Abstract
During a biodiversity survey of undiscovered taxa in Korea, two zygomycetous fungal strains were isolated. The first strain, EML-FSDY6-1 was isolated from a soil sample collected at Dokdo Island in the East Sea of Korea in 2013, and the second strain, EML-DG-NH3-1 was isolated from a rat dung sample collected at Chonnam National University garden, Gwangju, Korea in 2014. Based on the morphological characteristics and phylogenetic analysis of the internal transcribed spacer, 18S and 28S rDNA, actin and translation elongation factor-1α genes. EML-FSDY6-1 and EML-DG-NH3-1 isolates were confirmed as zygomycete species, Absidia pseudocylindrospora and Absidia glauca, respectively. Neither species has previously been described in Korea.
Keywords: Absidia glauca, A. pseudocylindrospora, Dokdo, Undiscovered taxa, Zygomycete fungi
The Zygomycota is a group of related basal clades comprising the subphyla Mucoromycotina, Entomophthoromycotina, Mortierellomycotina, Zoopagomycotina, and Kickxellomycotina [1]. Members of the Zygomycota generally reproduce asexually via sporangiospores and sexually via the formation of zygospores. The Mucorales is the largest order of the Zygomycota and include 56 genera and approximately 300 species. Most mucoralean species (called pin moulds) live as saprophytes growing on different organic substrates such as fruit, dung, plant and soil. Some species are parasites or pathogens of animals, plants and fungi. A few species cause human and animal disease zygomycosis, as well as allergic reactions. Zygomycete fungi belong to undiscovered taxa in Korea, due to lack of the species diversity information about it. According to the Korean species list of fungi [2], approximately 25 species of zygomycetes were known in Korea.
The genus Absidia belongs to the order Mucorales within the Cunninghamellaceae family. This genus was originally described by van Tieghem [3] with the type species Absidia reflexa Tieghem. To date, 21 species of Absidia have been recognized [4]. Absidia species are characterized by the production of stolons and sporangiophores bearing pyriform columellate sporangia with deliquescent walls and a septum below the apophysis and by zygospores enveloped in appendages from the suspensors. Absidia is closely related to Rhizopus; however, unlike the sporangiophores of Rhizopus, those of Absidia never arise opposite the rhizoids. Absidia also resembles Phycomyces with regard to its manner of sexual reproduction. However, in Phycomyces, large rough-walled zygospores are formed between tong-like suspensors [5,6].
Subsequent to van Tieghem's original description of the genus Absidia, some species of this genus were transferred to other genera such as Tieghemella Berl. & De Toni, Mycocladus Beauverie, Pseudoabsidia Bainier, and Proabsidia Vuill. However, with the exception of Lichtheimia, all are regarded as synonyms of Absidia [4,5].
Absidia species typically exhibit rapid growth at temperatures ranging from 25℃ to 34℃. They are frequently isolated from soil, dung, and dead or dying plant tissue [7,8,9]. Several species of Absidia are implicated in diseases affecting humans and animals, such as mucormycosis [10,11]. Recently, Hoffmann et al. [7] revised the classification of Absidia based on physiological, phylogenetic, and morphological characteristics. They observed temperature-dependent growth differences and divided the species into three groups—thermotolerant (optimal growth between 37℃ and 45℃), mesophilic (optimal growth between 25℃ and 34℃), and mycoparasitic (species that are potentially parasitic to other fungi within the order Mucorales and which exhibit optimal growth between 16℃ and 20℃).
Twenty five zygomycetous fungi which have been described in Korea were assigned in 12 genera and 8 families. Within the Mucoraceae, only 8 species have been described [2]. Data regarding the Mucoraceae of Korea, especially with respect to Absidia species, are lacking. A new species, Absidia koreana, was isolated from a soil sample collected at Dokdo Island in 2013 [12]. Almost all species belonging to the Mucoraceae in Korea have been discovered from soil, and to our knowledge, there are no previous published literature records of this family from animal dung.
Recently, several studies have evaluated the phylogenetic relationships between species of mucoralean fungi and other zygomycetes, using combined sequences of internal transcribed spacer (ITS) region, 18S and 28S rDNA, actin (act-1), and translation elongation factor-1α (EF-1α) genes [13,14,15,16,17].
The objective of the present study was to characterize two unrecorded zygomycete species, Absidia pseudocylindrospora and Absidia glauca, based on the morphological characteristics and sequence analyses.
