Abstract
Tree peony (Paeonia suffruticosa) is a perennial plant indigenous to China known for its elegant and vibrantly colorful flowers. A few genes involved in petal pigmentation have been cloned in tree peony. However, to date, there have been few studies on the comparison and selection of stable reference genes for gene expression analysis by quantitative reverse-transcription PCR (qRT-PCR) in this species. In this study, 10 candidate reference genes were evaluated for the normalization of qRT-PCR in three tree peony cultivars. GAPDH and UBC were identified as the top two most stable reference genes in ‘Feng Dan’ and ‘Xi Shi,’ and EF-1α/UBC was recommended to be the best combination for ‘Que Hao.’ The expression stability of various reference genes differed across cultivars, suggesting that selection and validation of reliable reference genes for quantitative gene expression analysis was necessary not only for different species but also for different cultivars. The results provided a list of reference genes for further study on gene expression in P. suffruticosa. However, in any case, a preliminary check on the accuracy of the best performing reference genes is requested for each qRT-PCR experiment.
Keywords: tree peony, reference genes, gene expression normalization, qRT-PCR, flower development
Introduction
Gene expression analysis provides improved understanding and insights into the molecular basis underpinning various biological processes (Bustin et al., 2005). Quantitative real-time polymerase chain reaction (qRT-PCR) is one frequently used platform to quantify transcript abundance, for its sensitivity, accuracy and reproducibility (Gachon et al., 2004). However, the accuracy of qRT-PCR is heavily dependent on the stability of the internal reference genes used to normalize transcript abundances (Huggett et al., 2005). An ideal reference gene must be expressed steadily both in different tissues and at different developmental stages and remain unaffected by any experimental treatment (Czechowski et al., 2005). Many studies have confirmed the fact that the utilization of unstable reference genes may result in significant biases and misinterpretations of data (Ferguson et al., 2010; Mafra et al., 2012). It is thus critical to select suitable reference genes before quantifying the expression level via qRT-PCR.
Various software packages, most prominently geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), Bestkeeper (Pfaffl et al., 2004), and RefFinder (Xie et al., 2012) have been developed to identify appropriate reference genes in different experimental systems. They have been widely used in different plants, including the model plant Arabidopsis (Wang H. B. et al., 2014), ornamental plants (Wang et al., 2015; Zhang et al., 2015), vegetables and fruits (Campos et al., 2014; Wu et al., 2014), crops (Wei et al., 2013; Ling et al., 2014), grasses (Yang et al., 2015; Zhuang et al., 2015), and some wild plants grown in special habitats (Wang H. L. et al., 2014; Xiao et al., 2015). All these studies have proved the importance of reference gene screening and validation before gene expression analysis.
The tree peony (Paeonia suffruticosa Andr.) is an ornamental plant native to China with striking ornamental and medicinal value. It has a cultivation history going back more than 1600 years in China (Cheng and Li, 1998). Qualitative and quantitative analyses of anthocyanins have been conducted in P. suffruticosa (Zhang et al., 2007; Yin et al., 2012). A few genes involved in petal pigmentation have also been cloned in the tree peony (Zhou et al., 2013; Du et al., 2015; Shi et al., 2015). However, to date, there have been few studies on the comparison and selection of reference genes for qRT-PCR during flower development in tree peony. Only few studies focusing on the bud dormancy release (Zhang et al., 2011), various tissues (Liu et al., 2015), and different treatments in cut flowers (Wang et al., 2012) have been reported, showing that no gene has a constant expression profile under all developmental or experimental conditions.
To normalize the gene expression level during flower development in different cultivars of tree peony, 10 candidate reference genes were selected based on the floral transcriptome datasets of tree peony and the stability of their expression level was evaluated. The results provided a list of reference genes suitable for the accurate quantification of gene expression during flower development.
Materials and methods
Plant materials
Tree peonies were grown in the germplasms nursery of Shanghai Chenshan Botanical Garden. Three cultivars (‘Feng Dan,’ ‘Xi Shi,’ and ‘Que Hao’) were used as the plant materials. For each cultivar, petal samples were collected from three different plants at six opening stages (February to April, 2015). Samples from 9 plants (3 plants per cultivar) at 6 different developmental stages were taken in 2015: February 17, March 1, March 8, March 15, April 9, and April 14. The 54 petal samples were immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
RNA extraction and cDNA synthesis
Total RNA was extracted using an E.Z.N.A.™ Plant RNA Kit-R6827 (Omega) following its manufacturer instructions. To avoid DNA contamination, the RNA samples were treated with an RNase-free DNase Set (Omega). The total RNA concentration, purity and integrity were determined using a NanoDrop2000c Spectrophotometer (Thermo Scientific, U.S.) and visually assessed via 1.5% agarose gel electrophoresis. The RNA samples with absorption ratios of A260/A280 between 1.9 and 2.1 and A260/A230 ratios above 2.0 were used for subsequent analyses. First-strand cDNA was synthesized using 1 μg total RNA with the PrimeScript RT Master Mix (Perfect Real Time; Takara, Japan) according to the manufacturer's protocol.
