Abstract
Gram‐negative pathogens often use conserved type three secretion systems (T3SS) for virulence. The Shigella type three secretion apparatus (T3SA) penetrates the host cell membrane and provides a unidirectional conduit for injection of effectors into host cells. The protein Spa47 localizes to the base of the apparatus and is speculated to be an ATPase that provides the energy for T3SA formation and secretion. Here, we developed an expression and purification protocol, producing active Spa47 and providing the first direct evidence that Spa47 is a bona fide ATPase. Additionally, size exclusion chromatography and analytical ultracentrifugation identified multiple oligomeric species of Spa47 with the largest greater than 8 fold more active for ATP hydrolysis than the monomer. An ATPase inactive Spa47 point mutant was then engineered by targeting a conserved Lysine within the predicted Walker A motif of Spa47. Interestingly, the mutant maintained a similar oligomerization pattern as active Spa47, but was unable to restore invasion phenotype when used to complement a spa47 null S. flexneri strain. Together, these results identify Spa47 as a Shigella T3SS ATPase and suggest that its activity is linked to oligomerization, perhaps as a regulatory mechanism as seen in some related pathogens. Additionally, Spa47 catalyzed ATP hydrolysis appears to be essential for host cell invasion, providing a strong platform for additional studies dissecting its role in virulence and providing an attractive target for anti‐infective agents.
Keywords: type III secretion system (T3SS), virulence factor, protein export, ATP, ADP, ATPase, oligomerization
Abbreviations
- CBD
chitin binding domain
- CD
circular dichroism
- IPTG
isopropyl β‐D‐1‐thiogalactopyranoside
- LPS
lipopolysaccharide;
- SEC
size exclusion chromatography
- SV‐AUC
sedimentation velocity analytical ultracentrifugation
- T3SA
type III secretion apparatus
- T3SS
type III secretion system
- TEM
transmission electron microscopy
- spp.
species (plural).
Introduction
Shigella species cause a severe form of bacillary dysentery (shigellosis) throughout the world with an estimated 90 million annual infections responsible for greater than 100,000 deaths each year.1 Shigella require as few as 10–100 organisms to cause infection and are efficiently spread through the fecal oral route and/or contaminated drinking water.2 Most reported cases of shigellosis occur in developing regions where sanitation and access to clean drinking water are limited. Increasing reports in industrialized regions3 and the appearance of antibiotic resistant strains further emphasize the need to better understand the mechanism of Shigella virulence and specifically the means by which it invades targeted host cells.4
Shigella invasion and subsequent spread throughout the colonic epithelium relies on a complex type three secretion system (T3SS) including effector proteins that are directly injected into host cell cytoplasm and the needle‐like apparatus that facilitates effector transfer.5 The type three secretion apparatus (T3SA) is highly conserved and resembles a nano‐needle and syringe consisting of a cytoplasmic bulb, basal body that spans the inner and outer membranes of the bacterium, a hollow needle that extends beyond the thick LPS layer, and an associated tip complex that interacts with the host cell membrane (Fig. 1).6, 7 Effector proteins and those that make up the T3SA tip complex are secreted through the 2.5 nm diameter channel of the MxiH needle both as the complex matures and upon activation of the system.8, 9, 10, 11 Proper timing and levels of secretion are essential for virulence, making identification of the trigger mechanism and driving force of secretion of great interest.12, 13 Sequence and structural similarities between the β‐subunit of F1Fo ATP synthase,14 FliI of the bacterial flagellar system,15 and identified ATPases associated with the translocon‐associated T3SS apparatus of several pathogens16, 17, 18 suggest that many T3SSs contain an ATPase at the base of the needle which could provide the necessary energy source for protein secretion.19, 20
Figure 1.
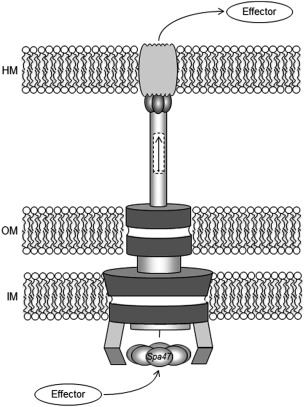
Organization of the Type III secretion system. Cartoon of the T3SS from Shigella is shown with the basal body spanning the inner membrane (IM) and outer membrane (OM) of the pathogen and the external needle inserting into the host membrane (HM). The Spa47 hexamer is depicted at the base of the needle assembly. Arrows show the path of the effector protein passage from the pathogen to the host through the apparatus.
The Shigella T3SS protein Spa47 shares 37.4% sequence identity and 53.6% similarity to the Salmonella enterica ATPase FliI which drives hook rotation responsible for flagellar motion and even higher levels of homology to identified T3SS ATPases from Escherichia, Yersinia, and Chlamydophila species (40.1%, 42.0% and 43.4% identity to Spa47, respectively) as determined by the alignment software Clustal Omega. Additionally, Spa47 knockout strains of S. flexneri do not properly secrete some T3SS proteins including MxiH which is necessary for construction of the needle.21 As a result, Spa47 is proposed to be a Shigella T3SS ATPase which may play a vital role in virulence by powering the T3SA machinery. Here, we describe the development of an expression and purification methodology for Spa47, allowing us to directly evaluate its ATPase activity in vitro. ATPase activity assays together with biophysical characterization of Spa47 found that the oligomerization state of the enzyme significantly impacts its activity, providing a potential mechanism for in vivo control over the associated T3SS. Specifically, the most active form identified purifies as a stable homo‐oligomer while the monomeric species are eight‐fold less active. A Spa47 point mutant was generated in the predicted catalytic Walker A domain to prevent ATP binding and provide an ATPase inactive Spa47 species. The oligomeric distribution of the mutant was similar to native Spa47 though it is unable to hydrolyze ATP and when used to complement a spa47 knockout Shigella strain, is unable to restore the invasion phenotype. The overall findings identify Spa47 as an oligomerization‐dependent active Shigella T3SS ATPase and suggest that its ability to hydrolyze ATP is key to the Shigella invasion phenotype and ultimately virulence.
