Abstract
Clinical oncology is being revolutionized by the increasing use of molecularly targeted therapies. This paradigm holds great promise for improving cancer treatment; however, allocating specific therapies to the patients who are most likely to derive a durable benefit continues to represent a considerable challenge. It is becoming increasingly clear that cancers are characterized by extensive intratumour genetic heterogeneity, and that patients being considered for treatment with a targeted agent might, therefore, already possess resistance to the drug in a minority of cells. Indeed, multiple examples of pre-existing subclonal resistance mutations to various molecularly targeted agents have been described, which we review herein. Early detection of pre-existing or emerging drug resistance could enable more personalized use of targeted cancer therapy, as patients could be stratified to receive the therapies that are most likely to be effective. We consider how monitoring of drug resistance could be incorporated into clinical practice to optimize the use of targeted therapies in individual patients.
Introduction
For the past seven decades, cancer therapy has been defined by nonselective, cytotoxic agents. Historically, choice of treatment was determined by histological features of the tumour and clinical characteristics of the patient, with limited or no focus on targeting the specific molecular aberrations that bestow tumour cells with the ability to proliferate abnormally and uncontrollably. Unsurprisingly, this untargeted cytotoxic approach all too frequently results in substantial toxicity with only marginal clinical benefit.
In the past decade, however, a dramatic change in emphasis has permeated medical oncology, driven by a rapidly growing number of rationally designed therapies that target specific molecular alterations in the tumour. Only a modest number of such drugs are currently available for use in routine clinical practice (Table 1), although many more are being evaluated in clinical trials. These targeted therapies are often paired with an associated diagnostic assay, which is used to test for the presence of a molecular alteration that indicates whether the patient is likely to respond to the specific drug. This approach is conceptually appealing, but response rates to targeted agents can be low, cures are infrequent, and drug resistance often develops rapidly. A targeted therapy will result in significant clinical improvement only if the target is both rate-limiting in terms of tumour growth and present in most or all of the tumour cells. Within any given patient, however, cancer can be extremely heterogeneous in nature, reflecting a continuously evolving population of tumour cells.1 Large-scale sequencing efforts have revealed that most human cancers have a substantial burden of clonal mutations, defined for the purposes of this manuscript as mutations that are shared by most or all of the malignant cells in the sequenced tumour sample—and thus arose in the founding clone.2,3 Growing evidence indicates that cancers also contain many subclonal mutations, defined as mutations that are present in a few cells, or perhaps a substantial minority of the tumour-cell population. These subclones are derived from the founding clone, and are defined by the additional mutations they carry, which are not present in the bulk population. Of note, many subclonal mutations are not detected using routine clinical assays because their abundance often falls below the lower limit of sensitivity; sampling issues can also lead to subclonal mutations being missed.
Table 1.
FDA-approved therapies with an associated companion diagnostic
| Target and drug | FDA-approved indications | Companion diagnostic test |
|---|---|---|
| ABL/KIT | ||
| Bosutinib | Ph+ CML | BCR–ABL fusion* |
| Dasatinib | Ph+ CML; Ph+ ALL | BCR–ABL fusion* |
| Imatinib | Ph+ CML; Ph+ ALL; KIT+ GIST | BCR–ABL fusion* (CML and ALL), KIT protein expression (GIST) |
| Nilotinib | Ph+ CML | BCR–ABL fusion* |
| Ponatinib | BCR–ABLT315I-mutated CML, or CML with no other TKI indicated | BCR–ABLT315I mutation*, or ABL mutation* and failure of other TKIs |
| EGFR | ||
| Cetuximab | KRAS-wild-type CRC | KRAS mutation, NRAS mutation* |
| Panitumumab | KRAS-wild-type CRC | KRAS mutation, NRAS mutation* |
| Afatinib | EGFR del19 or EGFRL858R NSCLC | EGFR mutation |
| Erlotinib | EGFR del19 or EGFRL858R NSCLC | EGFR mutation |
| Gefitinib | EGFR del19 or EGFRL858R NSCLC | EGFR mutation |
| BRAF | ||
| Dabrafenib | BRAFV600E melanoma | BRAF V600 mutation |
| Vemurafenib | BRAF V600 mutant melanoma | BRAF V600 mutation |
| ALK | ||
| Ceritinib | ALK+ NSCLC | ALK fusion |
| Crizotinib | ALK+ NSCLC | ALK fusion |
| MEK | ||
| Trametinib | BRAFV600E/K melanoma | BRAF V600 mutation |
| PARP | ||
| Olaparib | Ovarian cancer with deleterious germline BRCA mutation | BRCA mutation |
| HER2 | ||
| Ado-trastuzumab emtansine | HER2+ breast cancer | HER2 overexpression |
| Lapatinib | HER2+ breast cancer | HER2 overexpression |
| Pertuzumab | HER2+ breast cancer | HER2 overexpression |
| Trastuzumab | HER2+ breast cancer; HER2+ gastric cancer | HER2 overexpression |
Not an FDA-approved companion diagnostic, but a commercially-available test is in clinical use.
Abbreviations: ALL, acute lymphoblastic leukaemia; CML, chronic myeloid leukaemia; CRC, colorectal cancer; GIST, gastrointestinal stromal tumour; NSCLC, non-small-cell lung cancer; PARP, poly(ADP-ribose) polymerase; Ph+, Philadelphia chromosome positive; TKI, tyrosine-kinase inhibitor.
Targeted therapies need to be directed at the founding clonal mutations shared by all of the billions of cells in the cancer to be effective. For a few cancers that are heavily dependent on a single driver mutation, such treatment is potentially curative. For example, acute promyelocytic leukaemia is driven by the promyelocytic leukaemia protein (PML)–retinoic acid receptor α (RARA) fusion protein, which can be effectively targeted via treatment with all-trans-retinoic acid (ATRA) and arsenic.4 Subclonal mutations are frequently present in a variable proportion of the cancer cells in this disease,5 but these subclones remain susceptible to ATRA and arsenic because they are derived from the founding clone harbouring the treatment-sensitizing PML–RARA fusion protein; thus, the disease remains curable. Occasionally, point mutations in the PML and RARA genes can drive resistance to this standard treatment, and the presence of these genetic alterations in even a small fraction of the cancer cells precludes cure with ATRA–arsenic therapy alone.6 For most cancer types, therapies directed against a single molecular target are not durably curative owing to abundant similar forms of resistance; if subclones are present that bear mutations conferring resistance to therapy, these cells will rapidly expand and repopulate the tumour during treatment (Figure 1). Hence, if this pre-existing drug resistance could be identified, patients could avoid the toxicity of drugs that are destined to fail, and instead pursue alternate treatments with a higher probability of success.
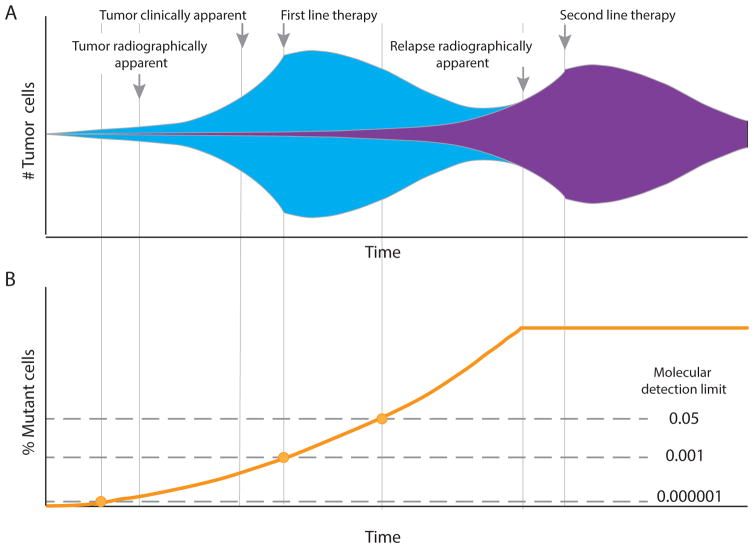
Figure 1.
The evolution and detection of drug-resistance in patients with cancer. a. DNA replication errors can introduce increasing genetic diversity at every cell division after the clonal founding of a tumour; thus, considerable genetic heterogeneity exists in the tumour at the time of diagnosis. As a consequence, a small subset of tumour cells with mutations that confer resistance to particular therapies will often be present (represented by purple shading). Initially, these drug-resistant cells have no specific growth advantage and expand at the same overall rate as the entire tumour; however, with introduction of a therapeutic pressure that hinders the growth of all but the resistant cells, the latter will rapidly takeover the tumour, becoming the predominant clone, until another non-cross-resistant treatment is applied. b. Until recently, a lack of sufficiently sensitive tools to detect these rare subclones meant that resistance could only be identified using clinical criteria, such as radiographic imaging, at a relatively late stage of disease. Newer molecular means for identifying resistance mutations (see ‘Detecting pre-existing drug resistance’ section) are enabling iteratively earlier detection of drug resistance as technical sensitivity improves, and are thus increasing the opportunity to better customize therapy.