Candidate reference genes selection and PCR primer design
An Illumina/Solexa library of tree peony was constructed in our preliminary study. A total of 15019 unigenes (NR database) were annotated with BLASTX (data was not shown). The indices of mean expression value (MV), standard deviation (SD), and coefficient of variation (CV) value of RPKM, were calculated using the method described by de Jonge et al. (2007) for mining candidate reference genes from transcriptome data. A total of 10 candidate genes were selected and assessed (Table 1).
Table 1.
Primer sequences and amplification characteristics for 10 candidate reference genes and 1 gene of interest.
| Unigene | Gene symbol | Gene name | Accession number | Arabidopsis Homolog locus | Primer [5′−3′] Forward/reverse | Product Size (bp) | TM (°C) | E (%) | R2 |
|---|---|---|---|---|---|---|---|---|---|
| comp39032_c0_seq1 | Actin | Actin | KU853024 | AT3G27000.1 | AAGGTGATGGAATGGCTGAC/CAAGAACTGCTCCTCCAAGG | 264 | 60 | 96.48 | 0.997 |
| comp72555_c2_seq1 | α-TUB | α-tubulin | KU853025 | AT1G50010.1 | AAAACTGTTGGTGGAGGCGA/TCTGGGTGAAAGAGTTGGCG | 152 | 60 | 95.71 | 0.999 |
| comp78934_c0_seq1 | β-TUB | β-tubulin | KU853026 | AT5G23860.2 | AGAACGCCGACGAGTGTATG/AGGGAAACGAAGGCAGCAAG | 149 | 60 | 90.51 | 0.994 |
| comp21853_c0_seq1 | UBC | Ubiquitin conjugating enzyme | KU853027 | AT3G46460.1 | TACCCAAACAGCCCTCCAAC/AGCAGCTTCAATGTTTGCCG | 234 | 59 | 98.82 | 0.994 |
| comp72593_c0_seq1 | GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | KU853028 | AT1G13440.1 | CTGGAGTTTTCACCGACAAGG/CCAATGGGGCAAGACAGTTAG | 186 | 60 | 92.78 | 0.998 |
| comp21491_c1_seq1 | PP2A | Protein phosphatase 2A | KU853029 | AT1G10430.1 | TGATTACTTGCCCCTCACAGCC/CATAACAGGTCGCACATTGGTC | 150 | 60 | 91.46 | 0.995 |
| comp62257_c0_seq1 | TIP41 | TIP41-like protein | KU853030 | AT4G34270.1 | ACGTTCCATTCTCACCTCCC/GCTTCCAACCAGTAAGAGCG | 164 | 59 | 101.45 | 0.993 |
| comp54677_c1_seq1 | SAMS | s-adenosylmethionine synthetase | KU853031 | AT4G01850.2 | TCTTCCACCTCAACCCATCTG/TAAGCACCACTCCTGTCCAC | 166 | 59 | 104.74 | 0.991 |
| comp82277_c2_seq1 | EF-1α | Elongation factor 1 alpha | KU853032 | AT5G60390.2 | TCCTGGGCATCGTGACTTTA/TTCATCATACCGAGCCTTTG | 232 | 60 | 94.57 | 0.997 |
| comp62008_c0_seq1 | CYP | cyclophilin | KU853033 | AT2G36130.1 | GTTCTTCATTACCTTGGCACCT/TGACCGTTGACCGTAGTATCTT | 161 | 58 | 99.23 | 0.995 |
| comp63583_c0_seq1 | F3H | flavanone 3-hydroxylase | KU853034 | AT3G51240.1 | GCTCTACCGCCTGATGACAAG/GTTACTCGCTTCCACCCCTCC | 191 | 60 | 98.58 | 0.996 |
Unigenes were selected from the transcriptome of tree peony. The mean efficiency (E) of PCR amplification and the regression coefficient (R2) for each primer pair were calculated by the LightCycler 96 SW Software.
Primers were designed with Primer Premier 5.0 (Lalitha, 2004), using the following criteria: targeting the 3′-untranslated region, no hairpin/dimer/false priming/cross dimer structures, Tm = 55–60°C, 18–25 bases in length, GC content between 40 and 60%, and amplicon length from 100 to 300 bp. The designed primer sets were BLASTed against the local floral transcriptional data of tree peony to verify primer specificity. Amplification efficiency (E) was evaluated using a standard curve generated by qRT-PCR using a 5-fold dilution series. The primer specificity was judged by melting-curve analysis and agarose gel electrophoresis analysis.
Real-time quantitative PCR
Real-time PCR reactions were carried out in 96-well plates with a LightCycler96 System (Roche, Switzerland) using SYBR Green I Master. The reaction mixture contained 10 μL of SYBR Green I Master, 2.0 μL of diluted cDNA (1:25), 0.4 μL of each of the forward and reverse primers (10 μM), and 7.2 μL of PCR-grade water in a final volume of 20 μL. The following reaction conditions were applied: 30 s at 95°C, 40 cycles of 10 s at 95°C and 30 s at 60°C, and a melting curve protocol (65–95°C with fluorescence measured every 0.5°C). Each reaction was performed in three biological replicates and three technical replicates. Three no-template controls (NTC) were used to monitor possible DNA contamination.
