Abstract
Individual differences in human temperament can increase risk for psychiatric disorders like depression and anxiety. Our laboratory utilized a rat model of temperamental differences to assess neurodevelopmental factors underlying emotional behavior differences. Rats selectively bred for low novelty exploration (Low Responders, LR) display high levels of anxiety- and depression-like behavior compared to High Novelty Responder (HR) rats. Using transcriptome profiling, the present study uncovered vast gene expression differences in the early postnatal HR versus LR limbic brain, including changes in genes involved in cellular metabolism. These data led us to hypothesize that rats prone to high (versus low) anxiety/depression-like behavior exhibit distinct patterns of brain metabolism during the first weeks of life, which may reflect disparate patterns of synaptogenesis and brain circuit development. Thus, in a second experiment we examined activity of Cytochrome C Oxidase (COX), an enzyme responsible for ATP production and a correlate of metabolic activity, to explore functional energetic differences in HR/LR early postnatal brain. We found that HR rats display higher COX activity in the amygdala and specific hippocampal subregions compared to LRs during the first 2 weeks of life. Correlational analysis examining COX levels across several brain regions and multiple early postnatal time points suggested desynchronization in the developmental timeline of the limbic HR versus LR brain during the first two postnatal weeks. These early divergent COX activity levels may reflect altered circuitry or synaptic activity in the early postnatal HR/LR brain, which could contribute to the emergence of their distinct behavioral phenotypes.
Keywords: Anxiety, Neurodevelopment, Transcriptome, Metabolism, Amygdala, Cytochrome C Oxidase
1. INTRODUCTION
Individual differences in temperament and emotional reactivity shape the ability of humans to cope with stress and predispose certain individuals to psychiatric disorders such as depression and anxiety (Cloninger et al., 2006). Our laboratory uses a rodent model of temperamental differences to study the neurobiological basis of differences in emotional behavior. Sprague Dawley rats that were selectively bred to display low behavioral response to novelty (Low Responders, LR) also exhibit high anxiety- and depression-like behavior compared to High Novelty Responder rats (HRs). These behaviors include diminished sociability and sexual motivation, increased behavioral inhibition and helplessness, as well as greater vulnerability to chronic stress (Stead et al., 2006a, Stedenfeld et al., 2011, Cummings et al., 2013, Clinton et al., 2014, Cohen et al., 2015). Furthermore, these behavioral traits emerge in early life, and a previous microarray study from our group revealed widespread gene expression differences in the early postnatal HR versus LR brain (Clinton et al., 2011). More specifically, numerous genes were altered in the developing hippocampus of HR versus LR rats during the first few weeks of life (i.e. postnatal days 7, 14 and 21), with marked changes in genes involved in synaptogenesis and cellular metabolism (Clinton et al., 2011).
While dysregulated monoamine neurotransmission has long been hypothesized to underlie the pathophysiology of depression (Hirschfeld, 2000), data also point to a possible role for metabolic dysfunction in the illness (Gardner and Boles, 2011). Numerous human neuroimaging studies have documented abnormal metabolism within several limbic brain regions of individuals suffering major depression (Fitzgerald et al., 2008, Anand et al., 2009), and studies using postmortem brain tissue from depressed patients have reported alterations in genes involved in a variety of metabolic processes (Sequeira et al., 2007, Kim and Webster, 2010, Regenold et al., 2012). Depression and other affective disorders are highly co-morbid with metabolic disorders, and the two classes of illness share many symptoms such as fatigue, cognitive deficits, and psychomotor retardation (Gardner and Boles, 2011). A recent study even showed that monoaminergic (specifically serotonin) and mitochondrial dysfunctions may be linked through monoamine-dependent regulation of phosphorylated glucocorticoid receptors and transcriptional control of mitochondrial genes for cytochrome c oxidase (Adzic et al., 2013). Whether these processes are overlapping or provide separate mechanisms of action, it is important to gain a deeper understanding of their roles in shaping emotional behavior.
The high energy demands of the brain are predominately fulfilled via oxidative phosphorylation (Hall et al., 2012), and perturbation of this pathway may contribute to metabolic abnormalities known to occur in emotional disorders. ATP is critical for myriad cellular processes in the brain (e.g., neurotransmission, intracellular signaling, calcium buffering, synaptic plasticity) (Babcock and Hille, 1998, Brodin et al., 1999, Li et al., 2004); its synthesis requires proper functioning of the electron transport chain, which consists of four enzyme complexes positioned within the inner membrane of mitochondria (Saddar et al., 2008). Cytochrome C oxidase (COX) is the terminal rate-limiting enzyme in the electron transport chain; COX's activity in the brain is directly proportional to ATP production and is commonly used to compare metabolic activity within different regions of the brain in human postmortem studies as well as studies using animal models of psychopathology (Harro et al., 2011, Wong-Riley, 2012, Rice et al., 2014).
Our previous work suggested that HR/LR differences in anxiety/depression-like behavior stem from disparate developmental trajectories in the early postnatal hippocampus and possibly amygdala (Clinton et al., 2011, Cohen et al., 2015). These early life HR/LR gene expression differences involved several functional classes of genes, including those relevant to synaptogenesis, synaptic plasticity, and cellular metabolism. The present study builds on that work, examining transcript and metabolic changes in the developing LR versus HR limbic brain that may contribute to a high anxiety/depression-like phenotype. We hypothesized that rats prone to high (versus low) levels of anxiety/depression-like behavior would exhibit distinct patterns of gene expression and metabolic activity in the early postnatal limbic brain. We first used transcriptome profiling to assess widespread gene expression in brains of early postnatal HR/LR pups (postnatal days 7, 14 and 21), focusing on key brain areas known to regulate emotional behavior: the amygdala, hippocampus and prefrontal cortex. A second experiment utilized COX histochemistry to map metabolic activity patterns in the developing HR/LR brain to test the hypothesis that developing anxiety/depression-prone LR rats exhibit altered COX activity in one or more brain regions compared to HR pups.
2. METHODS
All experiments were approved by the Committee on the Use and Care of Animals at the University of Alabama at Birmingham. This work was performed in accordance with the National Institutes of Health (USA, 2011) and National Research Council (UK, 1996) guidelines on animal research.
2.1 Animals and Tissue Collection
The animals used in this study were obtained from our in-house colony where the HR/LR bred lines were recently re-derived using a breeding strategy described previously in our original publication with the bred HR/LR rats (Stead et al., 2006a). Briefly, we screened 60 male and 60 female Sprague-Dawley rats purchased from Charles River Laboratory based on locomotor activity in a novel environment. Rats were individually placed into Noldus PhenoTyper® Boxes (45 × 45 × 60 cm clear Plexiglas chambers) equipped with video cameras and Ethovision® XT 8.0 software to monitor exploratory behavior (distance traveled) during a 30-min test session. Males and females with the highest and lowest exploration scores were bred together to generate the first generation of HR and LR lines, respectively. When progeny from each generation reached adulthood, we repeated the novelty-induced locomotion test and selected the top and bottom-scoring animals from HR and LR families, respectively, for subsequent breeding. The present experiments were conducted using tissue from HR/LR males from the 4th generation of our colony. All housing and testing facilities were maintained at 21-23°C and 50-55% humidity. For all experiments, rats were pair-housed in a 12:12 light-dark cycle (lights on/off at 6 AM/6 PM).
Adult male/female pairs were mated to produce HR and LR litters. At birth, litters were culled to six male and six female pups to control for litter size and gender composition. HR and LR male offspring were sacrificed by rapid decapitation to harvest brains at three early postnatal time points: postnatal day (P)7, P14 and P21 (n=15/phenotype/time point). Brains were removed, flash frozen in isopentane cooled to −30°C on dry ice, and then stored at −80°C until further use. A subset of brains (n=5/phenotype/time point, with each of the five rats per time point derived from an independent litter) were sectioned on a cryostat at −10 to −12°C, and alternating sections of 20 and 300 μm were collected. The 20 μm sections were stained with cresyl violet and compared to developing and adult rat brain atlases (Paxinos and Watson, 1986, Paxinos et al., 1994) to identify target anatomical regions in the 300 μm sections. Portions of the hippocampus, amygdala, and prefrontal cortex were removed from the 300 μm-thick sections using a 0.5 mm tissue punch (Harris Micro-Punch, Ted Pella, Redding, CA), stored at −20°C, and later homogenized. RNA was isolated (NucleoSpin RNA II), quantified using a Nanodrop ND-1000 (Wilmington, DE), and stored at −80°C. The remaining brains (n=10/phenotype/time point) were sectioned on a cryostat at 15 μm, mounted onto slides in parallel series of 10 slides, and stored at −80°C until processing for COX histochemistry.
2.2 Transcriptome profiling in the early postnatal HR versus LR limbic brain
RNA samples from the HR and LR P7/P14/P21 hippocampus, amygdala, and prefrontal cortex (n=5 samples/group) were shipped on dry ice for genome-wide expression profiling using NimbleGen Rat Gene Expression 12×135 Arrays (26,419 target genes, 5 probes/targets; Arraystar, Rockville, MD) as previously described (Glover et al., 2015). Briefly, RNA quality and quantity were assessed with Nanodrop ND-1000 (Thermo Scientific, Wilmington, DE) and the RNA integrity was evaluated through denaturing agarose gel electrophoresis. Total RNA from each sample was linearly amplified with Agilent's Low Input Quick Amp Kit (Agilent Technology). Double-stranded cDNA (ds-cDNA) was synthesized from the amplified cRNA using an Invitrogen SuperScript ds-cDNA synthesis kit, cleaned and labeled in accordance with the NimbleGen Gene Expression Analysis protocol (NimbleGen Systems, Inc., USA). The labeled ds-cDNA was hybridized, washed, and scanned using an Axon GenePix 4000B microarray scanner (Molecular Devices Corporation). Scanned images were aligned and analyzed using NimbleScan software (version 2.5). Expression data were normalized through quantile normalization and the Robust Multichip Average (RMA) algorithm included in the NimbleScan software. Probe and gene level files were generated after normalization.
