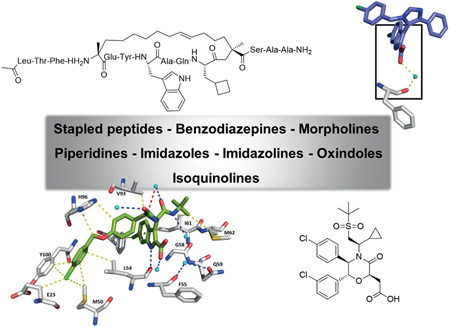
Abstract
A recent therapeutic strategy in oncology is based on blocking the protein-protein interaction between the murine double minute (MDM) homologues MDM2/X and the tumor-suppressor protein p53. Inhibiting the binding between wild-type (WT) p53 and its negative regulators MDM2 and/or MDMX has become an important target in oncology to restore the antitumor activity of p53, the so-called guardian of our genome. Interestingly, based on the multiple disclosed compound classes and structural analysis of small-molecule-MDM2 adducts, the p53-MDM2 complex is perhaps the best studied and most targeted protein-protein interaction. Several classes of small molecules have been identified as potent, selective, and efficient inhibitors of the p53-MDM2/X interaction, and many co-crystal structures with the protein are available. Herein we review the properties as well as preclinical and clinical studies of these small molecules and peptides, categorized by scaffold type. A particular emphasis is made on crystallographic structures and the observed binding modes of these compounds, including conserved water molecules present.
Keywords: cancer, conserved water, MDM2, MDMX, p53, protein–protein interactions
Graphical Abstract
1. Introduction
Protein–protein interactions (PPIs) are of utmost importance and are implicated in almost all biological processes. Proteins should not be considered to function as single isolated entities; indeed, they fulfill their roles by interacting with many other cellular components, and the various interaction patterns of a protein are at least as important as the intrinsic biochemical activity of the protein itself. Therefore, the biological role of a given protein is based heavily on the PPIs it undergoes. In particular, the majority of disease cases are ultimately governed by dysregulated PPIs. Identifying and successfully targeting these PPIs by finding inhibitors or activators is one the bases of drug discovery.[1]
The tumor-suppressor protein p53 is the principal regulator of cell division and growth,[2, 3] as it is able to control genes that are implicated in cell-cycle control, apoptosis, angiogenesis, senescence, and autophagia.[4] Mutations in this protein are present in ~50% of human cancers; altering the DNA binding domain impedes the activity of p53 as a transcription factor.[5] In the remaining tumors, the p53 pathway is inactivated by up-regulation of p53 inhibitors, such as the mouse double minute proteins (MDM2 and MDMX, also known as MDM4, HDM2, and HDMX in humans), or by downregulation of p53 cooperators, such as ARF.[6, 7]
The MDM2 gene was found to be upregulated in ~7% of tumors, with increased transcript levels and enhanced translation. Mutation of p53 and upregulation of MDM2 do not usually occur within the same tumor, indicating that MDM2 overexpression is an effective path for inactivation of p53 function in tumorigenesis.[8] MDM2 functions as an inhibitor of the N-terminal transactivation domain (TAD) of p53 and promotes p53 degradation through the ubiquitin–proteasome system (E3 ligase activity).[9, 10] On the other hand, MDMX can downregulate p53 by inhibition of the TAD domain and it can upregulate MDM2.[11] Moreover, it does not have E3 ligase activity, but its binding with MDM2 increases the rates of ubiquitination of p53 by MDM2.[11, 2] The C-terminal RING domains of both MDM2 and MDMX are involved in dimerization; MDM RING domains can form homodimers, and heterodimers can be formed by decreased auto-ubiquitination of MDM2 and increased p53 ubiquitination.[13] Consequently, MDM2 is stabilized by MDMX, keeping the levels of p53 low in healthy cells.[13–15] The use of dual-action MDM2/MDMX antagonists in cancer cells expressing wild-type (WT) p53 should activate p53 more significantly than agents that only inhibit MDM2, resulting in more effective antitumor activity.[16–18]
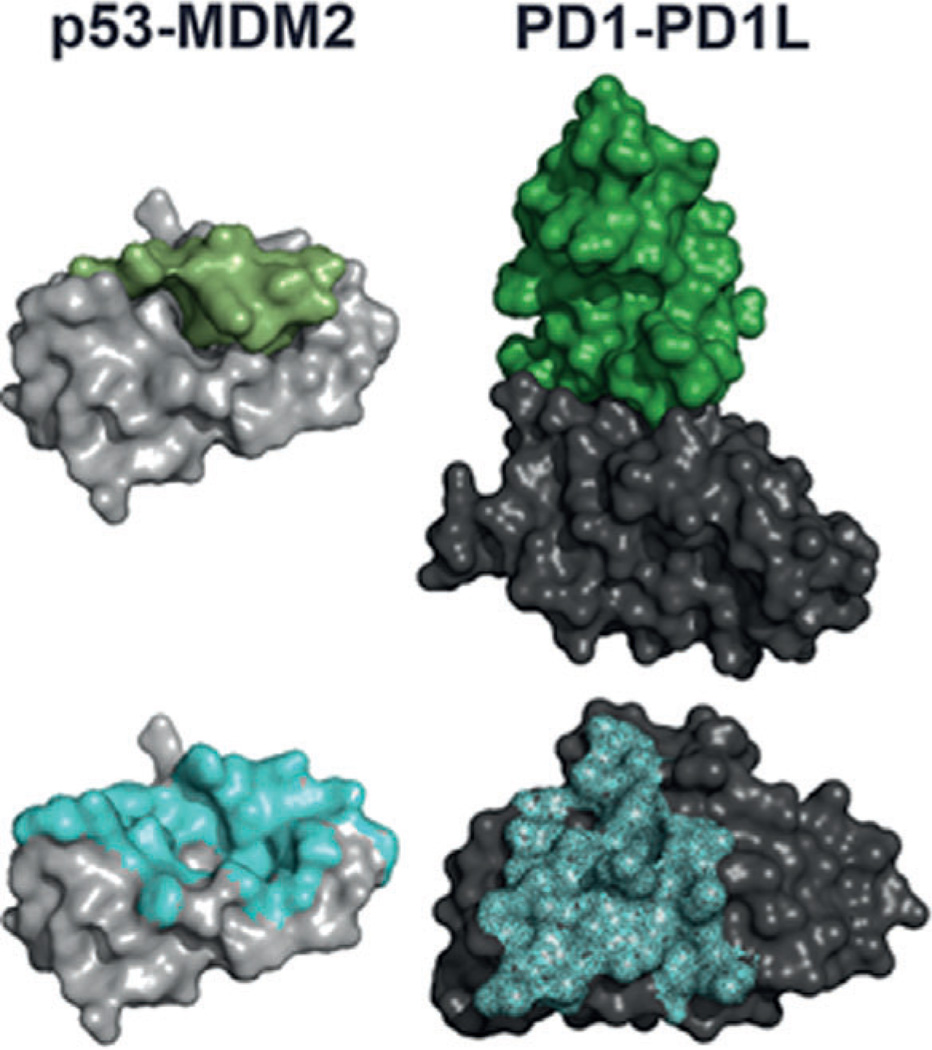
The p53–MDM2 interaction is druggable by small molecules based on a buried surface area of ~700 Å2 and a well-structured, deep, and hydrophobic binding site of similar dimensions to those of small molecules. For comparison, the recently solved immuno-oncology target protein–protein interaction PD1–PD1L has a buried surface area of ~2000 Å2, and is relatively flat and featureless (Figure 1).[19, 20]
Figure 1.
Two important protein–protein interaction targets. Left top: p53 (green) with MDM2 (grey), PDB ID: 1YCR; bottom: footprint of p53 on MDM2 shown as blue surface. Right top: PD1 (green) with PD1L (grey), PDB ID: 4ZQK; bottom: footprint of PD1 on PD1L shown as blue surface.
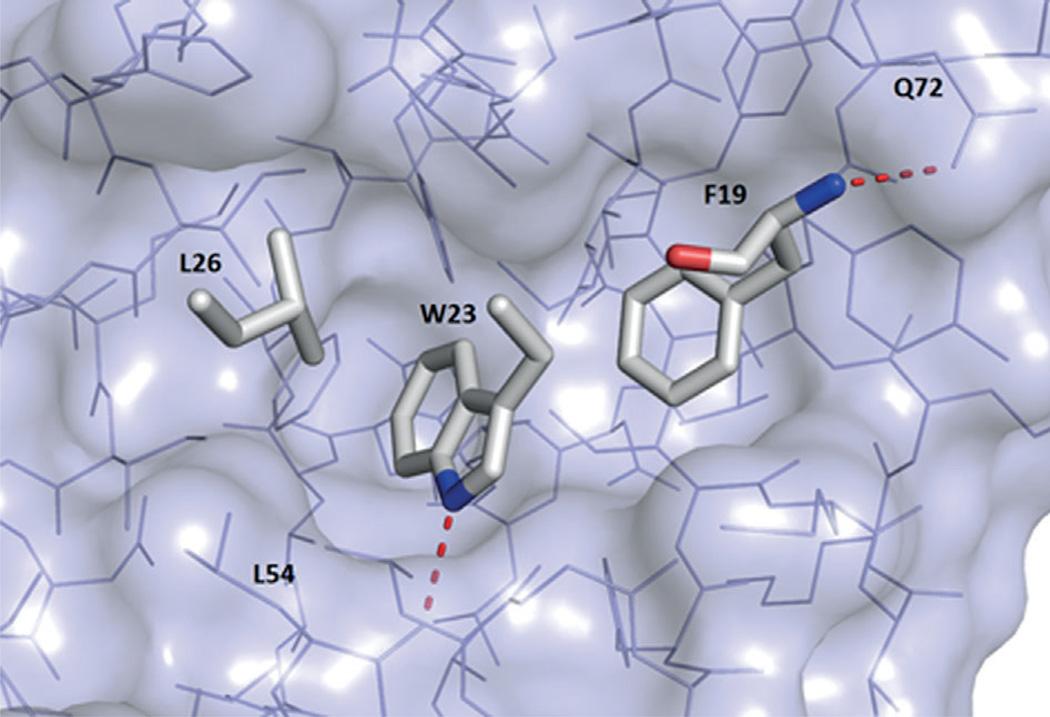
Notably, formation of the MDM2-based binding pocket is induced upon p53 or ligand binding. The NMR structure of apo-MDM2 does not show any pocket, expect a small indent anticipatory of the future p53Trp23 binding site (PDB ID: 1Z1M).[21 Nonetheless, all small-molecule ligand- or peptide-bound MDM2 structures are very similar in the induced binding pocket region and can be aligned with a very high RMSD. The binding site of p53 in MDM2 is formed by 14 residues: Leu54, Leu57, Ile61, Met62, Tyr67, Gln72, Val75, Phe86, Phe91, Val93, His96, Ile99, Tyr100, Ile101. The cleft at the surface of MDMX is highly similar; four out of 14 residues are, however, different: MDM2Leu54→MDMXMet53, MDM2Phe86→MDMXLeu85, MDM2His96→ MDMXPro95, and MDM2Ile99→MDMXLeu98.[18] MDM2 and MDMX have a deep hydrophobic pocket on which the p53 protein binds as an α helix. Three p53 residues are deeply buried in the MDM2 and MDMX clefts: Phe19, Trp23, and Leu26; the interaction is governed mostly by hydrophobic interactions (PDB ID: 1YCR, Figure 2).[22, 23]
Figure 2.
Key p53 residues Phe19, Trp23, and Leu26 bound to MDM2 (PDB ID: 1YCR). Red dotted lines indicate polar contacts.
Additionally, the Trp23 indole NH group forms a hydrogen bond with the backbone carbonyl group of MDM2Leu54. In MDMX, Met53 and Tyr99 are directed into the hydrophobic surface groove, making the binding pocket smaller and slightly different in shape (PDB IDs: 3DAB and 3DAC).[22] The known three-finger pharmacophore model for p53–MDM2 is now widely accepted to be responsible for the binding of small molecules and peptides to MDM2.[24–26] Recently, an extended four-finger model was proposed, taking into account the intrinsically disordered MDM2 N terminus, which can be ordered by certain small molecules, as shown by co-crystallization.[27, 28]
Given the importance of p53–MDM2/X PPI, several reviews have been published in the last few years.[29–32] More recently, however, several novel inhibitor classes have been disclosed, and new compounds have entered into clinical trials, thus justifying this updated review, which provides a broad recent overview of inhibitors of the p53–MDM2/MDMX protein–protein interaction, focusing mainly on the available structural data. Additionally, some insight into the rational design and optimization of MDM2/X binders, their in vitro and in vivo activities, and clinical trials are given. Moreover, a preliminary water analysis of the current crystal structures is presented.