Analysis of gene expression stability and validation of reference genes
Three software programs (GeNorm 3.5, BestKeeper and NormFinder) were used to rank the stability of the 10 selected reference genes in the study. Moreover, RefFinder (http://fulxie.0fees.us/?type=reference), a comprehensive web-based tool was used to integrate and rank comprehensively the tested candidate genes. The raw Ct values were converted into relative quantities using the formula: E−ΔCt (ΔCt = Ct value of each sample—the minimum Ct value) before data entry (Ramakers et al., 2003). All three of the software programs were run based on the software manuals to select suitable reference genes. RefFinder generated the final overall ranking of tested reference genes based on the geometric mean of the weights of every gene calculating by each program.
To validate the reliability of the reference genes, the relative expression levels of PsF3H in three cultivars, a key gene involved in the anthocyanin biosynthesis, was analyzed using the two top-ranking reference genes and the least stable reference gene, as determined by RefFinder, alone or in combination (calculated by geometric mean) for data normalization. The qRT-PCR amplification conditions were the same as described above. The primer specificity of PsF3H was verified as described for reference genes (Table 1). The relative expression data was calculated according to the 2−ΔΔCq method and presented as fold change (Livak and Schmittgen, 2001).
Results
Amplification specificity and efficiency
The primer sequences and amplicon characteristics of 10 candidate reference genes were summarized in Table 1. The primer specificities were assessed by melting-curves and agarose gel electrophoresis analysis, which showed only a single product of the expected size (Figure S1A) and a single peak melting curves (Figure S1B) for each primer set. The amplification efficiency (E) of each primer pair varied between 90.51% for β-TUB and 104.74% for SAMS, and the regression coefficient (R2) ranged from 0.991(SAMS) to 0.999 (α-TUB; Table 1). The amplicon size ranged from 100 to 264 bp.
To assess the expression stability of the reference genes in different samples, the transcript abundances of 10 candidate reference genes were presented as their mean Ct values (quantification cycles). The mean Ct values of these candidates varied from 20.9 ± 1.2 (SAMS) to 26.8 ± 1.0 (ACTIN) in different samples. Genes with greater SD of Ct values have more variable expression than those with lower SD. Of the 10 candidate genes, UBC showed the least variation in gene expression (23.7 ± 0.6), and the expression of SAMS was the most variable (20.9 ± 1.2). The candidate genes can be ranked as: UBC > EF-1α/TIP41 > PP2A/CYP > GAPDH > α-TUB/ ACTIN > β-TUB > SAMS (Table S1) based on the standard deviation of the Ct values.
Expression stability of the reference genes
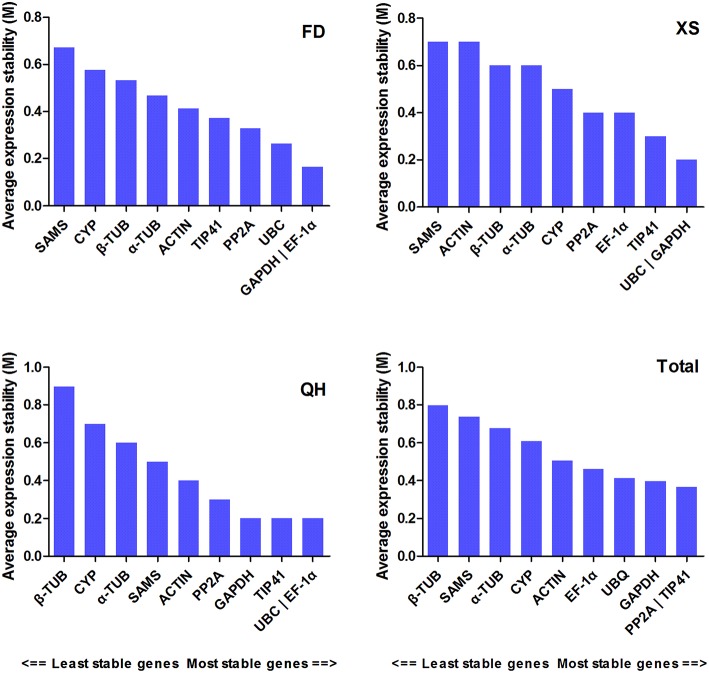
Different software programs (GeNorm, NormFinder, BestKeeper, and RefFinder) were used to assess and rank the expression stability of the candidate genes. The average expression stability value (M-value) and the average pairwise variation of template normalization factor (Vn∕n+1 value) were used to identify the most stable reference genes and obtain the required reference gene number in GeNorm. The M-value of a gene was inversely correlated with its expression stability, with the M-values less than 1.5 indicating stable expression (Hellemans et al., 2007). The cut-off value of 0.15 was used as a threshold of the Vn∕n+1 value. If Vn∕n+1 ≤ 0. 15, it is not necessary to introduce n+1 reference genes as the internal control. Figure 1 showed the ranking order of different candidate genes according to the M-value. For total samples, PP2A and Tip41 were identified as the most stable genes, while β-TUB and SAMS were the least stable reference genes. As depicted in Figure 2, the optional number of reference genes for qRT-PCR data normalization was two in each cultivar. Taking the results of the most stable gene selection into account, the combinations GAPDH/EF-1α, UBC/GAPDH, and UBC/EF-1α were predicted to deliver the most reliable level of normalization for ‘Feng Dan,’ ‘Xi Shi,’ and ‘Que Hao,’ respectively.
Figure 1.
