Maize Germplasm Conservation in Southern California’s Urban Gardens
Contemporary germplasm conservation studies largely focus on ex situ and in situ management of diversity within centers of genetic diversity. Transnational migrants who transport and introduce landraces to new locations may catalyze a third type of conservation that combines both approaches. Resulting populations may support reduced diversity as a result of evolutionary forces such as genetic drift, selection, and gene flow, yet they may also be more diverse as a result of multiple introductions, selective breeding and cross pollination among multiple introduced varietals. In this study, we measured the amount and structure of maize molecular genetic diversity in samples collected from home gardens and community gardens maintained by immigrant farmers in Southern California. We used the same markers to measure the genetic diversity and structure of commercially available maize varieties and compared our data to previously reported genetic diversity statistics of Mesoamerican landraces. Our results reveal that transnational dispersal creates an opportunity for the maintenance of maize genetic diversity beyond its recognized centers of diversity.
Key Words: Zea mays, biodiversity conservation, urban agriculture, germplasm erosion, genetic resources, crop evolution, home gardens, community gardens, immigrant farmers, ethnobotany
Introduction
Conserving landraces and crop wild relatives is crucial for maintaining genetic resources for future crop improvement (e.g., Ford–Lloyd et al. 2011; Maxted et al. 1997, 2012b). Landraces and crop wild relatives provide useful genetic material for breeding modern improved lines, minimizing the vulnerability of inbred crops to pathogens and pests, improving performance, and incorporating unique traits (e.g., Lopes et al. 2015; Mano and Omori 2007). Yet the sustainability of landraces is at risk, in both developing and developed countries. In developing countries, critical threats to genetic agrobiodiversity are policies and programs that encourage replacement of landraces with modern cultivars or encouraged migration to cities, often resulting in the abandonment of farming altogether (Nabhan 1985).
Maize genetic diversity in Mexico is eroding both in terms of the number of extant landraces as well as the amount of allelic diversity within creolized varieties (Dyer et al. 2014; van Heerwaarden et al. 2009a). Despite controversy regarding the speed of the decline (Brush et al. 2015; Dyer et al. 2015), “the question no longer is whether genetic erosion remains a presumption but how to respond to it” (Dyer et al. 2015).
Conservation strategies for minimizing germplasm loss in centers of diversity include both ex situ and in situ methods (Altieri and Merrick 1987; Brown et al. 1989; Brush 1991, 1995; Nabhan 1985; Oldfield and Alcorn 1987). Ex situ conservation in gene banks, botanical gardens, and reserves provides a controlled environment to conserve crop genetic resources. However, landraces and their wild and weedy ancestors are often underrepresented in such collections (Crop Wild Relatives and Climate Change 2013; Hammer 2004). Ex situ preservation also prevents evolutionary adaptive responses to abiotic and biotic changes (Maxted et al. 2012a; Simmonds 1962). In contrast, in situ or farmer–based conservation incorporates traditional agricultural breeding practices that permit crop evolution and adaptation to changing environments while preserving biological material and the social processes connected to them (Altieri and Merrick 1987; Brush 1995). However, in situ conservation is often difficult to sustain in the face of socioeconomic forces promoting the replacement of landraces with alternate varieties or crops, especially in developing countries (Frankel 1974; International Board for Plant Genetic Resources 1985; Maxted et al. 2012b; Oldfield and Alcorn 1987). Despite these difficulties, the consensus is that ex situ germplasm conservation should be complemented with an in situ farmer–based approach (Brush 1995; Hawkes et al. 2000; Maxted et al. 2002).
What has been largely ignored in previous studies of farmer–based germplasm conservation is whether human migration may provide a third opportunity for crop genetic diversity maintenance. At times, transnational migrants transport crop seeds from their homeland across international borders (e.g., Perales et al. 2003; Soleri et al. 2005). If they plant those seeds and introduce threatened varieties to new locations, they may create conditions for a unique kind of germplasm conservation that combines both in situ and ex situ approaches. Traditional crops, like maize, managed by migrant farmers in Southern California’s urban gardens mimic in situ conservation in that the traditional practices are built on first–hand knowledge of traits, management, and informal breeding techniques. Because those crops are not located in centers of diversity or in centers of crop origin, they are removed from the geographic foci of recognized in situ conservation. Maize cultivated by migrant farmers in Southern California’s urban gardens supports a novel environment in that their immediate origin parallels ex situ grow–out conditions.