2. Inhibitors of p53–MDM2/MDMX interactions
2.1. Nutlin-type compounds
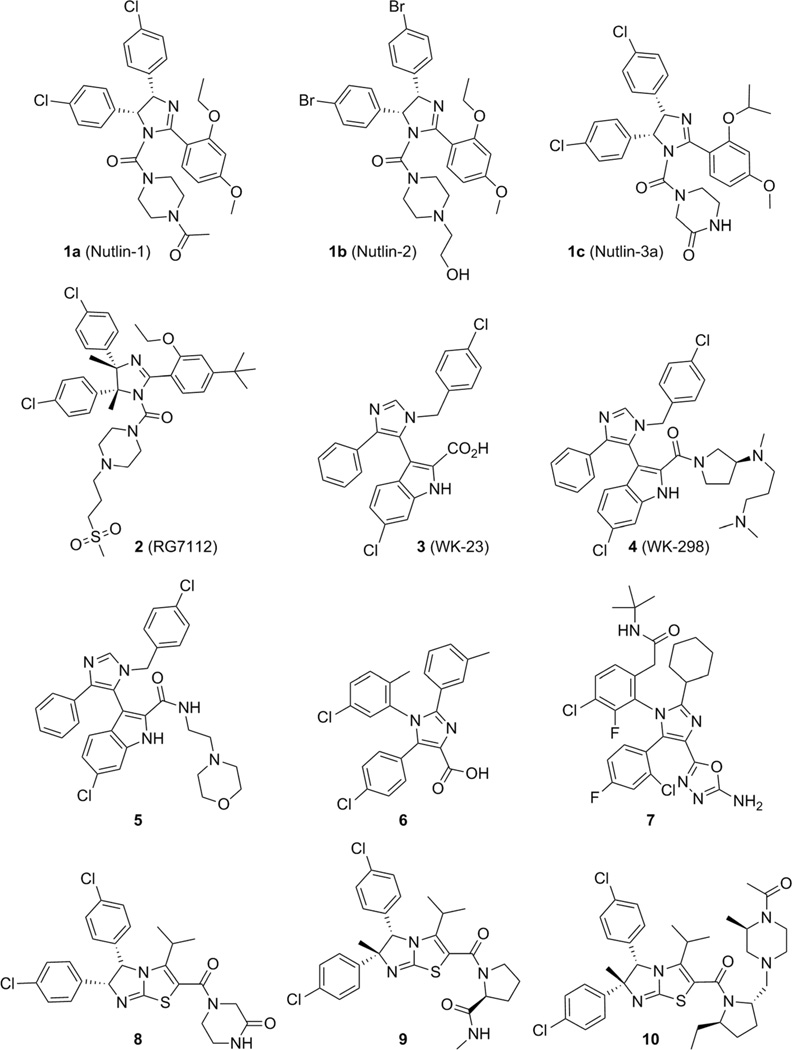
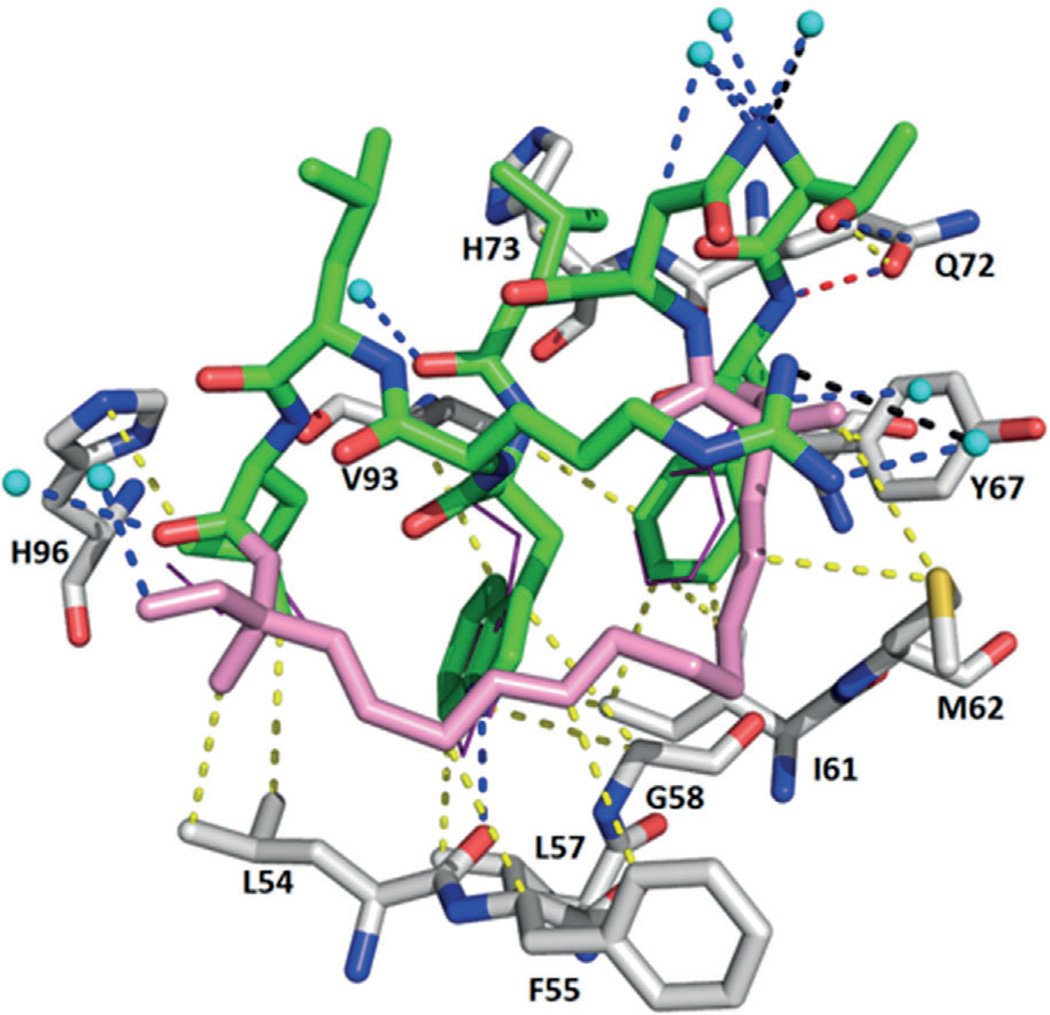
The first class of highly potent, specific, and orally active MDM2 inhibitors was disclosed in 2004 by scientists from Hoffmann-La Roche:[33] cis-diphenyl-substituted imidazolines, known as Nutlins (1). Modifications and optimizations resulted in derivatives named Nutlin-1 (1a), Nutlin-2 (1b), and Nutlin-3a (1c) (Figure 3). Nutlin compounds show 50% inhibitory concentration (IC50) values of 260, 140, and 90 nm, respectively. These compounds were observed to displace the recombinant p53 fragment (corresponding to residues 1–312 of human p53) from its complex with MDM2 in competition assays monitored by surface plasmon resonance (SPR).[33] A Nutlin-2– MDM2 complex co-crystal structure (PDB ID: 1RV1, Figure 4) marked the first structural elucidation of a non-peptide small molecule interacting with p53 protein.[33] Superimposition of the co-crystal structures MDM2–Nutlin-2 and MDM2–p53 (PDB ID: 1YCR) showed that the two bromo-substituted phenyl rings and the ethoxy group of Nutlin-2 mimic Trp23, Leu26, and Phe19, respectively: the key hydrophobic binding residues of the p53 peptide (Figure 1).[33] Nutlin-3a bound to MDM2 (PDB ID: 4J3E)[34] shows that both 4-chlorophenyl groups perfectly fill the Leu26 and Trp23 pockets, while the isopropoxy group reaches deep into the Phe19 pocket. It activates WT p53 and selectively kills cancer cells, with IC50 values of 1–2 µm in the SJSA-1 (osteosarcoma), HCT116 (colorectal), and RKO (colon carcinoma) cell lines, and showed 10-fold selectivity compared with the p53-mutated cell lines MDA-MB-435 (melanoma) and SW480 (colorectal adenocarcinoma).[33] Furthermore, in vivo studies demonstrate the capacity of Nutlin-3a to decrease tumor growth by 90% in an SJSA-1 mouse xenograft model.[33]
Figure 3.
Nutlins, imidazole and imidazothiazole derivatives.
Figure 4.
Crystal structure of Nutlin-2 bound to MDM2 (PDB ID: 1RV1). Nutlin-2 (green)–receptor (grey) interactions are indicated by colored dotted lines: yellow: hydrophobic, orange: π–π, blue: hydrogen bonding, black: cation dipolar, chocolate: halogen, red: dipolar. The receptor and compound are aligned with the p53 hot-spot residues (magenta lines, PDB ID: 1YCR; this color code and alignment are maintained throughout the remaining figures). The 3-(4-bromophenyl) group undergoes a π–π stacking interaction with His96 and is embedded in hydrophobic interactions with Leu54. The 2-(4-bromophenyl) residue makes multiple hydrophobic contacts with Leu54, Gly58, Ile61, and Val93.
In 2008, compound 2 (RG7112) based on Nutlins (PDB ID: 4IPF),[35] completed phase I clinical trials against advanced solid and soft-tissue tumors and hematological malignancies.[36,37]] Four key modifications were made to enhance the binding activity to MDM2, cell growth inhibition, and to improve pharmacokinetic properties: Two methyl groups were introduced in the imidazole ring to protect against metabolism, the isopropyl ether was replaced by ethyl ether to lower the molecular weight while maintaining hydrophobic interactions, the methoxy group was changed to a tert-butyl moiety to decrease metabolic liability, and lastly, a methylsulfonyl group was added to the piperidine ring to increase the binding affinity and to improve the pharmacokinetics by decreasing the logD value. After these modifications, compound 2 displayed enhanced binding activity for MDM2, with a Kd value of 10.7 nm and cell growth inhibition threefold more potent than Nutlin-3a.[34] The main drawback observed during clinical trials in patients with liposarcoma was thrombocytopenia, but the data obtained from biopsies in this preliminary trial suggested that the p53 pathway can be reactivated despite the presence of excess MDM2, resulting in cytostatic and possibly apoptotic effects in tumor cells (Figure 3).[36, 38]
2.2. Imidazoles and imidazothiazoles
Trisubstituted imidazole compounds 3–5 with a 6-chloroindole moiety as tryptophan mimic were discovered by a virtual Trp23-anchor-based pharmacophore screening approach and validated as MDM2 inhibitors (Figure 3).[39] The side chain of p53 Trp23 is embedded in a deep hydrophobic pocket formed by the MDM2 residues Leu57, Phe86, and Ile99, and it was noted early on that at the bottom of the Trp23 indole group, unfilled hydrophobic space is left which could be filled by a suitable hydrophobic group.[40] Thus substitution of the indole 6-position with methyl or alkynyl groups or a halogen atom can enhance the binding affinity by a factor up to 50. In addition, two other fragments were introduced to mimic leucine and phenylalanine residues, leading to compound 3 (WK-23), co-crystalized with MDM2 (PDB ID: 3LBK) with binding affinities of 916 nm and 36 µm for MDM2 and MDMX, respectively. Its analogue 4 (WK-298) with binding affinities of 109 nm and 11 µm for MDM2 and MDMX, respectively, was part of the first co-crystal structure of a small molecule bound to MDMX (PDB ID: 3LBJ).[41] These crystal structures clearly reveal that 1,3,5-trisubstituted 5-indolo-imidazoles can bind both to MDM2 and MDMX and thus provide a starting point for the design of dual-action MDM2/X antagonists. Compound 3 binds to MDM2 by filling its Trp23 sub-pocket with the 6-chloroindole group, the 4-phenyl group is located in the Phe19 pocket, and the 1-(4-chlorobenzyl) group resides in the Leu26 pocket. A hydrogen bond is formed between the indole of 3 and the Leu54 carbonyl oxygen atom of MDM2 similar to the Trp23– Leu54 hydrogen bond found in the endogenous p53–MDM2 interaction (Figures 3 and 5).[41] The inhibitor 4 binds to MDMX in a similar manner, despite the different shapes of the p53 binding sites in MDM2 and MDMX. The His96–Tyr100 region has the most pronounced differences in the shape of the Leu26 pocket, but the position of 1-(4-chlorobenzyl) is not altered. Two hydrogen bonds with MDMXMet53 and MDMXHis54 are formed, whereas the N,N-dimethylpropylamine portion of 4 folds over MDMXGly57 and MDMXMet61, forming an additional hydrophobic protection of the binding cleft, where MDMXTyr99 closes the p53Leu26 sub-pocket.[41]
Figure 5.
Crystal structure of compound 3 (WK-23) bound to MDM2 (PDB ID: 3LBK). A hydrogen bond between the indole NH group of 3 and the MDM2Leu54 carbonyl oxygen is depicted. The 1-(4-chlorobenzyl) group undergoes a T-shaped π–π stacking with His96. The compound makes multiple hydrophobic interactions with Leu57, Gli58, Ile61, Met62, Val93, His96, Ile99, and Tyr100.
Furthermore, the 3-imidazolyl indole inhibitor 5 (PDB ID: 4DIJ) was reported based on the central valine concept of Novartis by which a planar aromatic ring (imidazole) was placed in van der Waals contact with the side chain of Val93, occupying a central position in the upper part of the MDM2 pocket and providing different types of substitutions.[42] In this structure, an aromatic π–π stacking interaction between MDM2 residue His96 and the benzylic chlorophenyl ring of the compound was observed. Attempts to optimize compound 5 either by promoting the formation of a hydrogen bond with His96 and/or improving the cellular activity led to the tetrasubstituted imidazoles 6 (PDB ID: 4OQ3) and 7. Modeling showed that the plane of the inhibitor core imidazole ring and that of its chlorophenyl ring are nearly perpendicular, with an angle of 808 (Figure 3).[42, 43]
Novel dihydroimidazothiazole derivatives also containing a core imidazole heterocycle were developed as potent inhibitors of the p53–MDM2 interaction. As an initial lead, scientists from Daiichi Sankyo reported compound 8.[22] Further optimization by attaching a methyl group at the C6 position to avoid oxidation, and by modifying the C2 moiety of the additional proline motif, led to compound 9 (PDB ID: 3VZV) with an IC50 value of 8.3 nm in a homogeneous time-resolved fluorescence (HTRF) assay.[44, 45] The pyrrolidine moiety at C2 induced another hydrophobic interaction site with MDM2 (Met50 and Tyr67), as revealed by analysis of the co-crystal structure (Figure 6). Moreover, solubility was improved by introducing an alkyl group onto the pyrrolidine at the C2 substituent and by modifying the terminal substituent of the proline motif.
Figure 6.
Crystal structure of compound 9 bound to MDM2 (PDB ID: 3VZV). The pyrrolidine moiety induces a new hydrophobic interaction site with Met50 and Tyr67. Multiple hydrophobic interactions are formed with Thr26, Leu54, Ile61, Met62, Val93, Ile99, and Tyr100.
Compound 10 (PDB ID: 3W69), with an IC50 value of 59 nm (HTRF), also exhibited a good pharmacokinetic profile and significant antitumor efficacy via oral administration in a mouse xenograft model using MV4-11 cells bearing WT p53 (Figure 3).[44, 45]
2.3. Indoles and spirooxindoles
Using the pharmacophore search software ANCHOR.QUERY,[46] a series of 6-chloroindole derivatives were discovered, based on Ugi multicomponent reaction (MCR) chemistry (Figure 7).[47] A novel class of compounds derived from an Ugi MCR (Ugi four-component, five centers, U-5C-4CR) showed potent binding at MDM2 and MDMX, via the classical three-finger pharmacophore model, mimicking the hot-spot residues Phe19, Trp23, and Leu26. The most potent compounds are 11 (PDB ID: 3TJ2), 12 (PDB ID: 4MDQ), and 13 (PDB ID: 4MDN), with respective Ki values of 0.4, 1.2, and 0.6 µm in a fluorescence polarization (FP) assay, which was verified by a heteronuclear single quantum coherence (HSQC) experiment, in which 15N-labeled MDM2 was used, and thus the ligand-induced perturbations in NMR chemical shifts were observed.[27, 47] Compound 13 exhibited a novel fourth pharmacophore binding motif in MDM2 that is directly connected to the Leu26 sub-pocket, forming a deep binding spot around the fourth pharmacophore point of the molecule. A hydrogen bond is formed again between the indole of 13 and the Leu54 carbonyl oxygen atom of MDM2, mimicking the Trp23–Leu54 interaction. Furthermore, a halogen bond between the chlorine atom of the p-chlorobenzyl moiety and the carbonyl groups of Glu23 is possibly formed (Figure 8).[27, 47, 48]
Figure 7.
Indole and spirooxindole derivatives as inhibitors of the p53–MDM2/X interaction.
Figure 8.
Crystal structure of compound 13 bound to MDM2 (PDB ID: 4MDN). A hydrogen bond is formed again between the indole NH group of 13 and the carbonyl oxygen atom of Leu54. The compound makes hydrophobic interactions with Glu23, Met50, Leu54, Phe55, Gly58, Gln59, Ile61, Met62, Phe86, Val93, Ile99, and Tyr100.
