thus, taken together with our in vitro findings, IAIPs expression in different brain regions and cell types in vivo suggest the potential for diverse functions in healthy brain and potentially in neonatal related brain disorders. Moreover, our recent finding that exogenous blood derived IAIPs have important neuroprotective effects (Threlkeld et al., 2014, Gaudet et al., 2016) would appear to suggest that endogenous IAIPs could also exert neuroprotective effects analogous to other previously reported neuroprotective factors such as FGF-2 (Nozaki et al., 1993, Noda et al., 2014).
The source of the endogenous IAIPs cannot be discerned by the current study. However, their presence in cultured neurons, microglia, and astrocytes, along with the presence of genes of IAIPs within the cultured neurons, would tend to suggest that these molecules are locally produced in many CNS cell types. Therefore, we speculate that expression of these molecules in such a wide range of cell types suggest that they could also exert paracrine/autocrine modes of action. Although IAIPs are large molecules that are unlikely to cross the developing blood-brain barrier (Stonestreet et al., 1996), we cannot rule out the possibility that there could be transporters that could also facilitate their entry into the intact brain parenchyma. However, such a mechanism could not account for their presence in cultured cells.
The balance between the proteolytic activities of serine proteases and their natural inhibitors serves an important role in the brain development and synaptic plasticity. Impairment of this balance could lead to neurological injury such as hypoxia-ischemia-induced brain damage and neurodegenerative diseases (Sutton et al., 1994, Wang et al., 1998, Vivien and Buisson, 2000). As a canonical protein inhibitor of serine proteases, exogenous IAIPs are gaining increasing attention based upon their potential therapeutic benefits in sepsis and neonatal hypoxic-ischemic brain injury (Singh et al., 2010, Threlkeld et al., 2014, Gaudet et al., 2016).
Binding with hyaluronan (HA) to form a Serum-derived Hyaluronan-Associated Protein (SHAP)-HA complex is a critical step for the biological functions of IAIPs, particularly with regard to the stability and integrity of the extracellular matrix, and in IAIPs anti-inflammatory actions (Flahaut et al., 1998, Zhuo et al., 2004). Recent work also suggests that IAIPs and high molecular weight (HMW)-HA co-localized with histones in necrotic tissues suggesting that IAIPs, chondroitin sulfate, and HMW-HA are potential therapeutic agents to protect against histone-induced cytotoxicity, systemic inflammation, and organ damage during inflammatory conditions (Chaaban et al., 2015).
There are several limitations to the present study. We were not able to detect the HC-4 and HC-5 protein expression in CNS, because the pR21 antibody only recognizes the IaI and PaI complexes. The expression of IAIPs in multiple cell types and different regions of the brain that are derived from different embryonic origins within the telencephalon, suggest that it would be critical to examine the processes and mechanisms govern IAIPs expression. It would also be of interest to tract the developmental profiles of these proteins within specific cell types at different stages of brain maturation. Future studies would be required to answer fundamental questions regarding the endogenous roles and mechanisms of action of these critical proteins at the cellular and regional levels in the during brain development.
CONCLUSION
Neuroinflammation has been recognized as a critical contributor to both normal brain development and perinatal brain injury (Ferriero, 2004, Harry, 2012, Harry and Kraft, 2012). Exogenous IAIPs exhibit anti-inflammatory and neuroprotective effects in neonatal hypoxic-ischemic brain injury (Singh et al., 2010, Threlkeld et al., 2014, Gaudet et al., 2016). We have identified the wide spread existence of endogenous IAIPs in brain and other organs (Spasova et al., 2014), which has suggested the importance of examining the cellular expression and localization of IAIPs in brain as a first step to understand the physiological and molecular mechanisms of IAIPs in brain. The current study is the first to demonstrate systematically that IAIPs genes and proteins are broadly expressed in CNS including in cultured brain cells in vitro and different brain cells and regions in E18 mouse and P10 rat in vivo. Our results suggest the importance of further studies, designed to elucidate the mechanisms of IAIPs in the neurological disorders particularly as related to neonatal hypoxic-ischemic brain injury.
Proteins and mRNA of IAIP light and heavy chains are expressed in mouse neurons.
Proteins of IAIPs were detected in cultured rat neurons, microglia, and astrocytes.
IAIP immunoreactivity was detected in different brain regions.
IAIPs are expressed in a wide variety of brain cells and regions in rodent CNS.