Whether the crops of a diaspora warrant conservation attention depends on whether they harbor relatively high or unique diversity. The amount of genetic diversity in anthropogenically dispersed landraces depends on the combined action of the evolutionary forces of genetic drift, selection, and gene flow (Hancock 2004). Genetic drift via founder effect due to limited initial seed samples, bottlenecks, reduced gene flow from other sources, and/or chronically small crop population size results in depleted diversity. Likewise, strong selection for adaptation to new conditions can reduce genetic diversity. Migrants have made a deliberate effort to collect and transport genetically diverse maize seeds, the diversifying selection of which may lead to urban garden plots that are as much or more genetically diverse than source populations. Furthermore, gene flow in the form of repeated introduction of seeds from the same or different source populations, seed exchange among farmers, and cross–pollination between adjacent plots would be expected to increase diversity (e.g., van Heerwaarden et al. 2009b). Despite these varied expectations, to our knowledge, the relative within–population diversity of anthropogenically dispersed crops has not been studied.
Southern California provides an exemplary case study of joint human and plant migration. California immigrants from Mexico and Mesoamerica frequently maintain plots of crops from their homelands in urban home gardens and community gardens. Home gardens are typically small, multi–species agroecosystems located near their associated household (Galluzzi et al. 2010), whereas community gardens are characteristically larger in size and composed of individual plots with additional common areas where garden meetings and other social events take place. Such gardens have recently been hypothesized to serve as reservoirs of genetic as well as taxonomic diversity (Altieri and Merrick 1987; Corlett et al. 2003; Galluzzi et al. 2010; Hodgkin 2002; Hoogendijk and Williams 2002; Watson and Eyzaguirre 2002; Zaldivar et al. 2004).
In this study, we compared maize genetic diversity (Zea mays spp. mays) sampled from Southern Californian home gardens and community gardens to local commercial varieties for sale. Likewise, we used genetic diversity data for Mesoamerican maize reported in the literature to determine whether the gardens were more or less diverse relative to those in centers of diversity. We chose maize because the genetic erosion of maize diversity in Mexico, its center of domestication and diversity, has gained increased attention as farmers abandon the cultivation of traditional landrace varieties either in favor of migration to urban centers or in favor of relatively homogenous improved lines. Our results provide an examination of the population genetics of maize in Southern California gardens and reveal that maintaining maize germplasm diversity is not restricted to its traditionally recognized centers of diversity.
Materials and Methods
Plant Material
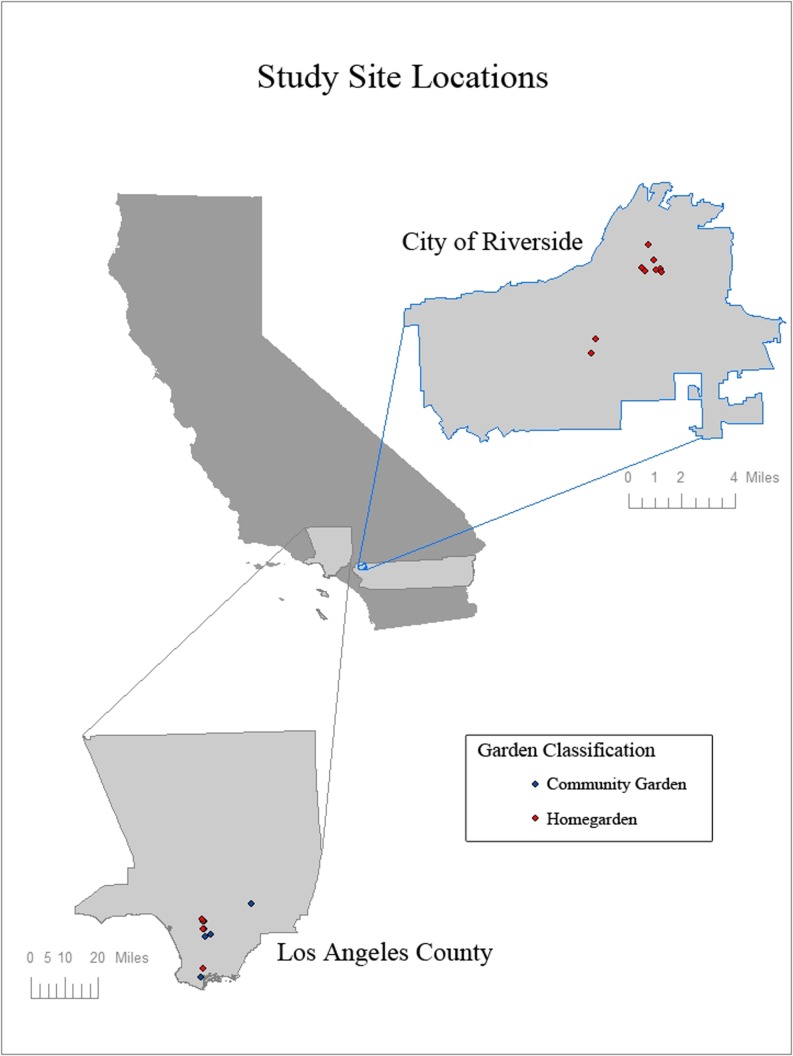
We collected maize samples between June and September 2008 in Los Angeles and Riverside, California. Populations and participants were selected at random in Mesoamerican enclave communities by visually surveying urban gardens in Los Angeles and Riverside and selecting households and gardens cultivating maize and willing to participate in the experimental study. Twenty total locations were sampled: six home gardens and four community gardens in Los Angeles and ten home gardens in Riverside. Locations of sampled populations are shown in Fig. 1. Table 1 gives detailed collection information for each site.