MATERIALS AND METHODS
Isolation of fungal strains from soil and rat dung sample
Soil samples were collected from Dongdo (eastern islet) of Dokdo (37°14'21.3'' N, 131°52'04.4'' E) Island in the East Sea of Korea in 2013. Samples were obtained at a depth of 10~15 cm, transported in sterile 50-mL conical tubes, and stored at 4℃ until examination. Fungi were isolated using serial dilution plating method. Briefly, 1 g of soil was mixed with 9mL of sterile distilled water and shaken for 15 min at room temperature; serial dilutions ranging from 10−1 to 10−4 were then made. An aliquot of 0.1mL from each dilution was transferred to potato dextrose agar (39 g PDA in 1 L of deionized water; Becton, Dickinson and Co., Sparks, MD, USA) and incubated at 27℃ for 3~7 days.
Rat dung sample was collected from the garden of Chonnam National University, Gwangju, Korea in 2014 using sterile forceps. The samples were transferred to the laboratory in plastic bags, placed onto sterile moist Whatman's filter paper in a Petri dish, and incubated in a moist chamber at 25℃ for 7 days. Hyphal tips were transferred to PDA plates using a stereomicroscope.
To isolate pure cultures, individual colonies of varied morphologies were transferred to PDA plates. Pure isolates such as EML-FSDY6-1 and -2, EML-DG-NH3-1 and -2 were maintained in PDA slant tubes and stored in 20% glycerol at −80℃ at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, Korea. The dry cultures (EML-FSDY6-1 and EML-DG-NH3-1) were preserved at Chonnam National University Fungal Collection (CNUFC), Division of Food Technology, Biotechnology and Agrochemistry, College of Agriculture and Life Sciences, Gwangju, Korea. The ex-type (EML-FSDY6-1 and EML-DG-NH3-1) strains were also deposited as glycerol stock at −80℃ at the Culture Collection of National Institute of Biological Resources (NIBR), Incheon, Korea as KOSPFGC000002026 and KOSPFGC000002025, respectively.
Morphological studies
To obtain samples for microscopic examination and growth rate determination, EML-FSDY6-1 and EML-DG-NH3-1 were cultured on each of three different media. The media used were PDA, synthetic mucor agar (SMA; 40 g dextrose, 2 g asparagine, 0.5 g KH2PO4, 0.25 g MgSO4·7H2O, 0.5 g thiamine hydrochloride, and 15 g agar, in 1 L of deionized water), and malt extract agar (33.6 g MEA in 1 L of deionized water; Becton, Dickinson and Co.). The plates were incubated at 20℃, 23℃, 25℃, 27℃, 30℃, 35℃, and 37℃ in the dark for 7 days. Samples were mounted in lactophenol solution (Junsei Chemical Co. Ltd., Tokyo, Japan) and examined under a light microscope (DFC290; Leica Microsystems, Wetzlar, Germany). Fine fungal structures were observed using scanning electron microscopy (Hitachi S4700; Hitachi, Tokyo, Japan). Samples were cultured on PDA medium in the dark at 27℃ for 7 days, fixed in 2.5% paraformaldehyde-glutaraldehyde in 0.05 M phosphate buffer (pH 7.2) for 2 hr, and then washed with cacodylate buffer (Junsei Chemical Co. Ltd.). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA, USA) diluted in cacodylate buffer for 1 hr, washing again in cacodylate buffer, dehydrating in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei Chemical Co. Ltd.), and drying in a fume hood. Finally, samples were sputter-coated with gold and observed under a Hitachi S4700 field emission scanning electron microscope at the Korea Basic Science Institute, Gwangju, Korea.
DNA extraction, PCR, and sequencing
Total genomic DNA was extracted directly from the mycelia of fungal isolates using the HiGene Genomic DNA Prep Kit (BIOFACT Co., Daejeon, Korea). The ITS region; small subunit of 18S rDNA, large subunit of 28S rDNA, actin (act-1), and elongation factor 1α (EF-1α) sequences were amplified with the primer pairs ITS1, ITS4 [18]; NL1, NS4 [14]; LROR, LR5F [19,20]; Act-1, Act-4R [21]; and MEF-11, MEF-4 [14], respectively. The PCR amplification mixture (total volume, 20 µL) contained 10 ng of fungal gDNA template, 5 pmol/µL of each primer, and Accupower PCR Premix (Taq DNA polymerase, dNTPs, buffer, and a tracking dye; Bioneer Co., Daejeon, Korea). PCR products were purified using the Accuprep PCR Purification Kit (Bioneer Co.) according to the manufacturer's instructions. DNA sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA).