Acknowledgments
We gratefully acknowledge the advice and technical support of Beth McGonnigal, M.S., Joan Stabila, M.S.; Yulian Wang, M.S.; Yitang, Zeng, Ph.D, Yanchun Xu, MD, Ph.D.; and Quanfu Mao, M.S.
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number 1R01-HD-057100 and R21 NS095130-01, by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 RR018728 and P20GM103537 and the Small Business Innovation Research Proposal 1R43NSO084575-01.
Abbreviations
- Ambp
α1-microglobulin/bikunin precursor light chain mRNA transcript
- Itih
alpha-trypsin inhibitor heavy chain mRNA transcripts
- AMBP
polypeptide precursors of light chain
- HP
polypeptide precursors of heavy chain
- LC
mature polypeptide of light chain
- HC
mature polypeptide of heavy chain,
- IaI
Inter-alpha Inhibitor
- PaI
Pre-alpha Inhibitor
- PIS
Pre-immune serum
Footnotes
Disclosure/Conflict of Interests
Yow-Pin Lim is employed by ProThera Biologics, which develops human plasma derived IAIPs as a therapeutic agent for hypoxic-ischemic brain injury. All other authors have no duality or conflicts of interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abney ER, Bartlett PP, Raff MC. Astrocytes, ependymal cells, and oligodendrocytes develop on schedule in dissociated cell cultures of embryonic rat brain. Dev Biol. 1981;83:301–310. doi: 10.1016/0012-1606(81)90476-0. [DOI] [PubMed] [Google Scholar]
- Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res. 1999;117:145–152. doi: 10.1016/s0165-3806(99)00113-3. [DOI] [PubMed] [Google Scholar]
- Banker GA, Cowan WM. Rat hippocampal neurons in dispersed cell culture. Brain Res. 1977;126:397–342. doi: 10.1016/0006-8993(77)90594-7. [DOI] [PubMed] [Google Scholar]
- Banker GA, Cowan WM. Further observations on hippocampal neurons in dispersed cell culture. J Comp Neurol. 1979;187:469–493. doi: 10.1002/cne.901870302. [DOI] [PubMed] [Google Scholar]
- Banks WA. Peptides and the blood-brain barrier. Peptides. 2015;72:16–19. doi: 10.1016/j.peptides.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer SA, Altman J. Neocortical Development. New York: Raven Press; 1991. [Google Scholar]
- Beaudoin GM, 3rd, Lee SH, Singh D, Yuan Y, Ng YG, Reichardt LF, Arikkath J. Culturing pyramidal neurons from the early postnatal mouse hippocampus and cortex. Nat Protoc. 2012;7:1741–1754. doi: 10.1038/nprot.2012.099. [DOI] [PubMed] [Google Scholar]
- Bottenstein JE, Hunter SF, Seidel M. CNS neuronal cell line-derived factors regulate gliogenesis in neonatal rat brain cultures. J Neurosci Res. 1988;20:291–303. doi: 10.1002/jnr.490200303. [DOI] [PubMed] [Google Scholar]
- Bratt T, Olsson H, Sjoberg EM, Jergil B, Akerstrom B. Cleavage of the alpha 1-microglobulin-bikunin precursor is localized to the Golgi apparatus of rat liver cells. Biochim Biophys Acta. 1993;1157:147–154. doi: 10.1016/0304-4165(93)90058-g. [DOI] [PubMed] [Google Scholar]
- Cameron RS, Rakic P. Glial cell lineage in the cerebral cortex: a review and synthesis. Glia. 1991;4:124–137. doi: 10.1002/glia.440040204. [DOI] [PubMed] [Google Scholar]
- Chaaban H, Keshari RS, Silasi-Mansat R, Popescu NI, Mehta-D’Souza P, Lim YP, Lupu F. Inter-alpha inhibitor protein and its associated glycosaminoglycans protect against histone-induced injury. Blood. 2015;125:2286–2296. doi: 10.1182/blood-2014-06-582759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Risler JL, Raguenez G, Salier JP. The three heavy-chain precursors for the inter-alpha-inhibitor family in mouse: new members of the multicopper oxidase protein group with differential transcription in liver and brain. Biochem J. 1995;306( Pt 2):505–512. doi: 10.1042/bj3060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P, Salier JP. Mouse alpha-1-microglobulin/bikunin precursor: cDNA analysis, gene evolution and physical assignment of the gene next to the orosomucoid locus. Biochim Biophys Acta. 1993;1174:195–200. doi: 10.1016/0167-4781(93)90115-t. [DOI] [PubMed] [Google Scholar]