Fig. 1.
Geographic map of locations of maize samples collected from home garden and community gardens in California. Credit L. M. Hayden.
Table 1.
Results from PopGene analysis by maize population sampled from home gardens, community gardens, and commercial varieties including collection ID, location, and number of accessions collected.*
| Population # | Classification ID | Classification | Location | # of accessions | # of polymophic loci | P | Na | Mean Na | StDev Na | He | StDev He | Fis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RIHG1 | HG | Riverside, CA | 10 | 5 | 1.00 | 16 | 3.20 | 1.79 | 0.53 | 0.23 | 0.05 |
| 2 | RIHG2 | HG | Riverside, CA | 10 | 4 | 0.80 | 10 | 2.00 | 0.71 | 0.37 | 0.22 | −0.15 |
| 3 | RIHG3 | HG | Riverside, CA | 10 | 5 | 1.00 | 13 | 2.60 | 0.89 | 0.45 | 0.21 | 0.12 |
| 4 | RIHG4 | HG | Riverside, CA | 10 | 5 | 1.00 | 15 | 3.00 | 1.22 | 0.48 | 0.03 | 0.00 |
| 5 | RIHG5 | HG | Riverside, CA | 10 | 5 | 1.00 | 11 | 2.20 | 0.45 | 0.47 | 0.04 | 0.08 |
| 6 | RIHG6 | HG | Riverside, CA | 10 | 4 | 0.80 | 13 | 2.60 | 1.14 | 0.35 | 0.22 | −0.21 |
| 7 | RIHG7 | HG | Riverside, CA | 10 | 5 | 1.00 | 12 | 2.40 | 0.55 | 0.57 | 0.95 | −0.08 |
| 8 | RIHG8 | HG | Riverside, CA | 10 | 5 | 1.00 | 14 | 2.80 | 2.15 | 0.56 | 0.05 | 0.06 |
| 9 | RIHG9 | HG | Riverside, CA | 10 | 5 | 1.00 | 13 | 2.60 | 0.89 | 0.47 | 0.26 | 0.07 |
| 10 | RIGH10 | HG | Riverside, CA | 10 | 5 | 1.00 | 12 | 2.40 | 0.55 | 0.32 | 0.18 | −0.16 |
| 11 | LAHG1 | HG | Los Angeles, CA | 10 | 5 | 1.00 | 10 | 2.00 | 0.00 | 0.38 | 0.12 | 0.23 |
| 12 | LAHG2 | HG | Los Angeles, CA | 10 | 5 | 1.00 | 13 | 2.60 | 0.55 | 0.45 | 0.25 | 0.10 |
| 13 | LAHG3 | HG | Los Angeles, CA | 10 | 5 | 1.00 | 17 | 3.40 | 2.32 | 0.57 | 0.14 | 0.22 |
| 14 | LAHG4 | HG | Los Angeles, CA | 10 | 4 | 0.80 | 10 | 2.50 | 0.58 | 0.47 | 0.10 | 0.00 |
| 15 | LAHG5 | HG | Los Angeles, CA | 10 | 4 | 0.80 | 11 | 2.20 | 1.10 | 0.45 | 0.28 | −0.08 |
| 16 | LAHG6 | HG | Los Angeles, CA | 10 | 4 | 0.80 | 10 | 2.00 | 0.71 | 0.41 | 0.24 | 0.61 |
| 17 | LAHG7 | CG | Los Angeles, CA | 10 | 5 | 1.00 | 10 | 2.00 | 0.00 | 0.39 | 0.18 | −0.40 |
| 18 | LACG1 | CG | Los Angeles, CA | 10 | 2 | 0.40 | 6 | 1.50 | 0.58 | 0.26 | 0.30 | −0.14 |
| 19 | LACG2 | CG | Los Angeles, CA | 10 | 4 | 0.80 | 9 | 1.80 | 0.45 | 0.25 | 0.19 | 0.28 |
| 20 | LACG3 | CG | Los Angeles, CA | 10 | 5 | 1.00 | 16 | 3.20 | 0.84 | 0.55 | 0.11 | 0.21 |
| 21 | LACG4 | CG | Los Angeles, CA | 10 | 4 | 0.80 | 9 | 1.80 | 0.45 | 0.24 | 0.20 | 0.13 |
| 22 | LACG5 | CG | Los Angeles, CA | 10 | 5 | 1.00 | 15 | 3.00 | 1.00 | 0.56 | 0.15 | 0.34 |
| 23 | HV1 | HV | 10 | 2 | 0.40 | 7 | 1.75 | 0.96 | 0.21 | 0.30 | 0.63 | |
| 24 | HV2 | HV | 10 | 1 | 0.20 | 6 | 1.20 | 0.45 | 0.04 | 0.08 | 1.00 | |
| 25 | IV1 | IV | 10 | 0 | 0.00 | 5 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| 26 | IV2 | IV | 10 | 2 | 0.40 | 7 | 1.40 | 1.32 | 0.19 | 0.27 | −0.67 | |