Results
Expression and purification of an active S. flexneri T3SS ATPase
T3SS ATPases from several organisms have been successfully expressed and purified, allowing them to be studied both structurally and biochemically.16, 22, 23, 24, 25, 26 While these findings have been key in understanding the role of ATPases in T3SSs, work with Shigella Spa47 has proven challenging and has limited the understanding of its role in the otherwise well‐defined Shigella system. Here, we have overcome the hurdle of Spa47 purification by generating a CBD‐tagged Spa47 fusion, resulting in E. coli expression of soluble Spa47 containing an N‐terminal CBD attached through an intein linker [Fig. 2(A)]. Affinity purification with chitin resin followed by DTT‐induced intein cleavage produced CBD‐less Spa47 at relatively high purity [Fig. 2(B)]. Negative selection using Q Sepharose anion exchange chromatography further removed contaminants and resulted in highly soluble full length recombinant Spa47 that was ≥85% pure (by SDS PAGE) with identity confirmed by in gel digestion followed by tandem mass spectrometry.
Figure 2.
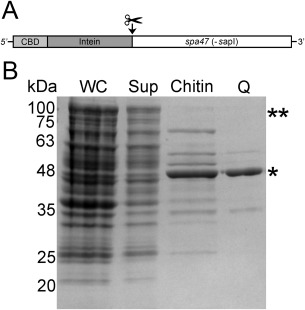
Purification of Spa47. (A) N‐terminal chitin binding domain (CBD) and Intein tagged version of Spa47 used for overexpression and purification. The site of cleavage is shown. (B) SDS‐PAGE of Spa47 at various stages during protein purification. Whole cell lysate (WC), clarified supernatant (Sup), cleaved protein after elution from the chitin column (Chitin) and, purified Spa47 from the Q‐sepharose (Q) are shown. ** and * denote the corresponding positions of the uncleaved CBD‐Intein‐Spa47 and cleaved Spa47, respectively.
Initial purification was followed by SEC to provide an additional purification step and as a means to characterize the oligomerization state of the recombinant Spa47. The elution profile identified multiple discrete species of Spa47 with the majority of the protein eluting later in the separation, suggesting that the recombinant Spa47 purified in multiple oligomeric states with the main peaks eluting at ∼0.4 and 0.6 column volumes, representing species III and I, respectively [Fig. 3(A,B)]. While we were not expecting such a distribution, ATPase oligomerization is not uncommon and is known to be essential for activity of the hetero‐hexameric F1Fo ATP synthase27 as well as suspected to play an important role in activation and control of hydrolysis for several T3SS ATPases,22, 25, 26, 28 providing a potential mechanism for regulation. We next tested if the purified Spa47 was active with respect to ATP hydrolysis which would not only validate the expression/purification method, but also confirm the hypothesis that Spa47 is a T3SS ATPase. As a first approach, the ATPase activity of each of the SEC separated Spa47 oligomers was tested. Radiolabeled ATP was mixed with a fixed volume of protein from each fraction and the formation of ADP was monitored. The results show a bimodal distribution of activity that precisely follows the protein elution profile, turning over nearly all of the 500 μM ATP in each of the peak fractions under the conditions tested [Fig. 3(C,D)]. Together, these results suggest that the recombinant Spa47 is not only active, but that it probably exists in multiple oligomeric forms.
Figure 3.
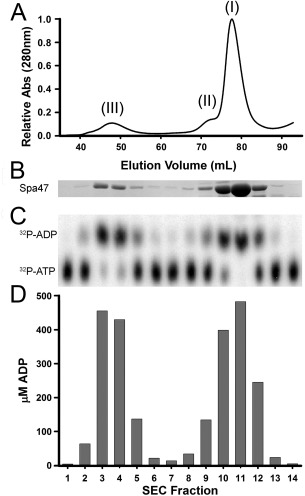
Oligomeric forms of Spa47. (A) Elution profile from size exclusion chromatographic (SEC) separation of Spa47 and (B) SDS‐PAGE analysis of protein in each fraction. (C) Thin layer chromatograph (TLC) showing the fraction of α32P‐ADP formed from α32P‐ATP after incubation with protein from the corresponding fraction in SEC separation (above) and (D) is the quantitation of total ADP formed in each SEC fraction.
Mass spectrometry identified the purified protein as Shigella Spa47 (data not shown), however since Spa47 has not previously been directly tested for activity, we felt it was important to generate an ATPase inactive Spa47 mutant as a negative control to ensure that the observed ATPase activity was in fact coming from Spa47 and not a co‐purifying contaminant. A Spa47 point mutant targeting the predicted Walker A motif was engineered by substituting an Alanine for Lysine 165. This residue was chosen based on sequence conservation and previous studies showing that the corresponding residues in FliI (Salmonella)29 and EscN (E. coli)26 are responsible for stabilizing the negative charge of ATP, supporting substrate interaction within the enzyme active site. The Spa47K165A mutant was expressed and purified exactly as was done for the native Spa47 construct with SEC analysis of the Spa47K165A maintaining a nearly identical oligomerization profile as native Spa47 (Fig. 4) with monomeric (I) and oligomeric (III) species evident in the chromatogram. When tested for ATP hydrolysis activity, Spa47K165A was inactive, with less than 4 μM total ADP observed following any reaction (Fig. 4 insert), compared to nearly 500 μM ADP for wild‐type Spa47. These data not only suggest that Spa47 belongs to the Walker type ATPase family,30 but also served as a control confirming that observed activity levels for native Spa47 were the direct result of Spa47 catalyzed ATP hydrolysis. Additionally, these results suggest that ATP hydrolysis by Spa47 is not a requirement for oligomerization however, we cannot rule out the possibility that perhaps transient higher order oligomers are driven by ATP hydrolysis.
Figure 4.
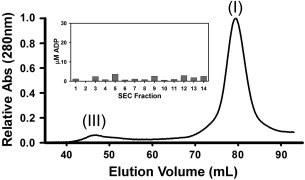
Size exclusion fractionation of Spa47K165A. Profile from SEC separation of the Walker A Spa47 mutant shows oligomeric distribution similar to wild type Spa47. Insert shows background levels of ATP hydrolysis by the mutant Spa47 protein.