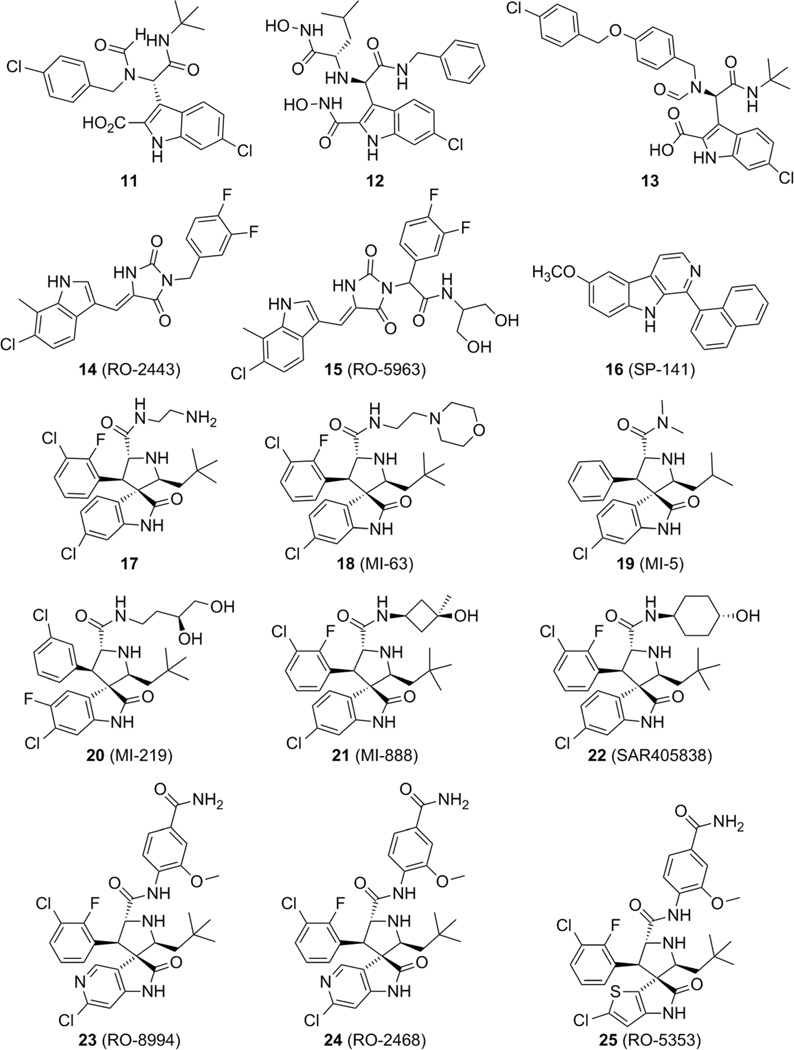
Researchers from Hoffmann-La Roche identified a series of indolylhydantoin compounds as potent dual MDM2/MDMX inhibitors. The most representative compound of the series, 14 (RO-2443, PDB ID: 3VBG) showed outstanding inhibitory activity against both MDM2 and MDMX, with IC50 values for blocking p53 binding of 33 and 41 nm, respectively. Additionally, RO-2443 binds to the p53 pocket of MDMX, and crystallographic and biochemical investigations suggested protein homodimerization and heterodimerization as mechanisms of action (PDB ID: 3U15).[49] Due to poor water solubility it was initially not possible to assess cellular activity; however, after supplementary optimization, compound 15 (RO-5963) was derived, which was found to inhibit the binding of p53 to MDM2 and MDMX and showed strong antiproliferative activity in cancer cell lines overexpressing MDMX (Figure 7).[49] These ‘dimerizer’ molecules comprise a Michael acceptor system, and thus the observed antiproliferative effects observed cannot be clearly assigned to a single mode of action.
A new class of pyrido[b]indole derivatives was recently discovered by a high-throughput virtual screening effort, and the most active of this class was compound 16 (SP-141), which has a Ki value of 28 nm for binding with MDM2 as determined in an FP-based assay. Significant inhibition of breast cancer cell growth, and decreased growth of tumors in two different breast cancer xenograft models were observed (Figure 7).[50] However, the binding mode of 16 is unclear and does not fit the three-point pharmacophore model shown by the rest of the compounds discussed herein.
Another privileged structure for p53–MDM2 inhibition is the spirooxindole. Based on the insight that an oxindole group can mimic the Trp23 moiety, spirooxindole-containing natural products were identified and docked into MDM2. A series of compounds, such as 17 (PDB ID: 4JVR), discovered by high-throughput screening assays with spirooxindoles, were developed with favorable results in HTRF assays (e.g., IC50: 9.4 nm for 17).[51] Compound 18 (MI-63) was also developed, having Ki values of 36 nm and 55 µm toward MDM2 and MDMX, respectively.[52] Analyzing the crystal structure of an MI-63 analogue (PDB ID: 3LBL), the compound also binds to MDM2 by nesting the chlorophenyl substructure of its 6-chlorooxindole into the Trp23 sub-pocket. The Leu26 sub-pocket is filled by the 2-fluoro-3-chlorophenyl ring, which is located similarly to the positioning in compounds 3 (WK-23) and 4 (WK-298), but its plane is rotated to allow the phenyl substituent atoms to fill the bottom of the MDM2 pocket. The neopentyl fragment fills the Phe19 pocket and causes a substantial induced-fit reshaping of the binding cleft, the Tyr67 side chain is rotated to close the binding region, and the whole Tyr67–His73 region acquires a different fold to allow the Tyr67 movement. The compound forms two hydrogen bonds with Leu54 and His96. The ethylmorpholino part of the compound does not take part in the binding and is not observed in the electron density (Figure 7).[41, 51]
Thorough optimization of the scaffold led to compounds with Ki values in the low-micromolar and nanomolar range (compounds 19–21), giving promising results in in vitro and in vivo assays, with compounds 20 (MI-219) and 21 (MI-888) showing Ki values of 8.5 µm and 5 nm (HTRF), respectively.[23, 51] An analogue of compounds 20 and 21 which has been advanced into phase I clinical trials is compound 22 (MI-773, SAR405838). This compound exhibits a Ki value of 0.88 nm (10-fold more potent than MI-219). As the co-crystal structure reveals, inhibitor 22 mimics the three important amino acids, captures additional interactions that were not observed in the p53–MDM2 complex, and induces refolding of the unstructured N-terminal region of MDM2 to achieve its high affinity. Furthermore, 22 activates WT p53 in vitro and in xenograft tumor tissue of leukemia and solid tumors, leading to p53-dependent cell-cycle arrest and/or apoptosis (Figure 7).[53]
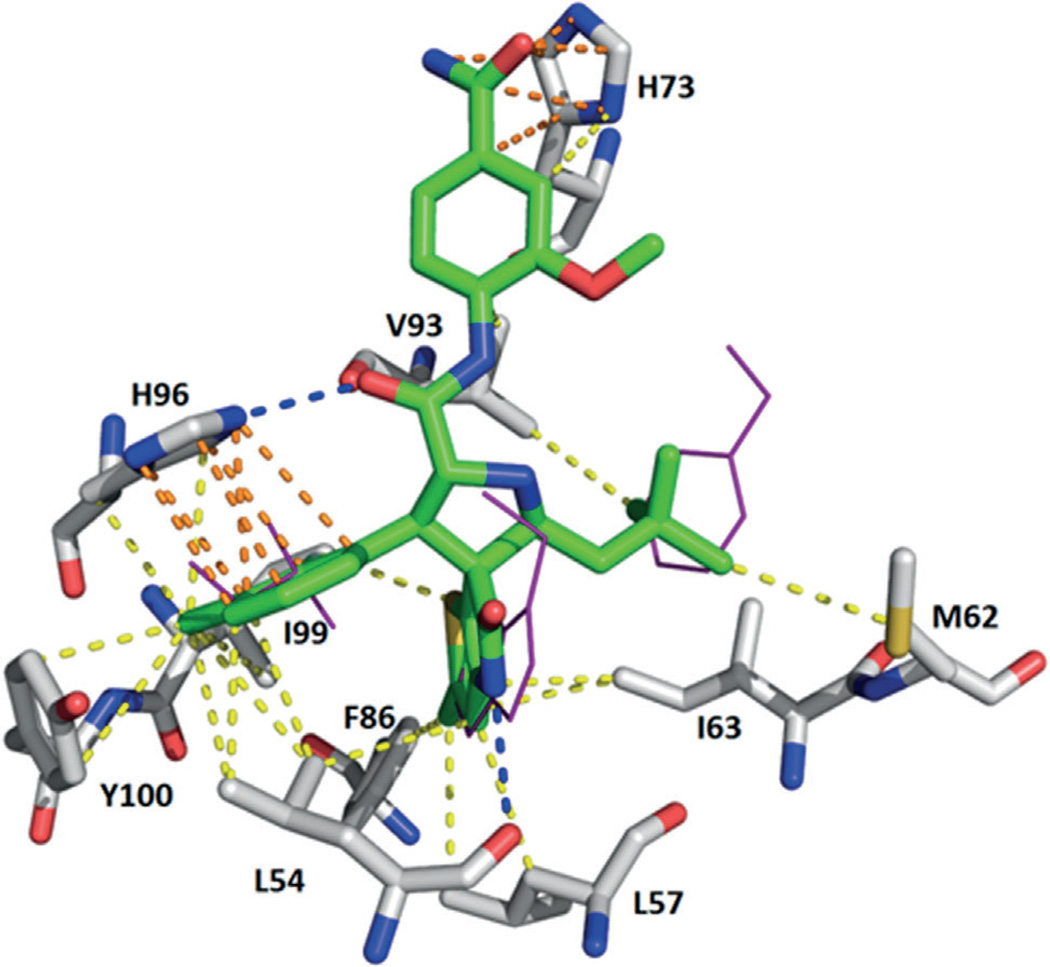
Based on the MI-series of compounds, scientists at Hoffman-La Roche developed modified spirooxindole small molecules using an additional phenyl moiety to fit into the Phe19 pocket and an isopropyl group to mimic Leu26 (23, RO-8994) with an IC50 value of 5 nm (HTRF).[54, 55] As expected, the 6-chloro-1,3-di-hydro-2H–pyrrolo[3,2-c]pyridin-2-one moiety in compound 23 is buried in the Trp23 pocket, and its NH group forms a hydrogen bond with a backbone carbonyl of MDM2 for enhanced binding affinity. Because the interaction in the Trp23 pocket appears to be the most critical, further exploration of bioisosteric replacements on the phenyl moiety of 6-chlorooxindole, while preserving other important architectural features in RO-8994 for optimal binding and pharmacological properties, led to 24 (RO-2468) and 25 (RO-5353) (PDB ID: 4LWV) with respective IC50 values of 6 and 7 nm (HTRF). In the latter, the 2-chloro-thienyl[3,2-b]pyrrol-5-one group is buried in the Trp23 pocket, whereas the 3-chloro-2-fluorophenyl group occupies the Leu26 pocket (Figure 9).[54] Examples 23–25 nicely show that bioisosteric replacement of the abundant p-halogen-substituted phenyl groups in the Trp23 pocket by heteroaromatic rings is possible and could be used to improve the overall very hydro-phobic properties of MDM2 antagonists.
Figure 9.
Crystal structure of compound 25 (RO-5353) bound to MDM2 (PDB ID: 4LWV). Hydrogen bonds are observed between both the pyrrole NH group and Leu54 and between the amide carbonyl group and His96. Two π–π stacking interactions are also formed between His73 with the 2-me-thoxy-4-carbamoyl amide aromatic ring, and His96 with the 2-fluoro-3-chlorobenzene ring. Hydrophobic interactions are observed with Leu54, Leu57, Ile61, Met62, Phe86, Val93, Ile99, and Tyr100.
2.4. Pyrrolidines
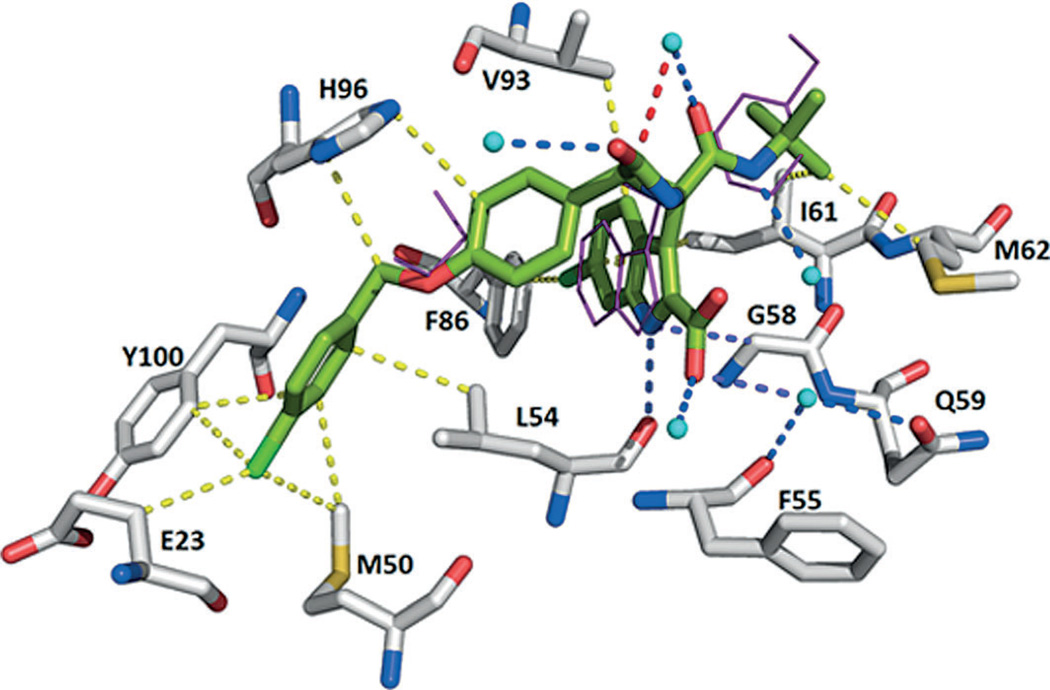
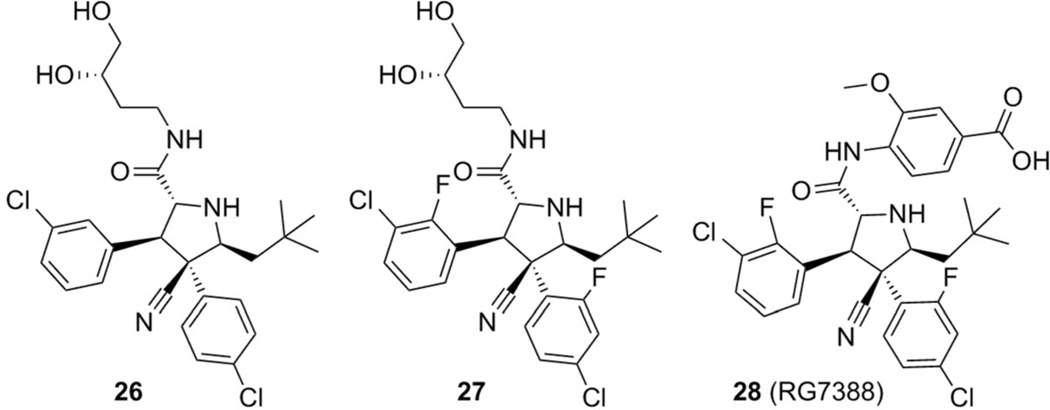
After the discovery of compounds 2 (RG7112) and 23 (MI-219), novel pyrrolidine derivatives were also developed as p53–MDM2/X inhibitors by Hoffman-La Roche scientists. The prototype compounds 26 and 27 (PDB IDs: 4JRG and 4JSC) showed good potency (IC50: 196 and 74 nm, respectively) in the HTRF assay.[55] The 4-chlorophenyl ring of compound 26 is buried in the Trp23 pocket, the 3-chlorophenyl group occupies the Leu26 pocket, and the neopentyl moiety binds to the Phe19 pocket (Figure 10).