| 27 | SV1 | SV | 10 | 4 | 0.80 | 11 | 2.20 | 0.84 | 0.40 | 0.23 | −0.32 |
*Key: HG, home garden; CG, community garden; HV, hybrid variety; IV, industrial variety; SV, supermarket variety. Number of polymorphic loci, P, proportion of polymorphic loci; Na, observed number of alleles (including mean±SD); He, mean expected heterozygosity ±SD; Fis, fixation index are reported.
Interviews were conducted with farmers at each site and are part of a larger ethnographic study exploring the origins, seed networks, and traditional agricultural practices of maize cultivation in these gardens. Details regarding those interviews and the associated methodology are available in Heraty (2010).
We sampled fresh leaf tissue from 10 plants per accession. In two cases (LAHG2, LAHG3 and LACG1, LACG2), respondents distinguished two distinct varieties of maize within their garden. In these instances, 10 plants from each of the two varieties were sampled and treated as separate accessions. Thus, we sampled 10 populations and 10 accessions from Riverside and 10 populations and 12 accessions from Los Angeles. Fresh leaf tissue was frozen at −80°C until DNA extraction could be performed.
We included five commercially available cultivars for comparison. Two sweet corn populations (HV1 and HV2) that are representative of locally available horticultural varieties for home gardeners were purchased from a Riverside garden retail store. We also obtained imported corn seed for human consumption from Mexico from a bulk food bin at Big Saver Foods supermarket in Riverside. The latter collection was motivated by interviews with respondents that revealed local ethnic markets supplying bulk seed for consumption were sometimes a source of seeds for their gardens. Finally, we included maize cultivars IV1 and IV2 to represent industrial maize cultivars. We germinated 10 randomly selected seeds from each cultivar in a temperature–controlled greenhouse. Fresh leaf tissue was harvested from germinated plants and stored at −80°C prior to extraction.
DNA was isolated from finely ground tissue by use of the DNeasy plant Mini Kit (Qiagen, Valencia, CA) with the following modifications: 3 μl Rnase A stock solution was added to the ground tissue; at the lysation phase, cells were incubated for 15 min; and at the final elution step, 50 μl Buffer AE was added to the DNeasy membrane. Extracted DNA was quantified with use of NanoDrop spectrophotometer (Thermo Fisher Scientific, Ealtham, MA) and stored at −20°C.
PCR (Polymerase Chain Reaction) Amplification
Five microsatellite markers previously described by Senior et al. (1998) were selected for analysis. The selected primer sequences are listed and detailed in Table 2. More information for each primer set can be found at http://www.maizegdb.org.