Recombinant Spa47 exists in discrete oligomeric states
SEC elution profiles [Fig. 3(A)] suggest that Spa47 exists as a monomer (I) and consistent homo‐oligomer (III); though a strong dependence of SEC on protein shape makes stoichiometry determination difficult. Sedimentation velocity analytical ultracentrifugation (SV‐AUC) measures the sedimentation coefficient for each species in a solution. The sedimentation coefficient is a function of both size and shape, and thus interpreting molecular weights from the peaks of a sedimentation coefficient distribution requires prior knowledge of the shape of each species. However, with the monomer peak identified, stoichiometric assignments can be made by comparing the sedimentation coefficient of the oligomer to that of the monomer. The sedimentation coefficient ratios expected for geometric oligomers up to octamer are outlined,31 and because sedimentation coefficients are compared directly to other sedimentation coefficients this analysis requires no knowledge of or deconvolution of the contribution of shape to the sedimentation coefficient. AUC analysis of purified recombinant Spa47 found two distinct peaks with sedimentation coefficients of 3.0 ± 0.05 and 5.5 ± 0.02 Svedbergs (Fig. 5). The 3.0 s peak was the most abundant, and the sedimentation coefficient is consistent with Spa47 monomer (I) with a moderately elongated shape (frictional ratio of 1.57). If the 3.0 s peak is Spa47 monomer, comparing its sedimentation coefficient to that of the 5.5 s peak yields a ratio of 1.83. As a ratio of 1.75 is expected of a linear trimer, and a ratio of 1.86 is expected of a triangular trimer, the 5.5s peak can be tentatively assigned as trimeric homo‐oligomers (III). A minor peak was also observed at 4.2 ± 0.40s (II) that is consistent with an intermediate species observed in the SEC profile (Fig. 3). Two additional peaks were observed in the sedimentation profiles and likely represent small contaminants that had not yet been removed as the AUC analysis was run prior to SEC to best determine species distribution in the population. The AUC results are consistent with the presence of three primary Spa47 species identified as a monomer (I) and two higher order oligomers (II and III) as seen by SEC. AUC sedimentation coefficient distributions were additionally obtained for Spa47 ranging in concentration from 0.1 to 1.0 mg mL−1 (Supporting Information Fig. S1), finding similar sedimentation profiles as observed in Figure 5. In these experiments the species previously identified as monomer sedimented consistently at ∼3.0 s and the species we believe to be a trimer consistently sedimented at 5.2 s, regardless of concentration, ensuring that the assigned peaks are the result of discrete species and not an artifact resulting from a reaction boundary.
Figure 5.
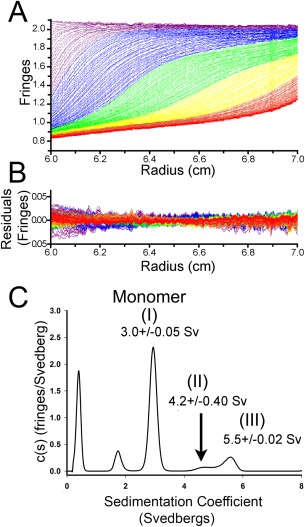
Sedimentation velocity analytical ultracentrifugation analysis of Spa47 oligomerization. (A) Representative interference scans of purified Spa47 monitored during sedimentation velocity analytical ultracentrifugation (SV‐AUC) and (B) representative residuals from fitting the data to a continuous c(s) distribution model as described in the methods. (C) Representative sedimentation coefficient distribution (c(s) versus S) showing the presence of distinct Spa47 species with sedimentation coefficients of 3.0 ± 0.05, 4.2 ± 0.40, and 5.5 ± 0.02 Svedbergs. Sedimentation coefficients represent mean ± SD from 3 independent measurements.
SEC purification isolated the predominant monomeric (I) and oligomeric (III) Spa47 species allowing further characterization of the individual oligomers. CD analysis was performed on the isolated species of Spa47 and Spa47K165A (Fig. 6). The Far‐UV CD spectra are nearly identical for each suggesting that major differences in structure are not driving the formation of oligomers or responsible for the observed increase in ATPase activity. Furthermore, the accompanying CD thermal unfolding curves exhibit very similar transition temperatures, suggesting that the protein structure is not significantly different between the two tested species. The monomeric species for both proteins do, however, exhibit sharper transitions than the oligomers (III), suggesting that the oligomer protein–protein interactions reduce the cooperativity of unfolding. Interestingly, the isolated species were both found to primarily maintain their original oligomeric state and activity levels after nearly 1 month at 4°C, suggesting that even though CD found their structure content to be similar, they remain stable under the tested in vitro storage conditions (data not shown).
Figure 6.
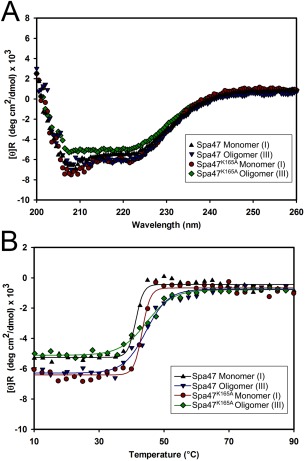
Circular dichroism (CD) analysis of Spa47. (A) Far‐UV CD spectra for monomeric (I) and oligomeric (III) forms of wild‐type Spa47 and Spa47K165A exhibit minima at 208 nm and 222 nm, characteristic of proteins containing high α‐helical secondary structure content. (B) Thermal unfolding profiles were collected by monitoring CD signal at 222 nm as the sample temperature increased from 10 to 90°C. The transition temperatures are similar for each species while the oligomeric (III) forms exhibit more gradual transitions compared to the monomers.
Spa47 ATPase activity is oligomer‐dependent
Together, the SEC and AUC profiles indicate that recombinant Spa47 exists in multiple states with monomer being the predominant species with lesser populations of homo‐oligomers (Figs. 3 and 5). While monomers, hexamers, and dodecamers of similar proteins have been captured before, the AUC results found that the largest observed Spa47 species (III) exhibits a sedimentation coefficient that is much lower than would be observed for a dodecamer or even homo‐hexamer of Spa47 warranting further characterization. A single time‐point activity assay of SEC fractions containing isolated Spa47 oligomers found that each of the oligomer species was able to hydrolyze ATP [Fig. 3(C,D)] although activity levels were not proportional to the quantity of protein in each fraction, suggesting that the formation of the oligomeric complex (III) leads to higher activity levels than seen in the monomeric and complex (II) species [Fig. 3(B)]. Reaction conditions were explored and it was found that consistent activity levels were observed at room temperature (21°C) and slightly basic pH (7.9) allowing a direct comparison of the SEC isolated wild‐type Spa47 oligomeric species as well as those for the Spa47K165A mutant. Reaction rates were determined by monitoring ATP hydrolysis through ADP product formation under equivalent enzyme and substrate concentrations (1.35 μM and 1.0 mM, respectively) as a function of time [Fig. 7(A)]. The results show that the Spa47K165A mutant essentially lacks the ability to hydrolyze ATP even after several minutes under these reaction conditions (k cat values ≤ 0.03 s−1) and identified significant differences in hydrolysis activity for the native Spa47 oligomers. Specifically, the Spa47 monomer and predicted homo‐dimeric species (II) hydrolyze ATP at rates of 0.21 ± 0.01 μM s−1 and 0.14 ± 0.01 μM s−1, respectively, while an equivalent concentration of oligomer (III) predicted to be a trimer exhibits a rate of 1.60 ± 0.05 μM s−1. This corresponds to an apparent k cat of 1.18 ± 0.03 s−1 for (III) which is similar to that of other T3SS ATPases including CdsN and FliI,23, 24, 29, 32 suggesting that Spa47 oligomerization plays a key role in its ATPase activity.