Subsequent data analysis was performed using Agilent GeneSpring GX software (version 12.6, Santa Clara, CA) and Metacore™ Data-Mining and Pathway Analysis software (Thomas Reuters, New York, NY). Probes with raw expression values less than 50 for any sample were excluded from analysis. Data were also filtered to exclude probes identified as “predicted mRNA model”, according to the most recent RefSeq database annotations. Following experimental grouping (i.e., HR/LR at a particular time point and brain region), expressed genes were considered statistically significant and included in downstream analysis if they displayed fold change >1.5, p-value < 0.05 after multiple test correction (Bonferroni family-wise error rate [FWER]). Principle Components analysis (PCA) was also performed on data from each microarray chip.
To gain a better understanding of top molecular pathways that differed between HR and LR in each brain region, we used GO (gene ontology) analysis (WebGestalt; http://bioinfo.vanderbilt.edu/webgestalt), considering genes with a fold change > 1.2, p-value of < 0.05. The pathway enrichment analysis was performed with these less stringent parameters to capture a broader scope of regulation. In the GO analysis of the amygdala, the following number of genes were included in the analysis at each developmental age: 1277 (P21), 798 (P14), and 1286 (P7). For the hippocampus analysis, the following numbers of genes were included for each age: 949 (P21), 908 (P14), and 676 (P7). Significantly enriched terms containing at least 15% of genes in each category were identified using a Benjamini-Hochberg false discovery rate and p< 0.05. All genes discussed in this paper are referenced using the Rattus norvegicus gene symbol. The microarray expression data will be provided on gene expression omnibus (GEO) database.
2.3 COX histochemistry
The COX enzymatic assay was performed as previously described by Melendez-Ferro et al. (Melendez-Ferro et al., 2013). From each rat, slides spanning the entire forebrain at intervals of 300 μm were removed from −80°C storage and warmed to room temperature for 15 min. Tissue sections were incubated in the reaction medium (described below) at 37°C in the dark for 120 min for the P7 group; 75 min for the P14 group; and 50 min for the P21 group. We performed preliminary experiments to find the length of incubation that was optimal for brain tissue samples at each developmental age. We found that there was no time length that allowed for accurate calculation of COX activity across all three ages due to oversaturation of the stain at the older ages. The reaction medium was a solution consisting of: 0.1792 g/L cytochrome c (Sigma-Aldrich, St. Louis, MO, USA; C3131), 0.92 g/L diaminobenzidine (DAB, Sigma-Aldrich, D5637), 36 g/L sucrose (Sigma-Aldrich, S0389), 0.1% nickel ammonium sulfate solution (Sigma-Aldrich, A1827), in 0.1M Hepes buffer (Sigma-Aldrich, H3375), pH 7.4 (Melendez-Ferro et al., 2013). To terminate the COX enzymatic reaction, sections were immersed in 4% paraformaldehyde in 0.1M phosphate buffer (PB) pH 7.4 for 1 h at room temperature. Sections were dehydrated and coverslipped using standard procedures. Negative controls were performed in adjacent tissue sections by immersing them in 4% paraformaldehyde in 0.1M phosphate buffer (PB) pH 7.4 for 60 min at room temperature, followed by incubation in a 10 mM sodium azide solution in PB for 17 h at room temperature. This process was performed to irreversibly inactivate COX in negative control tissue. After the COX enzyme was inactivated in negative control samples, the sections were incubated with the experimental slides in the COX incubation medium.
To more accurately assess differences in COX activity between experimental groups and between different brain regions at each developmental age, we created a standard dot blot of known COX protein concentrations (0.03-2 μg) as described in Melendez-Ferro et al. (Melendez-Ferro et al., 2013). These standards were loaded onto a nitrocellulose membrane using vacuum and a slot-blot microfiltration apparatus (Bio-Rad, Hercules, CA, USA; 170-6542). For all COX histochemical assays, negative controls and COX standards were incubated together with the brain tissue sections to be analyzed for COX activity.
2.4 Image acquisition and analysis
Slides containing rat brain tissue processed for COX histochemistry and the dot blot membrane containing COX standards were scanned on a MicroTek ScanMaker 9800XL in 16-bit grayscale settings without corrections. NIH Image J software (version 1.48v; http://rsbweb.nih.gov/ij/) was used to measure optical density. A separate set of slides from each subject was stained with cresyl violet in order to provide anatomical guides for the COX activity measurements and to create masks defining regions of interest on the corresponding COX-reacted slides. Sections were excluded from analysis if they showed damage due to tissue processing within a region of interest. An exponential standard curve was created using OD values from the COX standards on the dot blot for each experiment: P7, P14 and P21. The calculated standard curve for each experiment fitted an exponential curve where: R2= 0.97 for standards incubated for 120 min; R2= 0.95 for 75 min; and R2= 0.98 for 50 min.
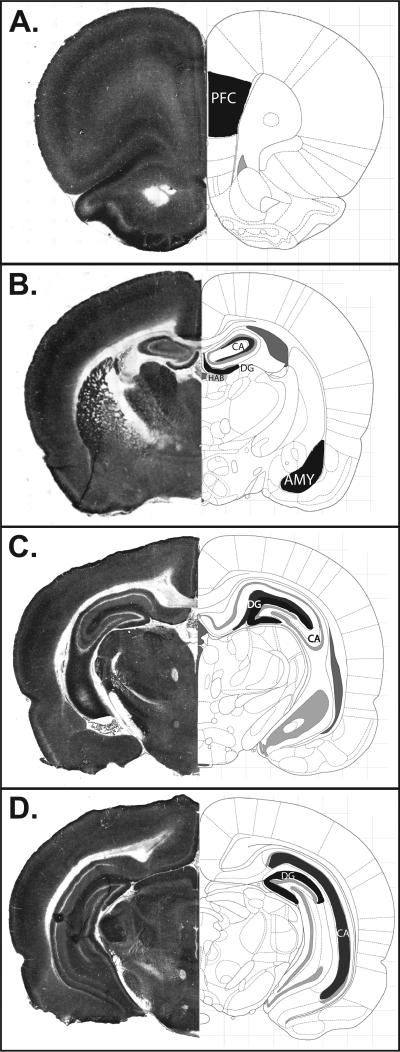
Using the COX-reacted sections, we measured optical density levels in three brain regions known to regulate emotional behavior: the amygdala, hippocampus, and prefrontal cortex. These measurements were made in at least 4 sections per rat that were spaced 300 μm apart across the rostro-caudal extent of each brain region. In the analysis of the hippocampus, we evaluated two layers of the cornu ammonis (CA) regions: the pyramidal cell layer and the innermost molecular layer, where the hippocampal neurons synapse. We chose to separately evaluate these two layers because they exhibited quite distinct patterns of COX labelling (as illustrated in Figure 3 and consistent with previous studies reporting especially high levels of COX activity and mitochondrial localization in the molecular layer, where afferent axons synapse onto principle neurons of the hippocampus (Kageyama and Wong-Riley, 1982)). For the analysis of the amygdala, we used two approaches. The first approach considered the amygdala as a whole. Because the amygdala is comprised of several functionally distinct nuclei, though, we were also interested to conduct a secondary analysis to evaluate COX levels on a nucleus-by-nucleus basis. We were able to reliably identify four sub-nuclei in the P21 amygdala: lateral, basolateral, central, and basomedial nuclei. Unfortunately, though, we were unable to reliably distinguish these nuclei in enough of our samples at P14 and P7. For each section measured, the Image J rectangle tool was used to create five non-overlapping boxes (0.002 cm square) that were randomly placed within a region of interest. Measurements from each subject from each brain region were averaged to yield a single value per animal for each brain region (or brain subregion). These values were then used for statistical comparison. Two separate blinded experimenters analyzed the tissue sections. Results were compared for consistency across experimenters and averaged together for subsequent analysis.
Figure 3. Representative Cytochrome c Oxidase (COX) staining and corresponding atlas images depicting brain regions analyzed.
Representative tissue sections processed for COX histochemistry at the level of the (A) prefrontal cortex (PFC; bregma +2.76 mm); (B) rostral hippocampus (HPC) and amygdala (AMY; bregma −2.04 mm); (C) mid-hippocampus (bregma −4.44 mm); and (D) caudal hippocampus (bregma −5.40 mm). Regions of interests are shown in black and dark grey on the atlas images, which were adapted from “The Rat Brain” (6th edition) by Paxinos and Watson (2006).
2.5 Statistical analysis
Data analysis for the microarray experiment is described above. Data from the COX histochemistry study were analyzed using GraphPad Prism Software (Version 6.0 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com). All data sets were first verified to be normally distributed. Student t-tests were used to compare HR versus LR COX activity within each brain region (or subregion) of interest. All tests were found to have equal variance.
We performed a secondary analysis of the COX data using correlation matrices to determine inter-correlations of COX activity within select brain regions of HR/LR pups across the three early postnatal time points. MATLAB software (R2015a, MathWorks, Inc., Natick, MA, USA) was used to display the results of this analysis in a colormap. Two-way ANOVA was used to compare average correlation coefficients (HR/LR phenotype and postnatal age as independent variables). In order to identify HR versus LR divergences in limbic region connectivity patterns, multidimensional scaling was implemented. Pair-wise differences were calculated between regional inter-correlations of HR versus LR at each developmental time point, and MATLAB software was used to configure these data on a 2-dimensional plot (as described in (Harro et al., 2014)).
3. RESULTS
3.1 Transcriptome profiling in the early postnatal HR/LR amygdala, hippocampus, and prefrontal cortex
Using genome-wide expression profiling, we examined HR/LR gene expression differences in the early postnatal brain, focusing on three key limbic regions (the amygdala, hippocampus, and prefrontal cortex) at three key early postnatal time points (P7, P14, and P21). We observed dramatic HR/LR gene expression differences in the early postnatal amygdala and hippocampus, with essentially no changes in the prefrontal cortex. Several hundred genes were differentially expressed in the LR versus HR P7 and P14 amygdala, with a surge in number of altered genes at P21. In contrast, the greatest numbers of HR/LR gene differences in the hippocampus occurred at P14 (Fig. 1A). Within the P7 amygdala, most of the altered genes were downregulated in HR versus LR; in the P21 amygdala and P14 hippocampus, more than two-thirds of the differentially expressed genes were upregulated in HR versus LR (Fig 1B). Based on our previous studies, we expected to find HR/LR differences in hippocampal gene expression, but these data highlighted stark HR/LR differences in the developing amygdala as well. Full lists of genes that were differentially expressed in the HR versus LR hippocampus and amygdala can be found on gene expression omnibus (GEO) database.