Figure 10.
Crystal structure of compound 26 bound to MDM2 (PDB ID: 4JRG). A π–π stacking interaction with His96 is depicted. Additionally, the pyrrolidine carbonyl group forms a hydrogen bond with the NH group of His96. Several hydrophobic interactions are observed with Leu54, Leu57, Ile61, Met62, Phe86, Phe91, Val93, Lys94, Ile99, and Tyr100.
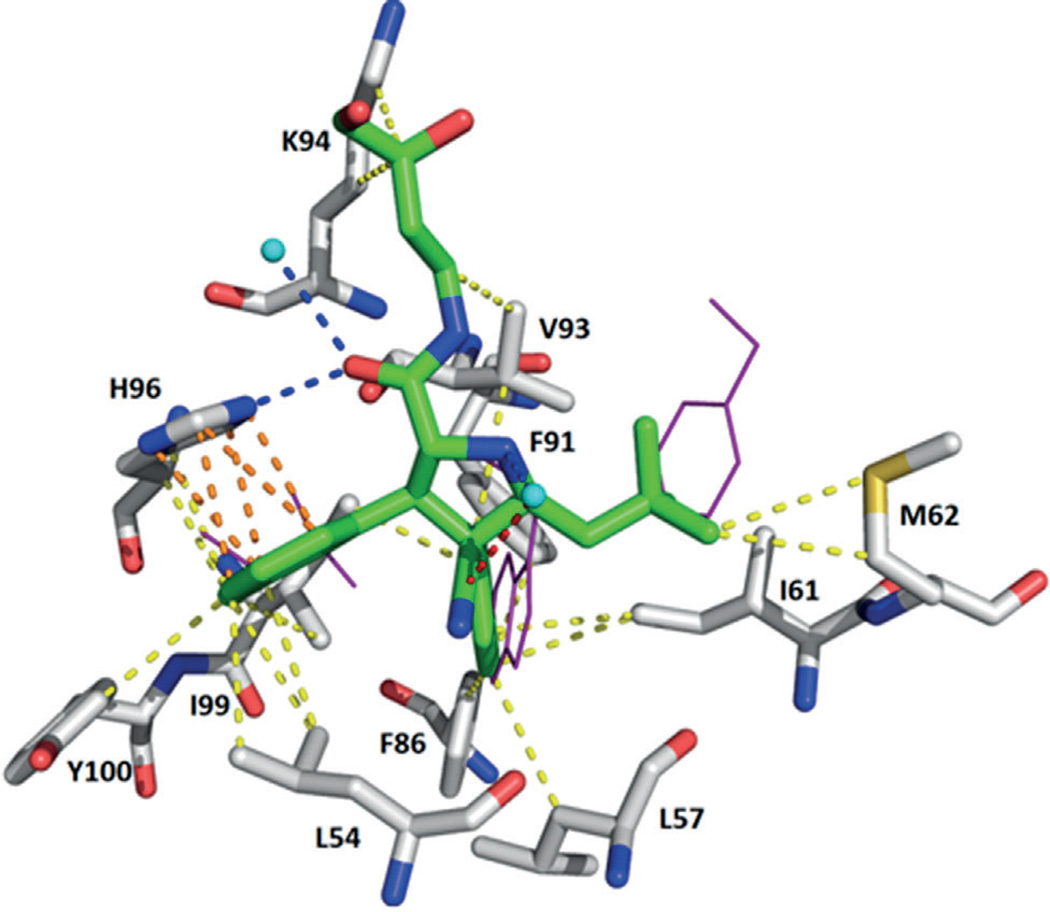
Further optimization led to the development of compound 28, with an IC50 value of 6 nm (RG7388), which effectively activates the p53 pathway, leading to cell-cycle arrest and/or apoptosis in cell lines expressing WT p53 and tumor growth inhibition or regression of osteosarcoma xenografts in nude mice. RG7388 is currently undergoing clinical investigation for the treatment of solid and hematological tumors (Figure 11).[55]
Figure 11.
Pyrrolidine derivatives screened for suppressing the p53–MDM2 interaction.
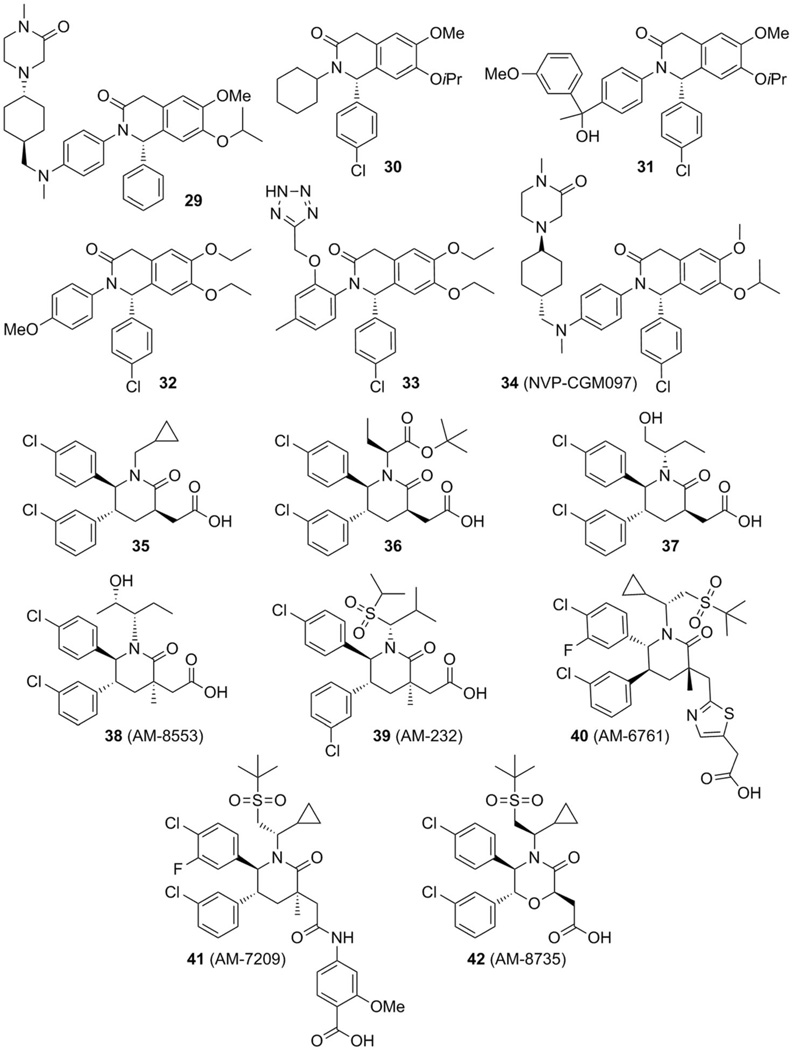
2.5. Isoquinolines, piperidinones, and morpholinones
In 2012 Novartis scientists described substituted isoquinolines and piperidinones as inhibitors of MDM2 and MDMX, with the most potent compound 29 having respective IC50 values of 0.8 and 2.1 µm for both proteins as determined in TR-FRET assays.[56] Two additional patents reported the cyclohexylisoquinoline compound 30 and hydroxyisoquinoline 31, which are also capable of inhibiting the interaction between p53 and MDM2/X with IC50 values of 0.894 µm (MDM2, 30), 44.25 µm (MDMX, 30), and 0.432 µm (MDM2, 31) (Figure 12).[57, 58] Recently, Novartis researchers described a new series of dihydroisoquinolinone derivatives derived from a virtual screening exercise using 2D and 3D presentations of ~50000 compounds from the Novartis compound collection, and compound 32 was identified with an IC50 value of 0.54 µm in a TR-FRET biochemical assay. The binding mode of compound 32 in the MDM2 pocket, obtained by docking, adheres to the central valine concept.[42] Further optimization yielded compound 33 (PDB ID: 4ZYC) with an IC50 value of 0.38 µm in a TR-FRET assay. Remarkably, the bicyclic core makes hydrophobic contacts with residues Ile54, Phe55, and Gly58 of MDM2 instead of the proposed interaction with Val93.[59] The development of this class of compounds led to compound 34 (NVP-CGM097, PDB ID: 4ZYI) with IC50 =1.7 nm against MDM2 and strong anti-proliferative effects in the HCT116 p53WT cell line compared with an isogenic HCT116 knockout for the expression of p53, showing 35-fold selectivity. Additionally, a 58-fold selectivity was observed between the SJSA-1 cell line and the p53-null osteosarcoma SAOS-2 cell line.[60] Presently, NVP-CGM097 is going through phase I clinical trials (Figure 12).[60, 61]
Figure 12.
Various isoquinolines, piperidinones, and morpholinones as potent p53–MDM2/X interaction inhibitors.
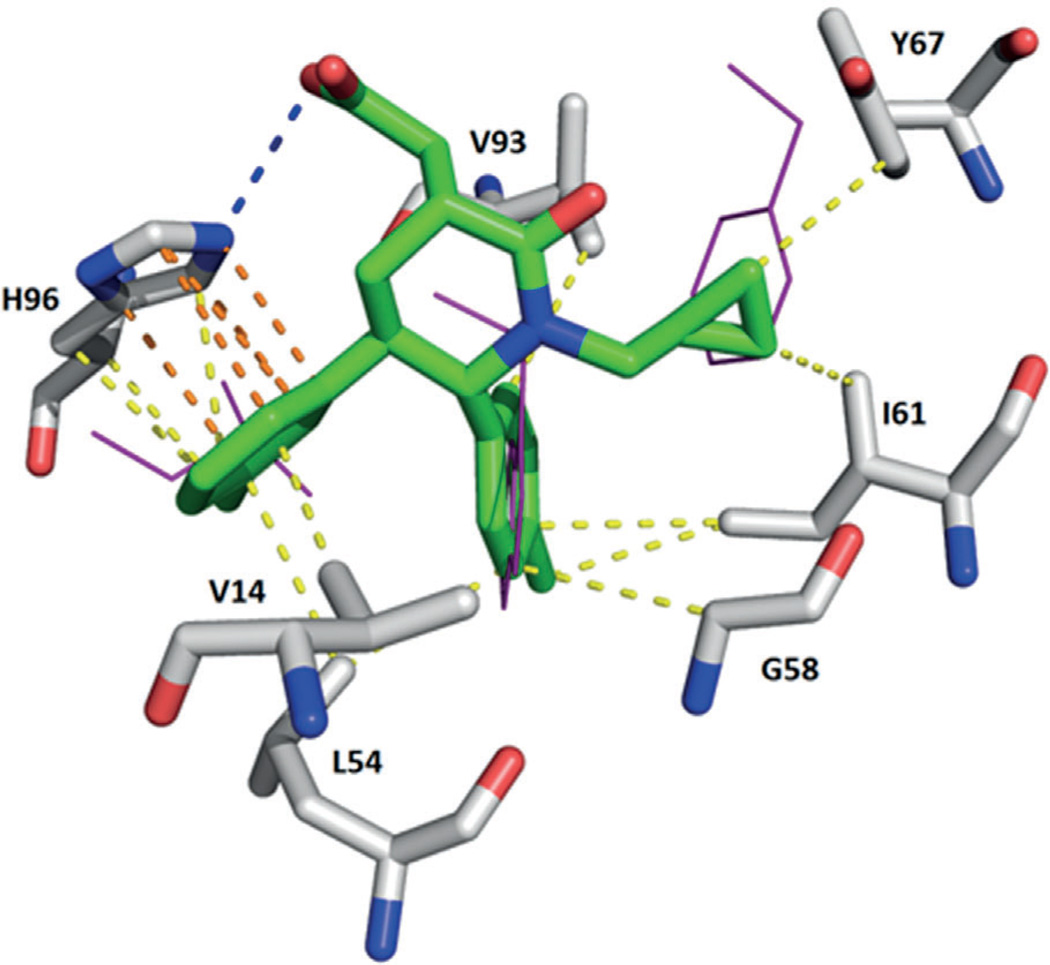
Scientists from Amgen described the structure-based design of new piperidinone compounds, based on yet another scaffold class.[62] In compound 35, with an IC50 value of 34 nm (PDB ID: 2LZG), the p-chloro-substituted phenyl ring occupies the Trp23 pocket, the m-chloro-substituted phenyl ring sits in the Leu26 pocket, and the cyclopropyl group fills the Phe19 pocket (Figure 13). The carboxylic acid moiety forms a hydrogen bond with the NH group of His96 (Figure 13).[62, 63]
Figure 13.
Crystal structure of compound 35 bound to MDM2 (PDB ID: 2LZG). The 3-chlorophenyl ring undergoes a π – π stacking interaction with His96; moreover, the carboxylic acid moiety forms a hydrogen bond with the NH group of His96. Numerous hydrophobic interactions with Val14, Leu54, Gly58, Ile61, Tyr67, and Val93 are shown.