Herein, we first review the evidence for extensive genetic heterogeneity within human cancers, which represents a pre-existing repository of drug-resistant subclones. We next consider the major molecularly targeted therapies in current use, and associated resistance mechanisms that arise from selection and expansion of pre-existing drug-resistant cells. Finally, we discuss the clinical potential for early detection of drug resistance, as well as important limitations, and describe ways in which to incorporate high-sensitivity detection of pre-existing drug resistance into the next generation of cancer therapies.
Tumour heterogeneity
At a fundamental level, cancer is a disease of somatic-cell evolution that is driven by successive waves of natural selection6b. Genetic and epigenetic heterogeneity within a tumour-cell population serves as the repository of selectable variation that fuels both disease progression and acquisition of resistance to therapy. The conceptual parallel between neoplastic evolution at a cellular level and Darwinian evolutionary processes among organisms was recognized more than 40 years ago,7,8 but until the past decade, remained largely a conceptual abstraction. Since the publication of the first tumour genome sequencing reports, mutational heterogeneity has come to the forefront as a recognized hallmark of cancer,9 and has garnered more widespread attention.
One of the earliest and most-striking findings of cancer genome sequencing efforts was the extraordinary variety of mechanisms by which different tumours, even within a given cancer type, acquire their neoplastic growth properties. The mutations identified by these approaches are predominantly clonal in origin—that is, they are shared by most of the malignant cells within an individual cancer. Heterogeneity in the overall burden of clonal mutations among different tumours highlights the role of stochastic mutational events early in tumour development prior to clonal outgrowth. A particularly large burden of clonal mutations has been noted in some tumour types, such as melanoma, which have historically been associated with rapid development of drug resistance.10 Whether this relationship reflects a relatively greater number of subclonal mutations—that, is mutations present in only a minority of cells—remains to be systematically examined for most cancers.
Using high-throughput DNA-sequencing methodologies, studies have begun to examine both intermixed and spatially distinct tumour-cell subclones within a variety of tumour types.11–13 These subclones carry distinct sets of oncogenic driver mutations and other phylogenetic signatures of their unique, and ongoing, evolutionary history.14,15 Subclonal mutations arise initially in a single cell; this cell can then expand to become a detectable minority population within the tumour. The mutations that confer a growth advantage—frequently referred to as ‘driver mutations’—can occur in concert with additional unselected mutations. Subclonal driver mutations can be clinically significant; for example, in patients with chronic lymphocytic leukaemia (CLL), the presence of subclonal driver mutations predicts more-rapid disease progression,16 and mutations in TP53, which when clonal are predictive of survival, are equally predictive if present in a minority subclone16b. The unselected ‘passenger mutations’ are also relevant, as these mutations—as well others that arise continually in single cells as the consequence of inevitable errors that occur during DNA replication—can function as a reservoir of genetic diversity from which resistance to subsequent therapies can emerge. Indeed, prevalent genetic changes found in relapsed tumours following initial therapy have been identified at low frequency prior to treatment (that is, in minority tumour subclones), implying that drug-resistance was pre-existing.15,17,18
Intuitively, a greater burden of subclonal mutations might be predicted to confer increased potential for rapid tumour evolution. In fact, greater intratumour genetic heterogeneity, as assessed by a variety of metrics, seems to portend worse outcomes for patients with several cancer types, including head and neck,19,20 cervical,21 and breast cancers,22 among others.23 Moreover, in patients with the premalignant condition Barrett oesophagus, clonal diversity has been demonstrated to be proportional to the risk of progression to oesophageal adenocarcinoma.24 The total mutation burden in a tumour is proportional to both the number of tumour cells and the frequency of mutations per cell genome. In aggressive triple-negative breast cancer (TNBC), the rate of mutation accumulation is more than an order of magnitude greater than that of the more-indolent oestrogen receptor (ER)-positive disease subtypes. 25 The high mutation rate could conceivably contribute to the worse outcomes of patients with TNBC, and the tendency of TNBCs to acquire the ability to metastasize at a smaller primary tumour size.26
Interestingly, a higher overall mutational burden does not invariably indicate a worse prognosis. For example, in colorectal cancer (CRC), mismatch repair (MMR) deficiency results in a ‘hypermutator’ phenotype, and can be associated with the presence of greater than 100-fold more somatic variants than are detected in their MMR-intact disease counterparts. Despite this increased mutational load, patients with MMR-deficient CRCs have a more-favourable prognosis than those with MMR-intact disease for reasons that are incompletely understood.27 One hypothesis is that the markedly elevated mutation frequency in the former might exceed some ceiling of somatic evolutionary benefit and leads to error catastrophe that hinders growth of the tumour.28 Alternatively, the high mutation rate might result in the generation of a larger number of immunogenic neoantigens, leading to more-effective immune control of MMR-deficient tumours. This latter concept has been supported by the observation that melanomas with the largest burden of clonal mutations are the most-likely to respond to immunotherapies,29 and more recently, that MMR-deficient colon cancers are uniquely responsive to immune-checkpoint blockade, as compared with MMR-intact tumours29b.
Genetic heterogeneity is an intrinsic feature of cancer;1,9 a higher level of genetic heterogeneity is associated with a more-aggressive disease course, at least in several cancer types,19–24 suggesting that this phenotype contributes to therapy resistance and disease progression. In the era of highly targeted antineoplastic agents, the presence of pre-existing variants that impart therapy resistance within these heterogeneous populations is of increased relevance and is now, arguably, the most substantial barrier to achieving durable cures.
Targeted agents and drug resistance
In the following sections we review selected targeted therapies that are associated with a specific, testable molecular alteration. This selection is not all-encompassing, and the list of such agents is certain to expand in the near future; our intent is to provide a conceptual overview based on the current clinical landscape.
Tyrosine kinase inhibitors targeting ABL
Nearly all patients with chronic myeloid leukemia (CML) have disease that is driven by the BCR–ABL1 gene fusion.30 ABL1 is a nonreceptor tyrosine kinase that is involved in regulation of multiple cellular processes, including cell division.31 ABL1 normally shuttles between the cytoplasm and nucleus; however, when fused with BCR (breakpoint cluster region protein), the ABL1 kinase is constitutively activated and becomes retained in the cytoplasm. Activated ABL1 then results in aberrant signalling and promotes uncontrolled cell proliferation through several routes, including the MAPK, JAK–STAT, and PI3K pathways.31 Inhibition of the ABL1 kinase with imatinib is the prototypical and arguably most-successful example of targeted cancer therapy: 98% of patients with CML responded to imatinib in initial trials of this agent, and 5-year survival rates for this disease improved from 30% among patients treated with interferon-α plus cytarabine to 89% in those treated with imatinib.32 In addition, approximately 20% of patients with primary acute lymphoblastic leukaemia (ALL) harbour the BCR–ABL1 fusion32b and these patients can also benefit from treatment with imatinib.33
Despite the remarkable effectiveness of imatinib, patients commonly develop resistance to therapy. Imatinib resistance is predominantly driven by point mutations in the ABL1 kinase that interfere with drug binding to the protein; more than 100 different mutations have been reported.34 Resistance mutations identified at the time of clinically observed treatment failure have been reported to frequently exist in rare tumour-cell subclones at the time of diagnosis, prior to the initiation of therapy.35–37 Resistance can thus arise via selective growth of cells with a single ABL1 mutation (monoclonal resistance), although simultaneous outgrowth of multiple drug-resistant subclones (polyclonal resistance) has also been reported.38
Additional ABL1 tyrosine-kinase inhibitors (TKIs) have now been approved by the FDA for the treatment of leukaemias harbouring the BCR–ABL1 fusion. Second-generation TKIs, including bosutinib, dasatinib and nilotinib, overcome many imatinib-resistance mutations in BCR–ABL1, and can be used either as initial therapy or following development of resistance to this agent.34 Importantly, specific mutations in BCR–ABL1 confer resistance to specific inhibitors. For example, the Y253H mutation results in resistance to imatinib and nilotinib, but BCR–ABL1 kinases with this alteration remain sensitive to dasatinib.34 Of note, the BCR–ABL1 T315I ‘gatekeeper’ mutation confers resistance to all currently approved ABL1 TKIs other than the newest of these agents, ponatinib.34
Sequencing of the ABL1 gene is typically carried out after failure of initial TKI therapy to help select an alternate non-cross-resistant TKI, based on the resistance mutation found.39 Genotyping of ABL1 is most-commonly performed using conventional Sanger DNA-sequencing methods, which can only detect mutations present in >10% of cells in the sampled population;40 mutations conferring TKI-resistance with an incidence below this limit will, therefore, go undetected. Mass spectrometry, which offers sensitivities of mutation detection of 0.05–0.5%, has been used to investigate subclonal TKI-resistance mutations at the time of relapse.41 Among 220 patients with CML studied after development of resistance to imatinib treatment in a retrospective analysis,41 55 mutations associated with resistance to second-line therapies (dasatinib and nilotinib) were identified in 50 patients by conventional Sanger sequencing. By contrast, mass spectrometry enabled 105 such resistance mutations to be identified in 71 patients. The majority (84%) of mutations detected in this retrospective analysis were found to rapidly become the dominant clones in patients who were treated with the drug to which the mutation conferred resistance. Among 100 patients with chronic phase CML, those found to have subclonal resistance mutations had a much worse outcome, with 0% failure-free survival after second-line therapy versus approximately 50% failure-free survival in patients who lacked subclonal resistance mutations;41 some of the unexplained treatment failures in this latter group could conceivably have resulted from mutations present below the detection limits of mass spectrometry.