Figure 1. HR/LR rats exhibit widespread gene expression differences in the early postnatal brain.
(A) Number of differentially expressed genes in the HR/LR hippocampus, amygdala, and prefrontal cortex at postnatal day (P) 7, P14, and P21. (B) Number of genes up-regulated or down-regulated in the early postnatal HR/LR amygdala (left) and hippocampus (right). Filtering for these gene lists was based on p-value < 0.05 fold change > 1.5 with multiple test corrections. (C) PCA of transcriptome in developing HR and LR amygdala and hippocampus. PCA was used to assess gene expression profiles from each amygdala or hippocampal sample dissected from HR and LR pup brains collected at P7, P14, and P21. The major component of variance in both brain regions separated animals by developmental age. Top panels depict samples colored based on developmental ages P7 (pink), P14 (blue), and P21 (orange). The second major component of variance was attributable to HR/LR phenotype. Thus, bottom panels depict samples colored based on HR (green) and LR (red) phenotypes. Green and red ellipses highlight the variablity of samples from each phenotype at each developmental age.
We next used principle component analysis (PCA) to further interrogate transcriptome profiles in the early postnatal HR/LR amygdala and hippocampus. PCA is a data reduction technique that apportions the major components of variance within a dataset into a limited number of dimensions, thus permitting visualization of global similarities/differences and variance among samples. Application of PCA to our data showed that the single greatest component of variance corresponded to developmental stage of the brain (Fig. 1C). For both the amygdala and hippocampus, there was a clustering of samples at each time point (top graphs in Fig. 1C display data points in different colors based on P7, P14, or P21 developmental age). In addition to these effects of developmental age on gene expression profiles, samples also clustered differently based on animals’ HR or LR phenotype, particularly in the amygdala (bottom graphs in Fig. 1C display HR data points in green and LR data points in red). The color-coded ellipses in the lower panel illustrate the degree of variability within each phenotype at the different developmental ages in the amygdala and hippocampus. HR and LR amygdala samples clustered distinct from each other at each developmental age. There was generally more overlap between HR and LR hippocampal samples; interestingly, though, the data implicate potentially distinct developmental trajectories for the HR versus LR hippocampus. In the HR hippocampal samples, there are distinct clusters at each timepoint. In the LR hippocampal samples, however, the later developmental ages (P14 and P21) cluster fairly close together and are quite separate from P7.
To better understand the biological processes altered in the early postnatal HR/LR brain, we performed enrichment analysis. Figure 2 displays several molecular pathways within the gene ontology (GO) cellular component that were found to differ in the HR/LR amygdala (Fig. 2A) and hippocampus (Fig. 2B). Interestingly, several components of the mitochondrion were enriched for altered expression within the developing amygdala (Fig. 2A; p< 0.05 FC > 1.5, BH correction) and hippocampus (Fig. 2B; p< 0.05 FC > 1.5, BH correction). Considering these apparent HR/LR differences in mitochondrion-related genes, we next wanted to look more closely at the expression of several classes of genes related to metabolic processes in the early postnatal HR versus LR brain. We therefore examined the number of genes significantly altered within several discrete metabolic pathways (e.g., carbohydrate metabolism, glycolysis, tricarboxylic acid cycle, fatty acid metabolism, electron transport chain, and lipid metabolism) in the early postnatal HR/LR amygdala (Fig. 2C) and hippocampus (Fig. 2D). Overall, the amygdala showed more robust HR/LR differences in metabolic genes compared to the changes observed in the hippocampus. In the amygdala, an average of 25% of genes within a given metabolic pathway (range 10-60%) were differentially expressed between HR and LR across the first three postnatal weeks. P7 was the time point when the greatest number of metabolic gene changes occurred in the HR/LR amygdala, particularly within pathways related to glycolysis, tricarboxylic acid cycle, fatty acid metabolism, and amino acid metabolism where 40-60% of genes within those pathways were altered between HR and LR (Fig. 2C). In the HR/LR hippocampus, an average of 10% of genes within a given metabolic pathway (range 3-27%) were significantly altered between HR and LR hippocampus across the first three postnatal weeks. P14 stood out as a time when the greatest number of changes occurred in the HR/LR hippocampus, particularly within pathways related to the tricarboxylic acid cycle, fatty acid metabolism, the electron transport chain, and lipid metabolism (Fig. 2D).
Figure 2. Pathways related to mitochondria and cellular metabolism are altered in the early postnatal HR/LR amygdala and hippocampus.
(A-B) Top gene ontology cellular component enrichment analysis identified top terms that were altered in the early postnatal HR versus LR amygdala (A) and hippocampus (B). (C) Percentage of genes within several metabolic pathways that were differentially expressed in HR versus LR amygdala at P7, P14, and P21. (D) Percentage of genes within each metabolic pathway that were differentially expressed in HR/LR hippocampus at P7, P14, and P21. Enrichment analysis and analysis of metabolic pathways were performed on altered genes across development ages that were filtered at p < 0.05 and FC > 1.2.
3.2 COX activity in HR versus LR early postnatal brain
Our transcriptome study revealed marked HR/LR gene expression differences within several metabolism-related genes in the early postnatal hippocampus and amygdala. We therefore hypothesized that such changes in metabolism-related genes would correspond with overall cellular energy changes. COX activity measurement is commonly used to estimate cellular metabolism as it is a terminal rate-limiting enzyme is ATP production through the electron transport chain. Consequently, we examined COX activity in the brains of P7/14/21 HR/LR rat pups to assess possible functional differences in cellular metabolism. Our analysis followed the same design as the first study by examining the amygdala and hippocampus and prefrontal cortex, another region relevant to regulating emotional behavior. Figure 3 shows representative tissue sections processed for COX activity, highlighting the brain regions that were analyzed, including the prefrontal cortex, amygdala, and subregions of the hippocampus. Sublayers of the hippocampus exhibited different activity; we therefore measured COX activity in the molecular (or synaptic) layer and cellular layers separately. In preliminary studies, we did not find differences in COX activity among the different CA regions (CA1, CA2 and CA3); therefore, these regions were measured as a single region in subsequent analyses.
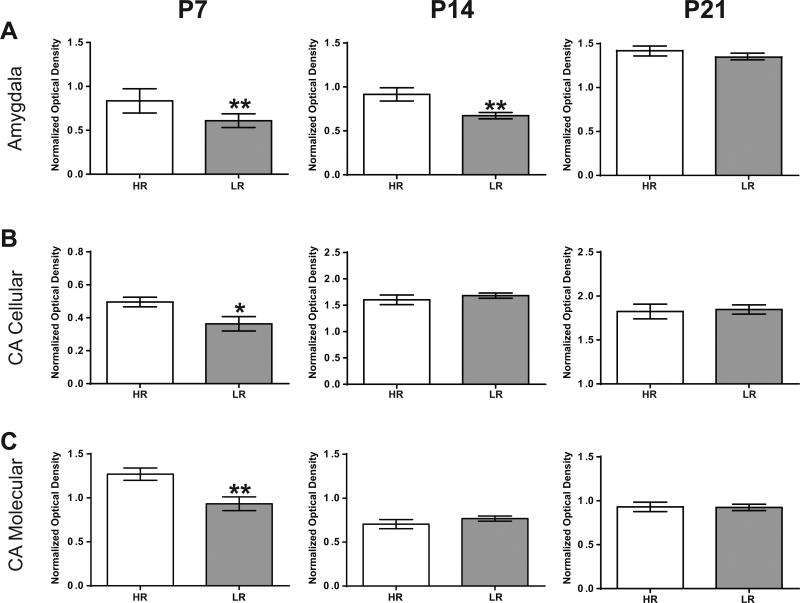
Figure 4 shows HR versus LR COX activity in the P7 (left column), P14 (middle column), and P21 (right column) amygdala (Fig. 4A), and CA region of the hippocampus (cellular and molecular layers; Fig. 4B-C). In the amygdala, HRs displayed higher COX activity compared to LRs at P7 (p= 0.0035, t= 3.70, df= 11, n= 6 HR, 7 LR). This HR/LR difference continued at P14 (p=0.0063, t=3.177, df=15, n= 7 HR, 11 LR), but was not present at P21 (Fig. 4A). In the CA region of the hippocampus, we found that HRs showed increased COX activity at P7 in both the pyramidal cell (p=0.0235, t=2.565, df=13, n= 8 HR, 7 LR; Fig. 4B) and molecular (p=0.0066, t=3.229, df=13, n=8 HR, 7 LR; Fig. 4C) layers compared to LRs. These HR/LR differences were absent at P14 and P21. In the dentate gyrus, we examined COX activity separately in the upper and lower blades. At P7, we found that HRs showed increased COX activity specifically in the lower blade/cellular layer (p=0.049, t=2.27; df=13, n=8 HR, 7 LR; Table 1), but these differences were not present at P14 and P21. We found no HR/LR differences in the upper blade of the dentate gyrus at any developmental age (Table 1). Likewise, we found that HR/LR rats showed similar COX activity in the P7/14/21 prefrontal cortex (Table 1).
Figure 4. Cytochrome c oxidase (COX) activity in the early postnatal HR versus LR limbic brain.
COX activity from tissue sections normalized to a dot blot of known COX protein amount was measured from amygdala (A), cellular and molecular layers of Cornu ammonis (CA; B-C) at three early postnatal time points: postnatal day (P) 7, 14 and 21. (A) At P7 and P14, HR pups exhibited greater COX activity in the amygdala compared to LR pups. (B-C) In the cellular and molecular layers of CA, HR showed increased COX activity at P7. ** indicates p-value < 0.01, * indicates p-value < 0.05.