- Choi BH, Suzuki M, Kim T, Wagner SL, Cunningham DD. Protease nexin-1. Localization in the human brain suggests a protective role against extravasated serine proteases. Am J Pathol. 1990;137:741–747. [PMC free article] [PubMed] [Google Scholar]
- Daveau M, Jean L, Soury E, Olivier E, Masson S, Lyoumi S, Chan P, Hiron M, Lebreton JP, Husson A, Jegou S, Vaudry H, Salier JP. Hepatic and extra-hepatic transcription of inter-alpha-inhibitor family genes under normal or acute inflammatory conditions in rat. Arch Biochem Biophys. 1998;350:315–323. doi: 10.1006/abbi.1997.0515. [DOI] [PubMed] [Google Scholar]
- Dobbing J, Sands J. Comparative aspects of the brain growth spurt. Early Hum Dev. 1979;3:79–83. doi: 10.1016/0378-3782(79)90022-7. [DOI] [PubMed] [Google Scholar]
- Enghild JJ, Thogersen IB, Pizzo SV, Salvesen G. Analysis of inter-alpha-trypsin inhibitor and a novel trypsin inhibitor, pre-alpha-trypsin inhibitor, from human plasma. Polypeptide chain stoichiometry and assembly by glycan. J Biol Chem. 1989;264:15975–15981. [PubMed] [Google Scholar]
- Faustino JV, Wang X, Johnson CE, Klibanov A, Derugin N, Wendland MF, Vexler ZS. Microglial cells contribute to endogenous brain defenses after acute neonatal focal stroke. J Neurosci. 2011;31:12992–13001. doi: 10.1523/JNEUROSCI.2102-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferriero DM. Neonatal brain injury. N Engl J Med. 2004;351:1985–1995. doi: 10.1056/NEJMra041996. [DOI] [PubMed] [Google Scholar]
- Flahaut C, Capon C, Balduyck M, Ricart G, Sautiere P, Mizon J. Glycosylation pattern of human inter-alpha-inhibitor heavy chains. Biochem J. 1998;333( Pt 3):749–756. doi: 10.1042/bj3330749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries E, Blom AM. Bikunin--not just a plasma proteinase inhibitor. Int J Biochem Cell Biol. 2000;32:125–137. doi: 10.1016/s1357-2725(99)00125-9. [DOI] [PubMed] [Google Scholar]
- Gaudet CM, Lim YP, Stonestreet BS, Threlkeld SW. Effects of age, experience and inter-alpha inhibitor proteins on working memory and neuronal plasticity after neonatal hypoxia-ischemia. Behav Brain Res. 2016;302:88–99. doi: 10.1016/j.bbr.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giussani DA, Winter JA, Jenkins SL, Tame JD, Abrams LM, Ding XY, Nathanielsz PW, Unno N, Wu WX, Li C, Hing WK, Collins JH, Derks JB, Owiny J, Sadowsky D, Wentworth R, Kelleman A, Binienda Z, Rittenhouse L, Mitchell M, Myers DA, Reimers T, Owiny JR, Jones MT, Massmann A. Changes in fetal plasma corticotropin-releasing hormone during androstenedione-induced labor in the rhesus monkey: lack of an effect on the fetal hypothalamo-pituitary-adrenal axis. Endocrinology. 1998;139:2803–2810. doi: 10.1210/endo.139.6.6044. [DOI] [PubMed] [Google Scholar]
- Grabrucker A, Vaida B, Bockmann J, Boeckers TM. Synaptogenesis of hippocampal neurons in primary cell culture. Cell Tissue Res. 2009;338:333–341. doi: 10.1007/s00441-009-0881-z. [DOI] [PubMed] [Google Scholar]
- Han X, Wu X, Chung WY, Li T, Nekrutenko A, Altman NS, Chen G, Ma H. Transcriptome of embryonic and neonatal mouse cortex by high-throughput RNA sequencing. Proc Natl Acad Sci U S A. 2009;106:12741–12746. doi: 10.1073/pnas.0902417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry GJ. Neuroinflammation: a need to understand microglia as resident cells of the developing brain. Neurotoxicology. 2012;33:558–559. doi: 10.1016/j.neuro.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Harry GJ, Kraft AD. Microglia in the developing brain: a potential target with lifetime effects. Neurotoxicology. 2012;33:191–206. doi: 10.1016/j.neuro.