A chiral tert-butyl 2-butanoate replaced the cyclopropyl group in compound 35 and led to piperidinone 36, eightfold more potent than its predecessor toward MDM2 (PDB ID: 4ERE).[62] Further modification of the tert-butyl ester of 36 yielded derivative 37 (PDB ID: 4HBM), which, after optimization, led to compound 38 (AM-8553, PDB ID: 4ERF), with an IC50 value of 1.1 nm toward MDM2 (Figure 12), showing satisfactory tumor regression in a mouse SJSA-1 tumor xenograft model, demonstrating its strong antitumor activity.[64] Supplementary development of AM-8553, with introduction of the sulfonamide moiety in order to search interactions between glycine in a shallow underused cleft of the MDM2 surface, yielded compound 39 (AM-232, PDB ID: 4OAS) with a Ki value of 0.045 nm, and IC50: 9.4, 11.3, and 23.8 nm in SJSA-1, HCT116, and ACHN (renal adenocarcinoma derived) cell lines, respectively.[65,66] Additional evaluations in seven p53-mutant cell lines showed no significant effects on cell growth, and in studies of SJSA-1, HCT116, A375 (malignant melanoma) and NCI-H460 (large-cell lung cancer) xenograft models, significant tumor regression was observed.[65] The evaluation of AM-232 in preclinical trials with rats, monkeys, and dogs showed differences in the pharmacokinetics with a good correlation between in vivo and in vitro studies. This methodology led to a prediction of low human plasma clearance and long half-life in humans for compound 39.[67] This compound is currently undergoing clinical trials (Figure 12).[65–68]
In 2014, additional efforts by Amgen researchers to enhance the biochemical and cellular potency, by using pyridine or thiazole as isosteres of the carboxylic acid moiety of AM-232, led to potent piperidinone inhibitors. Among them was compound 40 (AM-6761, PDB ID: 4ODE), a thiazolyl-containing inhibitor with IC50 values of 0.1 nm (HTRF), and 16 nm in SJSA-1 cells, and antitumor activity in the SJSA-1 osteosarcoma 9xenograft model with an ED50 value of 11 mg kg−1, along with promising pharmacokinetic properties.[69] Furthermore, an additional modification of AMG-232, replacing the carboxylic acid with 4-amidobenzoic acid, led to compound 41 (AM-7209, PDB ID: 4WT2), with a Kd value of 38 pm, IC50=1.6 nm in SJSA-1 cells, in vivo antitumor activity in SJSA-1 and HCT116 xenograft models, and outstanding pharmacokinetic properties.[70] Using the knowledge gained from the piperidinone series, changing to a morpholinone core had a significant impact on potency and metabolic stability. Morpholinone inhibitors are five- to tenfold less potent than their piperidinone counterparts; however, they are more stable in hepatocytes, a feature that can compensate for the decrease in potency. Compound 42 (AM-8735, PDB ID: 4OBA), the most representative compound of this series, showed an IC50 value of 25 nm in SJSA-1 cells, remarkable pharmacokinetic properties, and in vivo efficacy in a SJSA-1 osteosarcoma xenograft model (ED50 = 41 mg kg −1) [69,71]
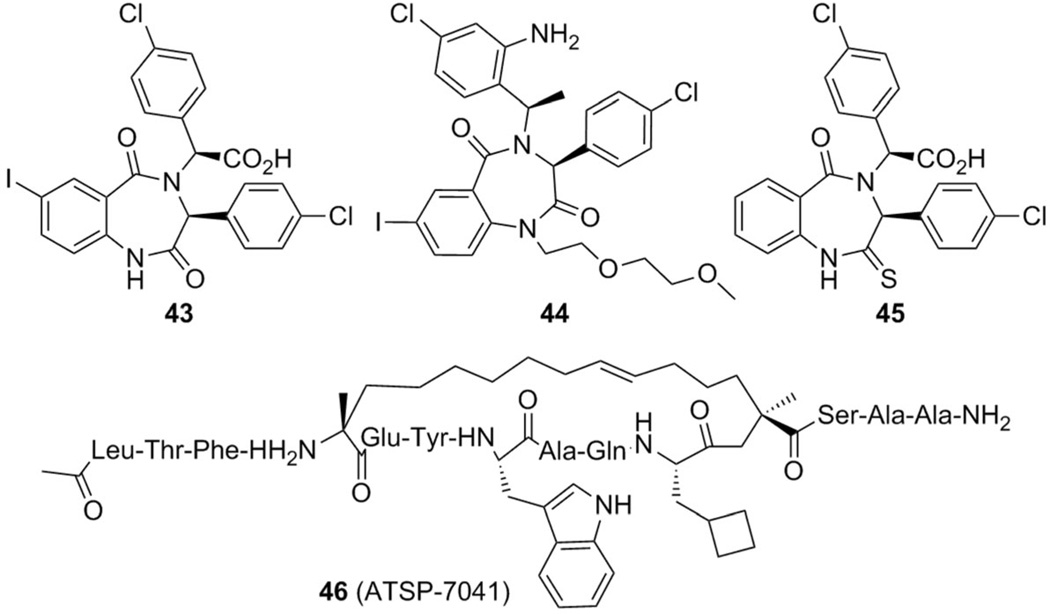
2.6. Benzodiazepines
In 2005 researchers at Johnson & Johnson reported a class of benzodiazepine compounds as MDM2 inhibitors (Figure 14).[72,73] With an initial high-throughput screening, two lead compounds were identified with a binding affinity for MDM2 between 15 and 30 µm. Further optimization of the substituents on the three phenyl rings resulted in compound 43, with IC50 = 0.22 µm and Kd = 80 nm.[74]
Figure 14.
Benzodiazepines and a stapled peptide that show potency as MDM2/X inhibitors.
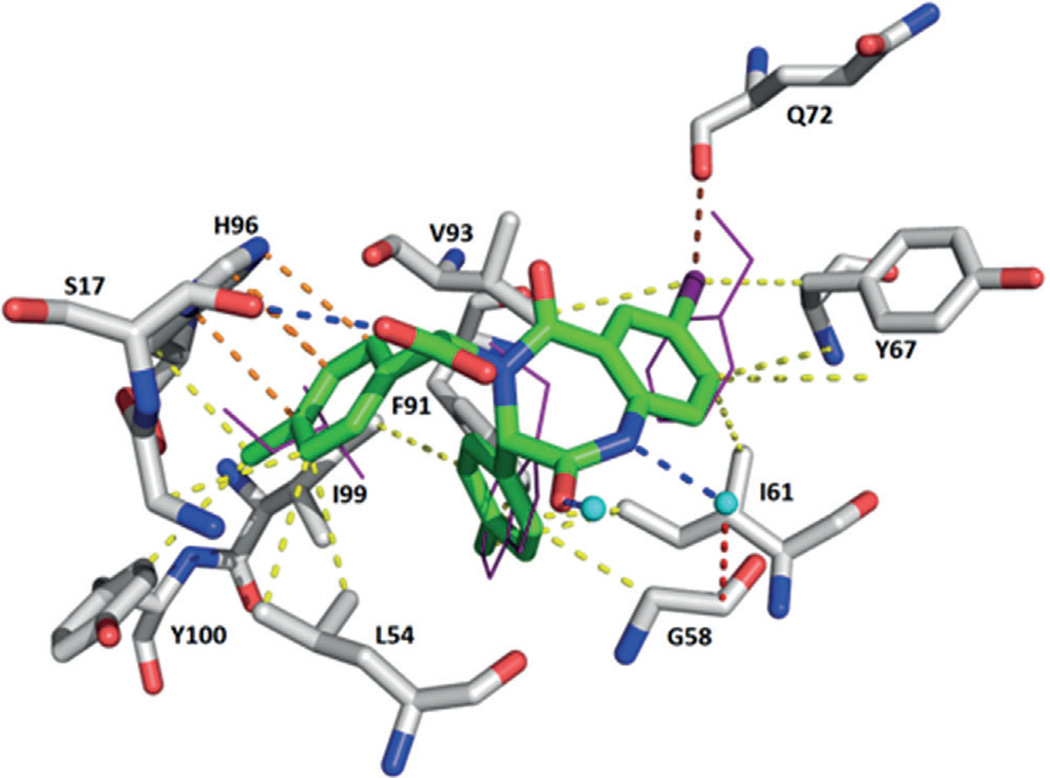
A co-crystal structure of MDM2 and S,S-43 at a resolution of 2.7 Å (PDB ID: 1T4E) showed that that the binding resembles the three key p53 residues for interaction with MDM2.[74] The Phe19 pocket is filled by the iodobenzene ring, the Trp23 pocket is occupied by the p-chlorophenyl group, and the Leu26 pocket is filled with the other p-chlorophenyl at the α-position corresponding to the carboxylic acid group (Figure 15). The iodo substituent of the benzodiazepinedione scaffold forms a strong halogen bond with the backbone carbonyl oxygen atom of Gln72. As expected, a steady decrease in affinity is observed with switches to smaller halogens.[72]
Figure 15.
Crystal structure of compound 43 bound to MDM2 (PDB ID: 1T4E). A halogen bond between the iodine and Gln72 is observed. Hydrogen bond between Ser17 and the carboxylic acid moiety and a π – π stacking interaction with His96 and the p-chlorophenyl group. Furthermore, various hydrophobic interactions with Leu54, Gly58, Ile61, Tyr67, Phe91, Val93, Ile99, and Tyr100 are shown.
Additional optimization resulted in compound 44 by reducing epimerization issues of the stereogenic center at the α-carbonyl position of 43. Compound 44 introduced a hydrogen bond with Val93, giving an IC50 value for binding with MDM2 of 394 nm,[75] compared with 0.49 µm for 43. However, none of this class of MDM2 inhibitors progressed into clinical development due to their unsatisfactory antitumor activity in human cancer cells, low cell permeability, rapid plasma clearance, and low bioavailability shown in vitro and in animal models.[29, 30, 75] In 2012, a 2-thiobenzodiazepine derivative (compound 45) was described in a patent as a MDM2 inhibitor, with an IC50 value of 3.18 µgmL−1 in SAOS-2 p53-null and 6.54 µgmL−1 in U-2-os WTp53 (osteosarcoma) cell lines (Figure 14).[76]
2.7. Peptides
Peptides and peptide derivatives can be designed to become potent p53–MDM2/X interaction inhibitors. In 2007, a 12-residue peptide (pDI) was identified by using phage display that selects for maximal inhibitory activity against MDM2 and MDMX, with respective IC50 values of 0.01 and 0.1 µm for binding as determined by ELISA.[77] The co-crystal structures of pDI with MDMX (PDB ID: 3JZO) and a single-mutant derivative (pDI6W, PDB IDs: 3JZP and 3JZR) bound to human MDMX and MDM2, served as templates to design 11 diverse pDI-derivative peptides that were tested for inhibitory potential. The best derivative (pDIQ, PDB ID: 3JZQ) exhibited a fivefold increase in potency over the parent peptide, with IC50 values for binding with MDM2 and MDMX of 8 and 110 nm, respectively.[78]
While peptidic inhibitors based on the modified p53 sequence offer very high affinity for MDM2, they suffer from low cell permeability and are unstable against proteolysis. A successful attempt to overcome these problems has been made by designing cyclic peptides that are closed by an all-hydrocarbon “staple”. The staple stabilizes the helical structure of the peptide, a feature which likely contributes to the enhanced affinity of the peptide for MDM2 relative to the WT peptide. The peptide SAH-p53–86 (PDB ID: 3V3B), which interestingly also targets MDMX, is the most effective stapled-peptide MDM2 inhibitor, using its Phe19, Trp23, and Leu26 residues to fill the binding site in a manner similar to that of the native p53 peptide. The Trp23 indole ring is bound to Leu54 by a hydrogen bond. The aliphatic staple protects this hydrogen bond and forms an extended hydrophobic network with Leu54, Phe55, Gly58, and Met62 of MDM2 (Figure 16).[79]
Figure 16.
Crystal structure of the stapled peptide SAH-p53-86 bound to MDM2 (PDB ID: 3V3B). The staple is shown in pink. The hydrogen bonds are the same as those present in the p53–MDM2 interaction, the NH group in SAH-p53-86Trp23 and the carbonyl group in MDM2Leu54, and between the NH of SAH-p53-86Phe19 and MDM2Gln72. Additionally, several hydrophobic interactions are depicted.
The PMI peptide (PDB ID: 3EQS bound to MDM2 and PDB ID: 3EQY bound to MDMX), a 12-mer peptide selected from a phage display peptide library, is highly soluble in aqueous media and has higher affinity (than pDI) for MDM2 and MDMX, with Kd values of 3.3 and 8.9 nm, respectively.[80] Aided by mirror-image phage display and native chemical ligation, several proteolysis-resistant 12-mer d-peptide antagonists of MDM2, termed DPMI-α, -β, -γ, and -δ were discovered.[81] The prototypic d-peptide inhibitor DPMI-α binds to MDM2 (PDB ID: 3LNJ) with an affinity of 220 nm, reactivates the p53 pathway in tumor cells in vitro, and inhibits tumor growth in vivo. Furthermore, the design of a superactive d-peptide antagonist of MDM2 was reported, named DPMI-δ (PDB ID: 3TPX), with a binding affinity for MDM2 improved over DPMI-α by three orders of magnitude (Kd =220 pm).[82]
In 2013, the stapled peptide 46 (ATSP-7041, PDB ID: 4N5T, co-crystallized with MDMX) was synthesized which activates the p53 pathway in tumors in vitro and in vivo, with Ki values of 0.9 and 6.8 nm for MDM2 and MDMX, respectively. This stapled peptide has shown improved bioavailability over previously described peptides and induces p53-dependent apoptosis and inhibits cell proliferation in multiple MDM2- and MDMX-overexpressing tumors in cell-based models. Interestingly, beyond the well-known triad of Phe19, Trp2, and Leu26, ATSP-7041 showed additional interactions between Tyr22 and the staple moiety itself with the MDMX protein. Tyr22 interacts with the MDMX binding pocket through van der Waals contact with Gln66, Arg67, Gln68, His69, Val89, and Lys90, as well as through water-mediated hydrogen bonds with the Ne atom of the Lys90 side chain and Nd1 of the His68 side chain. An extensive binding pocket for the staple exists on the MDMX protein, and a number of van der Waals contacts (with Lys47, Met50, His51, Gly54, Gln55, and Met58) were also observed (Figure 14).[83]
The MDM2-binding stapled peptide, M06, showed high affinity for binding with mutant (Nutlin-resistant) MDM2. The M06 stapled peptide forms interactions very similar to those made by the SAH-8 peptide, including reorientation of the Leu26 side chain that appears to be associated with increased helicity (PDB ID: 4UMN). Apart from attenuating the binding of Nutlin, the M62A mutation in MDM2 also causes a significant change in the conformation of the aliphatic staple. In the SAH-8 structure, the hydrocarbon chain packs predominantly against Leu54, Phe55, Gly58, and Met62. The absence of the methionine side chain in MDM2-M62A causes the conformation of the staple to change as it packs more closely against Gly58. The plasticity of the new designed stapled peptide enables it to respond to the M62A mutation by making compensatory contacts.[84]
3. Conclusions and Outlook
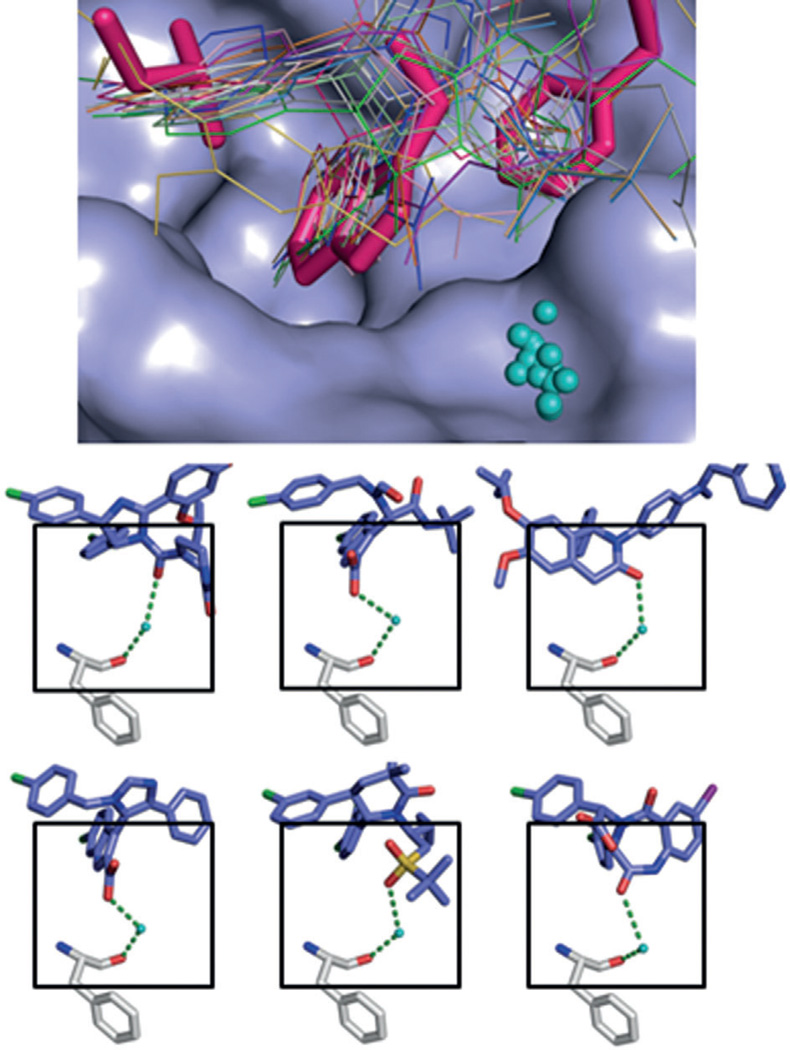
Recently, many novel small molecules with promising activity have been published as MDM2 inhibitors, including a new synthetic approach to obtain stapled peptides,[85] chlorofusin-in-spired class of analogues,[86] fluorescent triazolylpurines,[87] sulfamide and triazole benzodiazepines,[88] oxazoloisoindolinones,[89] dispiroindolinones,[90] a camptothecin analogue (FL118),[91] and an inauhzin analogue,[92] among others. Also, a 2,3’-bis(1H-indole) scaffold was recently published showing additional hydrophobic interactions with the p53Val93 as indicated by 2D NMR and modeling studies.[93] These compounds often possess modest affinities for MDM2 or MDMX in the micromolar range. In contrast, all potent MDM2 antagonistic scaffolds currently undergoing early clinical trials have been structurally characterized, and structure-based drug discovery played an important role in their optimization. Thorough analysis of the available structure and structure–activity relationship studies can provide some guiding principles for the design of novel and potent p53–MDM2 antagonists. The endogenous interaction of p53 with MDM2 and MDMX reveals a clear three-point pharmacophore model formed by the side chains of p53: Phe19, Trp23, Leu26, which competitive inhibitors must mimic. Multiple scaffolds have been designed that are capable of doing so and are discussed in this review. All structurally characterized MDM2 binders have a T-shaped topology in which the ends of the ‘T’ comprise the three moieties that address the pharmacophore points. The central scaffold is usually a heterocyclic ring, annulated rings, or in some cases an acyclic linear moiety. Amongst the three buried hydrophobic side chains, Trp23 contributes most to the interaction energy and thus must be closely mimicked in terms of hydrophobicity and shape. This has been accomplished with phenyl groups, indoles, oxindoles, and 2-oxobenzimidazoles. The choice of the proper anchor residue to mimic Trp23 may be important in terms of compound selectivity. A considerable increase in binding affinity can be reached by the suitable introduction of a hydrophobic halogen atom at the bottom of the Trp23 binding pocket. A per-p-halophenyl-substituted inhibitor scaffold might bind into other similar α-helix-mediated protein–protein interactions, whereas an anchor residue closely mimicking the Trp23–Leu54 hydrogen bond might be more selective and able to differentiate between similar PPI binding interfaces based on hydrogen bonding. Additional interactions with receptor residues observed in several co-crystal structures involve hydrogen bonding and π–π interactions with His96, hydrogen bonding with Ser17, halogen bonding or hydrogen bonding to backbone or side chain Glu72, respectively, and dipolar interactions with His73. In general, the MDM2 crystal structures show a high degree of crystallographic water presence, but a water on top of the indole ring of p53Trp23 seems to be highly conserved amongst many structures. In 17 out of 25 MDM2 small-molecule structures (~70%) a highly conserved water molecule is observed (Figure 17). This water is involved in a tight network of interactions between a polar inhibitor substituent on top of the central scaffold and the main-chain Phe55 carbonyl oxygen atom in the MDM2 receptor. The inclusion of this water network could be an important aspect to consider in the future design of MDM2 inhibitors.