Table 2.
Primer information.
| MARKER | CHROMOSOME | REPEAT | PRIMER SEQUENCE | Min/MAX Allele |
|---|---|---|---|---|
| Phi079 | 4 | AGATG | TGGTGCTCGTTGCCAAATCTACGA | 179/196 |
| GCAGTGGTGGTTTCGAACAGACAA | ||||
| Phi115 | 8 | AT/ATAC | GCTCCGTTTTCGCCTGAA | 291/312 |
| ACCATCACCTGAATCCATCACA | ||||
| Phi102228 | 3 | AAGC | ATTCCGACGCAATCAACA | 122/131 |
| TTCATCTCCTCCAGGAGCCTT | ||||
| Phi427424 | 2 | ACC | CAACTGACGCTGATGGATG | 123/139 |
| TTGCGGTGTTAAGCAATTCTCC | ||||
| Phi093 | 4 | AGCT | AGTGCGTCAGCTTCATCGCCTACAAG | 283/295 |
| AGGCCATGCATGCTTGCAACAATGGATACA |
DNA amplification involved the use of a PTC–100 Thermo Cycler (MJ Research Inc., Watertown, MA) in a 10 μl volume containing approximately 5.0 to 10.0 ng/μl template DNA, 10 μm each forward and reverse primer, 1 mM dNTPs, 10x Promega PCR Buffer, 25 μm magnesium chloride, and 5 units/μl Taq DNA polymerase. PCR conditions were 94°C for 5 min, 34 cycles of 94°C for 1 min, 55°C for 4 min, 72°C for 2.5 min, and a final cycle of 72°C for 15 min.
Polymorphism Detection
Polymorphisms in the amplified PCR product were detected by electrophoresis in a 2% Agarose Super Fine Resolution (SFR) gel (Amresco, Solon, OH). Gels were prepared with 1X TBE Buffer and were stained with ethidium bromide before electrophoresis in a submarine gel system (Wide Mini–sub Cell GT, Bio–Rad, Hercules, CA). Each well contained 4 μl PCR product and 1 μl 6X blue loading dye. Bands were visualized by use of GelDOC (Bio–Rad, Hercules, CA). Fragments were estimated by comparison with a 100–bp DNA ladder (New England Biolabs, Ipswich, MA). Each gel was run with a control sample of a known fragment size.
DNA for several individuals failed to amplify at certain loci, despite repeated amplifications, particularly B73. These data are treated as missing in our analyses.
Data Analysis
For each locus, bands were scored as heterozygous or homozygous, depending on the presence of a single or double band. Allele frequencies were calculated for each accession. Observed number of alleles (Na), proportion of polymorphic alleles, percentage of polymorphic loci, expected heterozygosity (He), and fixation index (Fis) were calculated for each population and population group by use of the computer program PopGene v1.32 (Yeh et al. 1997).
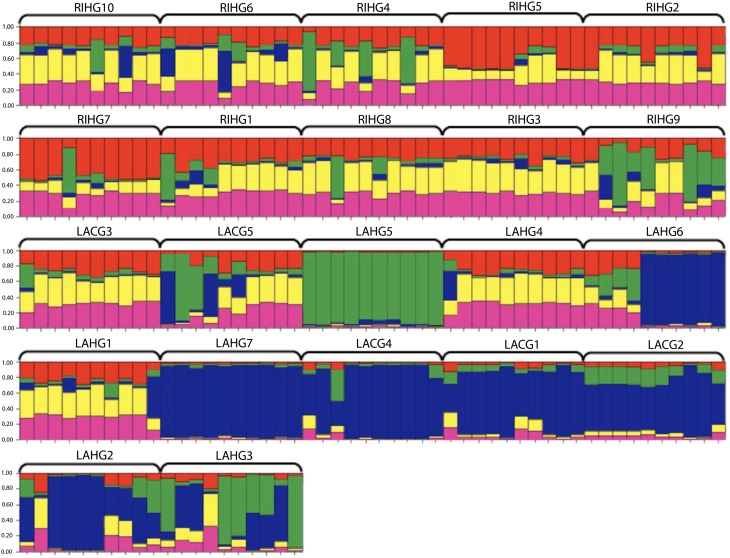
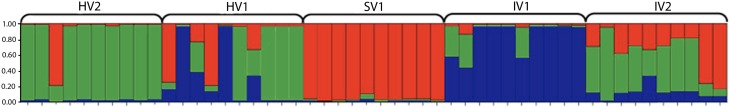
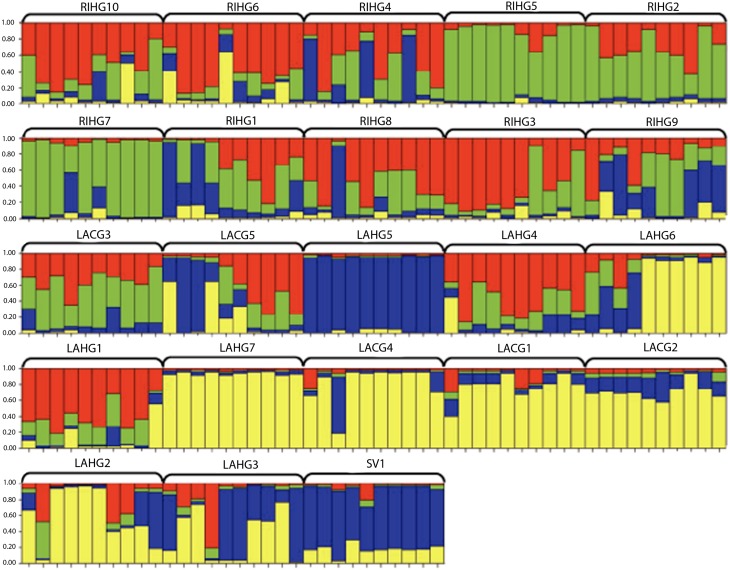