Table 1.
Measurements of Cytochrome C oxidase activity in select brain regions of early
| Brain region | Postnatal day(P) | HR | LR | student's t-test statistics |
|---|---|---|---|---|
| PFC | P7 | 0.54 ± 0.04, n=7 | 0.57 ± 0.067, n=7 | p=0.76 t=0.31 |
| Upper blade-cellular | 0.86 ± 0.04, n=5 | 0.82 ± 0.07, n=6 | p=0.67 t=0.45 | |
| Upper blade-molecular | 0.42 ± 0.03, n=5 | 0.34 ± 0.04, n=6 | p=0.12 t=1.67 | |
| Lower blade-cellular | 0.37 ± 0.03, n=5 | 0.28 ± 0.03, n=6 | *p= 0.05 t=2.27 | |
| Lower blade-molecular | 0.69 ± 0.05, n=5 | 0.55 ± 0.05, n=6 | p= 0.06 t=2.00 | |
| PFC | P14 | 1.11 ± 0.08, n=7 | 1.18 ± 0.04, n=7 | p=0.47 t=0.74 |
| Upper blade-cellular | 1.43 ± 0.07, n=10 | 1.51 ± 0.05, n=10 | p=0.35 t=0.95 | |
| Upper blade-molecular | 0.71 ± 0.05, n=10 | 0.76 ± 0.03, n=10 | p=0.40 t=0.86 | |
| Lower blade-cellular | 1.16 ± 0.06, n=10 | 1.23 ± 0.03, n=10 | p= 0.31 t=1.05 | |
| Lower blade-molecular | 0.62 ± 0.05, n=10 | 0.67 ± 0.02, n=10 | p= 0.32 t=1.05 | |
| PFC | P21 | 1.61 ± 0.05, n=8 | 1.48 ± 0.03, n=6 | p=0.08 t=1.94 |
| Upper blade-cellular | 1.80 ± 0.08, n=9 | 1.77 ± 0.05, n=9 | p= 0.734 t=0.34 | |
| Upper blade-molecular | 0.91 ± 0.05, n=9 | 0.90 ± 0.03, n=9 | p= 0.78 t=0.29 | |
| Lower blade-cellular | 1.66 ± 0.07, n=10 | 1.63 ± 0.04, n=9 | p=0.74 t= 0.33 | |
| Lower blade-molecular | 0.85 ± 0.05, n=9 | 0.85 ± 0.03, n=9 | p=0.97 t= 0.04 | |
| Lateral Amygdala | 1.38 ± 0.05, n=6 | 1.23 ± 0.03, n=9 | *p=0.03 t= 0.02 | |
| Basolateral Amygdala | 1.59 ± 0.067, n=6 | 1.40 ± 0.05, n=9 | *p=0.04 t=2.28 | |
| Central Amygdala | 1.45 ± 0.07, n=6 | 1.29 ± 0.05, n=9 | p=0.08 t=1.91 | |
| Basomedial Amygdala | 1.47 ± 0.08, n=6 | 1.27 ± 0.05, n=9 | *p=0.04 t=2.23 |
postnatal HR/LR rat brains.
Values represent average COX activity in normalized optical density ± standard errors for each experimental group: HR and LR at postnatal days (P) 7, 14, 21. Results of statistical analyses are shown with p-values and t-test statistics. Bolded statistics have a p-value < 0.05. Abbreviations: Prefrontal Cortex (PFC); dentate gyrus (DG).
3.3 Inter-correlations of limbic region COX activity in the early postnatal HR versus LR brain
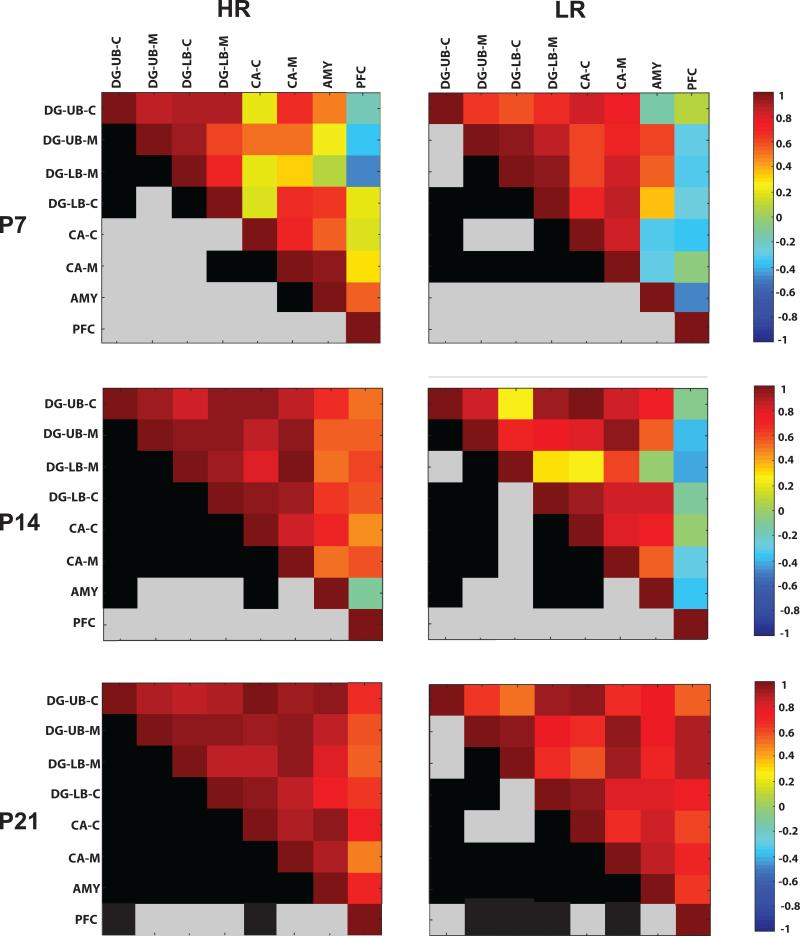
We extended our analysis of the COX activity data using a correlation matrices as described in Harro et al, 2014 in order to examine activity across brain regions at P7/14/21, which can, in turn, implicate connectivity across brain areas. Figure 5 shows colormaps of COX activity correlations among select limbic regions in HRs (left column) versus LRs (right column) at the three developmental time points. The top half of each square depicts a colormap showing how COX activity within select brain regions (i.e., sublayers of the dentate gyrus, CA region of the hippocampus, amygdala, or prefrontal cortex) correlate with the other measures. The mirrored bottom half of each square is color-coded in black and gray. A black square indicates that the correlation for that measure was statistically significant (p<0.05) whereas a gray square indicates that the correlation was not significant.
Figure 5. Inter-correlations of COX activity within limbic brain regions of HR (left) and LR (right) at postnatal day (P) 7, 14 and 21 (shown top to bottom).
The top half of each square depicts a colormap showing how COX activity within select brain regions (i.e., sublayers of the dentate gyrus, CA region of the hippocampus, amygdala, or prefrontal cortex) correlate with the other measures. The mirrored bottom half of each square is color-coded in black and gray. A black square indicates that the correlation for that measure was statistically significant (p<0.05) whereas a gray square indicates that the correlation was not significant. Abbreviations: DG-UB-C, dentate gyrus upper blade cellular layer; DG-UB-M. dentate gyrus upper blade molecular layer; DG-LB-M, dentate gyrus lower blade molecular layer; DG-LB-C, dentate gyrus lower blade cellular layer; CA-C, cornu ammonis cellular layer; CA-M, cornu ammonis molecular layer; AMY, amygdala; PFC, prefrontal cortex.
We compared the average correlation coefficients in HR/LR groups across the three developmental ages using two-way ANOVA. This analysis revealed a main effect of developmental age (F(2,210)=30.60, p<0.0001) since regardless of HR/LR phenotype, inter-correlations of limbic region COX activity strengthened as development progressed; there was less correlation among brain regions at the earliest developmental stage (P7; average correlation coefficient 0.49±0.03), increased correlation at P14 (average correlation coefficient 0.65±0.03), and the greatest inter-region correlation at P21 (average correlation coefficient 0.78±0.02; post-hoc comparison p<0.05 for each of the ages compared to one another). Although there was no main effect of HR/LR phenotype, there was a significant developmental age × phenotype interaction (F(2,210)=5.42, p<0.01). Post-hoc analysis showed that HR/LR groups exhibited similar average correlation coefficients at P7 (HR: 0.45±0.05; LR 0.54±0.04). At P14, HRs exhibited a higher average correlation coefficient compared to LRs (HR: 0.73±0.03; LR: 0.58±0.04; p<0.05). At P21, the groups did not significantly differ (HR: 0.80±0.02; LR: 0.77±0.02). These data suggest that stronger associations among these limbic regions are established earlier within the HR versus LR developing brain (Fig. 5).
3.4 Multidimensional scaling to visualize HR/LR COX activity differences in the P7/14/21 brain
Following calculation of region specific pair-wise differences between correlation matrices of HR/LR brain regions at P7/14/21, multidimensional scaling was used to express the data on a 2-dimensional plot where the distance between regions represented the degree of similarity of their metabolic demands (Fig. 6). This qualitative analysis, similar to PCA analysis used in the first study, highlighted the variance in COX activity within specific brain regions and developmental ages. The highest degree of HR/LR inter-regional divergence occurred at P7, where the prefrontal cortex, CA molecular layer, and amygdala exhibited the lowest levels of COX activity association with other limbic regions. Additionally, at P7, the cellular layer of CA and the cellular layer of the dentate gyrus showed less divergence in COX activity association in HR versus LR. Dissimilarity between regional inter-correlations in the brains of HR/LR rats diminished at later developmental time points. At P14, HR/LR COX activity inter-correlation differences were apparent at only a few regions: the molecular layer of the lower blade of the dentate gyrus, amygdala, and prefrontal cortex. At P21, all brain regions examined clustered, indicating similar associations in inter-regional metabolic demands in HR/LR animals (Fig. 6).
