SUMMARY
MicroRNAs (miRNAs) are important regulators of cell fate decisions in immune responses. They act by coordinate repression of multiple target genes, a property that we exploited to uncover regulatory networks that govern T helper-2 (Th2) cells. A functional screen of individual miRNAs in primary T cells uncovered multiple miRNAs that inhibited Th2 cell differentiation. Among these were miR-24 and miR-27, miRNAs coexpressed from two genomic clusters, which each functioned independently to limit interleukin-4 (IL-4) production. Mice lacking both clusters in T cells displayed increased Th2 cell responses and tissue pathology in a mouse model of asthma. Gene expression and pathway analyses placed miR-27 upstream of genes known to regulate Th2 cells. They also identified targets not previously associated with Th2 cell biology which regulated IL-4 production in unbiased functional testing. Thus, elucidating the biological function and target repertoire of miR-24 and miR-27 reveals regulators of Th2 cell biology.
INTRODUCTION
Polarized helper T cell differentiation both orchestrates beneficial anti-pathogen immunity and drives immunologic disease. Helper T cells (Th) differentiate from activated CD4+ T cells under the influence of cytokines and inducible transcription factors into effector cell subsets including Th1, Th2, Th17, T follicular helper (Tfh) and regulatory T (Treg) cells (Zhu et al., 2010). Th2 cell differentiation is driven by interleukin-4 (IL-4) which signals via the transcription factor STAT6. STAT6 induces the expression of GATA3, and together these transcription factors drive a gene expression program that includes the key Th2 cell lineage-defining cytokines IL-4, IL-13 and IL-5 (Ansel et al., 2006). Positive feedback loops amplify cell fate decisions to generate a pool of Th2 cells capable of orchestrating robust immune responses. Th2 cells support basophil, mast cell, and eosinophil survival, induce alternatively activated macrophages, and influence local stromal and epithelial cells (Pulendran and Artis, 2012). These functions effectively control parasitic infections. They also drive asthma and allergic disease pathogenesis (Fahy, 2014).
miRNAs influence helper T cell differentiation and function by modulating programs of gene expression through the inhibition of target mRNAs (Baumjohann and Ansel, 2013). The ability to globally process and generate functional mature miRNAs limits the differentiation of helper T cells into cytokine-producing effectors (Chong et al., 2008; Muljo et al., 2005; Steiner et al., 2011). However, individual miRNAs can either restrict or enhance helper T cell differentiation and function. In Th2 cells, miR-19 enhances and miR-155 limits effector cell differentiation and cytokine production (Rodriguez et al., 2007; Simpson et al., 2014; Thai et al., 2007). miR-27 has also been implicated in suppression of Th2 cell cytokine production in patients with multiple sclerosis (Guerau-de-Arellano et al., 2011; Guo et al., 2014).
Identifying miRNAs that regulate T cell differentiation and function is necessary to place them within the regulatory framework that governs immune responses. Furthermore, the intrinsic properties of miRNA biology can be leveraged to discover relevant gene networks through the identification of direct miRNA targets. A key feature of miRNA action is that the quantitative effect on an individual direct mRNA target is modest. Yet every miRNA targets tens or hundreds of mRNAs, and the collaborative inhibition of numerous direct targets in gene networks can have large biological consequences (Ebert and Sharp, 2012). This property can be used to connect miRNAs to previously described pathways as well as uncover novel regulators of cell fate decisions by empirical determination of direct miRNA targets and their effects on T cell responses.
In this study, we undertook to identify miRNAs that inhibit Th2 cell differentiation and cytokine production, and then use combined experimental and bioinformatic approaches to uncover target networks. We found that miR-24 and miR-27 both act to inhibit IL-4 production in T cells in vitro, and that mice lacking expression of these miRNAs in T cells show enhanced allergic airway hypersensitivity inflammatory responses. Global expression analysis uncovered target networks connecting predicted miR-24 and miR-27 targets to IL-4. A screen of candidate targets inhibited by these miRNAs and not biased by a priori knowledge of gene function uncovered known and novel regulators of IL-4 production in T cells. Therefore miR-24 and miR-27 regulate Th2 cell differentiation and cytokine production, targeting nodes in known Th2 cell network architecture and revealing novel players in allergic inflammation.
RESULTS
miRNAs limit the generation of Th2 effector cells
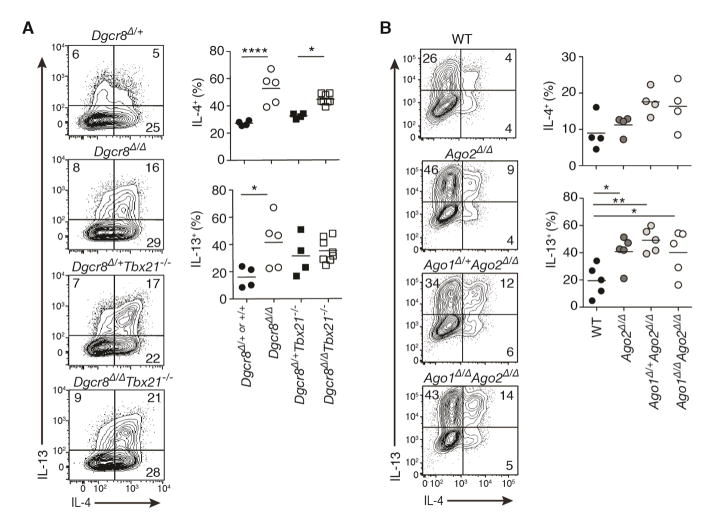
In non-polarizing culture conditions, globally miRNA-deficient CD4+ T cells lacking the critical miRNA processing factors Dgcr8, Drosha or Dicer exhibit aberrantly early and enhanced Th1 cell differentiation and interferon-γ (IFN-γ) cytokine production (Chong et al., 2008; Muljo et al., 2005; Steiner et al., 2011). To determine whether miRNAs also regulate Th2 cell generation and function, we examined production of the canonical Th2 cell-associated cytokines IL-4 and IL-13 under experimental conditions that prevent Th1 cell differentiation. Culturing Dgcr8f/fCd4-cre (Dgcr8Δ/Δ) CD4+ T cells in robust Th2 cell polarizing conditions with IFN-γ blocking antibodies increased the already high frequency of IL-4+ and IL-13+ cells observed in miRNA-sufficient control cells (Figure 1A). IL-4 production was similarly increased among Dgcr8Δ/Δ CD4+ T cells that also lack Tbx21, which encodes the lineage-defining Th1 cell transcription factor T-bet (Figure 1A).
Figure 1. miRNA-deficient T cells have enhanced cytokine production in Th2 cell culture conditions.