Figure 17.
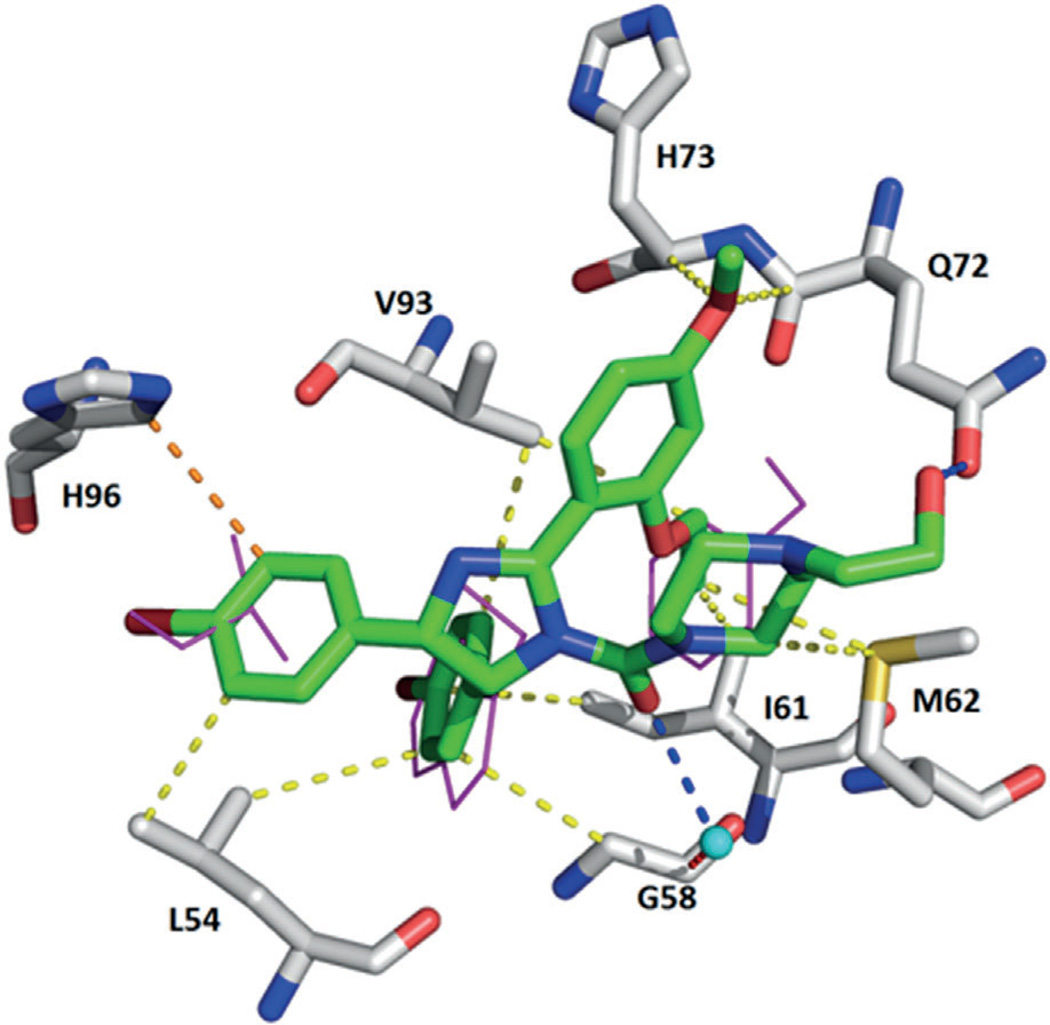
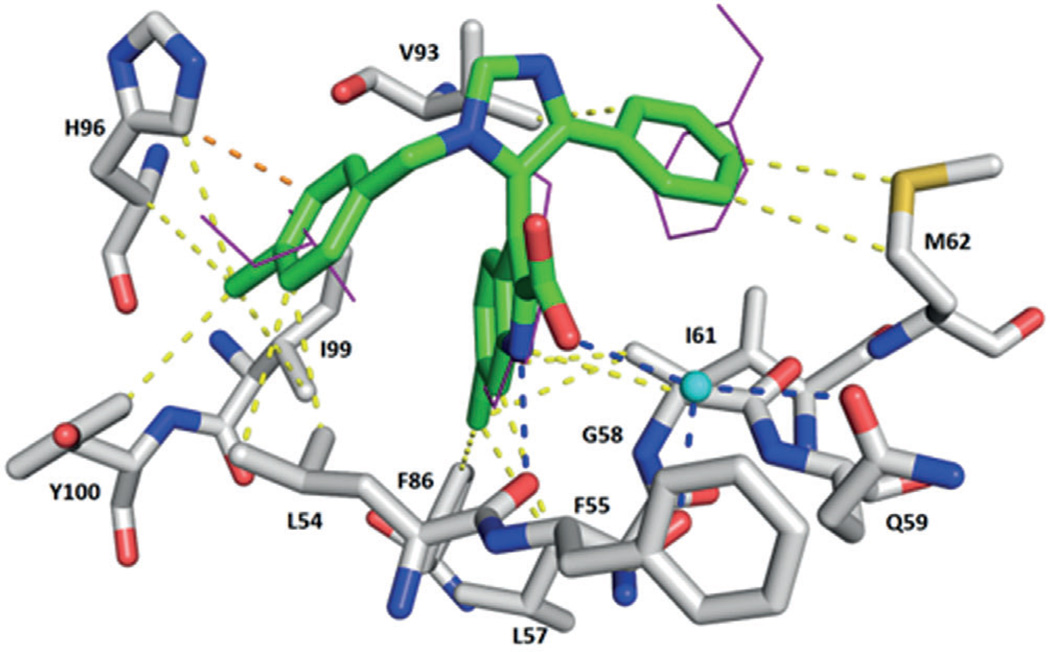
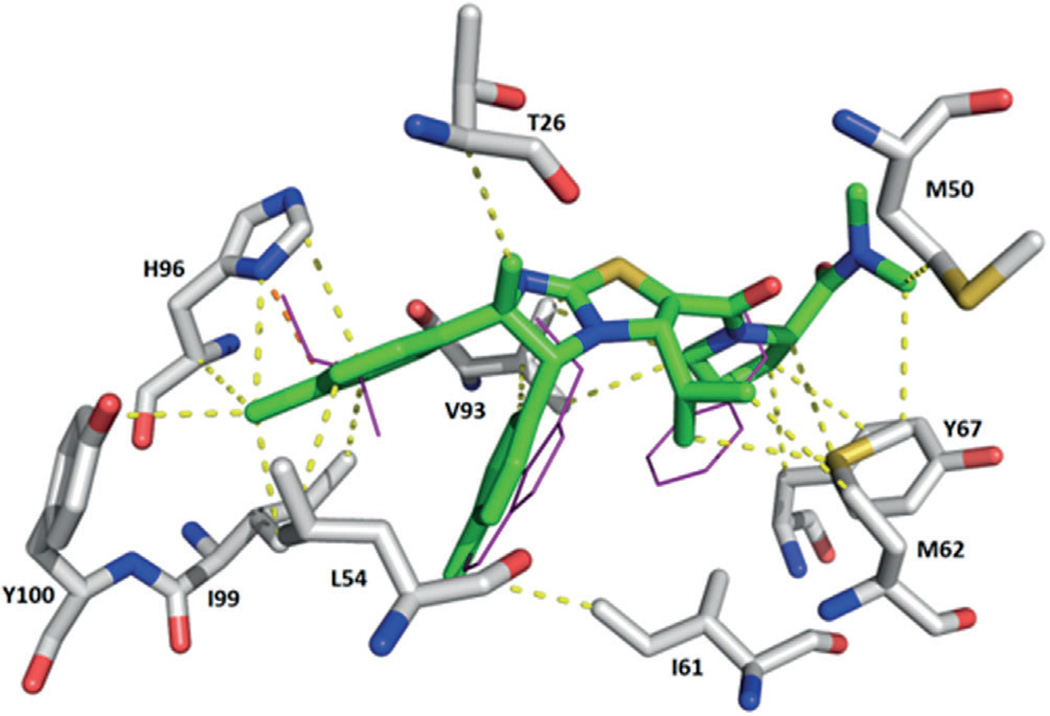
Highly conserved surface water in ligand–MDM2 structures. Above: superimposition of the co-crystal structures including the main cluster of waters (shown in cyan) located in the groove formed by Phe55, Tyr56, Lys57, Gly58, and Gln59. Below: examples of the interactions of the small molecules, water, and Phe55 of MDM2. First row: compounds 1c (Nutlin-3, PDB ID: 4J3E) and 3 (WK-23, PDB ID: 3LBK); second row: compounds 11 (PDB ID: 3TJ2) and 40 (AM-6761, PDB ID: 4ODE); third row: compounds 34 (NVP-CGM097, PDB ID: 4ZYI) and 43 (PDB ID: 1T4E).
Dual activity both in MDM2 and MDMX is essential for further development. However, current small molecules do not address the duality, nor are potent MDMX inhibitors known. In contrast, peptidic inhibitors can be designed with dual potency for MDM2 and MDMX. In addition, the intrinsically disordered protein states of MDM2 should be taken into account and the resulting fourth pharmacophore point, for the discovery of novel scaffolds as shown in previous studies.[27, 28]
Drug combinations and cyclotherapy had been studied as supplementary opportunities to treat cancer, exerting synergistic effects or to selectively protect normal tissues.[30] Representatives are the evaluation of Nutlin with actinomycin D, gemcitabine, vincristine, roscovitine, and doxorubicin in cell-based models.[94–99] Another highly interesting and complementary p53 reactivation approach not discussed herein is based on small-molecule restoration of certain p53 mutants.[100, 101]
Research efforts over the past decade have led to several compounds that are currently in clinical trials for the treatment of various types of cancer and also in combination with other classes of chemotherapeutics (Table 1). The available structural information and knowledge gathered in recent years have helped in the design of better compounds with greater affinity for MDM2, but not in the same degree for MDMX, improved pharmacokinetic profiles, oral bioavailability, and selectivity. During studies with animals as well as in clinical trials, it is imperative to determine proper doses, schedules, and possible combination therapies to circumvent the potential for acquired resistance to MDM2 inhibitors as well as unwanted side effects.
Table 1.