Phylogenetic analysis
Fungal sequences (Table 1) were subjected to phylogenetic analysis using Clustal_X v.1.83 [22] and Bioedit v. 5.0.9.1 software [23]. Phyologenies were assessed using MEGA 6 software [24]. Maximum likelihood (ML) phylogenetic trees were constructed for individual and combined datasets of the 18S and 28S rDNA, act-1 and EF-1α gene sequences. The nearest-neighbor-interchange was selected for the ML heuristic method, and the initial ML tree was set automatically. Umbelopsis isabellina and Umbelopsis nana sequences were used as outgroups. The percentage of sequence identity was obtained by applying the National Center for Biotechnology Information (NCBI) BLASTn search tool to each isolate.
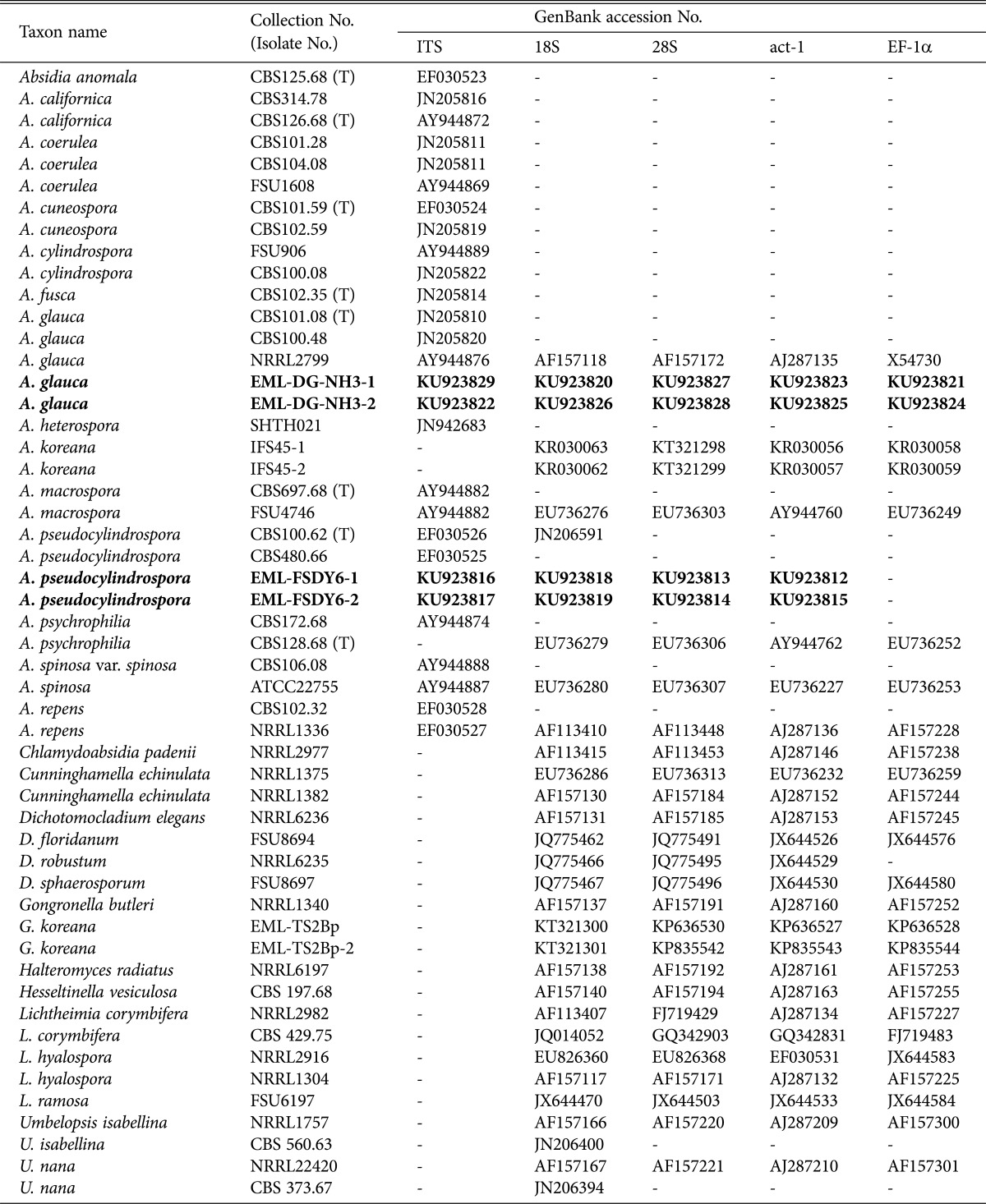
Table 1. Sequences used in this study, including GenBank accession numbers.
Bold letters indicate isolates and accession numbers determined in our study.
ITS, internal transcribed spacer; EF-1α, elongation factor-1α; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; T, ex-type strain; FSU, Friedrich Schiller University, Jena, Germany; NRRL, ARS Culture Collection, Peoria, Illinois, USA; EML, Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju, South Korea; ATCC, Biological Resource Center, Manassas, Virginia, USA.