The ranking order of reference genes. Expression stability values (M) of candidate reference genes calculated by GeNorm. Lower M-value indicates more stable expression. FD, ‘Feng Dan’; XS, ‘Xi Shi’; QH, ‘Que Hao.’
Figure 2.
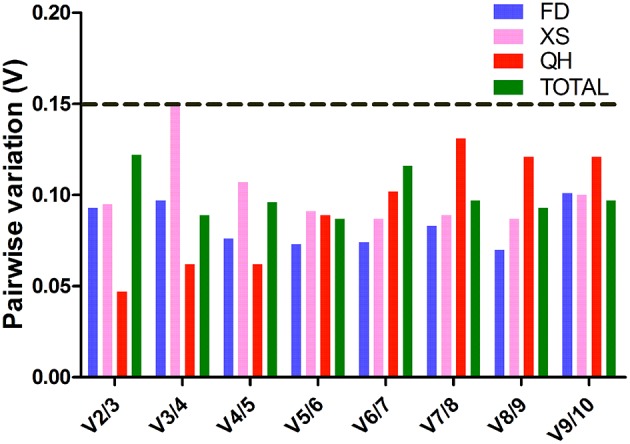
Pairwise variation (V) of 10 candidate reference genes calculated by GeNorm. The cutoff value to determine the optional number of reference genes for normalization is 0.15. FD, ‘Feng Dan’; XS, ‘Xi Shi’; QH, ‘Que Hao.’
NormFinder was used to determine the stability of reference genes based on inter- and intra-group variance in expression level. The stability value (SV) was calculated, with the lower stability value indicating the higher stability (Andersen et al., 2004). The results of NormFinder analysis were shown in Table 2. For total samples, UBC and TIP41 were the most stable genes, while SAMS and β-TUB being the least stable. The top two most stably expressed genes were GAPDH and UBC in ‘Feng Dan,’ GAPDH and TIP41 in ‘Xi Shi,’ and TIP41 and EF-1α in ‘Que Hao,’ respectively. The ranking order was slightly different from that of GeNorm.
Table 2.
Expression stability of the candidate reference genes calculated by NormFinder.
| Rank | FD | XS | QH | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | Stability | Gene | Stability | Gene | Stability | Gene | Stability | |
| 1 | GAPDH | 0.082 | GAPDH | 0.1 | TIP41 | 0.0 | UBC | 0.219 |
| 2 | UBC | 0.131 | TIP41 | 0.1 | EF-1α | 0.1 | TIP41 | 0.222 |
| 3 | EF-1α | 0.149 | UBC | 0.2 | UBC | 0.1 | GAPDH | 0.243 |
| 4 | PP2A | 0.236 | EF-1α | 0.3 | GAPDH | 0.3 | PP2A | 0.326 |
| 5 | TIP41 | 0.386 | PP2A | 0.4 | PP2A | 0.3 | EF-1α | 0.464 |
| 6 | ACTIN | 0.439 | CYP | 0.6 | ACTIN | 0.7 | ACTIN | 0.58 |
| 7 | α-TUB | 0.517 | ACTIN | 0.7 | α-TUB | 0.8 | CYP | 0.719 |
| 8 | β-TUB | 0.68 | α-TUB | 0.7 | CYP | 0.9 | α-TUB | 0.747 |
| 9 | CYP | 0.719 | β-TUB | 0.9 | SAMS | 1.0 | β-TUB | 0.918 |
| 10 | SAMS | 1.019 | SAMS | 1.0 | β-TUB | 1.2 | SAMS | 0.949 |
FD, ‘Feng Dan’; XS, ‘Xi Shi’; QH, ‘Que Hao.’
Coefficient of variance (CV) and the standard deviation (SD) were calculated by Bestkeeper analysis. The gene with the lowest CV and SD was considered to be the most stable reference gene (Chang et al., 2012). As shown in Table 3, PP2A and α-TUB were suggested to be the most stable genes in ‘Feng Dan.’ UBC and GAPDH had the lowest SD values in ‘Xi Shi,’ and CYP and UBC performed the best in ‘Que Hao.’
Table 3.
Expression stability of 10 candidate reference genes calculated by BestKeeper.
| Rank | FD | XS | QH | Total | ||||
|---|---|---|---|---|---|---|---|---|
| Gene | CV ± SD | Gene | CV ± SD | Gene | CV ± SD | Gene | CV ± SD | |
| 1 | PP2A | 1.42 ± 0.34 | UBC | 0.60 ± 0.10 | CYP | 1.70 ± 0.50 | UBC | 2.03 ± 0.48 |
| 2 | α-TUB | 1.96 ± 0.45 | GAPDH | 1.00 ± 0.20 | UBC | 2.40 ± 0.60 | EF-1α | 2.22 ± 0.52 |
| 3 | TIP41 | 1.75 ± 0.45 | TIP41 | 1.00 ± 0.30 | EF-1α | 2.60 ± 0.60 | TIP41 | 2.16 ± 0.56 |
| 4 | GAPDH | 2.25± 0.48 | PP2A | 1.50 ± 0.40 | TIP41 | 2.30 ± 0.60 | CYP | 2.37 ± 0.62 |
| 5 | UBC | 2.28 ± 0.53 | EF-1α | 1.70 ± 0.40 | α-TUB | 3.10 ± 0.80 | PP2A | 2.64 ± 0.66 |
| 6 | ACTIN | 2.04± 0.53 | ACTIN | 2.10 ± 0.60 | GAPDH | 3.50 ± 0.80 | GAPDH | 3.34 ± 0.74 |
| 7 | EF-1α | 2.46 ± 0.58 | α-TUB | 2.40 ± 0.60 | β-TUB | 3.30 ± 0.80 | α-TUB | 3.55 ± 0.85 |
| 8 | CYP | 2.77 ± 0.71 | CYP | 2.20 ± 0.60 | PP2A | 3.30 ± 0.80 | ACTIN | 3.17 ± 0.85 |
| 9 | SAMS | 3.67 ± 0.76 | β-TUB | 2.60 ± 0.70 | ACTIN | 3.70 ± 1.00 | β-TUB | 3.53 ± 0.86 |
| 10 | β-TUB | 3.84 ± 0.90 | SAMS | 3.20 ± 0.70 | SAMS | 5.70 ± 1.20 | SAMS | 4.37 ± 0.91 |
FD, ‘Feng Dan’; XS, ‘Xi Shi’; QH, ‘Que Hao.’