Interestingly, the presence of low-frequency mutations following imatinib failure is associated with worse clinical outcomes in patients treated with second-generation TKIs, even if the mutations were not predicted to confer resistance to the inhibitor used.42 Thus, subclonal diversity itself might be a marker of the potential to evolve drug resistance, and therefore could represent an important prognostic indicator.
Tyrosine kinase inhibitors targeting KIT
KIT is a receptor tyrosine kinase, also targeted by imatinib (Table 1), which is overexpressed in >90% of gastrointestinal stromal tumours (GISTs).43 KIT is typically activated only when bound by KIT ligand (also known as stem cell factor or mast cell growth factor), which leads to activation of several cell growth pathways, including the MAPK and PI3K pathways.44 KIT overexpression can lead to unregulated cell growth, as can constitutively activating point mutations affecting this protein, which are found in approximately 80% of GISTs.43 KIT mutations predominantly confer sensitivity—rather than resistance—to imatinib.43 Patients who lack KIT mutations frequently harbour mutations in the related receptor tyrosine kinase platelet-derived growth factor receptor α (PDGFRA), which can also confer sensitivity to imatinib.43b Historically, patients with GIST had a low response rate to chemotherapy, but treatment with imatinib induces marked clinical responses and has improved the median survival of patients with advanced-stage GIST from 18 months to 57 months.45 Unfortunately, most patients develop imatinib resistance within 2 years of starting therapy, predominately as a result of secondary mutations in the kinase domain of KIT or PDGFRA.46 Alternate TKIs can be used in the setting of imatinib resistance, and differential sensitivity to second-line TKIs depends on the specific imatinib-resistance mutation involved.47
Mutational profiling of KIT at the time of diagnosis can have prognostic and predictive value. For example, patients with exon 11 mutations generally have good responses to imatinib and improved survival; patients with exon 9 mutations, by contrast, are relatively resistant to imatinib, although resistance can be overcome to some extent by treatment with an elevated dose of imatinib.48 Interestingly, the KIT protein has equivalent sensitivity to imatinib in vitro regardless of whether the activating mutation is located in exon 9 or exon 11.49 The differential imatinib sensitivity of tumour cells harbouring these mutations in vivo has been hypothesized to reflect altered apoptotic thresholds that arise as a consequence of downstream signalling from the various mutant proteins.49
Results from mathematical modelling investigations suggest that imatinib-resistant subclones probably pre-exist in many GISTs prior to the initiation of therapy.50 As in patients with CML, if these resistant subclones could be detected early in the disease course, use of a higher dose of imatinib or of an alternative TKI could be considered in the first-line setting. Moreover, signalling downstream of KIT and PDGFRA is largely through the PI3K–mammalian target of rapamycin (mTOR) and MAPK pathways, and therapeutic targeting of these cascades in GIST is an area of active investigation.51
MAPK pathway inhibitors
The MAPK pathway is a key cellular signalling circuit that is involved in sensing and responding to extracellular stimuli, such as growth factors or inflammatory mediators (Figure 2). The stimulus (ligand) binds to and promotes dimerization of a receptor tyrosine kinase, such as KIT, PDGFRA, and EGFR, resulting in activation of the receptor’s intracellular kinase domain. The kinase domain mediates phosphorylation of a RAS protein (HRAS, KRAS, or NRAS), which then phosphorylates RAF kinases (ARAF, BRAF, or CRAF), causing their dimerization and activation. Dimeric RAF then phosphorylates MEK, which in turn phosphorylates ERK, which can subsequently enter the cell nucleus and regulate the activity of a variety of transcription factors to modulate gene expression. In this manner, the MAPK pathway drives cell growth, and thus constitutive activation of any of its components can contribute to cancer. Multiple FDA-approved targeted drugs are specifically directed at proteins in this pathway (Table 1), with many more in development. To date, the RAS family, despite being the most-mutated class of oncogenes in human cancers, has proved to be extremely difficult to selectively target.52
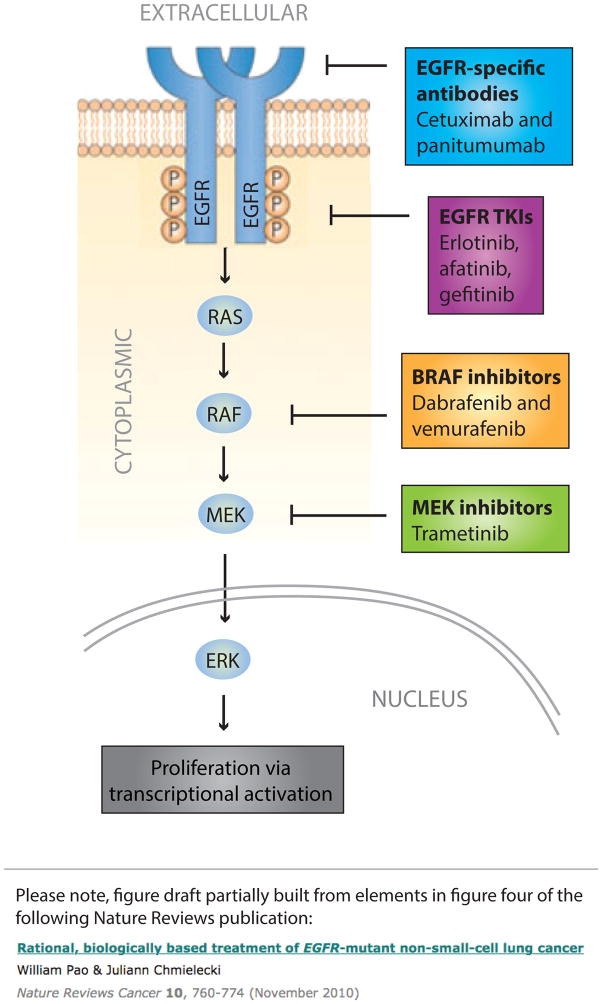
Figure 2.
Core elements of the human EGFR–MAPK pathway. The schematic representation shows the EGFR receptor tyrosine kinase and the MAPK signalling cascade activated downstream of this receptor. The current FDA-approved molecularly targeted agents with indications that are determined by a companion molecular diagnostic assay are shown at their main point of activity. The presence of particular mutations assessed using the companion diagnostic tests can determine whether the patient is eligible or ineligible for therapy, depending on the setting. RAS mutations, for example, rule out the use of EGFR monoclonal antibodies in patients with colorectal cancer. By contrast, the presence of activating mutations in EGFR (such as deletions in exon 19 or the single-nucleotide polymorphisms that result in the Leu858Arg mutation) indicate eligibility of patients with non-small-cell lung cancer for treatment with small-molecule TKIs of EGFR. Similarly, activating Val600 mutations in BRAF confer sensitivity, rather than resistance to BRAF and/or MEK inhibitors. Abbreviations: EGFR, epidermal growth factor receptor; TKIs, tyrosine-kinase inhibitors.