Figure 7.
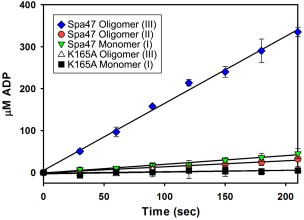
ATPase activity of Spa47. Kinetics of ATP hydrolysis by various oligomeric forms of Spa47. The active oligomeric form (III) of Spa47 hydrolyzes ATP with a k cat of 1.18 ± 0.03 s−1, which is 8‐ to 10‐fold faster than the monomer and smaller oligomeric form (II) (k cat = 0.15 ± 0.01 s−1 and 0.11 ± 0.01 s−1, respectively). All forms of the Spa47K165A mutant protein are essentially inactive for ATP hydrolysis with k cat values ≤0.03 ± 0.01 s−1. Each data point represents the mean ± SD of three independent measurements from two separate protein preparations.
ATPase active Spa47 is necessary for cellular invasion
The full spectrum of T3SS ATPase functions remains unclear though several key studies have demonstrated their importance in vivo and have provided insight into some specific functions.21, 33, 34 While this represents the first direct evidence that Spa47 is an ATPase, its importance in Shigella virulence was shown previously when a knockout strain failed to properly secrete several essential T3SA structural proteins as well as effectors associated with virulence.21 We suspected that this secretion handicap would result in a loss of ability for S. flexneri to invade cultured HeLa cells and directly tested this using a gentamycin protection assay. As expected, we found a spa47 null S. flexneri strain to be essentially noninvasive, exhibiting only 2% invasion compared to the wild‐type strain 2457T. Complementation of the spa47 null strain with a pWPsf4 vector containing the native spa47 gene restored invasion properties to >90% of wild‐type, showing that the presence of Spa47 is essential for cellular invasion and ultimately the ability of S. flexneri to cause infection (Table 1). Because Spa47 has been shown to interact with several T3SA proteins within the basal body of the apparatus,21, 35, 36, 37 the observed phenotype for spa47 null strains may result from a lack of necessary structural responsibilities and/or work directly provided by ATP hydrolysis. We used Spa47K165A developed and characterized in this study to provide an ATPase inactive form of Spa47 to complement the spa47 null strain. This strain containing a single Spa47 point mutation exhibited an identical noninvasive phenotype as the spa47 null strain, suggesting that Spa47 ATPase activity is necessary for cellular invasion. This does not rule out the possibility that Spa47 contributes key structural aspects to the basal body. However, uncoupling the structural and functional roles of Spa47 will require a dedicated set of phenotype and structure studies involving Spa47 mutants such as the ATPase deficient Spa47K165A mutant described in this study.
Table 1.
Invasion Phenotype of Spa47 Mutants
| S. flexneri strain | Complementation | Relative invasion (% ± SD) |
|---|---|---|
| 2457T (WT) | none | 100 ± 0 |
| spa47 null | none | 2 ± 3 |
| spa47 null | spa47/pWPsf4 | 91 ± 14 |
| spa47 null | spa47K165A/pWPsf4 | 2 ± 1 |
Gentamycin protection data outlining the effects of Spa47 mutations on virulence. This assay measures the ability of S. flexneri to invade host cells. The results represent a percentage of colonies formed relative to a wild‐type 2457T S. flexneri strain ± S.D. n = 3 independent experiments each performed in triplicate with an average of 182 colonies for WT.
Discussion
Shigella spp. rely on a T3SS to promote uptake by the epithelial cells lining the large intestine of their human hosts, initiate apoptosis of resident macrophages, escape endocytic vacuoles, and provide actin‐based motility.5, 38, 39 Providing these critical roles at the correct point during infection requires that the T3SS is under strict control at many levels including transcriptional and translational regulation as well as control over the maturation and activation of the injectosome itself.13 The secretion of T3SS translocator proteins and their incorporation into the maturing T3SA have been linked to environmental stimuli by small molecules such as the bile salt deoxycholate38, 40, 41 and lipid membrane components including cholesterol and sphingomyelin.11 This stimulation has been suggested to provide signals which essentially monitor progress as the pathogen enters the host and passes through the gastrointestinal tract, preparing the Shigella T3SA for invasion of the colonic epithelium. What has remained unclear, however, is what provides the driving force for protein secretion through the Shigella T3SA and ultimately how secretion is controlled.
Sequence homology between the Shigella protein Spa47 and identified ATPases including the β‐subunit of the hetero‐hexameric F1Fo ATP synthase, FliI of the bacterial flagellar system, and ATPases associated with the translocon T3SS of several pathogens suggests that Spa47 is a T3SS‐associated ATPase in Shigella and that it may provide the energy necessary for T3SS‐associated secretion. Much of this stems from work showing that a spa47 null Shigella strain is unable to construct the T3SA and as a result fails to secrete T3SS proteins, including several Ipas and the phosphatase effector IpgD,21 though challenges purifying Spa47 have prevented a direct correlation between Shigella T3SS activity and Spa47 ATP hydrolysis. In this study, we developed an expression and purification method for recombinant Spa47 that allowed us to provide the first direct characterization of its enzymatic activity and identify a link between Spa47 oligomer state and ATPase activity. Furthermore, we captured and characterized the active Spa47 homo‐oligomer (III), finding that it likely represents a smaller T3SS ATPase complex than has been observed for other systems and provide evidence suggesting that Shigella invasion of host cells requires Spa47 catalyzed ATP hydrolysis.