Intracellular cytokine staining of (A) Dgcr8Δ/+ and Dgcr8Δ/Δ and (B) wildtype (WT), Ago2 Δ/Δ, Ago1+/ΔAgo2 Δ/Δ and Ago1Δ/ΔAgo2 Δ/Δ CD4+ T cells cultured in Th2 cell conditions for 5 days. Also included are control and miRNA-deficient cells lacking T-bet (Dgcr8Δ/+Tbx21−/− and Dgcr8Δ/ΔTbx21−/−)(A). Numbers are percent of live CD4+ T cells singlets. For Dgcr8 cells, YFP+ pre-gating was performed to use a Rosa-YFP reporter as a measure of CRE activity and exclude cells that escaped deletion. Each graphed point represents an individual mouse. (n=4–8 mice from 2–3 independent experiments, one-way ANOVA with Bonferroni’s multiple comparison test for (A) (Dgcr8Δ/+or+/+ vs. Dgcr8Δ/Δ and Dgcr8Δ/+Tbx21−/− vs. Dgcr8Δ/ΔTbx21−/−) or Dunnett’s multiple comparison test for (B) (all samples compared to WT)). See also Figure S1
Dysregulated Th2 cell differentiation was also seen in T cells with reduced expression of Argonaute (Ago), the core effector protein of the miRNA-induced silencing complex. Four Ago are encoded in the genome (Ago1, Ago2, Ago3 and Ago4), and they mediate miRNA-dependent inhibition of gene expression. Ago2f/fCd4-cre (Ago2 Δ/Δ), Ago1+/fAgo2f/fCd4-cre (Ago1+/ΔAgo2 Δ/Δ) and Ago1f/fAgo2f/fCd4-cre (Ago1Δ/ΔAgo2 Δ/Δ) CD4+ T cells that lacked one or both of the most abundant Ago species in T cells (Figure S1A) displayed progressively reduced miRNA activity (Figure S1B) and enhanced IL-4 and IL-13 production in Th2 cell cultures (Figure 1B). Consistent with previous reports of T cells lacking critical miRNA machinery (Chong et al., 2008; Muljo et al., 2005; Steiner et al., 2011), the near complete loss of miRNA activity in Ago1Δ/ΔAgo2 Δ/Δ T cells resulted in enhanced IFN-γ production in non-polarizing (ThN) culture conditions and reduced peripheral T cell numbers (Figure S1C–S1G). Ago1Δ/ΔAgo2 Δ/Δ CD4+ T cell cultures also showed compromised proliferation and survival, allowing accumulation of cells that escaped gene deletion or induced compensatory Ago3 or Ago4 expression as indicated by pan-Ago protein immuno blots (Figure S1A). Importantly, enhanced Th1 and Th2 cell differentiation occurred independently of these defects, since Ago2 Δ/Δ and Ago1+/ΔAgo2 Δ/Δ displayed progressive increases in IFN-γ and IL-4 production with normal or even slightly increased proliferation rates (Figure S1H). Thus, a deficiency in the ability to generate mature miRNAs or have them function in post-transcriptional repression of gene expression results in enhanced Th2 cell differentiation. And in at least two helper T cell lineages (Th1 and Th2), the net effect of miRNA activity in the cell is to restrict differentiation into cytokine-producing effectors.
Identification of individual miRNAs that regulate Th2 cell cytokine production
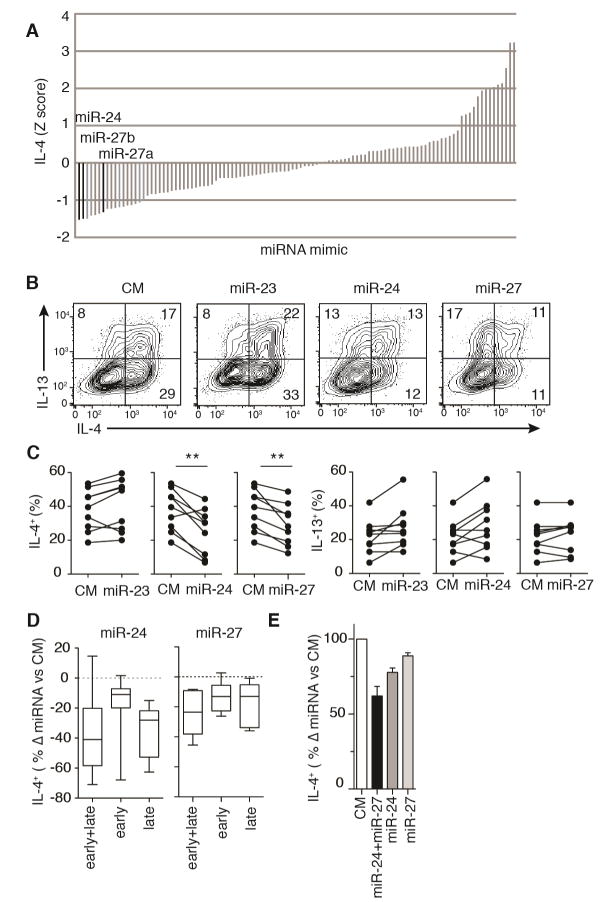
To define the impact of all individual miRNAs on Th2 cell differentiation and function in vitro, we employed a screening approach in primary mouse CD4+ T cells. We interrogated 108 of the most abundant miRNAs in activated CD4+ T cells (Steiner et al., 2011) by electroporating individual miRNA mimics into Dgcr8Δ/ΔTbx21−/− T cells twice over a 5 day culture period in the presence of low amounts of exogenous IL-4 (10U/ml) and no neutralizing antibodies. Using this protocol, transfections are >90% efficient with only minor effects on cell viability (Steiner et al., 2011). miRNA-deficient CD4+ T cells that also lacked Tbx21 were chosen for this screen to limit confounding effects of IFN-γ production, since Dgcr8Δ/Δ and Dgcr8Δ/ΔTbx21−/− cells show similar enhancements in IL-4 production in Th2 cell cultures (Figure 1A). Seventeen miRNAs reduced the frequency of IL-4 producing cells by greater than one standard deviation when compared to the median of all individually transfected miRNAs (Table S1 and Figure 2A). A subset of these top IL-4 inhibitors also substantially reduced IL-13 production in CD4+ T cells and included miR-140-5p, miR-101b, let-7a, miR-17 and miR-27b (Table S1). Among the miRNAs that enhanced IL-4 production was miR-19a (Table S1), which promotes Th2 cell-associated cytokine production in mouse and human CD4+ T cells (Simpson et al., 2014). Thus multiple individual miRNAs are sufficient to inhibit Th2 cell differentiation and cytokine production in vitro.
Figure 2. miR-24 and miR-27 inhibit IL-4 production in effector T cells.
(A) Z scores for the frequency of IL-4+ Dgcr8Δ/ΔTbx21−/− T cells among live YFP+ CD4+ T cell singlets by intracellular staining. Each vertical bar represents a singlicate measure of an individual miRNA mimic transfected twice over a 5 day culture. (B,C) IL-4 and IL-13 production by Dgcr8Δ/ΔTbx21−/− T cells transfected with miR-23, miR-24, miR-27 or control mimic (CM). Numbers in quadrants are percent of live YFP+ CD4+ singlets and each dot represents an individual mouse (n=9 from 8 independent experiments, two-tailed paired t-test with Bonferroni’s multiple comparison test). (D) Change in percent of IL-4+ cells after transfection of Dgcr8Δ/ΔTbx21−/− CD4+ T cells with miR-24 or miR-27 versus CM at early (day 1), late (day 4) or early and late (day 1 and day 4) culture timepoints (n≥7 mice from at least 5 independent experiments, box interquartile with whiskers minimum and maximum). (E) Change in percent of IL-4+ cells versus CM after transfection of Dgcr8Δ/ΔTbx21−/− CD4+ T cells with miR-24, miR-27, or miR-24 plus miR-27 (n=4 mice, representative of 2 independent experiments, linear model with the two miRNAs as binary code predictors and an interaction term). See also Figure S2 and S3.