Comprehensive ranking of the reference genes by RefFinder
RefFinder was used to calculate the geometric mean of weights for the comprehensive ranking order recommended by geNorm, NormFinder, and BestKeeper. The comprehensive ranking order recommended by RefFinder was shown in Table 4. In total samples, the ranking order recommended by RefFinder was similar to those of NormFinder and GeNorm: TIP41> UBC> GAPDH> PP2A> EF-1α> CYP> ACTIN> α-TUB> β-TUB >SAMS (from the most stable to the least stable). GAPDH and UBC were ranked as the top two stable genes in ‘Feng Dan’ and ‘Xi Shi.’ EF-1α and UBC were the most stable genes in ‘Que Hao.’ β-TUB and SAMS were supposed to be the least stable genes in three cultivars.
Table 4.
Expression stability ranking of 10 candidate reference genes.
| Method | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| RANKING ORDER IN FD (BETTER—GOOD—AVERAGE) | ||||||||||
| geNorm | GAPDH/EF-1α | UBC | PP2A | TIP41 | ACTIN | α-TUB | β-TUB | CYP | SAMS | |
| NormFinder | GAPDH | UBC | EF-1α | PP2A | TIP41 | ACTIN | α-TUB | β-TUB | CYP | SAMS |
| Bestkeeper | PP2A | α-TUB | TIP41 | GAPDH | UBC | ACTIN | EF-1α | CYP | SAMS | β-TUB |
| RefFinder | GAPDH | UBC | EF-1α | PP2A | TIP41 | α-TUB | ACTIN | β-TUB | CYP | SAMS |
| RANKING ORDER IN XS (BETTER—GOOD—AVERAGE) | ||||||||||
| geNorm | UBC/GAPDH | TIP41 | EF-1α | PP2A | CYP | α-TUB | β-TUB | ACTIN | SAMS | |
| NormFinder | GAPDH | TIP41 | UBC | EF-1α | PP2A | CYP | ACTIN | α-TUB | β-TUB | SAMS |
| Bestkeeper | UBC | GAPDH | TIP41 | PP2A | EF-1α | ACTIN | α-TUB | CYP | β-TUB | SAMS |
| RefFinder | GAPDH | UBC | TIP41 | EF-1α | PP2A | CYP | ACTIN | α-TUB | β-TUB | SAMS |
| RANKING ORDER IN QH (BETTER—GOOD—AVERAGE) | ||||||||||
| geNorm | UBC/EF-1α | TIP41 | GAPDH | PP2A | ACTIN | SAMS | α-TUB | CYP | β-TUB | |
| NormFinder | TIP41 | EF-1α | UBC | GAPDH | PP2A | ACTIN | α-TUB | CYP | SAMS | β-TUB |
| Bestkeeper | CYP | UBC | EF-1α | TIP41 | α-TUB | GAPDH | β-TUB | PP2A | ACTIN | SAMS |
| RefFinder | EF-1α | UBC | GAPDH | TIP41 | CYP | PP2A | ACTIN | α-TUB | SAMS | β-TUB |
| RANKING ORDER IN TOTAL SAMPLES (BETTER—GOOD—AVERAGE) | ||||||||||
| geNorm | PP2A/TIP41 | GAPDH | UBC | EF-1α | ACTIN | CYP | α-TUB | SAMS | β-TUB | |
| NormFinder | UBC | TIP41 | GAPDH | PP2A | EF-1α | ACTIN | CYP | α-TUB | β-TUB | SAMS |
| Bestkeeper | UBC | EF-1α | TIP41 | CYP | PP2A | GAPDH | α-TUB | ACTIN | β-TUB | SAMS |
| RefFinder | TIP41 | UBC | GAPDH | PP2A | EF-1α | CYP | ACTIN | α-TUB | β-TUB | SAMS |
FD, ‘Feng Dan’; XS, ‘Xi Shi’; QH, ‘Que Hao.’