MDM2/X inhibitors in clinical trials.
| Drug | Condition | Phase | Sponsor | Reference |
|---|---|---|---|---|
| RG7112 (RO5045337) | Solid Tumors, advanced solid tumors, leukemia, hematological neoplasms, liposarcomas |
I (completed) | Hoffmann-La Roche | [36– [38,102] |
| RG7112 (RO5045337) with doxorubicin | Soft tissue sarcoma | I (completed) | Hoffmann-La Roche | [103] |
| RG7112 (RO5045337) with cytarabine | Acute myelogenous leukemia | I (completed) | Hoffmann-La Roche | [104] |
| RG7775 (RO6839921) | Advanced cancers, including acute myeloid leu- kemia |
I (recruiting) | Hoffmann-La Roche | [105] |
| RO5503781 | Advanced malignancies, except leukemia | I (completed) | Hoffmann-La Roche | [106] |
| RO5503781 with posaconazole | Solid tumors | I (completed) | Hoffmann-La Roche | [107] |
| RO5503781 with cytarabine | Acute myelogenous leukemia | I (recruiting) | Hoffmann-La Roche | [108] |
| RG7388 (alone and with Pegasys) | Polycythemia vera and thrombocythemia | I (recruiting) | Hoffmann-La Roche | [109] |
| MK-8242 | Advanced solid tumors | I (completed) | Merck Sharp & Dohme Corp. |
[110] |
| MK-8242 -cytarabine | Acute myelogenous leukemia | I (completed) | Merck Sharp & Dohme Corp. |
[111] |
| DS-3032b | Hematological malignancies, advanced tumors or lymphomas |
I (recruiting) | Daiichi Sankyo | [112] |
| HDM201 | Advanced tumors WT TP53 | I (recruiting) | Novartis | [113] |
| HDM201 with LEE011 | Liposarcoma | I (recruiting) | Novartis | [114] |
| CGM097 | Solid tumor with WT p53 status | I (recruiting) | Novartis | [61], [115] |
| SAR405838 (MI-888 analogue) | Malignant neoplasm | I (active) | Sanofi | [116] |
| SAR405838 (MI-888 analogue) with pimasertib |
Malignant neoplasm | I (recruiting) | Sanofi | [117] |
| AMG232 | Advanced solid tumors or multiple myeloma | I (recruiting) | Amgen | [68] |
| AMG232 | Metastatic melanoma | Ib/IIa (recruit- ing) |
Amgen | [118] |
| AMG232 with trametinib | Acute myeloid leukemia | Ib (recruiting) | Amgen | [119] |
Acknowledgments
Research in the area of p53–MDM2/X interactions in the Dçmling research group was supported by the US National Institutes of Health (NIH) (1R01M097082). N.E.O. was supported by the Department of Sciences, Technology, and Innovation—COLCIENCIAS (Colombia).
Biographies

Natalia Estrada-Ortiz obtained her MSc in Biotechnology at the National University of Colombia in 2012, where her project involved the study of natural products from marine sponges and plants with promising anticancer activity. In September 2013 she started her PhD work at the University of Gronin-gen in the groups of Drug Design and Pharmacokinetics, Toxicology, and Targeting under the supervision of Profs. Alexander Dömling, Geny Grootehuis, and Angela Casini. Her research concerns the development and biological evaluation of compounds as possible anticancer agents.

Constantinos G. Neochoritis was born in Thessaloniki, Macedonia, Greece in 1982. He received his PhD (2011) in organic chemistry under the guidance of Professors J. Stephanidou Stephanatou and C. Tsoleridis in the Department of Chemistry at Aristotle University of Thessaloniki. In June 2012 he joined the research group of Prof. Alexander Dömling in the Drug Design Group at the University of Groningen. His research interests include bioactive heterocycles, drug design, multicomponent reactions, and high-throughput synthesis. He has published more than 25 peer-reviewed papers.

Alexander Dömling has held the chair for Drug Design at the University of Groningen since 2011. He studied chemistry and biology at the Technische Universität Munchen (Germany) and obtained his PhD under the guidance of Prof. Ivar Ugi. After postdoctoral research under a Humboldt fellowship in the group of Nobel laureate Barry Sharpless, he founded the biotech companies Morphochem and later Carmolex Inc. After his habilitation he worked as full professor at the University of Pittsburgh in the School of Pharmacy. His interests are centered around multi-component reaction (MCR) chemistry and its application to problems in drug design. His special focus is on MCR-centered pharmacophore methods, structure-based drug design, and MCR-centered fragment-based drug design. He is the author of more than 150 scientific articles, reviews, and book contributions. He has applied for more than 30 patents. His long-term vision is to bring a novel drug to patients in an indicated area of unmet medical need.
References
- 1.Ottmann C. In: Protein-Protein Interactions in Drug Discovery: Methods and Principles in Medicinal Chemistry. Dömling A, editor. Vol. 56. Weinheim: Wiley-VCH; 2013. pp. 1–19. [Google Scholar]
- 2.Lane DP. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 3.Levine AJ. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 4.Klein C, Vassilev LT. Br. J. Cancer. 2004;91:1415–1419. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hainaut P, Hollstein M. In: Advances in Cancer Research. Van-de Woude GF, Klein G, editors. San Diego: Elsevier; 1999. pp. 81–137. [Google Scholar]
- 6.Shangary S, Wang S. Annu. Rev. Pharmacol. Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherr CJ, Weber JD. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 8.Momand J, Zambetti GP. J. Cell. Biochem. 1997;64:343–352. [PubMed] [Google Scholar]
- 9.Momand J, Wu HH, Dasgupta G. Gene. 2000;242:15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 10.Haupt Y, Maya R, Kazaz A, Oren M. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 11.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. Proc. Natl. Acad. Sci. USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang L, Yan Z, Liao X, Li Y, Yang J, Wang Z-G, Zuo Y, Kawai H, Shadfan M, Ganapathy S, Yuan Z-M. Proc. Natl. Acad. Sci. USA. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 14.Sharp DA, Kratowicz SA, Sank MJ, L George D. J. Biol. Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 15.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 16.Khoury K, Popowicz GM, Holak TA, Dömling A. MedChemComm. 2011;2:246–260. doi: 10.1039/C0MD00248H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, Goydos J, Haigh JJ, Haupt Y, Larue L, Jochemsen A, Shi H, Moriceau G, Lo RS, Ghanem G, Shackleton M, Bernal F, Marine J-C. Nat. Med. 2012;18:1239–1247. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toledo F, Wahl GM. Int. J. Biochem. Cell Biol. 2007;39:1476–1482. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dömling A, Holak TA. Angew. Chem. Int. Ed. 2014;53:2286–2288. doi: 10.1002/anie.201307906. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2014;126:2318–2320. [Google Scholar]
- 20.Zak KM, Kitel R, Przetocka S, Golik P, Guzik K, Musielak B, Dömling A, Dubin G, Holak TA. Structure. 2015;23:2341–2348. doi: 10.1016/j.str.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhrinova S, Uhrin D, Powers H, Watt K, Zheleva D, Fischer P, McInnes C, Barlow PN. J. Mol. Biol. 2005;350:587–598. doi: 10.1016/j.jmb.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Popowicz GM, Czarna A, Holak TA. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 23.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y, Parrish DA, Deschamps JR, Wang S. J. Am. Chem. Soc. 2005;127:10130–10131. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 24.Moll UM, Petrenko O. Mol. Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
- 25.Chen J, Marechal V, Levine AJ. Mol. Cell. Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picksley SM, Vojtesek B, Sparks A, Lane DP. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- 27.Bista M, Wolf S, Khoury K, Kowalska K, Huang Y, Wrona E, Arciniega M, Popowicz GM, Holak TA, Dömling A. Structure. 2013;21:2143–2151. doi: 10.1016/j.str.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauer MR, Boeckler FM. Structure. 2013;21:2095–2097. doi: 10.1016/j.str.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Bernard D, Wang S. BioDiscovery. 2013;4 [Google Scholar]
- 30.Khoo KH, Verma CS, Lane DP. Nat. Rev. Drug Discovery. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 31.Neochoritis C, Estrada-Ortiz N, Khoury K. In: A. Dömling in Annual Reports in Medicinal Chemistry. Desai MC, editor. London: Elsevier; 2014. pp. 167–187. [Google Scholar]
- 32.Zhao Y, Aguilar A, Bernard D, Wang S. J. Med. Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 34.Tovar C, Graves B, Packman K, Filipovic Z, Higgins B, Xia M, Tar-dell C, Garrido R, Lee E, Kolinsky K, To K-H, Linn M, Podlaski F, Wovkulich P, Vu B, Vassilev LT. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 35.Vu B, Wovkulich P, Pizzolato G, Lovey A, Ding Q, Jiang N, Liu J-J, Zhao C, Glenn K, Wen Y, Tovar C, Packman K, Vassilev L, Graves B. ACS Med. Chem. Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray-Coquard I, Blay J-Y, Italiano A, Le Cesne A, Penel N, Zhi J, Heil F, Rueger R, Graves B, Ding M, Geho D, Middleton SA, Vassilev LT, Nichols GL, Bui BN. Lancet Oncol. 2012;13:1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 37.Andreeff M, Kelly KR, Yee KW, Assouline SE, Strair R, Popplewell L, Bowen D, Martinelli G, Drummond MW, Vyas P, Kirschbaum M, Iyer SP, Ruvolo V, Nogueras González GM, Huang X, Chen G, Graves B, Blotner S, Bridge P, Jukofsky L, Middleton S, Reckner M, Rueger R, Zhi J, Nichols G, Kojima K. Clin. Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iancu-Rubin C, Mosoyan G, Glenn K, Gordon RE, Nichols GL, Hoffman R. Exp. Hematol. 2014;42 doi: 10.1016/j.exphem.2013.11.012. 137 – 145.e5. [DOI] [PubMed] [Google Scholar]
- 39.Czarna A, Beck B, Srivastava S, Popowicz GM, Wolf S, Huang Y, Bista M, Holak TA, Dömling A. Angew. Chem. Int. Ed. 2010;49:5352–5356. doi: 10.1002/anie.201001343. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2010;122:5480–5484. [Google Scholar]
- 40.García-Echeverría C, Chène P, Blommers MJ, Furet P. J. Med. Chem. 2000;43:3205–3208. doi: 10.1021/jm990966p. [DOI] [PubMed] [Google Scholar]
- 41.Popowicz GM, Czarna A, Wolf S, Wang K, Wang W, Dömling A, Holak TA. Cell Cycle. 2010;9:1104–1111. doi: 10.4161/cc.9.6.10956. [DOI] [PubMed] [Google Scholar]
- 42.Furet P, Chène P, De Pover A, Valat TS, Lisztwan JH, Kallen J, Masuya K. Bioorg. Med. Chem. Lett. 2012;22:3498–3502. doi: 10.1016/j.bmcl.2012.03.083. [DOI] [PubMed] [Google Scholar]
- 43.Vaupel A, Bold G, De Pover A, Stachyra-Valat T, Hergovich Lisztwan J, Kallen J, Masuya K, Furet P. Bioorg. Med. Chem. Lett. 2014;24:2110–2114. doi: 10.1016/j.bmcl.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki M, Naito H, Sugimoto Y, Kawato H, Okayama T, Shimizu H, Miyazaki M, Kitagawa M, Seki T, Fukutake S, Aonuma M, Soga T. Bioorg. Med. Chem. Lett. 2013;23:728–732. doi: 10.1016/j.bmcl.2012.11.091. [DOI] [PubMed] [Google Scholar]
- 45.Miyazaki M, Naito H, Sugimoto Y, Yoshida K, Kawato H, Okayama T, Shimizu H, Miyazaki M, Kitagawa M, Seki T, Fukutake S, Shiose Y, Aonuma M, Soga T. Bioorg. Med. Chem. 2013;21:4319–4331. doi: 10.1016/j.bmc.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 46.Koes D, Khoury K, Huang Y, Wang W, Bista M, Popowicz GM, Wolf S, Holak TA, Dömling A, Camacho CJ. PLoS ONE. 2012;7:e32839. doi: 10.1371/journal.pone.0032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Wolf S, Koes D, Popowicz GM, Camacho CJ, Holak TA, Dömling A. ChemMedChem. 2012;7:49–52. doi: 10.1002/cmdc.201100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dömling A. Int. PCT Pub. No. WO 2012033525 A3. University of Pittsburgh; 2012. [Google Scholar]
- 49.Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D, Lello PD, Fry D, Garvie C, Huang K-S, Lin Gao, Tovar C, Lovey A, Wanner J, Vassilev LT. Proc. Natl. Acad. Sci. USA. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang W, Qin J-J, Voruganti S, Srivenugopal KS, Nag S, Patil S, Sharma H, Wang M-H, Wang H, Buolamwini JK, Zhang R. Nat. Commun. 2014;5:5086. doi: 10.1038/ncomms6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez-Lopez de Turiso F, Sun D, Rew Y, Bartberger MD, Beck HP, Canon J, Chen A, Chow D, Correll TL, Huang X, Julian LD, Kayser F, Lo M-C, Long AM, McMinn D, Oliner JD, Osgood T, Powers JP, Y Saiki A, Schneider S, Shaffer P, Xiao S-H, Yakowec P, Yan X, Ye Q, Yu D, Zhao X, Zhou J, Medina JC, Olson SH. J. Med. Chem. 2013;56:4053–4070. doi: 10.1021/jm400293z. [DOI] [PubMed] [Google Scholar]
- 52.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, Roller PP, Wang S. J. Med. Chem. 2006;49:3432–3435. doi: 10.1021/jm051122a. [DOI] [PubMed] [Google Scholar]