RESULTS
Phylogenetic analysis
The strains were identified based on morphological characteristics, ITS region, and combined gene sequences. The EML-FSDY6-1, EML-FSDY6-2 and EML-DG-NH3-1, EML-DG-NH3-2 sequences were deposited in the NCBI database under accession numbers as shown in Table 1.
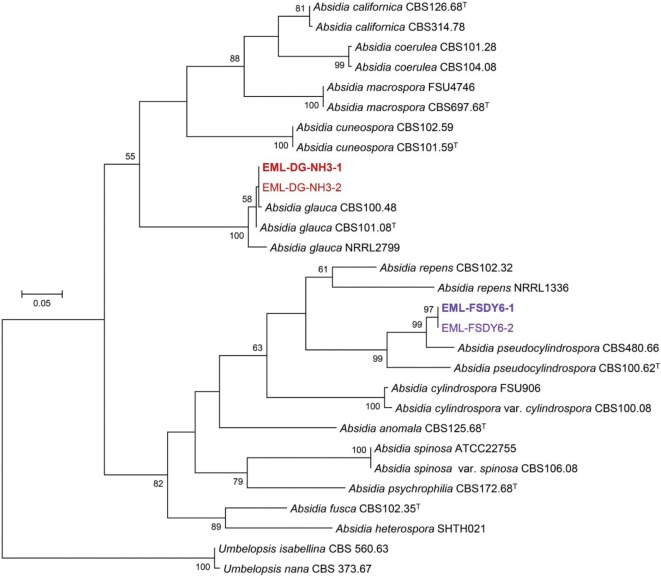
A BLASTn search was performed against the ITS sequences of EML-FSDY6-1 and EML-DG-NH3-1. The sequence similarities revealed 96.1% (469/488 bp) and 97.7% (606/620 bp) homologies with the two most closely related species, namely, A. pseudocylindrospora CBS480.66 (GenBank accession No. EF030525) and A. glauca FSU1615 (GenBank accession No. AY944875), respectively. Based on the 28S rDNA sequence analysis, EML-FSDY6-1 and EML-DG-NH3-1 strains showed 98.1% (599/611 bp) and 99.4% (694/698 bp) homologies with A. pseudocylindrospora CBS100.62 (GenBank accession No. JN206591) and A. glauca NRRL2799 (GenBank accession No. AF157172), respectively. In addition, the 18S rDNA sequence similarities against EML-FSDY6-1 and EML-DG-NH3-1 isolates were 98.8% (927/938 bp) and 99.9% (926/927 bp) homologous with A. repens SHTH004 (GenBank accession No. JQ004930) and A. glauca FSU660 (GenBank accession No. EU736275), respectively. The BLASTn search results for the act-1 and EF-1α gene sequences indicated that EML-DG-NH3-1 is closely to A. glauca FSU660 (GenBank accession No. EU736225); the identity values for the act-1 and EF-1α gene sequences were 98.7% (614/622 bp) and 99.6% (725/728 bp), respectively. The act-1 gene sequence of isolate EML-FSDY6-1 showed an identity value of 98.5% (599/608 bp) with A. pseudocylindrospora FSU5893 (GenBank accession No. EF030534). Based on the ITS sequence analysis, the two isolates were identical to A. pseudocylindrospora CBS 100.48 and A. glauca CBS 480.66 (Fig. 1). Moreover, analysis of the combined genes placed the two strains within the Cunninghamellaceae family, similar to other Absidia species, with well-supported branch values (Fig. 2).
Fig. 1. Phylogenetic tree based on maximum likelihood analysis of internal transcribed spacer rDNA sequences for Absidia pseudocylindrospora EML-FSDY6-1, A. pseudocylindrospora EML-FSDY6-2, A. glauca EML-DG-NH3-1, and A. glauca EML-DG-NH3-2. Umbelopsis isabellina and U. nana were used as outgroups. Bootstrap support values of ≥ 50% are indicated at the nodes. The bar indicates the number of substitutions per position. Ex-type strains are marked as bold purple and red letters.
Fig. 2. Phylogenetic tree based on maximum likelihood analysis of multiple loci including 18S and 28S rDNA, actin (act-1), and translation elongation factor-1α for Absidia pseudocylindrospora EML-FSDY6-1, A. pseudocylindrospora EML-FSDY6-2, A. glauca EML-DG-NH3-1, and A. glauca EML-DG-NH3-2. Umbelopsis isabellina and U. nana were used as outgroups. Bootstrap support values of ≥ 50% are indicated at the nodes. The bar indicates the number of substitutions per position. Ex-type strains are marked as bold purple and red letters.
Taxonomy of EML-FSDY6-1.