miR-24 and miR-27 act independently to reduce IL-4 production in helper T cells
The two miRNA mimics that most inhibited IL-4 production were miR-24 and miR-27 (Figure 2A). miR-24 and miR-27 family miRNAs reside together in polycistronic clusters with a third miRNA family, miR-23. miR-24 and miR-27, but not miR-23, consistently reduced the frequency of Dgcr8Δ/ΔTbx21−/− CD4+ T cells producing IL-4 by 20–40% in cells cultured with low levels of exogenous IL-4 (Figure 2B and 2C). Production of a second canonical Th2 cell-associated cytokine, IL-13, was not reduced by miR-24 or miR-27 (Figure 2B and 2C). Reductions in IL-4 but not IL-13 production were also observed in Dgcr8Δ/ΔTbx21−/− CD4+ T cells transfected with miR-24 and miR-27 and cultured in full Th2 cell polarizing conditions (500U/ml IL-4 and anti-IFN-γ neutralizing antibodies) (Figure S2A). In contrast, all let-7 family miRNAs inhibited their direct target IL-13 (Polikepahad et al., 2010) with variable effects on IL-4 production (Figure S2B). Specific activity of miRNA mimics was confirmed by downregulation of GFP expression from retroviral sensors carrying miRNA target sequences (Figure S3A). Comparison of sensor activity in wildtype and Dgcr8Δ/ΔTbx21−/− CD4+ T cells demonstrated that mimic transfection results in at most physiologic levels of miR-23, miR-24 or miR-27 activity in miRNA-deficient cells (Figure S3B). Reduced IL-4 production was not secondary to compromised cell survival (Figure S3C).
To determine at what timepoints during the in vitro differentiation of effector cells these miRNAs might be most active, miRNA mimic transfections were performed early (day 1), late (day 4), or early and late (day 1 and day 4). miR-24 inhibited IL-4 production predominantly late in culture, whereas miR-27 acted cumulatively over the culture period (Figure 2D). Co-transfecting miR-24 with miR-27 late in culture further suppressed IL-4 production (Figure 2E). These data are consistent with additive or partially additive, but not synergistic, effects of these two co-expressed miRNAs. Taken together, these results suggest that miR-24 and miR-27 act collaboratively through different downstream pathways to inhibit Th2 cell differentiation and IL-4 production. They imply that miR-24 can inhibit cytokine production by effector cells while miR-27 inhibits the differentiation of activated T cells into Th2 cells.
Mirc11 and Mirc22 cluster expression limit type 2 inflammation and tissue pathology in a mouse model of asthma
To determine whether miR-24 and miR-27 limit effector T cell differentiation and cytokine production during type 2 immune responses, we developed a mouse model in which T cells lacked expression of these miRNAs. miR-24 and miR-27 are encoded twice in the genome in the Mirc11 cluster (which encodes miR-23a, miR-27a and miR-24-2) and the Mirc22 cluster (which encodes miR-23b, miR-27b and miR-24-1). In these clusters, the ‘a’ and ‘b’ members of the miR-23 and miR-27 families vary only by a single nucleotide, and both clusters encode an identical mature miR-24. Since both the Mirc11 and Mirc22 clusters are expressed in mouse and human T cells (Barski et al., 2009; Steiner et al., 2011) and they likely share the majority of their mRNA targets, we eliminated both clusters to examine the role of endogenous miR-24 and miR-27 in T cells. We generated mice from embryonic stem cells with targeted alleles for each cluster (Park et al., 2012; Prosser et al., 2011) and intercrossed them to obtain Mirc11−/−Mirc22f/fCd4-cre (Mirc11−/−Mirc22ΔT/ΔT) mice with T cells that lack miR-23, miR-24 and miR-27 expression and activity (Figure S4A and S4B). Mirc11−/−Mirc22ΔT/ΔT mice had normal numbers and proportions of peripheral naïve, memory, activated, and regulatory T cell subsets, and normal T cell proliferation in vitro (Figure S4C–S4F).
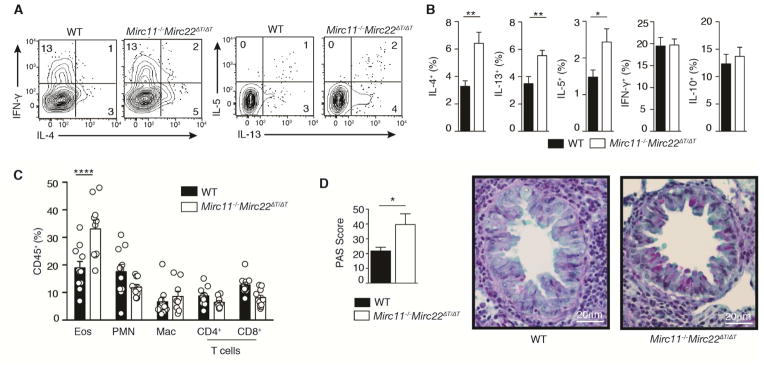
Given the critical role of type 2 immune responses in the pathogenesis of asthma, we used a mouse model of allergic airway inflammation to test the generation and function of Th2 cells in vivo. Mice were sensitized with a single intraperitoneal injection of ovalbumin (OVA), then challenged one week later with OVA in the airways on three consecutive days. Mirc11−/− Mirc22ΔT/ΔT mice had twice as many bronchoalveolar CD4+ T cells capable of producing IL-4 ex vivo (Figure 3A and 3B). Concomitant increases in IL-13 and IL-5 were also observed (Figure 3A and 3B). These changes were specific to cytokines that define type 2 responses, as there were no differences in the proportion of cells producing IFN-γ or IL-10 (Figure 3A and 3B). This increase in the capacity to produce Th2 cell-associated cytokines correlated with an increase in eosinophil recruitment to the lung (Figure 3C), a hallmark of allergic lung inflammation in both mice and humans. Mirc11−/−Mirc22ΔT/ΔT mice also displayed enhanced tissue pathology with increased goblet cell metaplasia in lung epithelium (Figure 3D). These data show that the Mirc11−/−Mirc22ΔT/ΔT clusters are endogenous inhibitors of Th2 cell-associated cytokine production and limit allergic responses in vivo.
Figure 3. Loss of the Mirc11 and Mirc22 clusters in T cells enhances allergic airway inflammation and tissue pathology.
(A,B) Intracellular cytokine staining of CD4+ T cells from WT and Mirc11−/−Mirc22ΔT/ΔT mice restimulated ex vivo 11 days after intraperitoneal OVA sensitization and 4 days after daily OVA airway challenge. Numbers in quadrants are percent of live CD4+ singlets (n≥10 mice from 2 independent experiments, two-tailed t-test). (C) Flow cytometry phenotyping of inflammatory cells in BAL from the same experiments quantifying the frequency of eosinophils (Eos, CD11b+SiglecF+), neutrophils (PMN, CD11b+Ly6G+), alveolar macrophage (Mac, CD11c+CD11blo), CD4+ T cells (CD4, CD11b−CD11c−CD4+) and CD8+ T cells (CD8, CD11b−CD11c−CD8+) among live hematopoietic cells (n≥10 mice from 2 independent experiments, one-way ANOVA with Bonferroni’s multiple comparison test). (D) Periodic Acid Shift (PAS) histologic staining for epithelial cell mucus metaplasia (pink) in lungs from the same experiment. PAS Score is the summed product of the percent of bronchioles with 0% (x0), 1–25% (x1), 26–50% (x2), 51–75% (x3) and 76–100% (x4) PAS positive cells (n≥10 mice from 2 independent experiments, 38–91 bronchioles counted per mouse, two-tailed t-test). See also Figure S4.