Figure 6. Multidimensional scaling analysis of HR/LR COX activity differences in the first weeks of life.
Following calculation of region specific pair-wise differences between correlation matrices of HR/LR brain regions at P7/14/ 21, multidimensional scaling expresses the data on a 2-dimensional plot where the distance between two given brain regions represents their association strength. Abbreviations: DG-UB-C, dentate gyrus upper blade cellular layer; DG-UB-M. dentate gyrus upper blade molecular layer; DG-LB-M, dentate gyrus lower blade molecular layer; DG-LB-C, dentate gyrus lower blade cellular layer; CA-C, cornu ammonis cellular layer; CA-M, cornu ammonis molecular layer; AMY, amygdala; PFC, prefrontal cortex.
4. DISCUSSION
To elucidate neural and molecular changes in the developing brain that may contribute to an individual developing a highly anxious/depressive-like phenotype, the present study utilized a rat model of individual differences in temperament. As noted earlier, LR rats selectively-bred for low behavioral response to novelty also display high levels of anxiety- and depression-like behaviors from early life through adulthood relative to high novelty responding HR rats (Clinton et al., 2011, Clinton et al., 2014). The current study used transcriptome profiling to examine widespread gene expression in the early postnatal limbic brains of LR vs. HR rats, focusing on the hippocampus, amygdala, and prefrontal cortex – 3 key brain regions that regulate emotional behaviors known to differ in HR/LR rats. These experiments asked: (1) when do HR/LR brains begin to develop differently; and (2) what brain regions vary the most in developing HR/LR rats. The most prominent differences occurred in the developing hippocampus and amygdala, with hundreds of genes differing at P7 and P14. By P21, the amygdalar gene expression differences dramatically increased (>1000 genes altered) whereas hippocampal changes dropped to zero. There were surprisingly few changes in the prefrontal cortex at any age, with no genes remaining after multiple test corrections. Among the many interesting functional classes of genes that differed in the early postnatal HR vs. LR brain were molecules critical for metabolism (Fig. 6), synaptogenesis, and neuroplasticity (data not shown). Thus, a follow-up experiment examined COX activity in the developing HR/LR brain to explore functional energetic differences that may correlate to HR/LR differences in the development of hippocampal-amygdala circuits.
4.1 Neurodevelopment in the amygdala, hippocampus, and prefrontal cortex and how perturbations of these regions impacts emotional behavior
The rodent amygdala, hippocampus and prefrontal cortex each undergo unique patterns of neurodevelopment during late gestation and the early postnatal period. Peak neurogenesis in the amygdala occurs at embryonic days (E)11-16 (Berger et al., 2002) and neuronal migration continues through P7 when subnuclear organization begins (Berdel et al., 1997). Cell number and density increases during the second postnatal week (stabilizing around P14), and amygdalar volume continues to increase during the third postnatal week until stabilizing at P28 (Berdel et al., 1997, Chareyron et al., 2012). This is accompanied by dendritic expansion and increased synaptic number from P7-P28 (Escobar and Salas, 1993, Morys et al., 1998, Ryan et al., 2014) . These changes reflect increased connectivity within amygdalar subregions and increased inputs to the amygdala from areas like the prefrontal cortex (Bouwmeester et al., 2002). Our transcriptome study identified marked HR/LR gene expression changes in the amygdala at P7, P14, and P21. Although previous work from our group and others have suggested that the distinct HR/LR behavioral phenotypes stem from differences in the developing (Clinton et al., 2011) and adult (Lemaire et al., 1999, Rosario and Abercrombie, 1999, Kabbaj et al., 2000, Isgor et al., 2004) hippocampus, our present data indicate a role for the amygdala as well
A number of studies by Sullivan et. al. and others have elegantly described the ontogeny of amygdalar circuits, amygdala-regulated fear and stress responses, and how perturbations of the developing rodent amygdala elicit lasting changes in emotional behavior (Thompson et al., 2008, Landers and Sullivan, 2012, Tallot et al., 2016). For example, excitotoxic lesions of the P7 rat amygdala impairs juvenile play and adult social behavior, increases novelty-induced ambulation (Wolterink et al., 2001, Daenen et al., 2002), and exacerbates amphetamine-induced locomotion (Solis et al., 2009). Lasting neural consequences of P7 amygdala lesions include altered dopamine receptor density in the adult nucleus accumbens, olfactory tubercle, substantia nigra and central grey (Bouwmeester et al., 2007), as well as decreased cerebral glucose utilization in several limbic brain regions, including the amygdala itself, the cingulate cortex, lateral septum, and anterior hippocampus (Gerrits et al., 2006). It is interesting to consider whether HR/LR differences in amygdala development contribute to their disparate biobehavioral phenotypes. Although our findings in the HR/LR model do not perfectly align with the P7 amygdala lesion literature, HR/LR rats show divergent social behavior, as well as novelty- and psychostimulant-induced locomotion (Stead et al., 2006a, Clinton et al., 2012, Cohen et al., 2015). They also exhibit a number of dopamine system anomalies, including altered expression of dopamine receptor transcripts in the nucleus accumbens and striatum, and greater number of spontaneous dopamine release events in the nucleus accumbens (for review, see (Flagel et al., 2014)). New studies should examine anatomical and functional differences in the developing HR/LR amygdala that may contribute to their disparate behavioral phenotypes. Future experiments should also examine the impact of manipulating the amygdala of HR/LR rats during early postnatal development to determine its effects on their adult phenotypes.
Within the hippocampus, the pyramidal neurons of Ammon's horn (fields CA1-3) are generated between E16- E20. The volume of the CA region rapidly expands from E21 through P1 (Bayer, 1980a, b), with its dendritic system rapidly expanding throughout the first three postnatal weeks (Pokorny and Trojan, 1986). In the dentate gyrus, a fraction of granule cells are generated around E16-17, but the majority (85%) of the cells are generated from P0-19 and migrate from P10-P25. Although this initial phase of neurogenesis in the dentate gyrus tapers around P21, it persists at low levels through adulthood (Bayer, 1980a). Our current transcriptome study as well as a prior microarray experiment (Clinton et al., 2011) revealed dramatic HR/LR gene expression differences in the hippocampus at P7 and P14, with minimal changes at P21. These data suggest, then, that there is a critical developmental window when hippocampal circuits may diverge in HR/LR rats, which may, in turn, contribute to life-long differences in behavior and hypothalamic pituitary adrenal axis stress reactivity. Indeed, the hippocampus has the potential to contribute to a number of well-established HR/LR behavioral differences given its contributions to detecting novelty (Roullet and Lassalle, 1990, Lever et al., 2006, Jeewajee et al., 2008), regulating neural stress circuits (Sapolsky et al., 1984, Jacobson and Sapolsky, 1991), and controlling fear and anxiety-like behavior (Gray, 1982, McNaughton and Gray, 2000, Bannerman et al., 2002).
The prefrontal cortex, like other neocortical and limbic cortical regions, undergoes peak neurogenesis between E14-20 (Bayer and Altman, 1991). Most of these newborn cells migrate within the first postnatal week, although cytoarchitectual organization in the PFC continues through P18 (van Eden et al., 1990). We were somewhat surprised to find no transcriptome or COX HR/LR differences in early postnatal prefrontal cortex, since some of the known HR/LR behavioral differences (such as impulsivity (Flagel et al., 2009)) may be related to divergent prefrontal cortical function. It is possible that the neurobiological factors contributing to HR/LR prefrontal cortex-associated behavioral differences may emerge later in life (i.e. post-weaning/puberty), which may explain why we saw minimal differential prefrontal cortex gene expression from P7-P21.
4.2 Examining COX activity as an indicator of metabolic activity in the early postnatal HR/LR brain
Our transcriptome analysis pointed to significant alterations of mitochondrial and metabolism-related genes in HR/LR developing amygdala and hippocampus. While genes from a number of metabolism pathways were altered in HR versus LR brain, we choose to focus on COX activity in our follow-up experiment because it serves as the terminal rate-limiting enzyme in ATP production and is thought to correlate with total neuronal activity (Harro et al., 2011). For example, early work by Wong-Riley et. al. began to describe COX localization in the brain and its relationship to synaptic activity (Wong-Riley, 1979, Wong-Riley, 1989). Among this early work was a study describing COX localization in the hippocampus, showing that the highest levels corresponded to synaptic terminal fields of the hippocampus (stratum moleculare of CA1-3 regions and outer molecular layer of the dentate gyrus); they also reported differential COX staining of distinct hippocampal cell populations, which corresponded with spontaneous firing rates of those neurons (Kageyama and Wong-Riley, 1982). Other studies mapped COX distribution throughout the adult rat brain, demonstrating the significant heterogeneity across several cortical and subcortical brain regions, white versus gray matter, and neuron types (e.g.,(Hevner et al., 1995)). In addition, several research groups have assessed COX activity in the early postnatal rat brain (Bilger and Nehlig, 1991, Diaz et al., 1994, Gonzalez-Pardo et al., 1996, Nair et al., 1999, Liu and Wong-Riley, 2003). Not surprisingly, the developmental patterns of COX activity vary across different brain regions. For instance, white matter tracts in the hippocampus showed relatively low levels of COX activity at P7, with steadily increasing levels at P12 and P17 before dramatically tapering at P30 through adulthood. Other white matter pathways exhibited different developmental patterns, with COX levels peaking at P7 in cerebellar white matter and declining with age, but the corpus callosum and anterior commissure showing peak COX levels at P17 and remaining fairly high through adulthood (Nair et al., 1999).