Absidia pseudocylindrospora C. W. Hesseltine & J. J. Ellis, Mycologia 53: 406 (1961) (Table 2, Fig. 3).
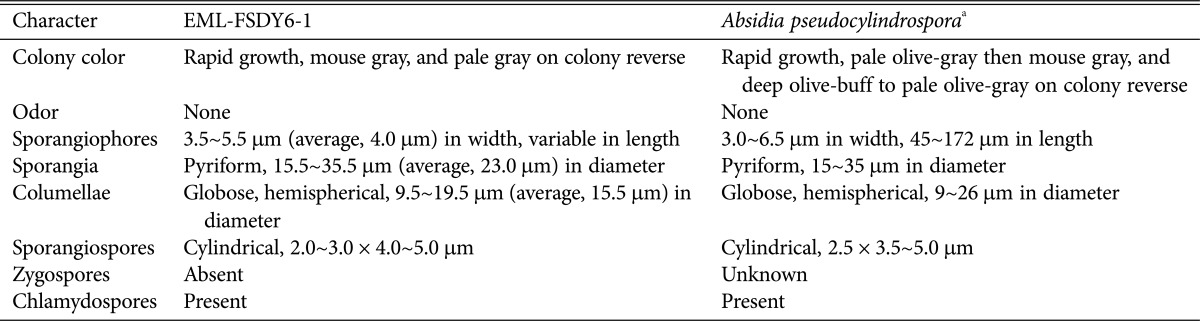
Table 2. Morphological characteristics of EML-FSDY6-1 and the reference species Absidia pseudocylindrospora on synthetic mucor agar medium at 25℃.
aFrom the description Hesseltine and Ellis [25].
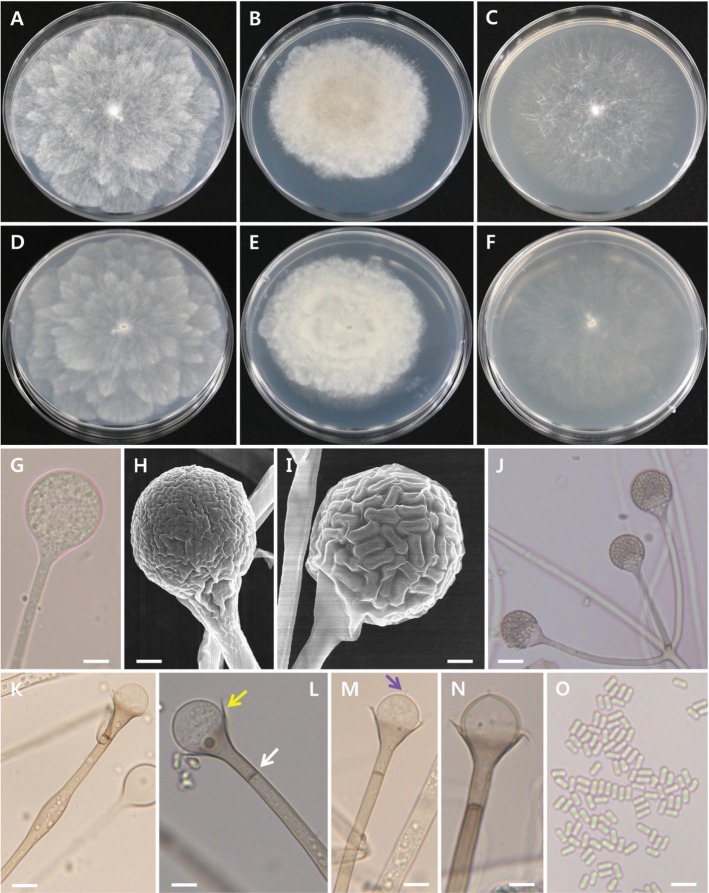
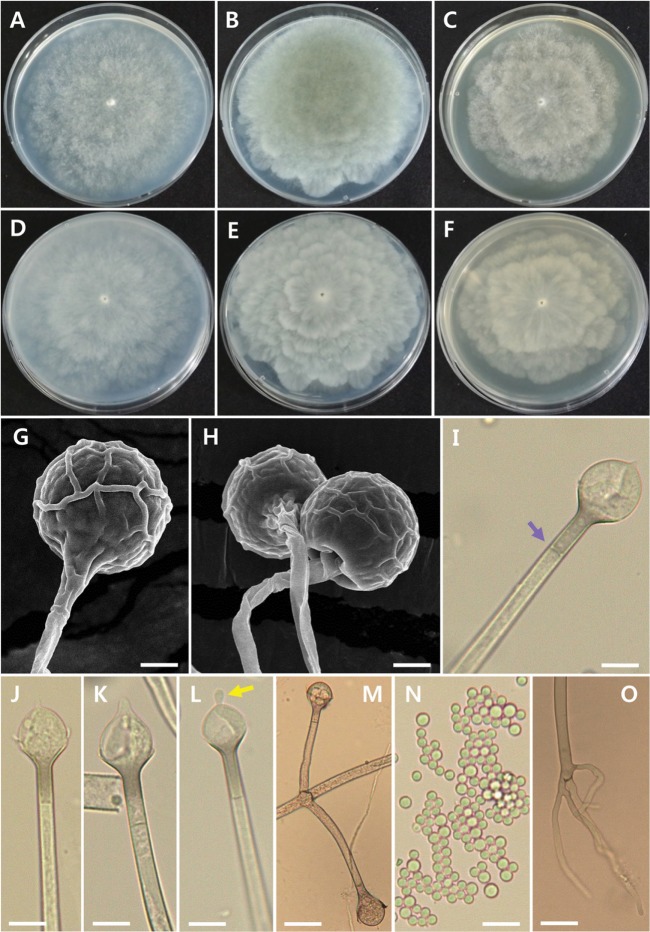
Fig. 3. Morphology of Absidia pseudocylindrospora EML-FSDY6-1. Colonies on synthetic mucor agar (A, D), potato dextrose agar (B, E), and malt extract agar (C, F). A~C, Obverse view; D~F, Reverse view; G, Young sporangium; H, I, Young and mature sporangia; J, Attachment of sporangiophores; K~N, Columella with collarette (yellow arrow), sporangial septum (white arrow), and projection (purple arrow); O, Sporangiospores (scale bars: G, K~O = 20 µm, H, I = 10 µm, J = 50 µm).
Description
Colonies exhibited rapid growth on SMA, attaining a diameter of 82~85 mm after 4 days at 25℃. The colony color was initially white, later turning to mouse gray. The colony reverse was pale gray and regularly zonate. Sporangiophores were 3.5~5.5 µm (average, 4.0 µm) wide, appearing upright, in groups of 1~10 (average, 3~5) per whorl from a single point in the stolons, and consistently had a septum under the sporangium. Sporangia measured 15.5~35.5 µm (average, 23.0 µm) in diameter and were pyriform, deep gray, and multispored. Columellae measured 9.5~19.5 µm (average, 15.5 µm) in diameter and were globose and hemispherical. Sporangiospores were mostly cylindrical, 2.0~3.0 µm × 4.0~5.0 µm. Chlamydospores were present in the aerial mycelia. Zygospores were not observed on this medium.
Taxonomy of EML-DG-NH3-1