The Mirc11 and Mirc22 clusters inhibit Th2 cell differentiation
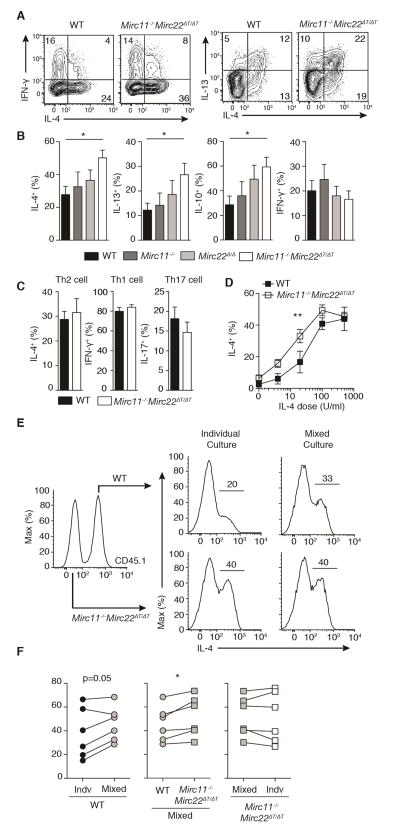
To define the cellular mechanism by which the Mirc11 and Mirc22 clusters restrict type 2 immune responses, we utilized well-characterized helper T cell cultures. In non-polarizing T helper (ThN) conditions where no exogenous cytokines or neutralizing antibodies are added to the culture, Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells exhibited increased production of Th2 cell-associated cytokines including IL-4 and IL-13 (Figure 4A and 4B). IL-10 production was also increased in vitro (Figure 4B), though no difference in the proportion of IL-10 producing T cells was observed in the bronchoalveolar lavage in our allergic airway model (Figure 3B). Cells lacking only one of the two clusters showed a non-significant trend toward increased Th2 cell-associated cytokine production. IFN-γ production was unchanged (Figure 4B). In strong polarizing conditions where saturating levels of exogenous cytokines are provided, Th1, Th2 and Th17 cells were generated at normal frequency (Figure 4C). However, Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells displayed enhanced IL-4 production over a wide range of sub-saturating IL-4 doses, demonstrating the robust capacity of these miRNAs to regulate Th2 cell differentiation and cytokine production (Figure 4D). In mixed culture experiments, Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells enhanced IL-4 production by congenically marked wildtype cells, supporting a partially cell-extrinsic mechanism of action (Figure 4E and 4F). These results are consistent with the ability of the Mirc11 and Mirc22 clusters to regulate IL-4, which is both a product of Th2 cells and the major instructing cytokine for their differentiation..
Figure 4. Mirc11−/−Mirc22ΔT/ΔT T cells have enhanced Th2 cell-associated cytokine production in cultures with limiting exogenous cytokines.
Intracellular cytokine staining of wildtype (WT) and Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells cultured in vitro for 5 days in ThN (A,B) (n=6–9 mice from 4 independent experiments, one-way ANOVA with Dunnett’s multiple comparison test), and Th1, Th2 and Th17 cell conditions (C) (n≥5 mice from 3 independent experiments, two-tailed t-test). Numbers in quadrants are percent of live CD4+ singlets. (D) Dose response of WT and Mirc11−/−Mirc22ΔT/ΔT T cells cultured in vitro for 5 days over a range of exogenous IL-4 doses and measured by intracellular cytokine staining (n=6 mice from 2 independent experiments, least squares four parameter nonlinear regression with logEC50 F-test). (E, F) Representative data and quantitation of intracellular cytokine staining of CD45.1+ WT and CD45.2+ Mirc11−/−Mirc22ΔT/ΔT T cells cultured individually or together in mixed cultures (n=6 pairs of mice from 3 independent experiments, two-tailed paired t-tests).
miR-24 and miR-27 act through downstream mRNA target networks
To determine the molecular mechanisms by which miR-24 and miR-27 inhibit Th2 cell differentiation and cytokine production, we investigated the regulation of their target gene networks in Th2 cells. To this end, Dgcr8Δ/ΔTbx21−/− CD4+ T cells transfected with miR-23, miR-24, miR-27 or control mimic were sorted for RNA sequencing expression analysis. Ingenuity Pathway Analysis (IPA) confirmed significant changes in canonical pathways associated with helper T cell differentiation and cytokine signaling (Table S2). IL-4 and STAT6 were identified as major upstream regulators of observed expression changes (Table S2). These analyses identified broad effects on gene expression that are consistent with our primary observation that miR-24 and miR-27 inhibit IL-4 production in T cells.
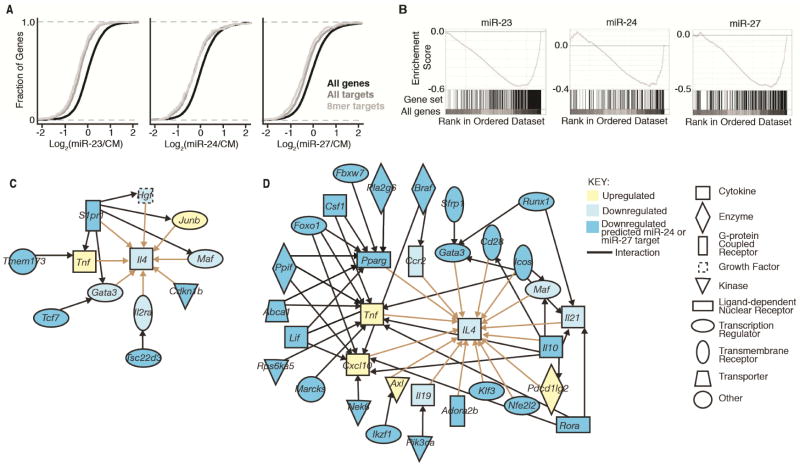
Expression analysis was also used to investigate direct effects on bioinformatically predicted miRNA targets. Cumulative distribution frequency plots and gene set enrichment analysis demonstrated that predicted targets were downregulated by their corresponding miRNAs (Figure 5A and 5B, and (Garcia et al., 2011)). Leading edge analyses revealed significant enrichment for miR-23, miR-24 and miR-27 predicted targets at 23%, 30% and 30% decreased expression, respectively. 68 of 414 predicted miR-24 targets and 118 of 813 predicted miR-27 targets expressed in T cells were downregulated by at least 30% after miRNA mimic transfection with a raw P-value of <0.05 among RNA sequencing replicates. These results highlight the quantitatively small but statistically significant effects miRNAs have on their direct targets.
Figure 5. miR-24 and miR-27 affect networks of genes upstream of IL-4.