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelfarb M, Klopocki E, Grube S, Staub E, Klaman I, Hinzmann B, Kristiansen G, Rosenthal A, Durst M, Dahl E. ITIH5, a novel member of the inter-alpha-trypsin inhibitor heavy chain family is downregulated in breast cancer. Cancer Lett. 2004;204:69–77. doi: 10.1016/j.canlet.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Itoh H, Tomita M, Kobayashi T, Uchino H, Maruyama H, Nawa Y. Expression of inter-alpha-trypsin inhibitor light chain (bikunin) in human pancreas. J Biochem. 1996;120:271–275. doi: 10.1093/oxfordjournals.jbchem.a021409. [DOI] [PubMed] [Google Scholar]
- Jacobson M. Developmental Neurobiology. New York: Plenum Press; 1991. [Google Scholar]
- Jansen EM, Low WC. Long-term effects of neonatal ischemic-hypoxic brain injury on sensorimotor and locomotor tasks in rats. Behav Brain Res. 1996;78:189–194. doi: 10.1016/0166-4328(95)00248-0. [DOI] [PubMed] [Google Scholar]
- Jessen TE, Faarvang KL, Ploug M. Carbohydrate as covalent crosslink in human inter-alpha-trypsin inhibitor: a novel plasma protein structure. FEBS Lett. 1988;230:195–200. doi: 10.1016/0014-5793(88)80670-7. [DOI] [PubMed] [Google Scholar]
- Kaumeyer JF, Polazzi JO, Kotick MP. The mRNA for a proteinase inhibitor related to the HI-30 domain of inter-alpha-trypsin inhibitor also encodes alpha-1-microglobulin (protein HC) Nucleic Acids Res. 1986;14:7839–7850. doi: 10.1093/nar/14.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S. Identification of Neuronal Nuclei (NeuN) as Fox-3, a New Member of the Fox-1 Gene Family of Splicing Factors. Journal of Biological Chemistry. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Suzuki M, Hirashima Y, Terao T. The protease inhibitor bikunin, a novel anti-metastatic agent. Biol Chem. 2003;384:749–754. doi: 10.1515/BC.2003.083. [DOI] [PubMed] [Google Scholar]
- Koga Y, Fujita M, Tsuruta R, Koda Y, Nakahara T, Yagi T, Aoki T, Kobayashi C, Izumi T, Kasaoka S, Yuasa M, Maekawa T. Urinary trypsin inhibitor suppresses excessive superoxide anion radical generation in blood, oxidative stress, early inflammation, and endothelial injury in forebrain ischemia/reperfusion rats. Neurol Res. 2010;32:925–932. doi: 10.1179/016164110X12645013515133. [DOI] [PubMed] [Google Scholar]
- Levitt P, Rakic P. Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J Comp Neurol. 1980;193:815–840. doi: 10.1002/cne.901930316. [DOI] [PubMed] [Google Scholar]
- Lim YP, Bendelja K, Opal SM, Siryaporn E, Hixson DC, Palardy JE. Correlation between mortality and the levels of inter-alpha inhibitors in the plasma of patients with severe sepsis. J Infect Dis. 2003;188:919–926. doi: 10.1086/377642. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Appolloni I, Calzolari F. Radial glia and neural stem cells. Cell Tissue Res. 2008;331:165–178. doi: 10.1007/s00441-007-0481-8. [DOI] [PubMed] [Google Scholar]
- Milland J, Tsykin A, Thomas T, Aldred AR, Cole T, Schreiber G. Gene expression in regenerating and acute-phase rat liver. Am J Physiol. 1990;259:G340–347. doi: 10.1152/ajpgi.1990.259.3.G340. [DOI] [PubMed] [Google Scholar]
- Mission JP, Takahashi T, Caviness VS., Jr Ontogeny of radial and other astroglial cells in murine cerebral cortex. Glia. 1991;4:138–148. doi: 10.1002/glia.440040205. [DOI] [PubMed] [Google Scholar]
- Noda M, Takii K, Parajuli B, Kawanokuchi J, Sonobe Y, Takeuchi H, Mizuno T, Suzumura A. FGF-2 released from degenerating neurons exerts microglial-induced neuroprotection via FGFR3-ERK signaling pathway. J Neuroinflammation. 2014;11:76. doi: 10.1186/1742-2094-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki K, Finklestein SP, Beal MF. Delayed administration of basic fibroblast growth factor protects against N-methyl-D-aspartate neurotoxicity in neonatal rats. Eur J Pharmacol. 1993;232:295–297. doi: 10.1016/0014-2999(93)90788-j. [DOI] [PubMed] [Google Scholar]