Size exclusion chromatography of recombinant Spa47 resulted in multiple resolved species, suggesting it exists in discrete oligomeric states [Fig. 3(A,B)] which were determined by AUC to be primarily Spa47 monomer (I) with a lesser population of oligomers (II and III) which we predict to be Spa47 homo‐dimers and homo‐trimers, respectively, based on sedimentation coefficients obtained by AUC (Fig. 5). While the implication of this distribution was initially unclear, Spa47 activity was found to be highly dependent on oligomeric state, suggesting it may be an important component of a regulation mechanism. Many ATPases including members of the AAA and AAA+ classes undergo oligomerization to form active complexes, often serving as a means of regulation and providing an amplification of the mechanical force generated by ATP hydrolysis.42 One such example is the highly studied flagellar ATPase FliI (Spa47 homolog) whose oligomerization to an active hexamer is negatively influenced by interaction with the regulatory protein FliH and is stimulated by ATP and E. coli phospholipid binding.43, 44 Regulation of the ATPase FliI through oligomerization provides tight control over export of flagellar proteins and may serve a similar role in the evolutionarily related Shigella T3SS.
Activity profiles of the isolated Spa47 oligomers found that the activated oligomer (III) exhibited 8 fold higher activity levels than the monomer (I) (k cat = 1.18 ± 0.03 s−1) [Fig. 7(A)], suggesting that Spa47 activation may be controlled by oligomeric state and specifically formation of the observed complex. In contrast to FliI, however, Spa47 formed oligomers in the absence of ATP (Figs. 3 and 5) as has been demonstrated for the T3SS ATPase HrcN from the plant pathogen Pseudomonas syringae.26 Although active hexameric, dodecameric, and even higher order oligomeric states have been reported, we believe that the observed active Spa47 complex (III) studied here represents a novel trimeric T3SS ATPase though additional work is required to confirm the precise stoichiometry of the active species. Additionally, while we cannot rule out the possibility of ATP driven higher order oligomerization of the active oligomer (III) (e.g., hexamer as seen in the transmission electron microscopy generated structure),35 the ATP hydrolysis deficient Spa47K165A mutant displayed oligomeric behavior similar to wild‐type Spa47 suggesting that ATP is not required for Spa47 oligomer formation as the equivalent lysine in FliI and EscN was needed to stabilize ATP binding.26, 29
CD analysis of the monomeric and oligomeric (III) forms of wild‐type and the K165A Spa47 mutant found that each result in spectra characteristic of highly α‐helical proteins (Fig. 6) consistent with the known structures of related T3SS ATPases.26, 33, 45 Together these results suggest that neither the oligomeric state nor the K165A mutation drastically affect the protein secondary structure. Additionally, the thermal unfolding curves exhibit similar transition temperatures while the active species (III) demonstrating shallower transitions reflecting the additional protein interactions of the multimeric state. Without atomic resolution structural data, it is impossible to entirely rule out protein misfolding as the reason for the appearance of the less active monomeric species. However, insight from related systems together with the data presented here support the hypothesis that Spa47 oligomerization is involved in activation and regulation of the Shigella T3SS.
Recent transmission electron microscopy electron density maps provided new insight into the architecture of the Shigella T3SA sorting platform and confirmed the relevance of Spa47 oligomer formation by identifying a symmetric homo‐hexameric Spa47 ring at the base of the intact T3SA. The Spa47 hexamer is positioned directly below the MxiA export gate35, 46 suggesting that secreted proteins interact with Spa47 prior to entering the apparatus needle for export. The structure further suggests that Spa47 is positioned via symmetric hexameric linkage to six individual Spa33 units of the C ring through direct interaction with MxiN, confirming yeast two‐hybrid and coimmunoprecipitation assays that first described these interactions.21, 36, 37 The observed interaction between Spa47 and MxiN is particularly interesting as MxiN is homologous to the regulatory protein FliH in the flagellar export apparatus which appears to prevent formation of the active FliI homo‐hexamer, preventing the wasteful hydrolysis of ATP before the flagellar export apparatus is prepared for productive transport.43, 44 A similar mechanism would be beneficial to the Shigella T3SS although the association of MxiN and Spa47 appears to be necessary for proper formation of the sorting platform35 and a mxiN null Shigella strain less efficiently secretes MxiH and MxiI, preventing proper T3SA needle formation.21
We suspect that the role of Spa47 in the Shigella T3SS is multifaceted, serving as both a structural component within the sorting platform and as an ATPase that provides the energy necessary for secretion, perhaps through recognition, chaperone release, and partial unfolding of transported proteins as has been demonstrated for the Salmonella enterica ATPase InvC.34 In Shigella, it has been shown that a spa47 null Shigella strain is unable to form the external needle structure of the T3SA and as a result is secretion and invasion deficient.21, 47 We tested the invasion phenotype of a Shigella spa47 null strain complemented with the engineered ATPase inactive Spa47K165A mutant finding that it resulted in the same noninvasive phenotype as the null strain even though invasion was restored when complemented with native spa47. Together with recent structural findings35 and the enzymatic activity characterization provided in this study, it appears that Spa47 may play critical structural roles in the T3SA and is in fact a functional ATPase whose activity may be tied directly to Shigella virulence. The findings presented here are the result of overcoming the long‐standing hurdle of recombinant expression and purification of the Shigella ATPase Spa47 and together have uniquely positioned us to begin answering a number of key questions regarding Spa47 activation, T3SS secretion regulation, and potential structural/organizational roles of Spa47 within the sorting platform of the T3SA. Ultimately, answering these questions will require a direct comparison of protein interactions, phenotype assays, and further characterization of enzyme activity and structure. Together, the methods developed in this study to produce active recombinant Spa47, the generation of a key ATPase inactive Spa47 point mutant, and the link identified between Spa47 oligomeric state and ATPase activity will provide a strong platform for this additional work dissecting the mechanism of Spa47 catalyzed ATP hydrolysis and its role in the Shigella T3SS.
Materials and Methods
Materials
Wild‐type (WT) S. flexneri corresponds to the serotype 2a 2457T strain originally isolated in 1954.48 The S. flexneri spa47 null strain was engineered by Abdelmounaaïm Allaoui as described in Jouihri et al.21 E. coli strains and 2X ligation mix were from Novagen (Madison, WI). Restriction enzymes, the pTYB21 protein expression plasmid, PCR buffer, Phusion High‐Fidelity polymerase, and chitin resin were purchased from New England Biolabs (Ipswich, MA). Oligonucleotide primers and the synthesized Spa47 gene were from Integrated DNA Technologies (Coralville, IA). The Superdex 16/600 size exclusion and 5 mL Q FF columns were purchased from General Electric (Pittsburgh, PA). ATP was from Sigma–Aldrich (St. Louis, MO) and α‐32P‐ATP was from Perkin Elmer (Boston, MA). DTT and ampicillin were from Gold Biotechnology (St. Louis, MO). All other solutions and chemicals were of reagent grade.