(A) Cumulative distribution frequency plots depicting global mRNA expression by RNA sequencing as a log2 fold ratio of Dgcr8Δ/ΔTbx21−/− CD4+ T cells transfected with miR-23, miR-24, or miR-27 versus control mimic (CM) plotted against the cumulative fraction of all genes (black), miRNA TaregtScan targets (dark grey), and miRNA 8mer match TargetScan targets (light grey). (B) Gene Set Enrichment Analysis of same data for miRNA 8mer match TargetScan targets. Plotted are the enrichment curve for and positional location of each target in the total gene set arrayed in ranked order from most upregulated to most downregulated (left to right) (Nominal p-value <0.001 and FDR q-value <0.001 for each) (C,D) IPA pathway analysis of gene and target networks regulated upstream of IL-4 after transfection with miR-24 (C) or miR-27 (D) mimic versus control mimic in Dgcr8Δ/ΔTbx21−/− CD4+ T cells. Interactions between genes or targets and IL-4 (tan) and targets with genes or targets upstream of IL-4 (black) are depicted. RNA sequencing was performed on 3–4 biologic replicates for each sample. See also Figure S5.
We next hypothesized that miR-24 and miR-27 must regulate IL-4 production through direct inhibition of a subset of these regulated target genes. We used IPA to build pathways upstream of IL-4 to investigate potential miRNA-dependent regulatory networks. We first assembled a network of all genes significantly changed (P<0.05 in RNA sequencing replicates) and upstream of IL-4. Then all downregulated predicted miR-24 or miR-27 targets upstream of genes in this network were mapped, and all genes not connected to a target were pruned. For miR-24 transfected T cells, only 5 predicted targets connected to IL-4 (Figure 5C). A single miR-24 target, Sphingosine-1-phosphate receptor 1 (S1pr1) was a highly connected node in this network. For miR-27 transfected T cells, 24 predicted targets were connected to IL-4 (Figure 5D). Several of these genes had isolated linear connections to IL-4, while others formed highly interconnected nodes upstream of IL-4. In comparison, miR-23 transfected T cells, which do not demonstrate statistically significant changes in IL-4, had only three connected miRNA targets in this analysis (Figure S5). Thus using expression data to mine previously identified gene relationships provided candidate targets that may regulate Th2 cell differentiation and cytokine production.
miR-24 and miR-27 target known and unexpected regulators of IL-4 production
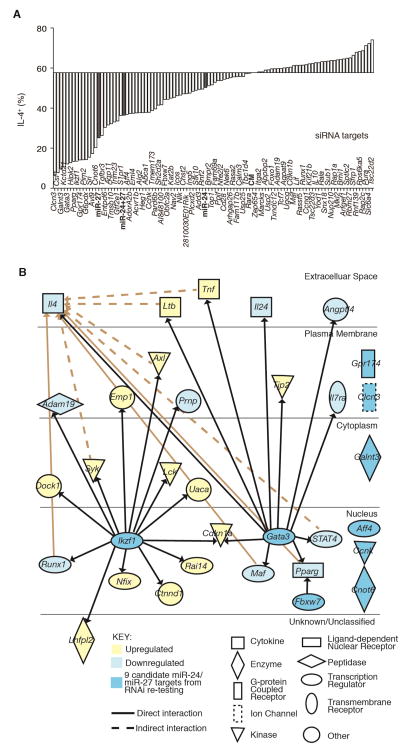
To determine the network of miR-24 and miR-27 targets that mediate miRNA regulation of IL-4 production, we tested the function of a set of candidate targets genes. In order to capture previously unknown regulators of Th2 cell differentiation and cytokine production, we narrowed the list of predicted targets for further testing without bias for previously established gene function. All downregulated predicted targets were ordered by a scoring system, which weighted robust miRNA-induced expression changes, connectivity among changed genes, and Ago binding in activated T cells (Loeb et al., 2012) (Figure S6A and S6B). The top 96 candidate miR-24 and miR-27 targets were tested for effects on IL-4 production by RNAi screening in Mirc11−/− Mirc22ΔT/ΔT CD4+ T cells. Pools of siRNAs against genes downstream of miR-24 and/or miR-27 that regulate IL-4 production should “rescue” (decrease) the proportion of IL-4+ Mirc11−/− Mirc22ΔT/ΔT CD4+ T cells. Overall, siRNAs against candidate targets were more likely to decrease than increase IL-4 production, consistent with the Mirc11 and Mirc22 clusters acting on a network of genes to limit IL-4 production in helper T cells (Figure 6A).
Figure 6. miR-24 and miR-27 candidate targets include known and novel genes that support Th2 cell differentiation and cytokine production.
(A) siRNA screen of candidate miRNA target genes. Pools of siRNAs were transfected twice over a 5 day culture period in Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells and IL-4 production measured by intracellular staining. Each bar represents a singlicate measurement. (B) IPA pathway analysis of 9 candidate direct targets of miR-24 and miR-27 that inhibit IL-4 production after re-testing and their connection to downstream expression changes in respective RNAseq expression data sets. All connections to genes that are upstream of IL-4 were also mapped (tan). See also Figure S6.
Re-testing was performed for the 30 candidate targets that most decreased IL-4 production. siRNAs against 9 genes reduced IL-4-producing cells in equal or greater magnitude than miR-24 or miR-27 transfection without affecting cell proliferation (Figure S6C and S6D). This set included two transcription factors previously associated with Th2 cell differentiation, Gata3 (Zheng and Flavell, 1997; Zhu et al., 2004) and Ikzf1 (a.k.a. Ikaros, (Grogan et al., 2003; Quirion et al., 2009)). Both are predicted targets of miR-27, and both also directly regulated a group of genes affected by miR-27 transfection (Figure 6B). Pathway analysis independently identified Ikzf1 as a critical upstream regulator of the gene expression changes induced by miR-27 (Table S2). Together, Gata3 and Ikzf1 anchor a network that converges on IL-4.
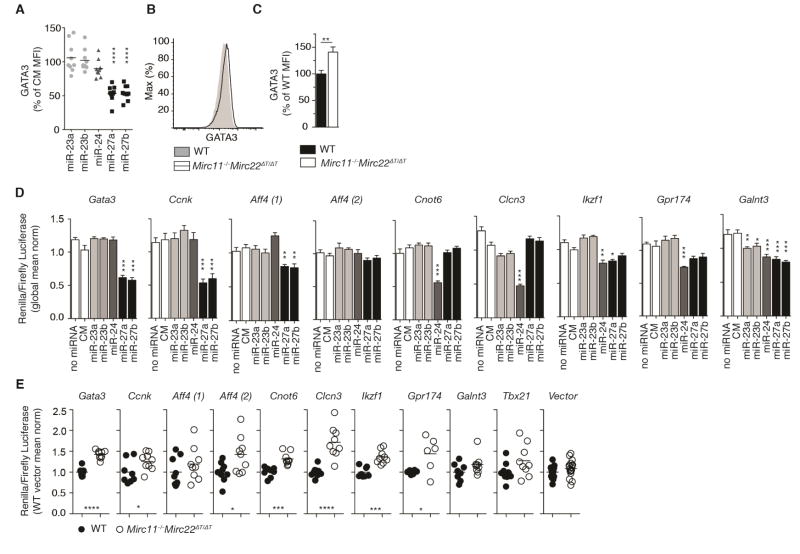
We further investigated the likely possibility that Gata3 is a functionally relevant part of the target network downstream of miR-27. Transfection with miR-27 reduced endogenous GATA3 protein abundance by 50% in Dgcr8Δ/ΔTbx21−/− CD4+ T cells within 24 hours (Figure 7A). Conversely, GATA3 expression was increased in Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells in ThN cultures (Figure 7B and 7C). Enhanced Th2 cell differentiation may account for part of the observed increase in Gata3 expression in Mirc11−/−Mirc22ΔT/ΔT cells. Indeed, a previous report indicates that Gata3 may be indirectly regulated by miR-27 (Guerau-de-Arellano et al., 2011). However, the Gata3 3′UTR contains multiple Ago binding peaks at predicted miR-27 target sites, suggesting direct targeting (Guerau-de-Arellano et al., 2011; Loeb et al., 2012). Dual luciferase assays in Dgcr8Δ/ΔTbx21−/− CD4+ T cells confirmed that miR-27 directly represses the 3′UTRs of Gata3 in primary T cells (Figure 7D). In addition, Gata3 3′UTR reporter assays showed decreased activity in wildtype cells compared with Mirc11−/−Mirc22ΔT/ΔT cells (Figure 7E). Taken together, these studies show that miR-27 directly regulates the Th2 cell transcription factor Gata3 in T cells. However, this single regulatory interaction cannot fully account for Mirc11 and Mirc22 cluster regulation of Th2 cell differentiation and IL-4 production. miR-24 did not directly regulate Gata3 (Figure 7A and 7D), and additional targets of both miR-24 and miR-27 likely contribute.