- Odum L, Halkier T, Hojrup P, Schousboe I. Characterization of urinary proteinase inhibitors with segments of amino acids sequences identical to sequences of pancreatic secretory trypsin inhibitor. Int J Biochem. 1989;21:1319–1327. doi: 10.1016/0020-711x(89)90151-1. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Academic Press; 2001. [Google Scholar]
- Paxinos G, Watson C. TThe Rat Brain in Stereotaxic Coordinates, Compact Third Edition. Academic Press; 1997. [Google Scholar]
- Potempa J, Kwon K, Chawla R, Travis J. Inter-alpha-trypsin inhibitor. Inhibition spectrum of native and derived forms. J Biol Chem. 1989;264:15109–15114. [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Ranasinghe S, McManus DP. Structure and function of invertebrate Kunitz serine protease inhibitors. Dev Comp Immunol. 2013;39:219–227. doi: 10.1016/j.dci.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Reuther C, Ganjam GK, Dolga AM, Culmsee C. The serine protease inhibitor TLCK attenuates intrinsic death pathways in neurons upstream of mitochondrial demise. Apoptosis. 2014;19:1545–1558. doi: 10.1007/s10495-014-1027-7. [DOI] [PubMed] [Google Scholar]
- Richardson WD. Encyclopedia of Neuroscience. Elsevier Ltd; 2009. Oligodendrocyte Specification. [Google Scholar]
- Rosenzweig S, Luu P. Adult Neurogenesis Workshop report. Frontiers in Neuroscience. 2010;4 doi: 10.3389/fnins.2010.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salier JP, Diarra-Mehrpour M, Sesboue R, Bourguignon J, Benarous R, Ohkubo I, Kurachi S, Kurachi K, Martin JP. Isolation and characterization of cDNAs encoding the heavy chain of human inter-alpha-trypsin inhibitor (I alpha TI): unambiguous evidence for multipolypeptide chain structure of I alpha TI. Proc Natl Acad Sci U S A. 1987;84:8272–8276. doi: 10.1073/pnas.84.23.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salier JP, Rouet P, Raguenez G, Daveau M. The inter-alpha-inhibitor family: from structure to regulation. Biochem J. 1996;315( Pt 1):1–9. doi: 10.1042/bj3150001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmaso N, Jablonska B, Scafidi J, Vaccarino FM, Gallo V. Neurobiology of premature brain injury. Nat Neurosci. 2014;17:341–346. doi: 10.1038/nn.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, Yang Y, Qiu W, Lu Z, Li Y, Bao J, Feng M, Hu X. Neuroprotection by ulinastatin in experimental autoimmune encephalomyelitis. Neurochem Res. 2011;36:1969–1977. doi: 10.1007/s11064-011-0520-4. [DOI] [PubMed] [Google Scholar]
- Singh K, Zhang LX, Bendelja K, Heath R, Murphy S, Sharma S, Padbury JF, Lim YP. Inter-alpha inhibitor protein administration improves survival from neonatal sepsis in mice. Pediatr Res. 2010;68:242–247. doi: 10.1203/PDR.0b013e3181e9fdf0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spasova MS, Sadowska GB, Threlkeld SW, Lim YP, Stonestreet BS. Ontogeny of inter-alpha inhibitor proteins in ovine brain and somatic tissues. Exp Biol Med (Maywood) 2014 doi: 10.1177/1535370213519195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stonestreet BS, Patlak CS, Pettigrew KD, Reilly CB, Cserr HF. Ontogeny of blood-brain barrier function in ovine fetuses, lambs, and adults. Am J Physiol. 1996;271:R1594–1601. doi: 10.1152/ajpregu.1996.271.6.R1594. [DOI] [PubMed] [Google Scholar]
- Sutton R, Keohane ME, VanderBerg SR, Gonias SL. Plasminogen activator inhibitor-1 in the cerebrospinal fluid as an index of neurological disease. Blood Coagul Fibrinolysis. 1994;5:167–171. doi: 10.1097/00001721-199404000-00002. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kobayashi H, Tanaka Y, Kanayama N, Terao T. Reproductive failure in mice lacking inter-alpha-trypsin inhibitor (ITI)--ITI target genes in mouse ovary identified by microarray analysis. J Endocrinol. 2004;183:29–38. doi: 10.1677/joe.1.05803. [DOI] [PubMed] [Google Scholar]