Microsatellite data were also analyzed for population genetic structure by use of STRUCTURE v2.2 (Pritchard et al. 2000) with a burn–in of 100,000 iterations run against a simulation of 250,000 iterations to estimate the genetic cluster (K) parameters. The optimal K–value can be estimated by graphing the output information for the log–transformed probability of K clusters [lnPr(X∣K)] from each run to display the greatest decrease in slope. Multiple runs at different K–values were conducted for accurate results. We conducted three separate analyses using STRUCTURE: one with all garden populations (Fig. 2), a second with all commercial populations (Fig. 3), and a third that assigned the imported bulk bin seed from Mexico to the garden populations group (Fig. 4).
Fig. 2.
STRUCTURE analysis of garden populations of maize. Most likely grouping K=5.
Fig. 3.
STRUCTURE analysis of commercial populations of maize. Most likely grouping K=3.
Fig. 4.
STRUCTURE analysis of garden populations of maize including imported supermarket variety SV1. Most likely grouping K=4.
Results
Microsatellite polymorphisms were detected for every locus. Population genetic properties for each individual population are in Table 1. Table 3 reports the distribution of genetic variation according to source: garden populations versus commercial populations, and commercial populations without the imported supermarket variety SV1. Table 3 also reports the distribution of genetic variation in garden populations by home gardens versus community gardens.
Table 3.
Results from PopGene analysis separated by garden populations (including a subanalysis between home gardens and community gardens) and commercial populations (including a separate analysis removing the imported supermarket variety SV1).
| Na | Mean Na | StDev Na | # of Polymorphic Loci | P | He | StDev He | Fis | |
|---|---|---|---|---|---|---|---|---|
| Garden Populations | 25 | 5.00 | 1.87 | 5 | 1.00 | 0.55 | 0.11 | 0.05 |
| **Homegardens | 25 | 5.00 | 1.87 | 5 | 1.00 | 0.55 | 0.10 | 0.05 |
| **Community Gardens | 19 | 3.80 | 1.30 | 5 | 1.00 | 0.51 | 0.16 | 0.09 |
| Commercial Populations | 14 | 2.80 | 1.10 | 4 | 0.80 | 0.31 | 0.29 | −0.23 |
| Commercial Populations without SV1 | 9 | 1.80 | 0.84 | 3 | 0.60 | 0.19 | 0.22 | 0.06 |
*Key: SV, supermarket variety. Number of polymorphic loci, P, proportion of polymorphic loci; Na, observed number of alleles (including mean±SD); He, mean expected heterozygosity ±SD; Fis, fixation index are reported.
All garden populations were polymorphic (100%). The total number of alleles observed was 25 (mean 5.00±1.87). The mean expected heterozygosity (He) for the group was 0.55±0.11, with mean Fis for the group of 0.05. A single population, LACG1 (one accession of two sampled from a Los Angeles community garden), had the lowest number of observed alleles, 6, and LAHG3 (one accession of two sampled from a Los Angeles home garden) had the highest, 17.