Figure 7. miR-24 and miR-27 regulate the 3′UTR of target genes.
(A) Intracellular staining for GATA3 one day after transfection of Dgcr8Δ/ΔTbx21−/− CD4+ T cells with miRNA mimic or control mimic (CM). Values are expressed as a percent of the CM mean fluorescence intensity (MFI). Each point represents an individual mouse (n=9 from 2 independent experiments, one sample t-test). (B,C) Intracellular staining for GATA3 in ThN conditions in WT and Mirc11−/− Mirc22ΔT/ΔT T cells. Values are expressed as a percent of wildtype (WT) mean fluorescence intensity (MFI) (n=9 from 4 independent experiments, two-tailed t-test). (D) Dual luciferase assay measuring the activity miR-23, miR-24, miR-27 or CM as a ratio of renilla to firefly luciferase on the 3′UTR of select target genes in Dgcr8Δ/ΔTbx21−/− CD4+ T cells one day after transfection. Values are normalized to the global mean across all transfection condition (n=6–12, 3 technical replicates each from 2–5 independent experiments, one-way ANOVA for reduced expression with Dunnett’s multiple comparison test). (E) Dual luciferase assay measuring regulation of the 3′UTR of select target genes in WT and Mirc11−/−Mirc22ΔT/ΔT T cells one day after transfection. Values are normalized to the mean of WT cells transfected with vector alone in each experiment (n=6–18 mice from 2–6 independent experiments, each dot represents the average of technical triplicates for an individual mouse, two-tailed t-test).
Seven additional genes encoding diverse proteins that have not been previously associated with Th2 cell biology were also identified as limiting factors for IL-4 expression (Figure 6B). They included a cyclin (Ccnk), a protein glycosyltransferase (Galnt3), an elongation complex scaffold (Aff4), a G protein-coupled receptor (Gpr174), a chloride channel (Clcn3), a ubiquitin ligase (Fbxw7) and a poly(A) deadenylase (Cnot6) (Table S3). Only one of these genes, Fbxw7, had been experimentally confirmed to be a direct target of mir-24 or miR-27 (Wang et al., 2011). Dual luciferase assays in Dgcr8Δ/ΔTbx21−/− CD4+ T cells demonstrated that Ccnk and Aff4 are direct targets of miR-27, and Cnot6 and Clcn3 are direct targets of miR-24 in primary T cells (Figure 7D). The remaining three genes, Ikzf1, Gpr174 and Galnt3, appear to be direct targets of both miR-24 and miR-27 (Figure 7D). These findings offer insight into the molecular mechanisms by which miR-24 and miR-27 may act collaboratively to regulate IL-4 production in T cells. Dual luciferase assays comparing wildtype and Mirc11−/−Mirc22ΔT/ΔT CD4+ T cells also showed that endogenous miRNAs from this cluster directly repress genes that are required to support IL-4 production (Figure 7E). Thus by combining bioinformatic and experimental approaches, we have identified both known and novel genes and gene networks capable of regulating IL-4 production directly downstream of miR-24 and miR-27.
DISCUSSION
Three decades of research have clearly established Th2 cells as central drivers of allergic inflammation. The core circuitry that defines and reinforces Th2 cell identity and polarizes immune responses in vivo, which consists of positive feedback mediated by GATA-3 and IL-4 signaling, is also well understood. Yet little is known about critical pathogenic determinants of allergic disease, especially the initiating and limiting factors that gate entry into this program and tip the balance toward Th2 cell responses. miRNAs are particularly effective at regulating cell fate decisions that are sensitive to small perturbations in environmental cues and intracellular signaling due to feedback loops that reinforce developmental outcomes. Therefore, we hypothesized and found that individual miRNAs were limiting factors that regulate Th2 cell differentiation. miRNA-directed pathway discovery established a direct connection with Gata3 and identified novel regulators of Th2 cell differentiation and IL-4 production.
T cells lacking miRNAs are hypersensitive to signals that induce Th1 cell differentiation (Chong et al., 2008; Muljo et al., 2005; Steiner et al., 2011). Aberrantly early and dominant Th1 cell differentiation in miRNA-deficient T cell cultures complicated previous analyses of miRNA regulation of Th2 cell differentiation. Our new results extend prior studies by demonstrating that miRNA deficiency also enhances the generation of Th2 cells.
Within the mouse T cell miRNAome, miR-24 and miR-27 were the most potent inhibitors of Th2 cell differentiation and cytokine production in reconstitution experiments. Our results show that endogenous miR-24 and miR-27 expression in T cells restricts Th2 cell differentiation over a wide range of limiting IL-4 doses. These culture conditions likely recapitulate in vivo conditions, where “pure” Th2 cell responses do not exist. Even in allergic and anti-helminth responses, T cells producing IFN-γ are typically present, and often even more prevalent than IL-4 producing cells. In addition, T cells themselves drive powerful autocrine/paracrine positive feedback mechanisms in helper cells in vivo (Ansel et al., 2006). Indeed, we found that Mirc11−/−Mirc22ΔT/ΔT deficient T cells are capable of enhancing the differentiation of co-cultured wildtype cells into IL-4 producing Th2 cells. This result is consistent with enhanced IL-4 production by Mirc11−/−Mirc22ΔT/ΔT T cells and explains why conditions that leave out or limit exogenous cytokines are useful in uncovering defects in helper T cell differentiation.
Loss of the Mirc11 and Mirc22 clusters in T cells enhances allergic inflammation and results airway pathology characterized by increased goblet cell metaplasia. Mucus production is a major determinant of asthmatic disease phenotype (Evans et al., 2015), and mucus plugs are a major cause of fatal asthma (Kuyper et al., 2003). Our findings demonstrate that unrelated families of miRNAs that are co-expressed from genomic clusters can evolve to collaborate in the coordination of cellular responses and contribute to tissue pathology. Previous work indicated that miR-19 and miR-155 promote type 2 immune responses and asthma (Malmhall et al., 2014; Okoye et al., 2014; Rodriguez et al., 2007; Simpson et al., 2014; Thai et al., 2007). Neither of these miRNAs nor the Mirc11 and Mirc22 clusters are differentially expressed in mouse or human Th2 cells as compared to other helper T cell subsets (Kuchen et al., 2010; Monticelli et al., 2005; Okoye et al., 2014; Rossi et al., 2011), yet these miRNAs clearly act as limiting positive and negative regulators of Th2 cell fate. This highlights the importance of studying targets downstream of miRNAs in the context of developing Th2 cells.