Methods
Cloning
The Spa47 gene was purchased as a gBlock gene fragment from Integrated DNA Technologies. A silent mutation was created within the gene to remove the native internal SapI restriction site. A PstI restriction site was included at the 3′ end of the gene immediately following the native stop codon and a SapI site followed by the sequence “CAAC” was introduced 5′ of the native start codon. The additional sequence maintains the proper reading frame and provides an N‐terminal Asparagine in Spa47 which aids in efficient intein cleavage during protein purification. Additional product was generated by amplifying the gBlock using complementary primers containing 5′‐GGTGGT sequences to aid in digestion (5′‐GGTGGT TGCTCTTCCAACATGAGCTATACAAAATTGCT‐3′ and 5′‐ GGTGGTCTGCAGTCATTATCTAATTGTTTCACCA ATA‐3′). The SapI/PstI digested gene was ligated into the expression plasmid pTYB21 which encodes an N‐terminal chitin binding domain (CBD) and intein linker. The ligated product was transformed into E. coli Nova blue cells by heat shock and screened for the presence of spa47 via PCR. Sequences were verified by Sanger sequencing (Genewiz, Inc., South Plainfield, NJ). spa47 was cloned into the plasmid pWPsf4 by introducing NdeI and BamHI restriction sites at the 5′ and 3′ ends, respectively, and ligating into the digested vector backbone. Sequences were again verified by Sanger sequencing prior to transformation into electro‐competent S. flexneri strains via electroporation. The Spa47K165A point mutation was made in both pTYB21 and pWPsf4 using inverse PCR followed by sequence verification and transformation into E. coli and S. flexneri, respectively.
Protein expression and purification
Spa47 and spa47K165A in pTYB21 were transformed into E. coli Tuner (DE3) cells and were grown overnight in LB Broth (Miller) containing 0.1 mg/mL ampicillin to ensure maintenance of the plasmid. A 5 mL L−1 volume of the starter culture was added to Terrific Broth media containing 0.1 mg mL−1 ampicillin and was grown to an OD600 of 0.8 at 37°C, 200 RPM. The culture was then cooled to 17°C before induction with 1 mM IPTG for ∼20 h (17°C, 200RPM). All subsequent steps were carried out at 4°C. Typical growths included 3 L of Spa47 expressing E. coli culture (1 L per 2.8‐L baffled Fernbach flask) Cells were pelleted by centrifugation, resulting in typical cell pellet masses of ∼27 g L−1, resuspended in binding buffer (20 mM Tris, 500 mM NaCl, pH 7.9), and lysed by sonication. The suspension was clarified by centrifugation and the supernatant run over a chitin affinity column (10 mL resin per liter of cultured cells) at 1 mL min−1 followed by washing with ∼20 bed volumes of Binding Buffer. The chitin beads were quickly equilibrated with 3 bed volumes of cleavage buffer (20 mM Tris, 500 mM NaCl, 50 mM DTT, pH 7.9) and incubated overnight to cleave Spa47 from the chitin binding domain (CBD). Column elutions were collected three times per day until no further elution of Spa47 was observed by SDS‐PAGE (∼4 days). Collected elutions were pooled and diluted to generate final buffer concentrations of 20 mM Tris, 100 mM NaCl, 5 mM DTT, pH 7.9, and further purified over a 5 mL Q Sepharose FF anion exchange column, consistently resulting in ∼10 mg of Spa47 per liter of expression culture. The Q column provided a negative selection and the purified Spa47 in the flow through was concentrated using an Amicon Ultra centrifugal filter unit with a 30‐kDa molecular weight cut off and further purified/characterized using a Superdex 16/600 size exclusion column equilibrated with 20 mM Tris, 100 mM NaCl, 5 mM DTT, pH 7.9. Spa47 contains no Tryptophan residues, resulting in a low molar A280 extinction coefficient that is below the limits of quantitation for the concentrations of Spa47 obtained and used throughout the study. All Spa47 concentrations were determined using a more sensitive Bradford protein assay with bovine serum albumin as a standard after verifying that comparison to a BSA standard curve provides accurate Spa47 quantitation results. All Spa47 concentrations are reported in monomer concentration units for consistency and clarity.
ATP hydrolysis activity assays
A single time point activity assay was used to observe ATP hydrolysis from collected size exclusion chromatography (SEC) fractions of Spa47 and Spa47K165A. The reactions were initiated by combining protein from the SEC fractions with a prepared ATP solution resulting in a final concentration of 0.5 mM ATP, 10 mM MgCl2, and 0.5 μCi (∼300 nM) α‐32P‐ATP. The reactions were allowed to proceed for 15 min before quenching with the addition of EDTA to a final concentration of 0.25 M. The level of ATP hydrolysis was quantified by first spotting 1 µL of the quenched reaction onto a TLC plate and developing in 0.6 M potassium phosphate buffer (pH 3.4) for 70 min. The α‐32P‐ATP and the α‐32P‐ADP were detected with a Storm PhosphorImager (Molecular Dynamics) and quantified using associated ImageQuant software (Molecular Dynamics). A multiple time point activity assay was used to determine apparent k cat values for Spa47 species and was carried out under similar conditions to the single time point assay described above. Specifically, the purified Spa47 concentration was held constant at 1.35 μM for each species, the ATP concentration was increased to 1 mM, and samples were removed from the reaction every 30 s for 3.5 min and rapidly quenched with 0.25 M EDTA prior to separation via TLC.
Far‐UV circular dichroism (CD)
Far‐UV CD spectra and thermal stability melts were obtained for isolated monomer (I) and oligomer fractions (III) of Spa47 and Spa47K165A. Measurements were obtained using a JASCO model J‐1500 spectropolarimeter with a Peltier temperature controller (Jasco, Easton, MD). Spectra were obtained from 200 to 260 nm at 10°C with a 0.1 cm path length, 0.5 nm spectral resolution, 50 nm min−1 scan rate and a 1‐s data integration time. Secondary structure thermal stability profiles were generated by monitoring CD signal at 222 nm while the temperature was increased from 10 to 90°C at 0.3°C min−1. All measurements were performed at 0.3 mg mL−1 protein concentration. CD signals were converted to mean residue molar ellipticity.