Targeting EGFR
Activating mutations in EGFR are found in approximately 15% of patient with non-small-cell lung cancers (NSCLCs) in the USA, and are particularly common in some Asian populations, in which they can be found in up to 62% of patients.53 Patients harbouring these mutations can be treated with TKIs that target this receptor (Table 1), such as erlotinib,54 afatinib,55 or gefitinib,56 which have been shown to greatly improve outcomes. For example, in the OPTIMAL trial,54 progression-free survival with erlotinib monotherapy in Chinese patients with advanced-stage NSCLC and activating mutations in EGFR was 13.1 months, relative to 4.6 months with gemcitabine plus carboplatin. Erlotinib is also approved for the treatment of patients with pancreatic cancer, although in this setting, use of this agent is not contingent on the presence of an activating mutation in EGFR. In a phase III study of erlotinib plus gemcitabine versus gemcitabine alone, the addition of erlotinib to therapy resulted in a small, but statistically significant, median overall survival benefit of approximately 2 weeks (HR 0.82; P = 0.038.)57 A more-recent randomized trial conducted in Taiwan reported a median overall survival of 7.2 months in patients treated with erlotinib plus gemcitabine versus 4.4 months among those treated with gemcitabine alone.58
In patients with NSCLC, detection of activating mutations in EGFR has generally been performed using the relatively low-sensitivity Sanger sequencing method. A study in which EGFR mutations were evaluated using the more-sensitive approach of next-generation sequencing (NGS) found that 22 of 87 patients with NSCLC who were considered EGFR wild-type based on the results of Sanger sequencing, in fact, harboured an EGFR mutation.59 In another study, the investigators used a mutation-specific PCR-based assay and demonstrated that patients carrying EGFR-activating mutations below the level of detection of traditional methods of genotyping also derived benefit from use of EGFR-targeted TKIs; the proportion of cells that were mutated was correlated with response rates and survival following EGFR blockade.60 Results of a meta-analysis have confirmed the improved ability to stratify patients who are likely to benefit from EGFR-TKI treatment with the use of higher-sensitivity genotyping assays.61 These empirical observations are biologically plausible: NSCLC is characterized by a high level of intratumour heterogeneity and thus key driver genes can be present in only a subset of tumour cells.62,63
As with other targeted therapies, acquired resistance to EGFR inhibitors is common. In a cohort of patients with NSCLC who became resistant to erlotinib, resistance was caused by a second active-site mutation in EGFR, the T790M ‘gatekeeper’ mutation, in approximately 50% of cases.64 Mutations or overexpression of downstream components of the MAPK pathway, such as KRAS, NRAS, BRAF, and MET, can also drive resistance to EGFR inhibitors, via constitutive pathway activation without dependence on EGFR. 65
In cell-line models, subclonal populations harbouring the EGFR T790M mutation have been found to pre-exist prior to erlotinib therapy, and clonally expand upon drug exposure.18 This pattern also seems to hold true in vivo; in one study that compared Sanger sequencing to mass spectrometry for mutational analysis of DNA from tumour tissue, the fraction of patients with a detectable pre-existing EGFR T790M mutation increased from 2.8% to 25.2% with the use of the latter technique66. An independent mass-spectrometry-based study also found a 25% incidence of pre-existing EGFR drug-resistance mutations in tumour samples from patients with NSCLC, and furthermore, investigators reported that the relative abundance of the mutation was proportional to the extent of both progression-free and overall survival.69
EGFR is also the target of therapeutic antibodies used in patients with CRC (panitumuab or cetuximab), and head and neck cancer (cetuximab). In these patients, the presence of an activating EGFR mutation is not a requirement for use of anti-EGFR antibody therapy, and resistance is not typically mediated by mutations in EGFR itself. However, pre-existing activating mutations in the KRAS gene, which encodes a small GTPase that mediates signalling downstream of the EGFR (Figure 2), are common in patients with CRC and functionally bypass EGFR blockade.70 For this reason, the anti-EGFR antibodies panitumumab and cetuximab are approved by the FDA for use only in patients with CRC who lack mutations in KRAS.71 The presence of KRAS mutations is also associated with a poor response to EGFR inhibition in patients with NSCLC, although tumour samples are not routinely tested for such mutations in this setting.72 As with EGFR-mutation testing in patients with NSCLC, the clinical benefit of KRAS-mutation detection is heavily dependent on assay sensitivity. In patients with CRC who are deemed KRAS-wild-type at the time of diagnosis, treatment with anti-EGFR-antibody therapy frequently results in emergence of KRAS mutations.73 These KRAS mutations might have been present at the time of diagnosis, below the detection limit of conventional DNA-sequencing assays. Indeed, as compared with conventional sequencing, the use of mass spectrometry,74 allele-specific PCR,75 or next-generation deep sequencing76 markedly improves the specificity of predicting responders to these agents.
Targeting BRAF and/or MEK
Activating substitution mutations of valine 600 (V600) in BRAF are detected in approximately 50% of patients with advanced-stage melanoma, and confer sensitivity to BRAF inhibitors, such as vemurafenib and dabrafenib (Figure 2).77 Most BRAF-mutant tumours are known to be responsive to the BRAF inhibitor vemurafenib; however, in an animal model, this inhibitor has been demonstrated to cause a paradoxical increase in MAPK activity and cell growth when an HRAS mutation is also present.78 Furthermore, development of cutaneous squamous-cell carcinomas and keratoacanthomas often occurs in patients treated with BRAF inhibitors,79 and might result from stimulation of the growth of skin cells that possess RAS mutations. Concomitant treatment with a MEK inhibitor, such as trametinib, which is also approved by the FDA for the treatment of BRAF V600-mutated melanoma alone or in combination with dabrafenib, abrogates this growth-stimulating effect in RAS-mutant cells and thus reduces the incidence of secondary skin cancers in patients with melanoma.80 Moreover, findings of a phase III trial have demonstrated superior survival with dual BRAF–MEK blockade (median overall survival in the combination group was 25.1 months versus 18.7 months in the BRAF-monotherapy group), and reduced risk of cutaneous squamous-cell carcinoma and keratoacanthoma (1% of patients in the combination therapy group versus 18% in the BRAF-monotherapy group).81
Unfortunately, approximately 30% of patients treated with dual BRAF–MEK inhibition experience disease progression within 6 months; in one study in 10 patients who developed rapidly progressive disease while receiving BRAF and MEK inhibitors, nine were found to have additional mutations in components of the MAPK pathway, most commonly BRAF amplification or activating mutations in NRAS or MEK2.82 These alterations were not found in the pretreatment tumour samples, but high-sensitivity assays were not used.82 Alternative RAF family inhibitors are in development that overcome these resistance mechanisms.82b Early detection of subclonal activating NRAS and MEK2 mutations could, therefore, enable stratification of the patients who would gain the greatest benefit from these inhibitors.
Detecting pre-existing drug resistance
Despite numerous demonstrations of excellent antineoplastic activity in some tumour types, targeted anticancer drugs almost uniformly select for drug-resistant subclones within a tumour, which eventually, and often rapidly, results in disease progression. For every molecular pathway discussed, we have highlighted examples in which pre-existing drug-resistance mutations can preferentially expand during exposure to the cognate drug and thus predict treatment failure. Such mutations can be found in the tumour itself, and sometimes in plasma cell-free DNA derived from the tumour or circulating tumour cells (CTCs), as discussed in a later section of this manuscript. The limited technical ability to accurately quantify rare genetic variants has been a substantial barrier to characterizing the extent to which pre-existing drug-resistant variants are present in different tumours, and precisely what clinical consequence the presence of these subclones foretells (Figure 3).
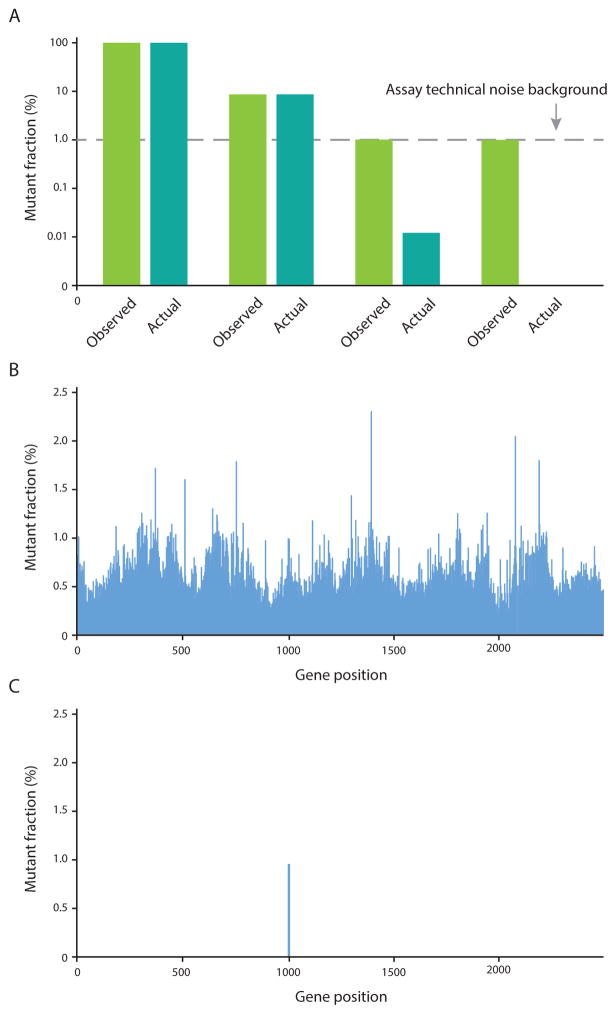
Figure 3.
The ability to detect mutations that are present at a low frequency depends on the assay error rate. a. Mutations present at a level substantially above technical background noise (error rate) of the assay (1% in this case) can be accurately quantified (i–ii), but those mutations with a prevalence below this threshold of detection (iii), or that are not present at all (iv), will be falsely assigned the background frequency. b. Genetic analysis via standard NGS has a background error rate of approximately 1%; at this error rate, with sufficiently deep sequencing, every genomic position would seem to be mutated near this level. In this example, exonic DNA encoding the active site region of ABL1 was enriched using a double-capture protocol and sequenced by conventional NGS on an Illumina® HiSeq® 2500 sequencer.68 c. Several biochemical error correction strategies have recently been developed that reduce this background noise and enable a lower the threshold of detection in order to capture extremely rare mutations. In this case, Duplex Sequencing of the same sample of tumour material across the same region of ABL1 reveals that only a single low-frequency mutation is actually present. 68 Abbreviations: NGS, next-generation sequencing. Permission obtained from Macmillan Publishers Limited © Schmitt, M. W. et al. Nat. Methods 12, 423–425 (2015)..