- 53.Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barrière C, Stuckey JA, Meagher JL, Bai L, Liu L, Hoffman-Luca CG, Lu J, Shangary S, Yu S, Bernard D, Aguilar A, Dos-Santos O, Besret L, Guerif St, Pannier P, Gorge-Bernat D, Debussche L. Cancer Res. 2014;74:5855–5865. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Chu X-J, Liu J-J, Ding Q, Zhang J, Bartkovitz D, Jiang N, Karnachi P, So S-S, Tovar C, Filipovic ZM, Higgins B, Glenn K, Packman K, Vassilev L, Graves B. ACS Med. Chem. Lett. 2014;5:124–127. doi: 10.1021/ml400359z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Q, Zhang Z, Liu J-J, Jiang N, Zhang J, Ross TM, Chu X-J, Bartkovitz D, Podlaski F, Janson C, Tovar C, Filipovic ZM, Higgins B, Glenn K, Packman K, T Vassilev L, Graves B. J. Med. Chem. 2013;56:5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 56.Berghausen J, Ren H, Novartis AG. Int. PCT Pub. No. WO 2012066095 A1. 2012 [Google Scholar]
- 57.Holzer P, Masuya K, Furet P, Kallen J, Stutz S, Buschmann N, Novartis AG. Int. PCT Pub. No WO 2012175520 A1. 2012 [Google Scholar]
- 58.Holzer P, Masuya K, Guagnano V, Furet P, Kallen J, Stutz S, Novartis AG. Int. PCT Pub. No. WO 2012175487 A1. 2012 [Google Scholar]
- 59.Gessier F, Kallen J, Jacoby E, Chène P, Stachyra-Valat T, Ruetz S, Jeay S, Holzer P, Masuya K, Furet P. Bioorg. Med. Chem. Lett. 2015;25:3621–3625. doi: 10.1016/j.bmcl.2015.06.058. [DOI] [PubMed] [Google Scholar]
- 60.Jeay S, Gaulis S, Ferretti S, Bitter H, Ito M, Valat T, Murakami M, Ruetz S, Guthy DA, Rynn C, Jensen MR, Wiesmann M, Kallen J, Furet P, Gessier F, Holzer P, Masuya K, Würthner J, Halilovic E, Hofmann F, R Sellers W, Graus Porta D. eLife. 2015:6498. doi: 10.7554/eLife.06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holzer P, Masuya K, Furet P, Kallen J, Valat-Stachyra T, Ferretti S, Berghausen J, Bouisset-Leonard M, Buschmann N, Pissot-Soldermann C, Rynn C, Ruetz S, Stutz S, Chène P, Jeay S, Gessier F. J. Med. Chem. 2015;58:6348–6358. doi: 10.1021/acs.jmedchem.5b00810. [DOI] [PubMed] [Google Scholar]
- 62.Rew Y, Sun D, Gonzalez-Lopez De Turiso F, Bartberger MD, Beck HP, Canon J, Chen A, Chow D, Deignan J, Fox BM, Gustin D, Huang X, Jiang M, Jiao X, Jin L, Kayser F, Kopecky DJ, Li Y, Lo M-C, Long AM, Michelsen K, Oliner JD, Osgood T, Ragains M, Saiki AY, Schneider S, Toteva M, Yakowec P, Yan X, Ye Q, Yu D, Zhao X, Zhou J, Medina JC, Olson SH. J. Med. Chem. 2012;55:4936–4954. doi: 10.1021/jm300354j. [DOI] [PubMed] [Google Scholar]
- 63.Michelsen K, Jordan JB, Lewis J, Long AM, Yang E, Rew Y, Zhou J, Yakowec P, Schnier PD, Huang X, Poppe L. J. Am. Chem. Soc. 2012;134:17059–17067. doi: 10.1021/ja305839b. [DOI] [PubMed] [Google Scholar]
- 64.Bernard D, Zhao Y, Wang S. J. Med. Chem. 2012;55:4934–4935. doi: 10.1021/jm3007068. [DOI] [PubMed] [Google Scholar]
- 65.Canon J, Osgood T, Olson SH, Saiki AY, Robertson R, Yu D, Eksterowicz J, Ye Q, Jin L, Chen A, Zhou J, Cordover D, Kaufman S, Kendall R, Oliner JD, Coxon A, Radinsky R. Mol. Cancer Ther. 2015;14:649–658. doi: 10.1158/1535-7163.MCT-14-0710. [DOI] [PubMed] [Google Scholar]
- 66.Sun D, Li Z, Rew Y, Gribble M, Bartberger MD, Beck HP, Canon J, Chen A, Chen X, Chow D, Deignan J, Duquette J, Eksterowicz J, Fisher B, Fox BM, Fu J, Gonzalez AZ, Gonzalez-Lopez De Turiso F, Houze JB, Huang X, Jiang M, Jin L, Kayser F, Liu J, Lo M-C, Long AM, Lucas B, McGee LR, McIntosh J, Mihalic J, Oliner JD, Osgood T, Peterson ML, Roveto P, Saiki AY, Shaffer P, Toteva M, Wang Y, Wang YC, Wortman S, Yakowec P, Yan X, Ye Q, Yu ┴ D, Yu M, Zhao X, Zhou J, Zhu J, Olson SH, Medina JC. J. Med. Chem. 2014;57:1454–1472. doi: 10.1021/jm401753e. [DOI] [PubMed] [Google Scholar]
- 67.Ye Q, Jiang M, Huang WT, Ling Y, Olson SH, Sun D, Xu G, Yan X, Wong BK, Jin L. Xenobiotica. 2015;45:681–692. doi: 10.3109/00498254.2015.1010632. [DOI] [PubMed] [Google Scholar]
- 68. ClinicalTrials.gov identifier for AMG232: NCT01723020 [Google Scholar]
- 69.Gonzalez AZ, Li Z, Beck HP, Canon J, Chen A, Chow D, Duquette J, Eksterowicz J, Fox BM, Fu J, Huang X, Houze J, Jin L, Li Y, Ling Y, Lo M-C, Long AM, McGee LR, McIntosh J, Oliner JD, Osgood T, Rew Y, Saiki AY, Shaffer P, Wortman S, Yakowec P, Yan X, Ye Q, Yu D, Zhao X, Zhou J, Olson SH, Sun D, Medina JC. J. Med. Chem. 2014;57:2963–2988. doi: 10.1021/jm401911v. [DOI] [PubMed] [Google Scholar]
- 70.Rew Y, Sun D, Yan X, Beck HP, Canon J, Chen A, Duquette J, Eksterowicz J, Fox BM, Fu J, Gonzalez AZ, Houze J, Huang X, Jiang M, Jin L, Li Y, Li Z, Ling Y, Lo M-C, Long AM, McGee LR, McIntosh J, Oliner JD, Osgood T, Saiki AY, Shaffer P, Wang YC, Wortman S, Yakowec P, Ye Q, Yu D, Zhao X, Zhou J, Medina JC, Olson SH. J. Med. Chem. 2014;57:10499–10511. doi: 10.1021/jm501550p. [DOI] [PubMed] [Google Scholar]
- 71.Gonzalez AZ, Eksterowicz J, Bartberger MD, Beck HP, Canon J, Chen A, Chow D, Duquette J, Fox BM, Fu J, Huang X, Houze JB, Jin L, Li Y, Li Z, Ling Y, Lo M-C, Long AM, McGee LR, McIntosh J, L McMinn D, Oliner JD, Osgood T, Rew Y, Saiki AY, Shaffer P, Wortman S, Yakowec P, Yan X, Ye Q, Yu D, Zhao X, Zhou J, Olson SH, Medina JC, Sun D. J. Med. Chem. 2014;57:2472–2488. doi: 10.1021/jm401767k. [DOI] [PubMed] [Google Scholar]
- 72.Parks DJ, Lafrance LV, Calvo RR, Milkiewicz KL, Gupta V, Lattanze J, Ramachandren K, Carver TE, Petrella EC, Cummings MD, Maguire D, Grasberger BL, Lu T. Bioorg. Med. Chem. Lett. 2005;15:765–770. doi: 10.1016/j.bmcl.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 73.Raboisson P, Marugán JJ, Schubert C, Koblish HK, Lu T, Zhao S, Player MR, Maroney AC, Reed RL, Huebert ND, Lattanze J, Parks DJ, Cummings MD. Bioorg. Med. Chem. Lett. 2005;15:1857–1861. doi: 10.1016/j.bmcl.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 74.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, V LaFrance L, Milkiewicz KL, Calvo RR, Maguire D, Lattanze J, Franks CF, Zhao S, Ramachandren K, Bylebyl GR, Zhang M, Manthey CL, Petrella EC, Pantoliano MW, Deckman IC, Spurlino JC, Maroney AC, Tomczuk BE, Molloy CJ, Bone RF. J. Med. Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 75.Parks DJ, V LaFrance L, Calvo RR, Milkiewicz KL, Marugán JJ, Raboisson P, Schubert C, Koblish HK, Zhao S, Franks CF, Lattanze J, Carver TE, Cummings MD, Maguire D, Grasberger BL, Maroney AC, Lu Ti. Bioorg. Med. Chem. Lett. 2006;16:3310–3314. doi: 10.1016/j.bmcl.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 76.Zhang W, Miao Z, Zhuang C, Sheng C, Zhu L, Zhang Y, Yao J, Guo Z. Pat. Appl. CN 102321034. 2012 [Google Scholar]
- 77.Hu B, Gilkes DM, Chen J. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 78.Phan J, Li Z, Kasprzak A, Li B, Sebti S, Guida W, Schçnbrunn E, Chen J. J. Biol. Chem. 2010;285:2174–2183. doi: 10.1074/jbc.M109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Baek S, Kutchukian PS, Verdine GL, Huber R, Holak TA, Lee KW, Popowicz GM. J. Am. Chem. Soc. 2012;134:103–106. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 80.Pazgier M, Liu M, Zou G, Yuan W, Li C, Li C, Li J, Monbo J, Zella D, Tarasov SG, Lu W. Proc. Natl. Acad. Sci. USA. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhan C, Zhao L, Wei X, Wu X, Chen X, Yuan W, Lu WY, Pazgier M, Lu W. J. Med. Chem. 2012;55:6237–6241. doi: 10.1021/jm3005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu M, Li C, Pazgier M, Li C, Mao Y, Lv Y, Gu B, Wei G, Yuan W, Zhan C, Lu W-Y, Lu W. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang YS, Graves B, Guerlavais V, Tovar C, Packman K, To K-H, Olson KA, Kesavan K, Gangurde P, Mukherjee A, Baker T, Darlak K, Elkin C, Filipovic Z, Qureshi FZ, Cai H, Berry P, Feyfant E, Shi XE, Horstick J, Annis DA, Manning AM, Fotouhi N, Nash H, Vassilev LT, Tomi Sawyer K. Proc. Natl. Acad. Sci. USA. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chee SMQ, Wongsantichon J, Tng QS, Robinson R, Joseph TL, Verma C, Lane DP, Brown CJ, Ghadessy FJ. PLoS ONE. 2014;9:e104914. doi: 10.1371/journal.pone.0104914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Chou DH-C. Angew. Chem. Int. Ed. 2015;54:10931–10934. doi: 10.1002/anie.201503975. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015;127:11081–11084. [Google Scholar]
- 86.Cominetti MMD, Goffin SA, Raffel E, Turner KD, Ramoutar JC, O’Connell MA, Howell LA, Searcey M. Bioorg. Med. Chem. Lett. 2015;25:4878–4880. doi: 10.1016/j.bmcl.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 87.Pettersson M, Bliman D, Jacobsson J, Nilsson JR, Min J, Iconaru L, Guy RK, Kriwacki RW, Andréasson J, Grøtli M. PLoS ONE. 2015;10:e0124423. doi: 10.1371/journal.pone.0124423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu Z, Zhuang C, Wu Y, Guo Z, Li J, Dong G, Yao J, Sheng C, Miao Z, Zhang W. Int. J. Mol. Sci. 2014;15:15741–15753. doi: 10.3390/ijms150915741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soares J, Pereira NAL, Monteiro Â, Leäo M, Bessa C, dos Santos DJVA, Raimundo L, Queiroz G, Bisio Â, Inga A, Pereira C, Santos MMM, Saraiva L. Eur. J. Pharm. Sci. 2015;66:138–147. doi: 10.1016/j.ejps.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 90.Ivanenkov YA, Vasilevski SV, Beloglazkina EK, Kukushkin ME, Machulkin AE, Veselov MS, Chufarova NV, Chernyaginab ES, Vanzcool AS, Zyk NV, Skvortsov DA, Khutornenko AA, Rusanov AL, Tonevitsky AG, Dontsova OA, Majouga AG. Bioorg. Med. Chem. Lett. 2015;25:404–409. doi: 10.1016/j.bmcl.2014.09.070. [DOI] [PubMed] [Google Scholar]
- 91.Ling X, Xu C, Fan C, Zhong K, Li F, Wang X. Cancer Res. 2014;74:7487–7497. doi: 10.1158/0008-5472.CAN-14-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu H, Zeng S, Zhang Q, Ye Q, Ding D. Int. PCT Pub. No. WO 2014018953 A1. Indiana University Research & Technology Corp.; 2014. [Google Scholar]
- 93.Neochoritis CG, Wang K, Estrada-Ortiz N, Herdtweck E, Kubica K, Twarda A, Zak KM, Holak TA, Dömling A. Bioorg. Med. Chem. Lett. 2015;25:5661–5666. doi: 10.1016/j.bmcl.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kranz D, Dobbelstein M. Cancer Res. 2006;66:10274–10280. doi: 10.1158/0008-5472.CAN-06-1527. [DOI] [PubMed] [Google Scholar]
- 95.Rao B, Lain S, Thompson AM. Br. J. Cancer. 2013;109:2954–2958. doi: 10.1038/bjc.2013.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Michaelis M, Rothweiler F, Klassert D, von Deimling A, Weber K, Fehse B, Kammerer B, Doerr HW, Cinatl J. Cancer Res. 2009;69:416–421. doi: 10.1158/0008-5472.CAN-08-1856. [DOI] [PubMed] [Google Scholar]
- 97.Ribas J, Boix J, Meijer L. Exp. Cell Res. 2006;312:2394–2400. doi: 10.1016/j.yexcr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 98.Cheok CF, Dey A, Lane DP. Mol. Cancer Res. 2007;5:1133–1145. doi: 10.1158/1541-7786.MCR-07-0161. [DOI] [PubMed] [Google Scholar]
- 99.Coll-Mulet L, Iglesias-Serret D, Santidrián AF, Cosialls AM, de Frias M, Castaño E, Campàs C, Barragan M, de Sevilla AF, Domingo A, Vassilev LT, Pons G, Gil J. Blood. 2006;107:4109–4114. doi: 10.1182/blood-2005-08-3273. [DOI] [PubMed] [Google Scholar]
- 100.Joerger AC, Ang HC, Fersht AR. Proc. Natl. Acad. Sci. USA. 2006;103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Basse N, Kaar JL, Settanni G, Joerger AC, Rutherford TJ, Fersht AR. Chem. Biol. 2010;17:46–56. doi: 10.1016/j.chembiol.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 102. ClinicalTrials.gov identifiers for RG7112: NCT01164033, NCT01677780, NCT01143740, NCT00623870, and NCT00559533 [Google Scholar]
- 103. ClinicalTrials.gov identifier for RG7112/doxorubicin: NCT01605526 [Google Scholar]
- 104. ClinicalTrials.gov identifier for RG7112/cytarabine: NCT01605526 [Google Scholar]
- 105. ClinicalTrials.gov identifier for RG7775: NCT02098967 [Google Scholar]
- 106. ClinicalTrials.gov identifier for RO5503781: NCT01462175 [Google Scholar]
- 107. ClinicalTrials.gov identifier for RO5503781/posaconazole NCT01901172 [Google Scholar]
- 108. ClinicalTrials.gov identifier for RO5503781/cytarabine NCT01773408 [Google Scholar]
- 109. ClinicalTrials.gov identifier for RG7388/Pegasys: NCT02407080 [Google Scholar]
- 110. ClinicalTrials.gov identifier for MK-8242: NCT01463696 [Google Scholar]
- 111. ClinicalTrials.gov identifier for MK-8242/cytarabine: NCT01451437 [Google Scholar]
- 112. ClinicalTrials.gov identifiers for DS-3032b: NCT02319369 and NCT01877382 [Google Scholar]
- 113. ClinicalTrials.gov identifier for HDM201: NCT02143635 [Google Scholar]
- 114. ClinicalTrials.gov identifier for HDM201/LEE011: NCT02343172 [Google Scholar]
- 115. ClinicalTrials.gov identifier for CGM097: NCT01760525 [Google Scholar]
- 116. ClinicalTrials.gov identifier for SAR405838: NCT01636479 [Google Scholar]
- 117. ClinicalTrials.gov identifier for SAR405838/pimasertib: NCT01985191 [Google Scholar]
- 118. ClinicalTrials.gov identifier for AMG232: NCT02110355 [Google Scholar]
- 119. ClinicalTrials.gov identifier for AMG232/trametinib NCT02016729 [Google Scholar]