Sedimentation velocity analytical ultracentrifugation
Sedimentation velocity analytical ultracentrifugation (SV‐AUC) experiments were conducted using an Optima XL‐I (Beckman Coulter, Fullerton, CA) analytical ultracentrifuge equipped with an interference optical detection system. 1.5 mg mL−1 Spa47 and a buffer reference were loaded into Beckman charcoal‐epon two sector cells with 12‐mm path lengths and sapphire windows. The samples were analyzed at 20°C and 40,000 RPM, using interference detection and scanning until complete sedimentation was achieved. The data were regularized with a confidence interval of 0.95 and analyzed using the software program Sedfit49 with a continuous c(s) distribution and 500 scans. The Spa47 partial specific volume (0.7422 mL g−1) and the buffer density and viscosity used in the analysis (1.0031 g mL−1 and 0.01018 Poise, respectively) were calculated using Sednterp.50 Additional SV‐AUC distributions were collected for Spa47 concentrations of 1.0, 0.66, and 0.1 mg mL−1 Spa47 to ensure that the assigned peaks are the result of discrete species and not an artifact resulting from a reaction boundary (Supporting Information Fig. S1).
Bacterial invasion of epithelial cells
S. flexneri invasion of cultured HeLa cells was monitored by a gentamicin protection assay as previously described.51 Sterile 24‐well plates were seeded with passaged HeLa cells and grown overnight in DMEM supplemented with 10% fetal calf serum, penicillin, and streptomycin at 100% relative humidity, 37°C, and 5% CO2. Tested S. flexneri strains were streaked onto tryptic soy agar plates containing 0.025% Congo red and grown overnight at 37°C. Small cultures containing appropriate antibiotics to maintain the transformed plasmid were inoculated from the agar plates and grown to an OD600 ∼0.4 at 37°C and 200 RPM. Equivalent bacterial loads were then introduced to the cultured HeLa cells as described previously.51 The plates were centrifuged at 1000 g to synchronize contact between the bacteria and HeLa cells, incubated at 37°C for 30 min, and rinsed to remove most of the extracellular bacteria. Shigella that had not successfully invaded the HeLa cells were killed by treatment with 50 μg mL−1 gentamicin. Bacteria that had invaded were visualized by lysing the host cells with 1% agarose in water and overlaying with a 2× LB agar solution. Overnight incubation at 37°C resulted in colony formation that is quantified and used to provide relative levels of invasiveness between the tested S. flexneri strains.
Supporting information
Supporting Information
Acknowledgments
The authors would like to thank Robert Burgess for assistance with protein expression and purification and William and Wendy Picking (University of Kansas in Lawrence, KS) for access to the CD spectropolarimeter and for critical reading of the manuscript. They also acknowledge Andrew Keightley with the mass spectrometry and proteomics facility at UMKC in Kansas City, MO for providing mass spectrometry services.
References
- 1. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins‐Browne RM, Levine MM (2013) Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case‐control study. Lancet 382:209–222. [DOI] [PubMed] [Google Scholar]
- 2. DuPont HL, Levine MM, Hornick RB, Formal SB (1989) Inoculum size in shigellosis and implications for expected mode of transmission. J Infect Dis 159:1126–1128. [DOI] [PubMed] [Google Scholar]
- 3. Kotloff KL, Winickoff JP, Ivanoff B, Clemens JD, Swerdlow DL, Sansonetti PJ, Adak GK, Levine MM (1999) Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ 77:651–666. [PMC free article] [PubMed] [Google Scholar]
- 4. Dutta S, Ghosh A, Ghosh K, Dutta D, Bhattacharya SK, Nair GB, Yoshida S (2003) Newly emerged multiple‐antibiotic‐resistant Shigella dysenteriae type 1 strains in and around Kolkata, India, are clonal. J Clin Microbiol 41:5833–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carayol N, Nhieu GTV (2013) The inside story of Shigella invasion of intestinal epithelial cells. Csh Perspect Med 3:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schroeder GN, Jann NJ, Hilbi H (2007) Intracellular type III secretion by cytoplasmic Shigella flexneri promotes caspase‐1‐dependent macrophage cell death. Microbiology 153:2862–2876. [DOI] [PubMed] [Google Scholar]
- 7. Chatterjee S, Chaudhury S, McShan AC, Kaur K, De Guzman RN (2013) Structure and biophysics of Type III secretion in bacteria. Biochemistry 52:2508–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ashida H, Mimuro H, Sasakawa C (2015) Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front Immunol 6:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enninga J, Mounier J, Sansonetti P, Tran Van Nhieu G (2005) Secretion of type III effectors into host cells in real time. Nat Methods 2:959–965. [DOI] [PubMed] [Google Scholar]
- 10. Cordes FS, Komoriya K, Larquet E, Yang S, Egelman EH, Blocker A, Lea SM (2003) Helical structure of the needle of the type III secretion system of Shigella flexneri . J Biol Chem 278:17103–17107. [DOI] [PubMed] [Google Scholar]
- 11. Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD (2009) Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infection Immun 77:2754–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deane JE, Abrusci P, Johnson S, Lea SM (2010) Timing is everything: the regulation of type III secretion. Cell Mol Life Sci 67:1065–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marteyn B, Gazi A, Sansonetti P (2012) Shigella: a model of virulence regulation in vivo. Gut Microbes 3:104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pallen MJ, Bailey CM, Beatson SA (2006) Evolutionary links between FliH/YscL‐like proteins from bacterial type III secretion systems and second‐stalk components of the FoF1 and vacuolar ATPases. Protein Sci 15:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blocker A, Komoriya K, Aizawa S (2003) Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci USA 100:3027–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eichelberg K, Ginocchio CC, Galan JE (1994) Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol 176:4501–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Akeda Y, Galan JE (2004) Genetic analysis of the Salmonella enterica type III secretion‐associated ATPase InvC defines discrete functional domains. J Bacteriol 186:2402–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woestyn S, Allaoui A, Wattiau P, Cornelis GR (1994) YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol 176:1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee PC, Rietsch A (2015) Fueling type III secretion. Trends Microbiol 23:296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burkinshaw BJ, Strynadka NC (2014) Assembly and structure of the T3SS. Biochim Biophys Acta 1843:1649–1663. [DOI] [PubMed] [Google Scholar]