Absidia glauca Hagem, Skrifter udgivne af Videnskabs-Selskabet I Christiania. Mathematisk-Naturvidenskabelig Klasse 7: 43 (1908) (Table 3, Fig. 4).
Table 3. Morphological characteristics of EML-DG-NH3-1 and the reference species Absidia glauca on synthetic mucor agar medium at 25℃.
aFrom the description by Ellis and Hesseltine [26].
Fig. 4. Morphology of Absidia glauca EML-DG-NH3-1. Colonies on synthetic mucor agar (A, D), potato dextrose agar (B, E), and malt extract agar (C, F). A~C, Obverse view; D~F Reverse view; G, H, Sporangia with wall net; I~L, Columella with septum (purple arrow) and projection (yellow arrow); M, Attachment of sporangiophores; N, Sporangiospores; O, Young rhizoid (scale bars: G, I~O = 20 µm, H = 30 µm).
≡ Tieghemella glauca (Hagem) Naumov, Tab. Opred. Predst. Mucor: 42 (1915)
= Absidia septata Tiegh., Annales des Sciences Naturelles Botanique 4: 362 (1878)
= Absidia sphaerosporangioides Manka & Truszk., Acta Societatis Botanicorum Poloniae 27: 54 (1958)
Description
Colonies exhibited rapid growth on SMA, attaining a diameter of 80~85 mm after 5 days at 25℃. The colony color was initially white, later turning to storm gray. The colony reverse was light gray and irregularly zonate. Mycelial growth on SMA was sparse, but sporulation was extensive. Sporangiophores were 4.0~10.5 µm wide, appearing upright, in groups of 1~8 (average, 2~4) per whorl from a single point on the stolons, and consistently had a septum under the sporangium. Sporangia were 19.0~43.0 µm × 21.5~52 µm and were globose, pyriform, and brownish yellow. The surface was covered by wall net. Columellae measured 14.5~33 µm × 12~27 µm and were globose to hemispherical. Sporangiospores were globose and measured 2.5~4.5 µm in diameter. Zygospores were not observed on this medium. Rhizoids were not well developed.