We utilized a COX histochemistry experiment to map metabolic activity in the early postnatal HR versus LR brain, focusing on the amygdala, hippocampus and prefrontal cortex since those regions were studied in the microarray experiment. We found significantly decreased COX activity in the amygdala and several hippocampal subregions of LR versus HR pups at P7. Some of the HR/LR COX activity differences (particularly in the amygdala) persisted at P14, but were not statistically significant at P21, at least when we considered the amygdala as a whole. Interestingly, when we conducted a secondary analysis of the P21 amygdala on a nucleus-by-nucleus basis, three of the four nuclei that were examined (lateral, basolateral, and basomedial nuclei) showed significantly reduced COX levels in LR versus HR. (Mody et al., 2001, Stead et al., 2006b, Liscovitch and Chechik, 2013, Thompson et al., 2014)(Flagel et al., 2009)
Although prior studies have examined COX activity in brain tissue from rodent models of psychopathology, this previous work mainly focused on adult subjects (Gallo et al., 2002, Shumake and Gonzalez-Lima, 2003, Harro et al., 2011, Sampedro-Piquero et al., 2013, Harro et al., 2014) (for review see (Harro et al., 2011)). For example, one study mapped COX activity in adult brain tissue within five distinct rodent models of depression, such as exposure to environmental adversity via chronic variable stress or chronic social defeat (Harro et al., 2014). These authors found that vulnerability to a depression-like phenotype was generally associated with higher COX levels in the brain, particularly within the reticular thalamus, hippocampal CA3 region, granular and agranular retrosplenial cortices, and pontine nuclei. Conversely, stress exposure tended to decrease region-wide oxidative metabolism relative to controls, with the notable exception of the median raphe (Harro et. al., 2014). Another study using a congenitally helpless rat model identified elevated levels of COX activity within several hippocampal regions and the paraventricular hypothalamic nucleus in depression-prone animals (Shumake et al., 2001, 2002).
Our current findings suggest that these previously identified COX activity differences in brains of adult animals known to display high levels of depression-like behavior may be the result of divergent establishment of neuronal circuitry during development. We found that anxiety/depression-prone LR rats displayed reduced COX activity in the amygdala and some subregions of the hippocampus at P7 compared to their HR counterparts, with progressively fewer HR/LR differences at the later time points (P14 and P21). Our inter-correlational analysis of COX activity across the multiple brain regions illustrated how inter-correlation of COX activity between brain regions strengthened over time (with relatively less correlation between brain regions at P7, but progressively more at P14 and P21). Interestingly, HR/LR animals displayed divergent patterns of inter-regional correlation, particularly at P7 and to a lesser extent at P14. These data coupled with our current as well as previous microarray findings (Clinton et al., 2011) suggest that HR/LR limbic circuits develop at different rates, although additional work will be required to more fully explore this notion.
Our results are largely consistent with a previous study comparing COX activity in the brains of one-day-old congenitally helpless versus non-helpless rats. They found decreased COX activity in several newborn brain regions, including the paraventricular nucleus of the hypothalamus, hippocampus, and multiple frontal cortical regions of helpless versus non-helpless rats (Shumake et al., 2004). A striking observation from this previous report was that brain regions that were most underactive in helpless versus non-helpless rats at birth became the most hyperactive in adulthood. The authors hypothesized that this reversal may result from an abnormal flip of the hypothalamic-pituitary-adrenal axis during development of the rodent depression model (Shumake et al., 2004). From P4-14, rodents are known to exhibit a stress hypo-responsive period (SHRP) that is characterized by a reduced capacity to secrete corticosterone, which protects the young brain from elevated glucocorticoids (Levine, 2001). It is possible that rats prone to develop a depressive-like phenotype may exhibit differences in the ontogeny of limbic and stress circuits, and such differences could be manifested as differences in COX activity. Results from our study are consistent with this notion, since we found the greatest HR/LR COX activity differences at P7, a lower number of significant differences persisting at P14, and no apparent differences by P21. In the future, it would be interesting to expand the brain regions examined in our studies to determine whether HR/LR pups display COX activity differences in additional brain regions (as reported in helpless versus non-helpless rats). Moreover, it would be interesting to determine whether adult HR/LR animals also exhibit a reversal in COX activity differences, akin to what was found in the congenitally helpless rats.
4.3 Considering the role of environmental factors that may shape the developing transcriptome and limbic circuit neurodevelopment
Brain and behavioral abnormalities related to an anxiety/depression-like phenotype may be triggered by innate biological differences, environmental factors, or a combination of both (Roth and Sweatt, 2011, Doherty et al., 2016). As noted above, several previous studies as well as our present work show COX activity differences in brains of rats either a) selectively bred to display depression-like behavioral differences, or b) chronically stressed to elicit a depression-like phenotype (Harro et al., 2011, Harro et al., 2014). The HR/LR differences in COX activity reported here for the amygdala and hippocampal subregions may be related to naturally occurring differences in HR versus LR brain development. However, it is also important to consider whether these COX activity differences could also be influenced by early life factors like maternal care, given the critical role of maternal care in shaping the developing brain (Meaney, 2001, Bredy et al., 2003, Champagne et al., 2003, Weaver et al., 2006), and considering the fact that we previously reported significant differences in maternal style between HR and LR dams (Clinton et al., 2007, Clinton et al., 2010). Many aspects of the HR/LR phenotype appear to be strongly driven by genetic differences (Stead et al., 2006a, Flagel et al., 2014), although recent cross-fostering experiments showed that fostering LR pups to HR mothers improves aspects of their anxiety-like behavior, particularly social interaction (Cohen et al., 2015). This recent study also found that cross-fostering specifically shifts gene expression in the developing LR amygdala, while there was only minimal effect in the hippocampus (Cohen et al., 2015). Future experiments will combine cross-fostering with COX histochemistry to test whether brain metabolism in LR pups can be normalized to resemble more closely HR patterns by exposure to a different maternal style.
4.4 Conclusions
Our work-to-date highlights the role for the early postnatal hippocampus and amygdala in driving individual differences in depression- and anxiety-like behavior. Our prior and current findings are consonant with a growing body of literature suggesting that the roots of many mental illnesses like depression involve aberrant brain development. Future therapeutic strategies may be highly effective if applied during early life (i.e., while the brain is highly plastic) and before mental illness takes hold. Such therapeutic advances will require more experiments using animal models of emotional dysfunction (such as the HR/LR model and other similar models) to help shed light on neurobiological processes that give rise to emotional disorders in humans.
HIGHLIGHTS.
Rats prone to high anxiety display distinct transcriptome profiles in limbic brain.
Cellular metabolism genes are altered in early postnatal anxiety-prone rat brain.
COX activity is reduced in amygdala and hippocampus of week old high-anxiety pups.
Metabolic changes may reflect abnormal circuit development in anxiety-prone brain.
ACKNOWLEDGMENTS
The authors would like to thank Dr. Matthew W. Rice for technical guidance with the COX histochemistry studies. The study was funded by NIH 4R00MH085859-02 (SMC), NIH R01MH105447-01 (SMC), and 5T90DE022736-04 (covering CRM).
Abbreviations
- LR
Low Novelty Responder rat
- HR
High Novelty Responder rat
- AMY
Amygdala
- HPC
Hippocampus
- PFC
Prefrontal Cortex
- CA
Cornu Ammonis
- DG
Dentate Gyrus
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- P
Postnatal day
- COX
Cytochrome C Oxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
REFERENCES
- Adzic M, Lukic I, Mitic M, Djordjevic J, Elakovic I, Djordjevic A, Krstic-Demonacos M, Matic G, Radojcic M. Brain region- and sex-specific modulation of mitochondrial glucocorticoid receptor phosphorylation in fluoxetine treated stressed rats: effects on energy metabolism. Psychoneuroendocrinology. 2013;38:2914–2924. doi: 10.1016/j.psyneuen.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Anand A, Li Y, Wang Y, Lowe MJ, Dzemidzic M. Resting state corticolimbic connectivity abnormalities in unmedicated bipolar disorder and unipolar depression. Psychiatry Res. 2009;171:189–198. doi: 10.1016/j.pscychresns.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock DF, Hille B. Mitochondrial oversight of cellular Ca2+ signaling. Curr Opin Neurobiol. 1998;8:398–404. doi: 10.1016/s0959-4388(98)80067-6. [DOI] [PubMed] [Google Scholar]
- Bannerman DM, Deacon RM, Offen S, Friswell J, Grubb M, Rawlins JN. Double dissociation of function within the hippocampus: spatial memory and hyponeophagia. Behav Neurosci. 2002;116:884–901. doi: 10.1037//0735-7044.116.5.884. [DOI] [PubMed] [Google Scholar]
- Bayer SA. Development of the hippocampal region in the rat. I. Neurogenesis examined with 3H-thymidine autoradiography. J Comp Neurol. 1980a;190:87–114. doi: 10.1002/cne.901900107. [DOI] [PubMed] [Google Scholar]