- 21. Jouihri N, Sory MP, Page AL, Gounon P, Parsot C, Allaoui A (2003) MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri . Mol Microbiol 49:755–767. [DOI] [PubMed] [Google Scholar]
- 22. Chatterjee R, Halder PK, Datta S (2013) Identification and molecular characterization of YsaL (Ye3555): a novel negative regulator of YsaN ATPase in type three secretion system of enteropathogenic bacteria Yersinia enterocolitica . PLoS One 8:e75028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Andrade A, Pardo JP, Espinosa N, Perez‐Hernandez G, Gonzalez‐Pedrajo B (2007) Enzymatic characterization of the enteropathogenic Escherichia coli type III secretion ATPase EscN. Arch Biochem Biophys 468:121–127. [DOI] [PubMed] [Google Scholar]
- 24. Stone CB, Johnson DL, Bulir DC, Gilchrist JD, Mahony JB (2008) Characterization of the putative type III secretion ATPase CdsN (Cpn0707) of Chlamydophila pneumoniae . J Bacteriol 190:6580–6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pozidis C, Chalkiadaki A, Gomez‐Serrano A, Stahlberg H, Brown I, Tampakaki AP, Lustig A, Sianidis G, Politou AS, Engel A, Panopoulos NJ, Mansfield J, Pugsley AP, Karamanou S, Economou A (2003) Type III protein translocase: HrcN is a peripheral ATPase that is activated by oligomerization. J Biol Chem 278:25816–25824. [DOI] [PubMed] [Google Scholar]
- 26. Zarivach R, Vuckovic M, Deng W, Finlay BB, Strynadka NC (2007) Structural analysis of a prototypical ATPase from the type III secretion system. Nat Struct Mol Biol 14:131–137. [DOI] [PubMed] [Google Scholar]
- 27. Habersetzer J, Ziani W, Larrieu I, Stines‐Chaumeil C, Giraud MF, Brethes D, Dautant A, Paumard P (2013) ATP synthase oligomerization: from the enzyme models to the mitochondrial morphology. Int J Biochem Cell Biol 45:99–105. [DOI] [PubMed] [Google Scholar]
- 28. Claret L, Calder SR, Higgins M, Hughes C (2003) Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol 48:1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan F, Macnab RM (1996) Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium . J Biol Chem 271:31981–31988. [DOI] [PubMed] [Google Scholar]
- 30. Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha‐ and beta‐subunits of ATP synthase, myosin, kinases and other ATP‐requiring enzymes and a common nucleotide binding fold. Embo J 1:945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia de la Torre JG, Bloomfield VA (1981) Hydrodynamic properties of complex, rigid, biological macromolecules: theory and applications. Quart Rev Biophys 14:81–139. [DOI] [PubMed] [Google Scholar]
- 32. Stone CB, Bulir DC, Gilchrist JD, Toor RK, Mahony JB (2010) Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae . BMC Microbiol 10(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Allison SE, Tuinema BR, Everson ES, Sugiman‐Marangos S, Zhang K, Junop MS, Coombes BK (2014) Identification of the docking site between a type III secretion system ATPase and a chaperone for effector cargo. J Biol Chem 289:23734–23744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Akeda Y, Galan JE (2005) Chaperone release and unfolding of substrates in type III secretion. Nature 437:911–915. [DOI] [PubMed] [Google Scholar]
- 35. Hu B, Morado DR, Margolin W, Rohde JR, Arizmendi O, Picking WL, Picking WD, Liu J (2015) Visualization of the type III secretion sorting platform of Shigella flexneri . Proc Natl Acad Sci USA 112:1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johnson S, Blocker A (2008) Characterization of soluble complexes of the Shigella flexneri type III secretion system ATPase. FEMS Microbiol Lett 286:274–278. [DOI] [PubMed] [Google Scholar]
- 37. Morita‐Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, Sasakawa C (2006) Shigella Spa33 is an essential C‐ring component of type III secretion machinery. J Biol Chem 281:599–607. [DOI] [PubMed] [Google Scholar]
- 38. Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD (2007) Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun 75:2626–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Niebuhr K, Sansonetti PJ (2000) Invasion of epithelial cells by bacterial pathogens the paradigm of Shigella . Subcell Biochem 33:251–287. [DOI] [PubMed] [Google Scholar]
- 40. Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD (2011) Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry 50:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dickenson NE, Arizmendi O, Patil MK, Toth RTT, Middaugh CR, Picking WD, Picking WL (2013) N‐terminus of IpaB provides a potential anchor to the Shigella type III secretion system tip complex protein IpaD. Biochemistry 52:8790–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White SR, Lauring B (2007) AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic 8:1657–1667. [DOI] [PubMed] [Google Scholar]
- 43. Minamino T, Kazetani K, Tahara A, Suzuki H, Furukawa Y, Kihara M, Namba K (2006) Oligomerization of the bacterial flagellar ATPase FliI is controlled by its extreme N‐terminal region. J Mol Biol 360:510–519. [DOI] [PubMed] [Google Scholar]
- 44. Minamino T, MacNab RM (2000) FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol Microbiol 37:1494–1503. [DOI] [PubMed] [Google Scholar]
- 45. Imada K, Minamino T, Tahara A, Namba K (2007) Structural similarity between the flagellar type III ATPase Flil and F‐1‐ATPase subunits. Proc Natl Acad Sci USA 104:485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Abrusci P, Vergara‐Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM (2013) Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol 20:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tamano K, Aizawa S, Katayama E, Nonaka T, Imajoh‐Ohmi S, Kuwae A, Nagai S, Sasakawa C (2000) Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. Embo J 19:3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Formal SB, Dammin GJ, Labrec EH, Schneider H (1958) Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol 75:604–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lebowitz J, Lewis MS, Schuck P (2002) Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci 11:2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laue T, Shah B, Ridgeway T, Pelletier S, Analytical Ultracentrifugation in Biochemistry and Polymer Science In: Harding S, Rowe A, Horton JC, Eds. (1992). United Kingdom: Royal Society of Chemistry, 1–644. [Google Scholar]
- 51. Niesel DW, Chambers CE, Stockman SL (1985) Quantitation of HeLa cell monolayer invasion by Shigella and Salmonella species. J Clin Microbiol 22:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