Detection of mutations in tumour biopsy tissue has typically been performed using Sanger sequencing; however, this methodology is optimal for detecting mutations that are present in most or all of the cells, with detection of mutations present in fewer than 25% of cells being unreliable.40 Thus, assessments of heterogeneous tumours, or tumours intermixed with normal cells, can lead to false-negative results. Newer methods of DNA-sequence analysis, such as allele-specific PCR,83 mass spectrometry,84 Random Mutation Capture,85 and digital droplet PCR,86 afford greater sensitivity, but these techniques can only be used to survey a limited number of specified mutations. NGS offers the ability to determine the sequence of multiple genes simultaneously and can resolve mutations present in subpopulations of cells; however, this methodology is generally limited to detection of mutations present in >5% of cells, as errors during PCR amplification and sequencing generate background ‘noise’, which obscures the detection of lower-frequency variants (Figure 3).40,87
Molecular tagging methods have been developed that can lower the background error rate of NGS by approximately 20-fold.88,89 These approaches are limited in their ability to resolve lower-level mutations, as they depend on amplification of single-stranded DNA in which the presence of DNA damage (such as oxidative damage or abasic sites) can result in recurrent errors and miscalling of variants. To overcome this limitation, we have developed a technology termed ‘Duplex Sequencing’ that independently tags and sequences the two complementary strands of DNA,87 as well as methods for efficient targeted capture and sequencing of individual exons of human genes.68 This approach improves on the accuracy of NGS by >100,000-fold, and enables detection of a single mutated base among >10 million sequenced nucleotides.87 With Duplex Sequencing and other error-correction methods, additional DNA-sequencer capacity is consumed relative to conventional NGS, as a single molecule of DNA is effectively sequenced multiple times to allow for error correction. By focusing on targeted regions of the genome that are likely to reflect actionable loci, however, the extra sequencing requirement is fairly modest. Highly sensitive assays will be essential to accurately characterize the pattern and timing of resistance to targeted therapies, and will be a necessary aspect of future drug trials in order to optimize sequential ordering of therapies, inform the choice of combination therapies, and to enable early adjustments in therapy at the first sign of drug resistance and disease progression.
Representative sampling of tumours
With liquid tumours (that is, leukaemias and some lymphomas), sampling a homogeneous population of the cancer is relatively simple, given the inherent mixing of cells in peripheral blood. The subclones within the bone marrow are not usually directly sampled, although the abundance of these subpopulations of cells seems to be similar to their frequency in the peripheral blood, at least in patients with acute myeloid leukaemia (AML).90 Obtaining a representative sample of solid tumours is much more challenging, as the potential for spatial heterogeneity implies that any single biopsy could result in much of the diversity being missed. Furthermore, tumours evolve over time in response to treatment, but performing repeated biopsies to assess the associated molecular changes is generally impractical. Thus, treatment decisions are frequently made on the basis of mutations detected in biopsy samples taken at the initial time of diagnosis, despite the fact that many months might have passed. Analysis of tumour products present in the circulation, an approach sometimes termed ‘liquid biopsy’, is one way to circumvent these challenges relating to spatial heterogeneity and tumour evolution.
The majority of advanced-stage solid tumours release DNA into the systemic circulation, known as circulating tumour DNA (ctDNA).91 With sufficiently large tumours, nearly all mutations identified as clonal events in tumour biopsies are accurately represented using this liquid-biopsy approach.92,93 Subclonal mutations that confer resistance to targeted therapies have also been identified in ctDNA during treatment, and the abundance of drug-resistance mutations has been shown to change dynamically over the course of therapy.94 Indeed, the major subclones comprising the tumour population are likely to be represented in ctDNA, although a direct comparison of subclonal structure in a tumor sample versus matched ctDNA remains to be reported.
The potential clinical utility of screening for subclonal drug-resistance mutations in ctDNA is being increasingly demonstrated.95,96 For instance, KRAS mutations have frequently been found in ctDNA from patients with CRC—who were initially deemed KRAS wild-type—at the time of clinical failure of EGFR blockade;97 in fact, multiple independent KRAS mutations were detected in ctDNA from some of the patients who developed resistance to panitumumab.97 In another study in patients with CRC,73 mutations that confer resistance to cetuximab were identified in serum samples collected up to 10 months prior to radiographic disease progression. Similarly, pre-existing MET amplification as a mechanism of resistance to EGFR blockade has also been observed in ctDNA prior to clinical treatment failure.98
Intact CTCs can also be isolated, and analysis of these cells might be more informative than evaluation of cell-free ctDNA in some situations. For example, in patients with NSCLC who are known to harbour EGFR-activating mutations, the mutation could be detected in CTCs from 92% of the patients, but the same mutation could be detected in cell-free ctDNA from only 32%.99 In this study, genotyping of CTCs was also more sensitive than ctDNA for identifying the prototypical T790M resistance mutation in EGFR at, and prior to, disease progression.99 In one interesting hybrid application in patients with breast cancer, capture of CTCs offered the opportunity to predict drug sensitivity by both empirical testing in cell culture experiments and through direct sequencing of CTC DNA for resistance mutations.100 Finally, enrichment for certain cell subtypes prior to genotyping, such as CD34+ progenitor cells in samples from patients with CML, can also improve the predictive value of subclonal mutations.101
Given the well-established spatial and temporal heterogeneity of solid tumours,13,62,63,102,103 a reasonable question is whether individual biopsies, cell-free ctDNA, or CTCs can be truly representative of the full extent of genetic diversity within the tumour itself: biopsies are limited to a defined location within tumours and are taken at potentially restricted time points, and whether all subclonal cell populations within a tumour contribute equally to circulating cell-free DNA or are represented as CTCs remains unknown. Rigorously determining concordance of mutations among these sample sources would require deep sequencing of individual biopsy samples, cell-free DNA and CTCs, in conjunction with deep sequencing of the entire homogenized tumour. Fundamentally, encompassing the complete genetic diversity of a tumour will never be possible without sequencing every tumour cell individually; therefore, an important practical question to be addressed in the coming years is what level of sampling provides the most clinically relevant approximation.
Issues in early detection of resistance
History is rife with examples of well-intentioned diagnostics that have ultimately borne out no benefit in terms of patient survival or quality of life. In some instances, tests have led to harm either directly, as a result of the diagnostic assessment itself; indirectly, by leading to unnecessary interventions; or psychologically, by introducing worry among the patients about their future (without offering the ability to make changes to affect its course). Even seemingly intuitive tests supported by evidence from modern clinical trials, such as serum PSA screening for prostate cancer in men, have resulted in a complex mixture of conflicting guidelines based on different interpretations of the data. Health-care costs within oncology are growing rapidly and any additional sources of expenditure need to be considered critically in the context of their overall value in improving clinical outcomes. In the following sections we consider some of these poignant issues.