Validation of the reference genes
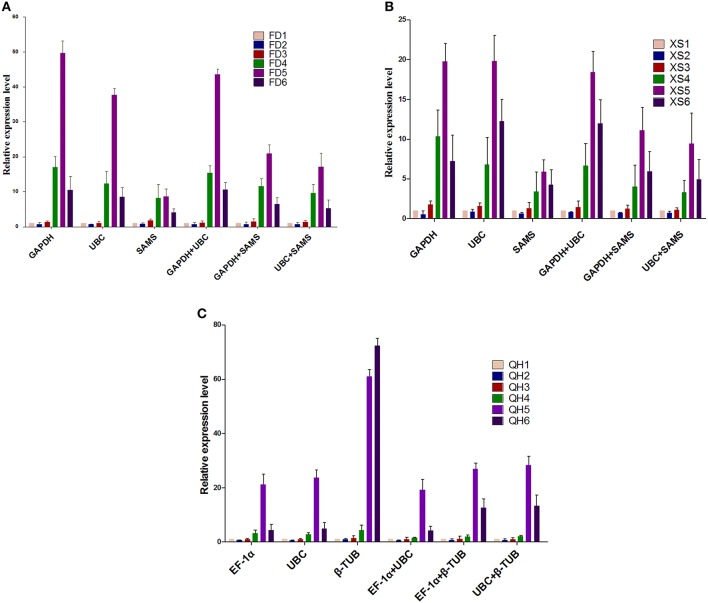
To assess the reliability of the selected candidate reference genes, the transcription levels of PsF3H were evaluated using different reference genes in three cultivars. The two most stable reference genes identified in each cultivar (UBC and GAPDH for ‘Feng Dan’ and ‘Xi Shi,’ EF-1α and UBC for ‘Que Hao’) and the least stable reference gene (SAMS or β-TUB) were used either singly or in combination in qRT-PCR analyses.
Although the overall expression patterns of PsF3H were similar, differences were found when normalized to different reference genes. In ‘Feng Dan’ and ‘Xi Shi,’ the normalized expression levels of PsF3H using the least stable gene SAMS were dramatically decreased compared to the stable genes GAPDH and UBC (Figures 3A,B). Conversely, in ‘Feng Dan’, ‘Xi Shi’ and ‘Que Hao,’ the expression level normalized by β-TUB significantly increased compared to EF-1α and UBC (Figure 3C). When the combinations of GAPDH/SMAS and UBC/SAMS were used as internal control, the expression patterns of PsF3H were similar to that of SAMS (Figures 3A,B). When the combinations of EF-1α/β-TUB and UBC/β-TUB were used, the expression level of PsF3H at stage 6 was significantly overestimated. The combination of GAPDH/UBC was recommended to be the optimum pairs of reference gene for ‘Feng Dan’ and ‘Xi Shi,’ and EF-1α/UBC was the suitable pairs of reference gene for accurate normalization in ‘Que Hao.’
Figure 3.
Normalized expression level of PsF3H in different cultivars. (A) Normalized expression level of PsF3H in ‘Feng Dan’ (FD). Different reference genes (GAPDH, UBC, and SAMS) and gene combinations (GAPDH + UBC, GAPDH + SAMS, and UBC + SAMS) were used. (B) Normalized expression level of PsF3H in ‘Xi Shi’ (XS). The reference genes and gene combinations similar to ‘Feng Dan’ were used. (C) Normalized expression level of PsF3H in ‘Que Hao’ (QH). Individual genes: EF-1α, UBC and β-TUB; Combination genes: EF-1α + UBC, EF-1α + β-TUB, UBC + β-TUB.
Discussion
To ensure the accuracy of analysis, it is important to validate suitable reference genes for gene expression analysis using qRT-PCR. In this study, 10 reference genes were evaluated for the normalization of qRT-PCR in tree peony cultivars. For the cultivars ‘Feng Dan’ and ‘Xi Shi,’ the top two most stable reference genes, GAPDH and UBC, were similar, while EF-1α was identified as the most stable gene in ‘Que Hao.’ The expression stability of various reference genes differed across cultivars. Similar results were reported in strawberries and Panax ginseng (Galli et al., 2015; Wang and Lu, 2016). The diverse genetic background and biological process between cultivars probably affected the expression stability of reference genes. The results suggested that selection and validation of reliable reference genes for quantitative gene expression analysis was necessary not only for different species but also for different cultivars.
Among the candidate genes evaluated, UBC and GAPDH were the two most stable reference genes in both ‘Feng Dan’ and ‘Xi Shi’ cultivars. This result was consistent with previous studies in Salicornia europaea (Xiao et al., 2015), Plukenetia volubilis (Niu et al., 2015), and navel oranges (Wu et al., 2014). They seemed unstable for normalization at different development stages in petunia hybrid (Mallona et al., 2010). Both ACTIN and β-TUB were involved in basic cellular processes, and have long been considered suitable reference genes in numerous species (Jin et al., 2013; Zhang et al., 2015). However, in this study, β-TUB was identified as the least stable genes in all three cultivars.
With the development of RNA sequencing, increasing amounts of transcriptome data have been successfully utilized to screen reference genes for non-model plants (Demidenko et al., 2011; Xiao et al., 2015; Zhuang et al., 2015). It has been demonstrated that the newly discovered reference genes could perform better than traditional reference genes in Brassica napus (Yang et al., 2014) and tea (Hao et al., 2014). The results of this study also showed that, compared to classic reference genes, the newly discovered ones were more stably expressed in tree peony cultivars.