- Bayer SA. Development of the hippocampal region in the rat. II. Morphogenesis during embryonic and early postnatal life. J Comp Neurol. 1980b;190:115–134. doi: 10.1002/cne.901900108. [DOI] [PubMed] [Google Scholar]
- Bayer SA, Altman J. Neocortical development. Raven Press; New York: 1991. [Google Scholar]
- Berdel B, Morys J, Maciejewska B. Neuronal changes in the basolateral complex during development of the amygdala of the rat. Int J Dev Neurosci. 1997;15:755–765. doi: 10.1016/s0736-5748(97)00022-1. [DOI] [PubMed] [Google Scholar]
- Berger MA, Barros VG, Sarchi MI, Tarazi FI, Antonelli MC. Long-term effects of prenatal stress on dopamine and glutamate receptors in adult rat brain. Neurochem Res. 2002;27:1525–1533. doi: 10.1023/a:1021656607278. [DOI] [PubMed] [Google Scholar]
- Bilger A, Nehlig A. Quantitative histochemical changes in enzymes involved in energy metabolism in the rat brain during postnatal development. I. Cytochrome oxidase and lactate dehydrogenase. Int J Dev Neurosci. 1991;9:545–553. doi: 10.1016/0736-5748(91)90015-e. [DOI] [PubMed] [Google Scholar]
- Bouwmeester H, Gerrits MA, Roozemond JG, Snapper J, Ronken E, Kruse CG, Westenberg HG, van Ree JM. Neonatal basolateral amygdala lesions affect monoamine and cannabinoid brain systems in adult rats. Int J Neuropsychopharmacol. 2007;10:727–739. doi: 10.1017/S1461145706007346. [DOI] [PubMed] [Google Scholar]
- Bouwmeester H, Smits K, Van Ree JM. Neonatal development of projections to the basolateral amygdala from prefrontal and thalamic structures in rat. J Comp Neurol. 2002;450:241–255. doi: 10.1002/cne.10321. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Grant RJ, Champagne DL, Meaney MJ. Maternal care influences neuronal survival in the hippocampus of the rat. European Journal of Neuroscience. 2003;18:2903–2909. doi: 10.1111/j.1460-9568.2003.02965.x. [DOI] [PubMed] [Google Scholar]
- Brodin L, Bakeeva L, Shupliakov O. Presynaptic mitochondria and the temporal pattern of neurotransmitter release. Philos Trans R Soc Lond B Biol Sci. 1999;354:365–372. doi: 10.1098/rstb.1999.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne FA, Francis DD, Mar A, Meaney MJ. Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiology and Behavior. 2003;79:359–371. doi: 10.1016/s0031-9384(03)00149-5. [DOI] [PubMed] [Google Scholar]
- Chareyron LJ, Lavenex PB, Lavenex P. Postnatal development of the amygdala: A stereological study in rats. Journal of Comparative Neurology. 2012;520:3745–3763. doi: 10.1002/cne.23132. [DOI] [PubMed] [Google Scholar]
- Clinton SM, Bedrosian TA, Abraham AD, Watson SJ, Akil H. Neural and environmental factors impacting maternal behavior differences in high- versus low-novelty-seeking rats. Horm Behav. 2010;57:463–473. doi: 10.1016/j.yhbeh.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Stead JD, Miller S, Watson SJ, Akil H. Developmental underpinnings of differences in rodent novelty-seeking and emotional reactivity. Eur J Neurosci. 2011;34:994–1005. doi: 10.1111/j.1460-9568.2011.07811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Turner CA, Flagel SB, Simpson DN, Watson SJ, Akil H. Neonatal fibroblast growth factor treatment enhances cocaine sensitization. Pharmacol Biochem Behav. 2012 doi: 10.1016/j.pbb.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Vazquez DM, Kabbaj M, Kabbaj MH, Watson SJ, Akil H. Individual differences in novelty-seeking and emotional reactivity correlate with variation in maternal behavior. Horm Behav. 2007;51:655–664. doi: 10.1016/j.yhbeh.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Watson SJ, Akil H. High novelty-seeking rats are resilient to negative physiological effects of the early life stress. Stress. 2014;17:97–107. doi: 10.3109/10253890.2013.850670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloninger CR, Svrakic DM, Przybeck TR. Can personality assessment predict future depression? A twelve-month follow-up of 631 subjects. J Affect Disord. 2006;92:35–44. doi: 10.1016/j.jad.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Cohen JL, Glover ME, Pugh PC, Fant AD, Simmons RK, Akil H, Kerman IA, Clinton SM. Maternal Style Selectively Shapes Amygdalar Development and Social Behavior in Rats Genetically Prone to High Anxiety. Dev Neurosci. 2015 doi: 10.1159/000374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JA, Clinton SM, Perry AN, Akil H, Becker JB. Male rats that differ in novelty exploration demonstrate distinct patterns of sexual behavior. Behav Neurosci. 2013;127:47–58. doi: 10.1037/a0031528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daenen EW, Wolterink G, Gerrits MA, Van Ree JM. The effects of neonatal lesions in the amygdala or ventral hippocampus on social behaviour later in life. Behav Brain Res. 2002;136:571–582. doi: 10.1016/s0166-4328(02)00223-1. [DOI] [PubMed] [Google Scholar]
- Diaz F, Villena A, Requena V, Perez de Vargas I. Cytochrome oxidase activity in the lateral geniculate nucleus of postnatal rats. Brain Res Bull. 1994;35:269–271. doi: 10.1016/0361-9230(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Doherty TS, Forster A, Roth TL. Global and gene-specific DNA methylation alterations in the adolescent amygdala and hippocampus in an animal model of caregiver maltreatment. Behav Brain Res. 2016;298:55–61. doi: 10.1016/j.bbr.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar C, Salas M. Neonatal undernutrition and amygdaloid nuclear complex development: an experimental study in the rat. Exp Neurol. 1993;122:311–318. doi: 10.1006/exnr.1993.1130. [DOI] [PubMed] [Google Scholar]
- Fitzgerald PB, Laird AR, Maller J, Daskalakis ZJ. A meta-analytic study of changes in brain activation in depression. Hum Brain Mapp. 2008;29:683–695. doi: 10.1002/hbm.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel SB, Akil H, Robinson TE. Individual differences in the attribution of incentive salience to reward-related cues: Implications for addiction. Neuropharmacology 56 Suppl. 2009;1:139–148. doi: 10.1016/j.neuropharm.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel SB, Waselus M, Clinton SM, Watson SJ, Akil H. Antecedents and consequences of drug abuse in rats selectively bred for high and low response to novelty. Neuropharmacology. 2014;76(Pt B):425–436. doi: 10.1016/j.neuropharm.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo A, Gonzalez-Lima F, Sadile AG. Impaired metabolic capacity in the perirhinal and posterior parietal cortex lead to dissociation between attentional, motivational and spatial components of exploration in the Naples High-Excitability rat. Behav Brain Res. 2002;130:133–140. doi: 10.1016/s0166-4328(01)00427-2. [DOI] [PubMed] [Google Scholar]
- Gardner A, Boles RG. Beyond the serotonin hypothesis: mitochondria, inflammation and neurodegeneration in major depression and affective spectrum disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:730–743. doi: 10.1016/j.pnpbp.2010.07.030. [DOI] [PubMed] [Google Scholar]
- Gerrits MA, Wolterink G, van Ree JM. Cerebral metabolic consequences in the adult brain after neonatal excitotoxic lesions of the amygdala in rats. Eur Neuropsychopharmacol. 2006;16:358–365. doi: 10.1016/j.euroneuro.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Glover ME, Pugh PC, Jackson NL, Cohen JL, Fant AD, Akil H, Clinton SM. Early-life exposure to the SSRI paroxetine exacerbates depression-like behavior in anxiety/depression-prone rats. Neuroscience. 2015;284:775–797. doi: 10.1016/j.neuroscience.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pardo H, Novelli A, Menendez-Patterson A, Arias JL. The development of oxidative metabolism in diencephalic structures of the rat: a quantitative study. Brain Res Bull. 1996;41:31–38. doi: 10.1016/0361-9230(96)00007-x. [DOI] [PubMed] [Google Scholar]
- Gray JA. The neuropsychology of anxiety: An enquiry into the functions of the septohippocampal system. Oxford University Press; Oxford, UK: 1982. [Google Scholar]
- Hall CN, Klein-Flugge MC, Howarth C, Attwell D. Oxidative phosphorylation, not glycolysis, powers presynaptic and postsynaptic mechanisms underlying brain information processing. J Neurosci. 2012;32:8940–8951. doi: 10.1523/JNEUROSCI.0026-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harro J, Kanarik M, Kaart T, Matrov D, Koiv K, Mallo T, Del Rio J, Tordera RM, Ramirez MJ. Revealing the cerebral regions and networks mediating vulnerability to depression: oxidative metabolism mapping of rat brain. Behav Brain Res. 2014;267:83–94. doi: 10.1016/j.bbr.2014.03.019. [DOI] [PubMed] [Google Scholar]
- Harro J, Kanarik M, Matrov D, Panksepp J. Mapping patterns of depression-related brain regions with cytochrome oxidase histochemistry: relevance of animal affective systems to human disorders, with a focus on resilience to adverse events. Neurosci Biobehav Rev. 2011;35:1876–1889. doi: 10.1016/j.neubiorev.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Liu S, Wong-Riley MT. A metabolic map of cytochrome oxidase in the rat brain: histochemical, densitometric and biochemical studies.x. Neuroscience. 1995;65:313–342. doi: 10.1016/0306-4522(94)00514-6. [DOI] [PubMed] [Google Scholar]
- Hirschfeld RM. History and evolution of the monoamine hypothesis of depression. J Clin Psychiatry. 2000;61(Suppl 6):4–6. [PubMed] [Google Scholar]
- Isgor C, Slomianka L, Watson SJ. Hippocampal mossy fibre terminal field size is differentially affected in a rat model of risk-taking behaviour. Behav Brain Res. 2004;153:7–14. doi: 10.1016/j.bbr.2003.10.039. [DOI] [PubMed] [Google Scholar]
- Jacobson L, Sapolsky R. The role of the hippocampus in feedback regulation of the hypothalamic-pituitary-adrenocortical axis. Endocr Rev. 1991;12:118–134. doi: 10.1210/edrv-12-2-118. [DOI] [PubMed] [Google Scholar]
- Jeewajee A, Lever C, Burton S, O'Keefe J, Burgess N. Environmental novelty is signaled by reduction of the hippocampal theta frequency. Hippocampus. 2008;18:340–348. doi: 10.1002/hipo.20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbaj M, Devine DP, Savage VR, Akil H. Neurobiological correlates of individual differences in novelty-seeking behavior in the rat: differential expression of stress-related molecules. Journal of Neuroscience. 2000;20:6983–6988. doi: 10.1523/JNEUROSCI.20-18-06983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama GH, Wong-Riley MT. Histochemical localization of cytochrome oxidase in the hippocampus: correlation with specific neuronal types and afferent pathways. Neuroscience. 1982;7:2337–2361. doi: 10.1016/0306-4522(82)90199-3. [DOI] [PubMed] [Google Scholar]