Clinical actionability and utility
How often early detection of drug-resistance mutations will be clinically actionable and how frequently such action will meaningfully improve patient care are important questions. At present, our ability to identify low-level resistance mutations exceeds the therapeutic tools available to prevent their outgrowth. A potential criticism of early-resistance testing is that it would add cost, while only offering the ability to present a patient with the somber information that the treatment they are receiving is likely to fail after a short amount of time. Increasingly, however, alternate treatments do exist and could be instituted early if the development of drug resistance could be predicted and assessed over time. Repeated assessment of the various ABL1 kinase mutations that confer differential sensitivities to the five currently approved TKIs that target this protein represents an important example of this approach; currently, switching rationally between these drugs according to the particular resistance mutation that arises at a clonal level is commonplace in the clinic.34 A further example in colon cancer involves the EGFR S492R mutation, which confers resistance to the EGFR inhibitor cetuximab, but not to panitumumab.104 With regard to the more common mode of resistance to anti-EGFR antibodies mediated by KRAS mutations, patients with resistant CRCs seem to retain sensitivity to targeting of downstream signalling with MEK inhibitors, and early detection of emerging KRAS mutations has been proposed as an indication for initiation of treatment with anti-MEK agents.73 Pre-existing MET amplification is another predictor of resistance to anti-EGFR agents in patients with CRC,98 and would theoretically be actionable with the MET/VEGFR2 inhibitor cabozantinib, which is currently approved for the treatment of medullary thyroid cancer, or one of several MET inhibitors that are under investigation in clinical trials.105
As the number of new targeted antineoplastic agents continues to grow, so too will the number of options for countering emerging resistance induced by a prior treatment. For example, the remarkably successful introduction within the past year of ibrutinib, which targets the Bruton tyrosine kinase (BTK) and is an effective treatment for patients with CLL and indolent lymphomas,106–108 has already led to recognition of specific resistance mechanisms. Mutation of the BTK target at cysteine 481 (C481) or gain-of-function mutations in phospholipase Cγ2 (PLCγ2), which is immediately downstream of BTK, results in ibrutinib resistance and disease progression.109,110 Inhibition of the downstream cyclin-dependent kinase 4 (CDK4)-signalling pathway with palbociclib, an agent now approved by the FDA for the treatment of breast cancer, restores sensitivity to ibrutinib.110 Likewise, second-generation BTK inhibitors are in development that maintain effectiveness in the presence of BTK C481 mutations.111
In scenarios in which emerging drug-resistance can be detected but no alternate therapy currently exists, sometimes other benefits to early recognition remain important. In patients with melanoma, tumours that have acquired resistance to BRAFV600E-targeted therapy seem to become dependent on the BRAF inhibitor for growth, and withdrawal of the failing drug has, in fact, been shown to lead to tumour regression in melanoma xenograft models;112 thus, early detection of emerging resistance could guide the decision on when to halt the use of a drug. More generally, nearly all drugs have adverse effects and an advanced warning of failure could, in some cases, improve a patient’s overall quality of life by enabling earlier discontinuation of therapy to avoid exposing patients to unnecessary toxicities of an ultimately futile treatment. Similarly, earlier detection of resistance might improve prognostication of disease trajectory and such information could be used to help patients to better prioritize life goals. In addition, detection of pre-existing resistance would enable clinical trials to be enriched for patients who lack detectable resistance, which would potentially decrease the number of patients that would be need to be enrolled in a trial and would, therefore, speed up the approval of novel therapeutics while decreasing costs.
Thresholds for treatment modification
The frequency of a drug-resistance mutation that should necessitate a change of treatment when an alternate therapy exists is another pertinent question. As discussed, we have only recently developed the capacity to readily detect subclonal drug-resistance mutations, and thus much information about the effects of such mutations on clinical outcomes remains to be established. For example, how a clinician should respond to the scenario in which 0.1% of tumour cells in a population that is otherwise sensitive to a targeted drug acquire a resistance mutation is unclear: a resistant clone is clearly emerging, but in this setting the majority of the tumour presumably continues to be suppressed by use of the current agent and the dilemma becomes how to balance future disease progression with prematurely abandoning one active drug among a finite pool of effective treatments.
Resistance mutations present in a small percentage of the cells in a tumour might be highly relevant if the mutation occurs in a cell that is capable of rapid growth. That tumours consist of multiple subpopulations of cells with differing growth rates is well established;90,113 thus, a minority population of ‘cancer stem cells’ might give rise to most of the cells in a bulk tumour. The hypothesis that cancer stem cells are drivers of resistance is controversial,114 although this concept indicates a mechanism by which resistance in a minor subclone could quickly expand and cause clinical disease progression. On the other hand, some mutant subclones are likely to be more indolent. For example, it has been reported that some BCR–ABL1 kinase domain resistance mutations can be present at low levels prior to TKI treatment without leading to clinical relapse.115 Likewise, in patients with AML, the AML–ETO fusion product, which is considered a driver of the disease, can remain detectable at low levels in the blood in patients who have been in complete remission for years.116
In the examples we have described in this Review, resistance mutations present at the lower limit of detection of the assays used, typically those present in 0.1–1% of cells, are clearly correlated with clinical outcome. A solid tumour that is detectable on imaging will typically consist of more than one billion cells, and 0.1% of the tumour thus comprises a population of at least 1 million cells. As higher-sensitivity techniques are more-widely adopted, the clinical relevance of a mutational burden below one in 1,000 needs to be explored. Ultimately, determining when and how to act on mutations that are present in a small fraction of cells is a considerable challenge that will require prospective clinical trials to evaluate actionability. The significance of rare subclonal mutations will likely depend on the specific disease, the magnitude of resistance conferred by a particular mutation and the effectiveness of subsequent second-line or third-line agents. In some cases, addition of another agent to the current regimen—rather than a complete switch to a different therapy—could be preferable, although this approach might be limited by multiplicative toxicity of the drugs.
Is genetic testing for resistance futile?
In a large, genetically unstable tumour, every drug-resistance mutation could potentially be pre-existing; with this in mind, one might ask: what is the benefit of testing? Indeed, acquisition of an elevated mutation rate has been proposed as a common feature of carcinogenesis,7,117 which implies that every possible mutation will be present in some subset of cells within a tumour. In this scenario, treatment with any targeted therapy might be expected to result in expansion of a drug-resistant population and clinical progression. Some data support the concept of a ‘mutator’ phenotype in specific cancers;118–120 however, whether this is a general phenomenon remains to be demonstrated.
Of note, not every mutation that theoretically confers drug resistance will be able to do so for a variety of reasons, including stochastic cell death, clonal interference from other tumour cells, or because the mutation is not carried by a long-lived tumour stem cell, which might entail only a small minority of a tumour population. The fact that thousands of different resistance mutations do not simultaneously expand to a detectable level upon exposure to a targeted therapy supports this reasoning. Thus, a very large number of resistance mutations might be present at an extremely low frequency, but those clones that have expanded to form a modest-sized subpopulation comprising thousands to millions of cells among the billions of cells within a cancer are likely to be the most-clinically relevant.
Independent of specific resistance mutations, quantifying subclonal heterogeneity itself is of clinical importance. The frequency of clonal mutations has been examined comprehensively for most major cancer types,2,3 whereas the extent of subclonal heterogeneity within the DNA-sequences of individual tumours has not. Greater subclonal diversity in a tumour might predict a higher likelihood of pre-existing resistance to any conceivable targeted therapy. Such information might be used to provide a rationale for accepting higher toxicity or increased costs of upfront targeted therapy combinations in certain settings. A high mutational load might also predict that a tumour is approaching an ‘error-catastrophe’ threshold, such that further mutagenesis would be lethal to the tumour,28 and thus might predict sensitivity to nontargeted cytotoxic chemotherapies that promote further mutagenesis. In the field of immunotherapy, patients with tumours bearing larger numbers of clonal mutations, and thus presumably more tumour-specific neoantigens, respond especially well to immune-checkpoint blockade.29,67 Whether a greater abundance of subclonal mutations similarly stimulates immune responses, or if instead such heterogeneity contributes to a pool of immune-evading resistance variants, merits examination.
Conclusions
Few other clinical disciplines have experienced such a foundation-shifting effect of molecular medicine on daily practice as felt in oncology. In patients with some malignancies, such as certain lymphomas and breast cancers, the addition of molecularly targeted drugs to cytotoxic chemotherapy regimens has increased the rate of definitive cure; for others, such as those with CML, rationally designed therapies have turned a life-ending diagnosis into a largely chronic disease. For many patients with cancers, however, the benefit of such agents remains limited by the invariable evolution of resistance via outgrowth of subclonal mutants.
The next frontier in cancer medicine will be developing methods of simultaneously suppressing the many mechanisms that neoplastic cells have at their disposal for circumventing the available therapies. A more immediate objective should be early identification of drugs that are failing or are likely to fail using the high-sensitivity mutation-detection tools that already exist. In the short term, this approach will expedite the use of therapies that are more likely to succeed, prevent unnecessary toxicities, and limit the substantial costs of treatment—the latter of which is an unfortunate, and often underappreciated, adverse effect of targeted approaches in oncology.
Carrying out robust clinical trials of the large number of new targeted agents in development is an intimidating, and immediate challenge. One possible means of improving trial efficiency would be to screen for, and exclude patients with pre-existing low-level drug-resistance mutations, to enrich small study cohorts for those individuals who are most likely to benefit from the treatment. Another approach would be to capitalize on the currently unused interval between when a drug is introduced and when resistance becomes clinically apparent by using high-sensitivity methods to detect early molecular changes in CTCs or cell-free ctDNA. Avoiding the need to wait for radiographic evidence of disease relapse would enable more-rapid cycling of experimental therapies in humans, the most promising of which could then be validated in the context of traditional survival end points.
Despite many limitations, personalized cancer therapy remains the incontrovertible future of oncology, and is rapidly being implemented. Our current tools for addressing resistance remain imperfect, although it should be remembered that personalized medicine strives to deliver the best care to individual patients—not only in terms of identifying a drug we can use, but also regarding the harms we can avoid. Moving forward, several short-term actions could be implemented coordinately to forestall the onset of drug resistance. These include: early detection of subclonal drug-resistance mutations; routine implementation of high-sensitivity liquid biopsies; monitoring patients for early disease recurrence; development of effective protocols for simultaneous treatment with multiple drugs; and, most importantly, continued efforts to expand our repertoire of targeted therapeutic options.
Key points.