Mycelial growth
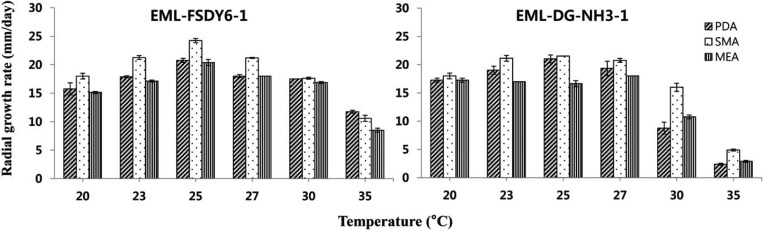
The isolates grew at different rates according to temperature and medium. The average growth rates of EML-FSDY6-1 on PDA, SMA, and MEA were 20.5 mm/day, 24 mm/day, and 20 mm/day, respectively; those of EML-DG-NH3-1 were 21 mm/day, 21.5 mm/day, and 16.5 mm/day, respectively. The optimal growth temperature range was 23~27℃. Slow growth was observed at 35℃ and no growth was visible at 37℃ (Fig. 5).
Fig. 5. Effect of temperature and culture medium on mycelial growth of Absidia pseudocylindrospora EML-FSDY6-1 and A. glauca EML-DG-NH3-1. Mycelia were grown on synthetic mucor agar (SMA), potato dextrose agar (PDA), and malt extract agar (MEA) at different temperatures, as indicated.
DISCUSSION
Until recently, we have reported some kinds of new and undescribed zygomycetous fungi from Dokdo Island of Korea based on current classification system using combined sequences [12, 27]. This study was also done to discover zygomycetous taxa on that unique island. Thus, the data with regard to such an undiscovered fungal taxa can be considerably meaningful in understanding of geographical distribution of fungal species diversity in Korean Peninsula.
Similar to many other Mucorales, the Absidia has been distinguished based on morphological features and growth temperature. With regard to phylogenetic and physiological studies on Absidia, Hoffmann et al. [7] showed that species of Absidia exhibited different growth patterns at different temperatures. The authors distinguished Absidia species based on the shape of the sporangiospores including cylindrical, conical, and globose. In our present study, the two investigated strains grew rapidly and were mesophilic (optimum growth temperature range of 23~27℃), but they responded differently at 30℃ and 35℃. The strain identified as A. glauca showed lower thermotolerance, suggesting that the culture medium influences the maximum growth temperatures of fungi. In addition, the colony morphology and culture characteristics of the two isolates were generally similar to those previously described by Hesseltine and Ellis [25] for A. pseudocylindrospora and by Ellis and Hesseltine [26] for A. glauca. However, some features differed. Our A. pseudocylindrospora isolate presented sporangiophores with swellings, which were not described by Hesseltine and Ellis [25]. Moreover, our A. glauca isolate produced columellae with a projection of variable shape and displayed more sporangiophores per whorl than the isolate described by Ellis and Hesseltine [26].
Despite the wide intraspecific variation found among some taxa, the ITS region is the barcode marker for mucoralean species identification. The results of our molecular data analysis of the two investigated species were consistent with the phylogeny presented by Walther et al. [17]. Furthermore, in the ITS tree, EML-FSDY6-1 belonged to the cylindrical Absidia group, whereas EML-DG-NH3-1 clustered within the globose Absidia group as defined by Hoffmann et al. [7]. Phylogenetic analyses of four genes showed a close relationship between A. pseudocylindrospora and A. glauca and other species of the family Cunninghamellaceae. The classification of the two investigated strains within this family is consistent with similarities in several morphological characters, including rapid growth and production of sporangia with multispores (except in the genus Hesseltinella).
Data regarding the diversity of zygomycetous fungi in Korea are lacking. Hence, further studies on the classification of different orders and families within the zygomycete fungi are required to expand our knowledge of undiscovered taxa in Korea.
ACKNOWLEDGEMENTS
This work was supported by the Project on Survey and Discovery of Indigenous Species of Korea and in part by Graduate Program for the Undiscovered Taxa of Korea funded by the National Institute of Biological Resources, Ministry of Environment (MOE), Republic of Korea.