- Kim S, Webster MJ. Correlation analysis between genome-wide expression profiles and cytoarchitectural abnormalities in the prefrontal cortex of psychiatric disorders. Mol Psychiatry. 2010;15:326–336. doi: 10.1038/mp.2008.99. [DOI] [PubMed] [Google Scholar]
- Landers MS, Sullivan RM. The development and neurobiology of infant attachment and fear. Dev Neurosci. 2012;34:101–114. doi: 10.1159/000336732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire V, Aurousseau C, Le Moal M, Abrous DN. Behavioural trait of reactivity to novelty is related to hippocampal neurogenesis. Eur J Neurosci. 1999;11:4006–4014. doi: 10.1046/j.1460-9568.1999.00833.x. [DOI] [PubMed] [Google Scholar]
- Lever C, Burton S, O'Keefe J. Rearing on hind legs, environmental novelty, and the hippocampal formation. Rev Neurosci. 2006;17:111–133. doi: 10.1515/revneuro.2006.17.1-2.111. [DOI] [PubMed] [Google Scholar]
- Levine S. Primary social relationships influence the development of the hypothalamic--pituitary--adrenal axis in the rat. Physiol Behav. 2001;73:255–260. doi: 10.1016/s0031-9384(01)00496-6. [DOI] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Liscovitch N, Chechik G. Specialization of gene expression during mouse brain development. PLoS computational biology. 2013;9:e1003185. doi: 10.1371/journal.pcbi.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wong-Riley MT. Postnatal changes in cytochrome oxidase expressions in brain stem nuclei of rats: implications for sensitive periods. J Appl Physiol (1985) 2003;95:2285–2291. doi: 10.1152/japplphysiol.00638.2003. [DOI] [PubMed] [Google Scholar]
- McNaughton N, Gray JA. Anxiolytic action on the behavioural inhibition system implies multiple types of arousal contribute to anxiety. J Affect Disord. 2000;61:161–176. doi: 10.1016/s0165-0327(00)00344-x. [DOI] [PubMed] [Google Scholar]
- Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- Melendez-Ferro M, Rice MW, Roberts RC, Perez-Costas E. An accurate method for the quantification of cytochrome C oxidase in tissue sections. Journal of neuroscience methods. 2013;214:156–162. doi: 10.1016/j.jneumeth.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody M, Cao Y, Cui Z, Tay KY, Shyong A, Shimizu E, Pham K, Schultz P, Welsh D, Tsien JZ. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci U S A. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morys J, Berdel B, Kowianski P, Dziewiatkowski J. The pattern of synaptophysin changes during the maturation of the amygdaloid body and hippocampal hilus in the rat. Folia Neuropathol. 1998;36:15–23. [PubMed] [Google Scholar]
- Nair HP, Collisson T, Gonzalez-Lima F. Postnatal development of cytochrome oxidase activity in fiber tracts of the rat brain. Brain Res Dev Brain Res. 1999;118:197–203. doi: 10.1016/s0165-3806(99)00149-2. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Ashwell KWS, Tork I. Atlas of the Developing Rat Nervous System. Academic Press; New York: 1994. [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. Acacemic Press Inc.; San Diego, CA: 1986. [Google Scholar]
- Pokorny J, Trojan S. The development of hippocampal structure and how it is influenced by hypoxia. Acta Univ Carol Med Monogr. 1986;113:1–79. [PubMed] [Google Scholar]
- Regenold WT, Pratt M, Nekkalapu S, Shapiro PS, Kristian T, Fiskum G. Mitochondrial detachment of hexokinase 1 in mood and psychotic disorders: implications for brain energy metabolism and neurotrophic signaling. J Psychiatr Res. 2012;46:95–104. doi: 10.1016/j.jpsychires.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Rice MW, Smith KL, Roberts RC, Perez-Costas E, Melendez-Ferro M. Assessment of cytochrome C oxidase dysfunction in the substantia nigra/ventral tegmental area in schizophrenia. PLoS One. 2014;9:e100054. doi: 10.1371/journal.pone.0100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario LA, Abercrombie ED. Individual differences in behavioral reactivity: correlation with stress-induced norepinephrine efflux in the hippocampus of Sprague-Dawley rats. Brain Res Bull. 1999;48:595–602. doi: 10.1016/s0361-9230(99)00040-4. [DOI] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Annual Research Review: Epigenetic mechanisms and environmental shaping of the brain during sensitive periods of development. J Child Psychol Psychiatry. 2011;52:398–408. doi: 10.1111/j.1469-7610.2010.02282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roullet P, Lassalle JM. Genetic variation, hippocampal mossy fibres distribution, novelty reactions and spatial representation in mice. Behav Brain Res. 1990;41:61–70. doi: 10.1016/0166-4328(90)90054-i. [DOI] [PubMed] [Google Scholar]
- Ryan SJ, Ehrlich DE, Rainnie DG. Morphology and dendritic maturation of developing principal neurons in the rat basolateral amygdala. Brain Struct Funct. 2014 doi: 10.1007/s00429-014-0939-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saddar S, Dienhart MK, Stuart RA. The F1F0-ATP synthase complex influences the assembly state of the cytochrome bc1-cytochrome oxidase supercomplex and its association with the TIM23 machinery. J Biol Chem. 2008;283:6677–6686. doi: 10.1074/jbc.M708440200. [DOI] [PubMed] [Google Scholar]
- Sampedro-Piquero P, Zancada-Menendez C, Begega A, Mendez M, Arias JL. Effects of forced exercise on spatial memory and cytochrome c oxidase activity in aged rats. Brain Res. 2013;1502:20–29. doi: 10.1016/j.brainres.2012.12.036. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Krey LC, McEwen BS. Glucocorticoid-sensitive hippocampal neurons are involved in terminating the adrenocortical stress response. Proceedings of the National Academy of Sciences of the U S A. 1984;81:6174–6177. doi: 10.1073/pnas.81.19.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, ffrench-Mullen J, Benkelfat C, Rouleau GA, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Shumake J, Conejo-Jimenez N, Gonzalez-Pardo H, Gonzalez-Lima F. Brain differences in newborn rats predisposed to helpless and depressive behavior. Brain Res. 2004;1030:267–276. doi: 10.1016/j.brainres.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Shumake J, Edwards E, Gonzalez-Lima F. Hypermetabolism of paraventricular hypothalamus in the congenitally helpless rat. Neurosci Lett. 2001;311:45–48. doi: 10.1016/s0304-3940(01)02142-5. [DOI] [PubMed] [Google Scholar]
- Shumake J, Edwards E, Gonzalez-Lima F. Dissociation of septo-hippocampal metabolism in the congenitally helpless rat. Neuroscience. 2002;114:373–377. doi: 10.1016/s0306-4522(02)00297-x. [DOI] [PubMed] [Google Scholar]
- Shumake J, Gonzalez-Lima F. Brain systems underlying susceptibility to helplessness and depression. Behavioral and cognitive neuroscience reviews. 2003;2:198–221. doi: 10.1177/1534582303259057. [DOI] [PubMed] [Google Scholar]
- Solis O, Vazquez-Roque RA, Camacho-Abrego I, Gamboa C, De La Cruz F, Zamudio S, Flores G. Decreased dendritic spine density of neurons of the prefrontal cortex and nucleus accumbens and enhanced amphetamine sensitivity in postpubertal rats after a neonatal amygdala lesion. Synapse. 2009;63:1143–1153. doi: 10.1002/syn.20697. [DOI] [PubMed] [Google Scholar]
- Stead JD, Clinton S, Neal C, Schneider J, Jama A, Miller S, Vazquez DM, Watson SJ, Akil H. Selective breeding for divergence in novelty-seeking traits: heritability and enrichment in spontaneous anxiety-related behaviors. Behav Genet. 2006a;36:697–712. doi: 10.1007/s10519-006-9058-7. [DOI] [PubMed] [Google Scholar]
- Stead JD, Neal C, Meng F, Wang Y, Evans S, Vazquez DM, Watson SJ, Akil H. Transcriptional profiling of the developing rat brain reveals that the most dramatic regional differentiation in gene expression occurs postpartum. Journal of Neuroscience. 2006b;26:345–353. doi: 10.1523/JNEUROSCI.2755-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedenfeld KA, Clinton SM, Kerman IA, Akil H, Watson SJ, Sved AF. Novelty-seeking behavior predicts vulnerability in a rodent model of depression. Physiol Behav. 2011;103:210–216. doi: 10.1016/j.physbeh.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallot L, Doyere V, Sullivan RM. Developmental emergence of fear/threat learning: neurobiology, associations and timing. Genes Brain Behav. 2016;15:144–154. doi: 10.1111/gbb.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee CK, Glattfelder K, Sunkin SM, Henry A, Lau C, Dang C, Garcia-Lopez R, Martinez-Ferre A, Pombero A, Rubenstein JL, Wakeman WB, Hohmann J, Dee N, Sodt AJ, Young R, Smith K, Nguyen TN, Kidney J, Kuan L, Jeromin A, Kaykas A, Miller J, Page D, Orta G, Bernard A, Riley Z, Smith S, Wohnoutka P, Hawrylycz MJ, Puelles L, Jones AR. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron. 2014;83:309–323. doi: 10.1016/j.neuron.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JV, Sullivan RM, Wilson DA. Developmental emergence of fear learning corresponds with changes in amygdala synaptic plasticity. Brain Res. 2008;1200:58–65. doi: 10.1016/j.brainres.2008.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eden CG, Kros JM, Uylings HB. The development of the rat prefrontal cortex. Its size and development of connections with thalamus, spinal cord and other cortical areas. Prog Brain Res. 1990;85:169–183. doi: 10.1016/s0079-6123(08)62680-1. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proceedings of the National Academy of Sciences of the USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink G, Daenen LEWPM, Dubbeldam S, Gerrits MAFM, Van Rijn R, Kruse CG, Van Der Heijden JAM, Van Ree JM. Early amygdala damage in the rat as a model for neurodevelopmental psychopathological disorders. European Neuropsychopharmacology. 2001;11:51–59. doi: 10.1016/s0924-977x(00)00138-3. [DOI] [PubMed] [Google Scholar]
- Wong-Riley M. Changes in the visual system of monocularly sutured or enucleated cats demonstrable with cytochrome oxidase histochemistry. Brain Res. 1979;171:11–28. doi: 10.1016/0006-8993(79)90728-5. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT. Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT. Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv Exp Med Biol. 2012;748:283–304. doi: 10.1007/978-1-4614-3573-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]