- Takano M, Mori Y, Shiraki H, Horie M, Okamoto H, Narahara M, Miyake M, Shikimi T. Detection of bikunin mRNA in limited portions of rat brain. Life Sci. 1999;65:757–762. doi: 10.1016/s0024-3205(99)00302-1. [DOI] [PubMed] [Google Scholar]
- Threlkeld SW, Gaudet CM, La Rue ME, Dugas E, Hill CA, Lim YP, Stonestreet BS. Effects of inter-alpha inhibitor proteins on neonatal brain injury: Age, task and treatment dependent neurobehavioral outcomes. Exp Neurol. 2014;261:424–433. doi: 10.1016/j.expneurol.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towfighi J, Yager JY, Housman C, Vannucci RC. Neuropathology of remote hypoxic-ischemic damage in the immature rat. Acta Neuropathol. 1991;81:578–587. doi: 10.1007/BF00310141. [DOI] [PubMed] [Google Scholar]
- Turgeon VL, Houenou LJ. The role of thrombin-like (serine) proteases in the development, plasticity and pathology of the nervous system. Brain Res Brain Res Rev. 1997;25:85–95. doi: 10.1016/s0165-0173(97)00015-5. [DOI] [PubMed] [Google Scholar]
- Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, Yamashita T. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci. 2013;16:543–551. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- Vivien D, Buisson A. Serine protease inhibitors: novel therapeutic targets for stroke? J Cereb Blood Flow Metab. 2000;20:755–764. doi: 10.1097/00004647-200005000-00001. [DOI] [PubMed] [Google Scholar]
- Wang YF, Tsirka SE, Strickland S, Stieg PE, Soriano SG, Lipton SA. Tissue plasminogen activator (tPA) increases neuronal damage after focal cerebral ischemia in wild-type and tPA-deficient mice. Nat Med. 1998;4:228–231. doi: 10.1038/nm0298-228. [DOI] [PubMed] [Google Scholar]
- Watkins RJ, Thomas MG, Talbot CJ, Gottlob I, Shackleton S. The Role of FRMD7 in Idiopathic Infantile Nystagmus. J Ophthalmol. 2012;2012:460956. doi: 10.1155/2012/460956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JR, Gold SJ, Cunningham DD, Gall CM. Cellular localization of thrombin receptor mRNA in rat brain: expression by mesencephalic dopaminergic neurons and codistribution with prothrombin mRNA. J Neurosci. 1995;15:2906–2919. doi: 10.1523/JNEUROSCI.15-04-02906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Anraku S, Nakayama R, Ushijima K. Neuroprotective effect of urinary trypsin inhibitor against focal cerebral ischemia-reperfusion injury in rats. Anesthesiology. 2003;98:465–473. doi: 10.1097/00000542-200302000-00028. [DOI] [PubMed] [Google Scholar]
- Yoshida E, Maruyama M, Sugiki M, Mihara H. Immunohistochemical demonstration of bikunin, a light chain of inter-alpha-trypsin inhibitor, in human brain tumors. Inflammation. 1994;18:589–596. doi: 10.1007/BF01535257. [DOI] [PubMed] [Google Scholar]
- Yoshida E, Sumi H, Tsushima H, Maruyama M, Mihara H. Distribution and localization of inter-alpha-trypsin inhibitor and its active component acid-stable proteinase inhibitor: comparative immunohistochemical study. Inflammation. 1991a;15:71–79. doi: 10.1007/BF00917911. [DOI] [PubMed] [Google Scholar]
- Yoshida E, Yoshimura M, Ito Y, Mihara H. Demonstration of an active component of inter-alpha-trypsin inhibitor in the brains of Alzheimer type dementia. Biochem Biophys Res Commun. 1991b;174:1015–1021. doi: 10.1016/0006-291x(91)91520-m. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Hascall VC, Kimata K. Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Kimata K. Structure and function of inter-alpha-trypsin inhibitor heavy chains. Connect Tissue Res. 2008;49:311–320. doi: 10.1080/03008200802325458. [DOI] [PubMed] [Google Scholar]