The current paradigm for miRNA function posits that they mediate their effects by tuning the expression of multiple direct targets in common biological networks. Therefore, we mapped gene pathways from our RNA sequencing expression set and found miR-27 targets in networks of highly interconnected nodes upstream of IL-4, consistent with the possibility that dozens of targets contribute to the regulatory effects of this miRNA. This analysis uncovered the well-known Th2 cell-associated transcription factor Gata3, which was confirmed as one of the direct target of miR-27 capable of regulating IL-4. miRNA inhibition of key lineage-defining transcription factors may be a common mechanism by which miRNAs impact cell fate, as thresholds for both Th1 and Th2 cell differentiation are regulated in this fashion by miR-29 and miR-24 or miR-27, respectively (Steiner et al., 2011). The effect of miR-27 on GATA3 is also a fine example of the quantitative effects, rather than switch-like behavior, typically exhibited by miRNAs. In addition, it highlights how both different quantitative thresholds and the activity of additional direct targets may be required to alter the expression of select downstream pathways. This likely explains why IL-4, but not IL-13, is changed by miR-27 transfection.
Pathway analysis depends on general curation of available literature. Not all relationships identified will be relevant in particular specialized cell types, such as Th2 cells. In fact, among the genes functionally tested by RNAi screening, many had no effect on IL-4 production in T cells including Cdkn1b, Tcf7, and Tsc22d3 for miR-24 and Foxo1, Il10, Lif, Marcks, Rps6Ka5, Runx1, and Sfrp1. Pathway analysis also cannot predict which factors within a network of genes are present at limiting concentration and therefore more likely to be critical nodes of regulation by miRNAs, which mediate only moderate repression of their targets.
Our findings demonstrate how miRNA-directed pathway discovery can reveal novel genes involved in immune responses. This was essential for miR-24, which had relatively few downregulated targets known to impact IL-4, but was also successful for miR-27 with its large target networks upstream of IL-4. Choosing candidate targets for testing without bias for previously established gene function enabled us to identify functionally relevant genes that were all validated as miR-24 and/or miR-27 targets, and that encode diverse proteins not previously implicated in IL-4 production. This includes genes involved in human disease, such as the lysophosphatidylserine receptor Gpr174, which is associated with autoimmune thyroid disease (Chu et al., 2013; Szymanski et al., 2014). It also includes two targets, Ccnk and Aff4, that both interact with the positive transcription elongation complex (p-TEFb, (Fu et al., 1999; Lin et al., 2002; Luo et al., 2012)), suggesting possible functional specialization in RNA polymerase II elongation machinery in the regulation of IL-4 production or Th2 cell differentiation. The miR-24 target Cnot6 encodes the catalytic component of the poly(A) deadenylase complex that interacts with mRNA-destabilizing proteins, including the miRNA-induced silencing complex (Behm-Ansmant et al., 2006; Piao et al., 2010), indicating that efficient post-transcriptional regulation is essential for IL-4 production in T cells. Thus, identifying functionally relevant miRNAs and vulnerable nodes in their target networks generates new information and novel hypotheses about the regulation of immune responses. Ultimately, these approaches hold promise to provide new therapeutic strategies for immune diseases.
EXPERIMENTAL PROCEDURES
Mice
Dgcr8Δ/ΔTbx21−/−, Ago2 Δ/Δ, Ago1+/ΔAgo2 Δ/Δ, Ago1Δ/ΔAgo2 Δ/Δ, Mirc11−/−, Mirc22ΔT/ΔT, Mirc11−/−Mirc22ΔT/ΔT, and CD45.1+ male and female mice were used at 5–12 weeks of age. Strain details are provided in the Supplemental Experimental Procedures. All mice were housed and bred in specific pathogen-free conditions in the Animal Barrier Facility at the University of California, San Francisco. Animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
In vitro cultures, transfections and transductions
CD4+ T cells from the spleen and lymph nodes of mice were enriched by Dynabead positive selection (Invitrogen, L3T4). Cells were stimulated with anti-CD3 and anti-CD28, in the presence of exogenous cytokines and neutralizing antibodies for polarization cultures, for 3 days then rested with 20 units/ml recombinant IL-2 (National Cancer Institute) for an additional 2 days (see Supplemental Experimental Procedures for details). For cytokine assays, T cells were restimulated on day 5 of culture for 5 hours with 20nM PMA and 1μM ionomycin. During the final 2.5hrs of restimulation, 5μg/ml brefeldin A was added. For proliferation assays, cells were labeled with 5μM CellTrace Violet (Invitrogen) per manufacturer’s instruction. Transfections with 500nM miRIDIAN miRNA mimics (Dharmacon) or siGENOME SmartPools (Dharmacon) were performed on day 1 and day 4 of culture using the Neon Transfection System (Invitrogen) as previous described (Steiner et al., 2011) unless otherwise specified. MSCV-PGK-hCD25 miRNA sensors for miR-23a, miR-23b, miR-24, miR-27a and miR-27b were constructed to express a GFP with four perfectly complementary binding sites for the miRNA in the 3′UTR. Cells were transduced on day 2 of ThN cultures by spin infection and hCD25+ CD4+ T cells analyzed on day 5 for GFP expression as previously described (Steiner et al., 2011).
Luciferase assays
3′UTR dual luciferase plasmids were constructed (see Supplemental Experimental Procedures), and CD4+ T cells transfected on day 4 of ThN culture with luciferase reporter constructs and/or miRNA mimics using the Neon Transfection System. Luciferase activity was measured 24 h after transfection with the Dual Luciferase Reporter Assay System (Promega) and a FLUOstar Optima plate-reader (BMG Labtech).
RNA sequencing
150,000–800,000 live RosaYFP+CD4+ Dgcr8Δ/ΔTbx21−/− cells were sorted directly into Trizol LS (Life Technologies) and RNA isolated using a miRNeasy Micro Kit (QIAGEN) with on-column DNase digestion after transfection at day 1 and day 4 of culture with control, miR-23, miR-24 or miR-27 mimic. RNA sequencing libraries were generated using 1ng of total RNA and the Ovation RNA-Seq System V2 and Ultralow Library Construction System sample prep kits (NuGEN). Libraries were multiplexed at a density of 4 per flow-cell lane and sequenced on the HiSeq 2500 to generate single end 50bp reads per the manufacturer’s instructions (Illumina). Of the 4 biologic replicates sorted for each condition, data from one control mimic transfected sample was excluded for low total read counts (203,495). The data pipeline and downstream analysis using cumulative distribution frequency plots, Gene Set Enrichment Analysis and Ingenuity Pathway Analysis are described in the Supplemental Experimental Procedures.