References
- 1.Muszewska A, Pawłowska J, Krzyściak P. Biology, systematics, and clinical manifestations of Zygomycota infections. Eur J Clin Microbiol Infect Dis. 2014;33:1273–1287. doi: 10.1007/s10096-014-2076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YS, Jung HY, Lee HB, Kim SH, Shin KS, Eom AH, Kim C, Lee SY Korean Society of Mycology. National list of species of Korea. Ascomycota, Glomeromycota, Zygomycota, Myxomycota, Oomycota. Incheon: National Institute of Biological Resources; 2015. [Google Scholar]
- 3.Van Tieghem MP. Sur le développement du fruit des Ascodesmis, genre Nouveau de L'ordre des Ascomycètes. Bull Soc Bot Fr. 1878;23:271–282. [Google Scholar]
- 4.Kirk PM, Cannon PF, Minter DW, Stalpers JA. Ainsworth and Bisby's dictionary of the fungi. 10th ed. Wallingford: CAB International; 2008. [Google Scholar]
- 5.Hesseltine CW, Ellis JJ. The genus Absidia: Gongronella and cylindrical-spored species of Absidia. Mycologia. 1964;56:568–601. [Google Scholar]
- 6.O'Donnell K. Zygomycetes in culture. Palfrey contribution in botany. No. 2. Athens (GA): Department of Botany, University of Georgia; 1979. [Google Scholar]
- 7.Hoffmann K, Discher S, Voigt K. Revision of the genus Absidia (Mucorales, Zygomycetes) based on physiological, phylogenetic, and morphological characters: thermotolerant Absidia spp. form a coherent group, Mycocladiaceae fam. nov. Mycol Res. 2007;111(Pt 10):1169–1183. doi: 10.1016/j.mycres.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Ho HM, Chuang SC, Chen SJ. Notes on Zygomycetes of Taiwan (IV): three Absidia species (Mucoraceae) Fungal Sci. 2004;19:125–131. [Google Scholar]
- 9.Benny GL. Methods used by Dr. R. K. Benjamin, and other mycologists, to isolate Zygomycetes. Aliso. 2008;26:37–61. [Google Scholar]
- 10.Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clin Microbiol Rev. 2000;13:236–301. doi: 10.1128/cmr.13.2.236-301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann K, Voigt K. Absidia parricida plays a dominant role in biotrophic fusion parasitism among mucoralean fungi (Zygomycetes): Lentamyces, a new genus for A. parricida and A. zychae. Plant Biol (Stuttg) 2009;11:537–554. doi: 10.1111/j.1438-8677.2008.00145.x. [DOI] [PubMed] [Google Scholar]
- 12.Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KW, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, et al. Fungal diversity notes 111-252: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. [Google Scholar]
- 13.O'Donnell K, Lutzoni FM, Ward TJ, Benny GL. Evolutionary relationships among mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia. 2001;93:286–297. [Google Scholar]
- 14.Voigt K, Wöstemeyer J. Phylogeny and origin of 82 Zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1α genes. Gene. 2001;270:113–120. doi: 10.1016/s0378-1119(01)00464-4. [DOI] [PubMed] [Google Scholar]
- 15.White MM, James TY, O'Donnell K, Cafaro MJ, Tanabe Y, Sugiyama J. Phylogeny of the Zygomycota based on nuclear ribosomal sequence data. Mycologia. 2006;98:872–884. doi: 10.3852/mycologia.98.6.872. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann K, Pawłowska J, Walther G, Wrzosek M, de Hoog GS, Benny GL, Kirk PM, Voigt K. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia. 2013;30:57–76. doi: 10.3767/003158513X666259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walther G, Pawłowska J, Alastruey-Izquierdo A, Wrzosek M, Rodriguez-Tudela JL, Dolatabadi S, Chakrabarti A, de Hoog GS. DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia. 2013;30:11–47. doi: 10.3767/003158513X665070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and application. San Diego (CA): Academic Press; 1990. pp. 315–322. [Google Scholar]
- 19.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Crytococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HB. Molecular phylogenetic status of Korean strain of Podosphaera xanthii, a causal pathogen of powdery mildew on Japanese thistle (Cirsium japonicum) in Korea. J Microbiol. 2012;50:1075–1080. doi: 10.1007/s12275-012-2618-z. [DOI] [PubMed] [Google Scholar]
- 21.Voigt K, Wöstemeyer J. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiol Res. 2000;155:179–195. doi: 10.1016/S0944-5013(00)80031-2. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 24.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesseltine CW, Ellis JJ. Notes on Mucorales, especially Absidia. Mycologia. 1961;53:406–426. [Google Scholar]
- 26.Ellis JJ, Hesseltine CW. The genus Absidia: globose-spored species. Mycologia. 1965;57:222–235. [Google Scholar]
- 27.Lee HW, Nguyen TT, Mun HY, Lee H, Kim C, Lee HB. Confirmation of two undescribed fungal species from Dokdo of Korea based on current classification system using multi loci. Mycobiology. 2015;43:392–401. doi: 10.5941/MYCO.2015.43.4.392. [DOI] [PMC free article] [PubMed] [Google Scholar]