In conclusion, this study evaluated the expression stability of 10 candidate reference genes in three tree peony cultivars. GAPDH/UBC was identified as the optimum pair of reference genes in ‘Feng Dan’ and ‘Xi Shi.’ EF-1α/UBC was recommended to be the best combination for ‘Que Hao.’ The results provided useful information for selection and validation of reference genes for transcript normalization in gene expression studies in P. suffruticosa. In any case, a preliminary check on the accuracy of the best performing reference genes is requested for each qRT-PCR experiment.
Author contributions
JL and JY conceived and designed the experiments. JL, JH, and YH performed the experiment and analyzed the data. JL wrote the paper. JY revised and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Shanghai Municipal Administration of Landscaping and City Appearance (Grant No. F122431, Grant No. G142417, Grant No. F132419 and Grant No. G142420), Science and Technology Commission of Shanghai Municipality (Grant No. 11ZR1436100).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00516
References
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Nolan T., Pfaffl M. W. (2005). Quantitative real-time RT-PCR-a perspective. J. Mol. Endocrinol. 34, 597–601. 10.1677/jme.1.01755 [DOI] [PubMed] [Google Scholar]
- Campos M. D., Frederico A. M., Nothnagel T., Schmitt B. A., Cardoso H. (2014). Selection of suitable reference genes for reverse transcription quantitative real-time PCR studies on different experimental systems from carrot (Daucus carota L.). Sci. Hortic. 186, 115–123. 10.1016/j.scienta.2014.12.038 [DOI] [Google Scholar]
- Chang E., Shi S., Liu J., Cheng T., Xue L., Yang X. (2012). Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS ONE 7:e33278. 10.1371/journal.pone.0033278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F. Y., Li J. J. (1998). Exportation of Chinese tree peonies (Mudan) and their development in other countries: cultivated. J. Northwest Norm. Univ. Nat. Sci. 34, 109–116. [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. 10.1104/pp.105.063743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge H. J., Fehrmann R. S., de Bont E. S., Hofstra R. M., Gerbens F., Kamps W. A., et al. (2007). Evidence based selection of housekeeping genes. PLoS ONE 2:e898. 10.1371/journal.pone.0000898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko N. V., Logacheva M. D., Penin A. A. (2011). Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS ONE 6:e19434. 10.1371/journal.pone.0019434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Wu J., Ji K. X., Zeng Q. Y., Su S., Shu Q. Y., Wang L. S., et al. (2015). Methylation mediated by an anthocyanin, O-methyltransferase, is involved in purple flower coloration in Paeonia. J. Exp. Bot. 66, 6563–6577. 10.1093/jxb/erv365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B. S., Nam H., Hopkins R. G., Morrison R. F. (2010). Impact of reference gene selection for target gene normalization on experimental outcome using Real-Time qRT-PCR in Adipocytes. PLoS ONE 5:e15208. 10.1371/journal.pone.0015208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon C., Mingam A., Charrier B. (2004). Real-time PCR: what relevance to plant studies?. J. Exp. Bot. 55, 1445–1454. 10.1093/jxb/erh181 [DOI] [PubMed] [Google Scholar]
- Galli V., Borowski J. M., Perin E. C., Messias R. S., Labonde J., Rombaldi C. V., et al. (2015). Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 554, 205–214. 10.1016/j.gene.2014.10.049 [DOI] [PubMed] [Google Scholar]
- Hao X., Horvath D. P., Chao W. S., Yang Y., Wang X., Xiao B. (2014). Identification and evaluation of reliable reference genes for quantitative realtime PCR analysis in Tea Plant (Camellia sinensis (L.) O.Kuntze). Int. J. Mol. Sci. 15, 22155–22172. 10.3390/ijms151222155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J., Dheda K., Bustin S., Zumla A. (2005). Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284. 10.1038/sj.gene.6364190 [DOI] [PubMed] [Google Scholar]
- Jin X. H., Fu J. X., Dai S. L., Sun Y., Hong Y. (2013). Reference gene selection for qPCR analysis in cineraria developing flowers. Sci Horti. 153, 64–70. 10.1016/j.scienta.2013.01.023 [DOI] [Google Scholar]
- Lalitha S. (2004). Primer Premier 5. Biotech. Softw. Inter. Rep. 1, 270–272. 10.1089/152791600459894 [DOI] [Google Scholar]
- Ling H., Wu Q., Guo J., Xu L., Que Y. (2014). Comprehensive selection of reference genes for gene expression normalization in Sugarcane by real time quantitative RT-PCR. PLoS ONE 9:e97469. 10.1371/journal.pone.0097469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. F., Gao L. X., Hu Y. H. (2015). Reference genes discovery and selection for quantitative real time PCR in tree peony seed and petal tissue of different development stages. J. Agr. Biotech. 23, 1639–1648. 10.3969/j.issn.1674-7968.2015.12.014 [DOI] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-11CT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mafra V., Kubo K. S., Alves-Ferreira M., Ribeiro-Alves M., Stuart R. M., Boava L. P. (2012). Reference genes for accurate transcript normalization in Citrus genotypes under different experimental conditions. PLoS ONE 7:e31263. 10.1371/journal.pone.0031263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I., Lischewski S., Weiss J., Hause B., Egea-Cortines M. (2010). Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrid. BMC Plant Biol. 10:4 10.1186/1471-2229-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu L., Tao Y. B., Chen S. M., Fu Q. T., Li C. Q., Dong Y. L., et al. (2015). Selection of reliable reference genes for gene expression studies of a promising oilseed crop, Plukenetia volubilis, by Real-Time quantitative PCR. Int. J. Mol. Sci. 16, 12513–12530. 10.3390/ijms160612513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., Tichopad A., Prgomet C., Neuvians T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- Ramakers C., Ruijter J. M., Deprez R. H. L., Moorman A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuro Sci. Lett. 339, 62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Shi Q., Zhou L., Wang Y., Li K., Zheng B., Miao K. (2015). Transcriptomic analysis of Paeonia delavayi wild population flowers to identify differentially expressed genes involved in purple-red and yellow petal pigmentation. PLoS ONE 10:e0135038. 10.1371/journal.pone.0135038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:reaearch0034. 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. B., Wang J. J., Jiang J. F., Chen S. M., Guan Z. Y., Liao Y., et al. (2014). Reference genes for normalizing transcription in diploid and tetraploid Arabidopsis. Sci. Rep. 4:6781. 10.1038/srep06781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. B., Chen S. M., Jiang J. F., Zhang F., Chen F. D. (2015). Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci. Rep. 5:8094. 10.1038/srep08094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. L., Chen J. H., Tian Q. Q., Wang S., Xia X. L., Yin W. L. (2014). Identification and validation of reference genes for Populus Euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol Plantarum. 152, 529–545. 10.1111/ppl.12206 [DOI] [PubMed] [Google Scholar]
- Wang M., Lu S. F. (2016). Validation of suitable reference genes for quantitative gene expression analysis in Panax ginseng. Front. Plant Sci. 6:1259. 10.3389/fpls.2015.01259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. J., Dong L., Zhang C., Wang X. Q. (2012). Reference gene selection for real-time quantitative PCR normalization in Tree Peony (Paeonia suffruticosa Andr.). J Agr Biotech. 5, 521–528. 10.3969/j.issn.1674-7968.2012.05.008 [DOI] [Google Scholar]
- Wei L., Miao H., Zhao R., Han X., Zhang T., Zhang H. (2013). Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta 237, 873–889. 10.1007/s00425-012-1805-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. X., Su S. Y., Fu L. L., Zhang Y. J., Chai L. J., Yi H. L. (2014). Selection of reliable reference genes for gene expression studies using quantitative real-time PCR in navel orange fruit development and pummelo floral organs. Sci. Hortic. 176, 180–188. 10.1016/j.scienta.2014.06.040 [DOI] [Google Scholar]
- Xiao X., Ma J., Wang J, Wu, X., Li P., Yao Y. (2015). Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 5:788. 10.3389/fpls.2014.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F. L., Xiao P., Chen D. L., Xu L., Zhang B. H. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. 10.1007/s11103-012-9885-2 [DOI] [PubMed] [Google Scholar]
- Yang H., Liu J., Huang S., Guo T., Deng L., Hua W. (2014). Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 538, 113–122. 10.1016/j.gene.2013.12.057 [DOI] [PubMed] [Google Scholar]
- Yang Z., Chen Y., Hu B., Tan Z., Huang B. (2015). Identification and validation of reference genes for quantification of target gene expression with quantitative real-time PCR for Tall Fescue under four abiotic stresses. PLoS ONE 10:e0119569. 10.1371/journal.pone.0119569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W. P., Duan W. L., Liu P., Deng R. X., Ren Y. L., Zhao S. (2012). GC-MS analysis of essential oil of Paeonia suffruticosa Andrews from ZhaoFen and RouFurong flowers in China. Nat Sci. 8, 552–554. 10.4236/ns.2012.48073 [DOI] [Google Scholar]
- Zhang C., Fu J. X., Wang Y. G., Bao Z. Y., Zhao H. B. (2015). Identification of suitable reference genes for gene expression normalization in the quantitative real-Time PCR analysis of Sweet Osmanthus (Osmanthus fragrans Lour.). PLoS ONE 10:e0136355. 10.1371/journal.pone.0136355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. J., Wang L. S., Shu Q. Y., Liu Z. A., Li C. H., Zhang J., et al. (2007). Comparison of anthocyanins in non-blotches and blotches of the petals of Xibei tree peony. Sci. Hortic. 114, 104–111. 10.1016/j.scienta.2007.05.009 [DOI] [Google Scholar]
- Zhang Y. X., Gai S. P., Liu C. Y., Mu P., Zheng G. S. (2011). Selection of control genes in real-time qPCR analysis during bud dormancy release in Tree Peony (Paeonia suffruticosa). Mol. Plant Breeding 9, 1052–1056. 10.5376/mpb.cn.2011.09.0007 [DOI] [Google Scholar]
- Zhou L., Wang Y., Ren L., Shi Q. Q., Zheng B. Q., Miao K., et al. (2013). Over expression of Ps-CHI1, a homologue of the chalcone isomerase gene from tree peony (Paeonia suffruticosa), reduces the intensity of flower pigmentation in transgenic tobacco. Plant Cell Tiss. Organ Cult. 3, 285–295. 10.1007/s11240-013-0403-2 [DOI] [Google Scholar]
- Zhuang H., Fu Y., He W., Wang L., Wei Y. (2015). Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 6:475. 10.3389/fpls.2015.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.