Airway hypersensitivity
Sex and age matched wildtype and Mirc11−/−Mirc22ΔT/ΔT mice were injected intraperitoneally with 50μg ovalbumin (OVA, Sigma) in 100μl of PBS plus 100μl of Imject alum (Thermo Scientific). Seven days later mice were challenged with 50μg OVA in 20 μl of PBS administered to the lungs by oropharyngeal aspiration daily for three consecutive days. BAL was performed by 4 consecutive washes of 1ml complete media, and cells used for cytokine production upon restimulation as described above. Inflammatory cells in BAL were quantified as the frequency of eosinophils (CD11b+SiglecF+), neutrophils (CD11b+Ly6G+), alveolar macrophage (CD11c+CD11blo), CD4+ T cells (CD11b−CD11c−CD4+) and CD8+ T cells (CD11b− CD11c−CD8+) among live CD45+ cells. Reactive epithelial changes were assessed by a blinded pathologists who scored PAS positive bronchioles in a complete cross-section of the right and left lung. The PAS score was defined as the summed product of the percent of bronchioles with 0% (x0), 1–25% (x1), 26–50% (x2), 51–75% (x3) and 76–100% (x4) PAS-positive cells.
Statistics and Data Analysis
Excel (Microsoft) and Prism (GraphPad) were used for data analysis. For all figures, bar graphs display mean+s.e.m. unless otherwise stated. Z-score was calculated from mean and s.d.. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001 for significance. For statistical tests of multiple groups, ANOVA was used with appropriate post hoc testing. All data was assumed to be normally distributed.
Supplementary Material
Highlights.
Multiple individual miRNAs inhibit Th2 cell differentiation and function
miR-24 and miR-27 limit IL-4 production in T cells
Mice lacking miR-24 and miR-27 in T cells have enhanced Th2 cell responses
miR-24 and miR-27 inhibit a network of direct targets that regulate IL-4 production
Acknowledgments
We thank Andrea Barczak, Rebecca Barbeau, and Joshua Pollack from the UCSF SABRE Functional Genomics Core Facility for help with RNA sequencing, Megan Laurance for help with pathway analysis, Chong Park and the Gladstone Transgenic Gene Targeting Core for help with generating mice, Nirav Bhakta for statistical analysis, Hannah Happ for help running flow cytometry experiments and Yelena Bronevetsky, Marisella Panduro, and Priti Singh for help with establishing and maintaining mutant mouse colonies. This work supported by the US National Institutes of Health (HL107202, HL109102, K08AI116949), the Sandler Asthma Basic Research Center, a Scholar Award (K.M.A.) and a Fellow Award (H.H.P.) from The Leukemia & Lymphoma Society, the W.M. Keck Foundation (M.T.M.) and the UCSF Diabetes Center (NIH P30 DK063720). The data reported in this paper has been archived in the GEO database. The authors have no conflicts of interest to report. An MTA with Genome Research Limited, Wellcome Trust Sanger Institute governs use of Ago1-deficient mice.
Footnotes
AUTHOR CONTRIBUTIONS
H.H.P. performed and analyzed most of the experiments; D.F.S, S.P, J.R.G, J.F.O.C, R.K., N.T.C., and A.G. performed and analyzed some of the experiments; D.K. and L.T.J. generated the Mirc22floxd chimeric mice. M.T.M. supervised the generation of the miR-Mirc22floxd mice in the Keck Consortium. D.J.E designed RNAseq experiments and pipeline analysis; H.H.P and K.M.A. designed the experiments, interpreted the data, and wrote the manuscript; and all authors discussed the results and read the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X, Shen M, Xie F, Miao XJ, Shou WH, Liu L, Yang PP, Bai YN, Zhang KY, Yang L, et al. An X chromosome-wide association analysis identifies variants in GPR174 as a risk factor for Graves’ disease. J Med Genet. 2013;50:479–485. doi: 10.1136/jmedgenet-2013-101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CM, Raclawska DS, Ttofali F, Liptzin DR, Fletcher AA, Harper DN, McGing MA, McElwee MM, Williams OW, Sanchez E, et al. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat Commun. 2015;6:6281. doi: 10.1038/ncomms7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy JV. Type 2 inflammation in asthma - present in most, absent in many. Nat Rev Immunol. 2014;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem. 1999;274:34527–34530. doi: 10.1074/jbc.274.49.34527. [DOI] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan JL, Wang ZE, Stanley S, Harmon B, Loots GG, Rubin EM, Locksley RM. Basal chromatin modification at the IL-4 gene in helper T cells. J Immunol. 2003;171:6672–6679. doi: 10.4049/jimmunol.171.12.6672. [DOI] [PubMed] [Google Scholar]
- Guerau-de-Arellano M, Smith KM, Godlewski J, Liu Y, Winger R, Lawler SE, Whitacre CC, Racke MK, Lovett-Racke AE. Micro-RNA dysregulation in multiple sclerosis favours pro-inflammatory T-cell-mediated autoimmunity. Brain. 2011;134:3578–3589. doi: 10.1093/brain/awr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Riley KJ, Iwasaki A, Steitz JA. Alternative capture of noncoding RNAs or protein-coding genes by herpesviruses to alter host T cell function. Mol Cell. 2014;54:67–79. doi: 10.1016/j.molcel.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper LM, Pare PD, Hogg JC, Lambert RK, Ionescu D, Woods R, Bai TR. Characterization of airway plugging in fatal asthma. Am J Med. 2003;115:6–11. doi: 10.1016/s0002-9343(03)00241-9. [DOI] [PubMed] [Google Scholar]
- Lin X, Taube R, Fujinaga K, Peterlin BM. P-TEFb containing cyclin K and Cdk9 can activate transcription via RNA. J Biol Chem. 2002;277:16873–16878. doi: 10.1074/jbc.M200117200. [DOI] [PubMed] [Google Scholar]
- Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol. 2012;13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- Malmhall C, Alawieh S, Lu Y, Sjostrand M, Bossios A, Eldh M, Radinger M. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol. 2014;133:1429–1438. 1438 e1421–1427. doi: 10.1016/j.jaci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye IS, Czieso S, Ktistaki E, Roderick K, Coomes SM, Pelly VS, Kannan Y, Perez-Lloret J, Zhao JL, Baltimore D, et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proc Natl Acad Sci U S A. 2014;111:E3081–3090. doi: 10.1073/pnas.1406322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, Richards H, Li Z, Adler D, Yoshinaga Y, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, Benham AL, Kim J, Soibam B, Harris RA, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Artis D. New paradigms in type 2 immunity. Science. 2012;337:431–435. doi: 10.1126/science.1221064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol. 2009;182:741–745. doi: 10.4049/jimmunol.182.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, et al. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- Simpson LJ, Patel S, Bhakta NR, Choy DF, Brightbill HD, Ren X, Wang Y, Pua HH, Baumjohann D, Montoya MM, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol. 2014;15:1162–1170. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K, Miskiewicz P, Pirko K, Jurecka-Lubieniecka B, Kula D, Hasse-Lazar K, Krajewski P, Bednarczuk T, Ploski R. rs3827440, a nonsynonymous single nucleotide polymorphism within GPR174 gene in X chromosome, is associated with Graves’ disease in Polish Caucasian population. Tissue Antigens. 2014;83:41–44. doi: 10.1111/tan.12259. [DOI] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li DC, Li ZF, Liu CX, Xiao YM, Zhang B, Li XD, Zhao J, Chen LP, Xing XM, et al. Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene. 2011;30:3875–3886. doi: 10.1038/onc.2011.103. [DOI] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.