All cancers probably contain an enormous number of co-existing subclonal mutations; in some cases, every possible mutation could exist in at least one cell in the tumour
Resistance to molecularly targeted therapies can arise from selective growth of pre-existing subclones within the bulk of the tumour that carry drug-resistance mutations and thus have a survival advantage
Drug-resistance mutations can be found in variable proportions of tumour cells prior to therapy; their early detection enables stratification of patients to more-effective treatments and avoidance of treatments that are destined to fail
Accurate identification of resistance mutations requires highly sensitive detection techniques and representative tumour sampling
Routine interrogation of the subclonal genetic structure of tumours, will be critical to the success of personalized cancer medicine
Acknowledgments
We gratefully acknowledge our many colleagues, collaborators, and patients for the stimulating discussions that lead to the conception of this manuscript. The work of the authors is funded by NIH grants: P50 CA097186 to M.W.S.; R01 CA160674 and R33 CA181771 to L.A.L.; and T32 HL007093 to J.J.S.
Biographies
Michael W. Schmitt is a medical scientist at the University of Washington Seattle, USA. He holds MD and PhD degrees; his PhD work was focused on the biochemistry of error-prone DNA polymerases. During a post-doctoral research fellowship, he developed a methodology known as Duplex Sequencing, which enables extremely sensitive detection of rare mutations in heterogeneous samples of DNA. Subsequently, Dr Schmitt completed an internal medicine residency at the University of Washington, and is currently undertaking a medical oncology fellowship at the Fred Hutchinson Cancer Research Center, Seattle, USA. He divides his time between research and patient care. His research is focused on accurate detection of subclonal mutations in cancers, with a goal of better stratifying patients to receive the most-effective therapies.
Lawrence A. Loeb is a Professor at the University of Washington, where he directs the Gottstein Memorial Cancer Research Laboratory in the Departments of Pathology and Biochemistry. His long scientific career has focused on the origins and consequences of DNA mutations in malignancy, particularly the roles of defects in DNA repair and DNA polymerases. He is credited with formulating the mutator phenotype hypothesis of cancer. He has previously served as President of the American Association for Cancer Research and President of the Environmental Mutagen Society.
Jesse J. Salk is a medical scientist at the University of Washington, where he received his MD and PhD degrees, followed by an internal medicine residency and ongoing medical oncology training. His research focuses on the dynamics of clonal evolution from normal cells to pre-neoplasia and cancer, and new technologies for deconstructing this process. He co-developed the ultrasensitive Duplex Sequencing method with Dr Schmitt, as well as an innovative approach for tumour clonal lineage mapping using passenger mutations in genomic hotspots. His clinical interests lie in translating the complex genetic clonal architecture of individual patient tumours to early detection of cancer, optimal selection of treatment, and monitoring.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
M.W.S and J.J.S. researched data for article. M.W.S., L.A.L., and J.J.S. all contributed to discussion of content and writing of the manuscript, and reviewed/edited the manuscript before submission.
References
- 1.Schmitt MW, Prindle MJ, Loeb LA. Implications of genetic heterogeneity in cancer. Annals of the New York Academy of Sciences. 2012;1267:110–116. doi: 10.1111/j.1749-6632.2012.06590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cull EH, Altman JK. Contemporary treatment of APL. Curr Hematol Malig Rep. 2014;9:193–201. doi: 10.1007/s11899-014-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welch JS, et al. The Origin and Evolution of Mutations in Acute Myeloid Leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu HH, Qin YZ, Huang XJ. Resistance to arsenic therapy in acute promyelocytic leukemia. N Engl J Med. 2014;370:1864–1866. doi: 10.1056/NEJMc1316382. [DOI] [PubMed] [Google Scholar]
- 6b.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Research. 1974;34:2311–2321. [PubMed] [Google Scholar]
- 8.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–506. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–509. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naxerova K, et al. Hypermutable DNA chronicles the evolution of human colon cancer. Proc Natl Acad Sci USA. 2014;111:E1889–E1898. doi: 10.1073/pnas.1400179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landau DA, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Rossi D, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–47. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong TN, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhang H-EC, et al. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nature medicine. 2015 doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- 19.Mroz EA, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mroz EA, Tward AM, Hammon RJ, Ren Y, Rocco JW. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the Cancer Genome Atlas. PLoS Med. 2015;12:e1001786. doi: 10.1371/journal.pmed.1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooke SL, et al. Intra-tumour genetic heterogeneity and poor chemoradiotherapy response in cervical cancer. 2010;104:361–368. doi: 10.1038/sj.bjc.6605971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 24.Merlo LMF, et al. A comprehensive survey of clonal diversity measures in Barrett’s Esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prevention Research. 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foulkes WD, Reis-Filho JS, Narod SA. Tumor size and survival in breast cancer-a reappraisal. Nature Reviews Clinical Oncology. 2010;7:348–353. doi: 10.1038/nrclinonc.2010.39. [DOI] [PubMed] [Google Scholar]
- 27.Sinicrope FA, Yang ZJ. Prognostic and predictive impact of DNA mismatch repair in the management of colorectal cancer. Future Oncology. 2011;7:467–474. doi: 10.2217/fon.11.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox EJ, Loeb LA. Lethal Mutagenesis: targeting the mutator phenotype in cancer. Seminars in Cancer Biology. 2010;20:353–359. doi: 10.1016/j.semcancer.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29b.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chereda B, Melo JV. Natural course and biology of CML. Ann Hematol. 2015;94(Suppl 2):S107–21. doi: 10.1007/s00277-015-2325-z. [DOI] [PubMed] [Google Scholar]
- 31.Cilloni D, Saglio G. Molecular pathways: BCR-ABL. Clin Cancer Res. 2012;18:930–937. doi: 10.1158/1078-0432.CCR-10-1613. [DOI] [PubMed] [Google Scholar]
- 32.Druker BJ, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 32b.Moorman AV, Harrison CJ, Buck GA, Richards SM, Secker-Walker LM, et al. Karyotype is an independent prognostic factor in adult acute lymphoblastic leukemia (ALL): analysis of cytogenetic data from patients treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial. Blood. 2007;109:3189–97. doi: 10.1182/blood-2006-10-051912. [DOI] [PubMed] [Google Scholar]
- 33.Lee KH, et al. Clinical effect of imatinib added to intensive combination chemotherapy for newly diagnosed Philadelphia chromosome-positive acute lymphoblastic leukemia. Leukemia. 2005;19:1509–1516. doi: 10.1038/sj.leu.2403886. [DOI] [PubMed] [Google Scholar]
- 34.Soverini S, et al. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leukemia Research. 2014;38:10–20. doi: 10.1016/j.leukres.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Roche-Lestienne C, et al. Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood. 2002;100:1014–1018. doi: 10.1182/blood.v100.3.1014. [DOI] [PubMed] [Google Scholar]
- 36.Roche-Lestienne C, Laï JL, Darré S, Facon T, Preudhomme C. A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N Engl J Med. 2003;348:2265–2266. doi: 10.1056/NEJMc035089. [DOI] [PubMed] [Google Scholar]
- 37.Pfeifer H, et al. Kinase domain mutations of BCR-ABL frequently precede imatinib-based therapy and give rise to relapse in patients with de novo Philadelphia-positive acute lymphoblastic leukemia (Ph+ ALL) Blood. 2007;110:727–734. doi: 10.1182/blood-2006-11-052373. [DOI] [PubMed] [Google Scholar]
- 38.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 39.Hughes T, et al. Impact of baseline BCR-ABL mutations on response to nilotinib in patients with chronic myeloid leukemia in chronic phase. Journal of Clinical Oncology. 2009;27:4204–4210. doi: 10.1200/JCO.2009.21.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox EJ, Reid-Bayliss KS, Emond MJ, Loeb LA. Accuracy of Next Generation Sequencing Platforms. Next Gener Seq Appl. 2014;1 doi: 10.4172/jngsa.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker WT, et al. Sensitive detection of BCR-ABL1 mutations in patients with chronic myeloid leukemia after imatinib resistance is predictive of outcome during subsequent therapy. Journal of Clinical Oncology. 2011;29:4250–4259. doi: 10.1200/JCO.2011.35.0934. [DOI] [PubMed] [Google Scholar]
- 42.Parker WT, Ho M, Scott HS, Hughes TP, Branford S. Poor response to second-line kinase inhibitors in chronic myeloid leukemia patients with multiple low-level mutations, irrespective of their resistance profile. Blood. 2012;119:2234–2238. doi: 10.1182/blood-2011-08-375535. [DOI] [PubMed] [Google Scholar]
- 43.Rubin BP, et al. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Research. 2001;61:8118–8121. [PubMed] [Google Scholar]
- 43b.Corless CL, Schroeder A, Griffith D, Town A, McGreevey L, et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol. 2005;23:5357–5364. doi: 10.1200/JCO.2005.14.068. [DOI] [PubMed] [Google Scholar]
- 44.Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev. 2012;92:1619–1649. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 45.Blanke CD, et al. Long-term results from a randomized phase II trial of standard- versus higher-dose imatinib mesylate for patients with unresectable or metastatic gastrointestinal stromal tumors expressing KIT. Journal of Clinical Oncology. 2008;26:620–625. doi: 10.1200/JCO.2007.13.4403. [DOI] [PubMed] [Google Scholar]
- 46.Heinrich MC, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. Journal of Clinical Oncology. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 47.Guo T, et al. Sorafenib inhibits the imatinib-resistant KITT670I gatekeeper mutation in gastrointestinal stromal tumor. Clinical Cancer Research. 2007;13:4874–4881. doi: 10.1158/1078-0432.CCR-07-0484. [DOI] [PubMed] [Google Scholar]
- 48.Heinrich MC, et al. Correlation of kinase genotype and clinical outcome in the North American intergroup phase III trial of imatinib mesylate for treatment of advanced gastrointestinal stromal tumor. Journal of Clinical Oncology. 2008;26:5360–5367. doi: 10.1200/JCO.2008.17.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinrich MC, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 50.Tomasetti C, Demetri GD, Parmigiani G. Why tyrosine kinase inhibitor resistance is common in advanced gastrointestinal stromal tumors. F1000Res. 2013 doi: 10.12688/f1000research.2-152.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel S. Exploring novel therapeutic targets in GIST: focus on the PI3K/Akt/mTOR pathway. Curr Oncol Rep. 2013;15:386–395. doi: 10.1007/s11912-013-0316-6. [DOI] [PubMed] [Google Scholar]
- 52.Baker NM, Der CJ. Cancer: Drug for an ‘undruggable’ protein. Nature. 2013;497:577–578. doi: 10.1038/nature12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Y, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncology. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 55.Yang JCH, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncology. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 56.Fukuoka M, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29:2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 57.Moore MJ, et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 58.Wang JP, et al. Erlotinib is effective in pancreatic cancer with epidermal growth factor receptor mutations: a randomized, open-label, prospective trial. Oncotarget. 2015;6:18162–18173. doi: 10.18632/oncotarget.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seo JS, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Research. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou Q, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. Journal of Clinical Oncology. 2011;29:3316–3321. doi: 10.1200/JCO.2010.33.3757. [DOI] [PubMed] [Google Scholar]
- 61.Lee JK, et al. Epidermal growth factor receptor tyrosine kinase inhibitors vs conventional chemotherapy in non–small cell lung cancer harboring wild-type epidermal growth factor receptor. JAMA: The Journal of the American Medical Association. 2014;311:1430. doi: 10.1001/jama.2014.3314. [DOI] [PubMed] [Google Scholar]
- 62.de Bruin EC, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346:256–259. doi: 10.1126/science.1256930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Balak MN, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 65.Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nature medicine. 2013;19:1389–1400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su KY, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. Journal of Clinical Oncology. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 67.Le DT, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schmitt MW, et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nature Methods. 2015;12:423–425. doi: 10.1038/nmeth.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee Y, et al. Clinical outcome according to the level of preexisting epidermal growth factor receptor T790M mutation in patients with lung cancer harboring sensitive epidermal growth factor receptor mutations. Cancer. 2014;120:2090–2098. doi: 10.1002/cncr.28711. [DOI] [PubMed] [Google Scholar]
- 70.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 71.Dolgin E. FDA narrows drug label usage. Nature. 2009;460:1069. doi: 10.1038/4601069a. [DOI] [PubMed] [Google Scholar]
- 72.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- 73.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molinari F, et al. Increased detection sensitivity for KRAS mutations enhances the prediction of anti-EGFR monoclonal antibody resistance in metastatic colorectal cancer. Clinical Cancer Research. 2011 doi: 10.1158/1078-0432.CCR-10-3137. [DOI] [PubMed] [Google Scholar]
- 75.Dono M, et al. Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: implications for treatment of metastatic colorectal cancer. Mol Med. 2012;18:1519–1526. doi: 10.2119/molmed.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tougeron D, et al. Effect of low-frequency KRAS mutations on the response to anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2013;24:1267–1273. doi: 10.1093/annonc/mds620. [DOI] [PubMed] [Google Scholar]
- 77.Long GV, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 78.Su F, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robert C, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 80.Flaherty KT, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long GV, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. The Lancet. 2015;386:444–451. doi: 10.1016/S0140-6736(15)60898-4. [DOI] [PubMed] [Google Scholar]
- 82.Long GV, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nature Communications. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 82b.Girotti MR, Lopes F, Preece N, Niculescu-Duvaz D, Zambon A, et al. Paradox-breaking RAF inhibitors that also target SRC are effective in drug-resistant BRAF mutant melanoma. Cancer Cell. 2015;27:85–96. doi: 10.1016/j.ccell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bottema CD, Sommer SS. PCR amplification of specific alleles: rapid detection of known mutations and polymorphisms. Mutat Res. 1993;288:93–102. doi: 10.1016/0027-5107(93)90211-w. [DOI] [PubMed] [Google Scholar]
- 84.Tost J, Gut IG. Genotyping single nucleotide polymorphisms by MALDI mass spectrometry in clinical applications. Clin Biochem. 2005;38:335–350. doi: 10.1016/j.clinbiochem.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 85.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nature Methods. 2005;2:285–290. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 86.Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods. 2013;59:101–107. doi: 10.1016/j.ymeth.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 87.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Nat Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Nat Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lou DI, et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Nat Acad Sci U S A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klco JM, et al. Functional Heterogeneity of Genetically Defined Subclones in Acute Myeloid Leukemia. Cancer Cell. 2014;25:379–392. doi: 10.1016/j.ccr.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science Translational Medicine. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zill OA, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discovery. 2015 doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lebofsky R, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol. 2015;9:783–790. doi: 10.1016/j.molonc.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Piotrowska Z, et al. Heterogeneity Underlies the Emergence of EGFRT790 Wild-Type Clones Following Treatment of T790M-Positive Cancers with a Third-Generation EGFR Inhibitor. Cancer Discovery. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Siravegna G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nature medicine. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thress KS, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non–small cell lung cancer harboring EGFR T790M. Nature medicine. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diaz LAJ, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bardelli A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discovery. 2013;3:658–673. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu M, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chu S, et al. Detection of BCR-ABL kinase mutations in CD34+ cells from chronic myelogenous leukemia patients in complete cytogenetic remission on imatinib mesylate treatment. Blood. 2005;105:2093–2098. doi: 10.1182/blood-2004-03-1114. [DOI] [PubMed] [Google Scholar]
- 102.Kim TM, et al. Regional biases in mutation screening due to intratumoural heterogeneity of prostate cancer. J Pathol. 2014;233:425–435. doi: 10.1002/path.4380. [DOI] [PubMed] [Google Scholar]
- 103.Kumar A, et al. Deep sequencing of multiple regions of glial tumors reveals spatial heterogeneity for mutations in clinically relevant genes. Genome Biol. 2014;15:530. doi: 10.1186/s13059-014-0530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Montagut C, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature medicine. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 105.Lee MS, Kopetz S. Current and Future Approaches to Target the Epidermal Growth Factor Receptor and Its Downstream Signaling in Metastatic Colorectal Cancer. Clin Colorectal Cancer. 2015 doi: 10.1016/j.clcc.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 106.Byrd JC, et al. Ibrutinib versus ofatumumab in previously treated chronic lymphoid leukemia. N Engl J Med. 2014;371:213–223. doi: 10.1056/NEJMoa1400376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Byrd JC, et al. Three-year follow-up of treatment-naive and previously treated patients with CLL and SLL receiving single-agent ibrutinib. Blood. 2015;125:2497–2506. doi: 10.1182/blood-2014-10-606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Advani RH, et al. Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J Clin Oncol. 2013;31:88–94. doi: 10.1200/JCO.2012.42.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Woyach JA, et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N Engl J Med. 2014;370:2286–2294. doi: 10.1056/NEJMoa1400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chiron D, et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discovery. 2014;4:1022–1035. doi: 10.1158/2159-8290.CD-14-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Daver N, et al. Secondary mutations as mediators of resistance to targeted therapy in leukemia. Blood. 2015;125:3236–3245. doi: 10.1182/blood-2014-10-605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thakur, Das M, et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jordan CT. Cancer stem cells: controversial or just misunderstood? Cell Stem Cell. 2009;4:203–205. doi: 10.1016/j.stem.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Willis SG, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106:2128–2137. doi: 10.1182/blood-2005-03-1036. [DOI] [PubMed] [Google Scholar]
- 116.Nucifora G, Larson RA, Rowley JD. Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood. 1993;82:712–715. [PubMed] [Google Scholar]
- 117.Beckman RA, Loeb LA. Efficiency of carcinogenesis with and without a mutator mutation. Proc Natl Acad Sci USA. 2006;103:14140–14145. doi: 10.1073/pnas.0606271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci USA. 2011;108:17087–17092. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shlien A, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]