In contrast, commercial populations showed less genetic diversity than garden populations, regardless of population genetic measurement. Only four loci were polymorphic (80%). The total number of observed alleles was 14 (mean 2.80±1.10). The mean He was 0.31±0.29 with mean Fis for the group of −0.23. The population B73 had the lowest number of observed alleles, 5. Interestingly, the population SV1 (supermarket variety of imported Mexican seed) had the highest number of total alleles, 11. Two–tailed t–test revealed significant difference in diversity (He) of individual garden populations and individual commercial populations (including SV1) (p≤0.016). When we removed the imported SV1 variety from the commercial group statistics, the total number of alleles for the commercial populations decreased to 9 (mean 1.80±0.84). The mean He was 0.19±0.22, with the mean Fis for the group of 0.06.
The contrast between home and community gardens was not nearly as strong. Both garden types were polymorphic at all five loci (100%). The total number of alleles in home gardens was 25 (mean 5.00±1.87), with 19 (mean 3.80±1.80) in community gardens. The mean He for home gardens was 0.55±0.10; for community gardens and 0.51±0.10 for community gardens. The mean Fis for home gardens was 0.05 and 0.09 for community gardens.
The program STRUCTURE assigned genetic affiliations to the sampled individuals within preselected populations. The most likely number of groups for all garden populations (unresolved amplifications treated as missing) was K = 5 (Fig. 2). Generally, most populations showed a moderate to high degree of admixture, except for population LAHG5, which grouped independently and uniformly from other garden populations. Also, populations LAHG6, LAHG7, LACG4, LACG1, LACG2, and LAHG2 showed some affiliation with each other.
Separate analysis of the five commercial varieties gave a group assignment of K = 3 (Fig. 3). The population SV1 (supermarket variety of imported Mexican seed) showed little genetic affiliation to other commercial populations. We reassigned SV1 to the garden populations to explore whether it had any affinity with them. Interestingly, adding SV1 to the garden populations gave a group assignment of K = 4 (Fig. 4). Furthermore, SV1 showed some genetic affiliation to some garden populations, especially LAHG5.
Discussion
As compared with commercially available maize from Southern California, maize collected from garden populations in Southern California showed a considerable amount of genetic variation in all five microsatellite loci investigated (Table 3). The results of this analysis assume that the samples of both the gardens and commercial populations are representative of the range of diversity of maize in Southern California. Several varieties of both cultivated and commercial maize could have served as comparative populations and varieties used in this study are assumed to be representative of overall varieties.
This analysis of the structure of diversity in the gardens gave several interesting results. First, PopGene revealed greater relative diversity in gardens than in commercial populations. In the STRUCTURE analysis, the imported SV1 corn seed showed greater allelic similarity to maize sampled from gardens than any of the commercial varieties, so the garden populations may have a closer genetic affiliation to this imported seed from Mexico. PopGene analysis showed less overall diversity in the commercial populations sampled.
Results of STRUCTURE analysis showed that the gardens exhibited greater admixture within and between populations than the commercial populations. While these results may indicate potential gene flow, it may also indicate a high degree of relatedness due to similar ancestry. When analyzing the commercial populations alone, SV1, the imported seed from Mexico, did not pair with the rest of the commercial varieties. The store labeling of this seed as an imported corn seed from Mexico suggests a potential landrace origin; if so, our finding of little genetic affiliation with other locally available commercial varieties is not surprising. On analyzing SV1 with the garden populations, SV1 showed a common affiliation with the populations LAHG5, LAHG7, LACG4, LACG1, LACG2, LAHG2, and LAHG3. The resulting affiliation suggests that the farmers who managed the populations might have used the supermarket variety in the past as a seed source or that some of the garden accessions share ancestry with the imported seed.
We considered that a community garden with multiple plots and accessions might have increased potential for gene flow and admixture, thus showing a more diverse germplasm profile. Results from grouped PopGene analysis of home garden and community garden populations do not support our original assumption of greater variation. These results could be attributed to the small number of community gardens we sampled as compared with the large number of home garden populations. Sampling from a larger set of community gardens in future studies would shed light on whether a small sample size affects the comparative outcome or whether other factors are involved.
Several existing studies have reported on the genetic diversity of landraces of maize in Mexico. Their results (Table 4) on fixation index, allele number, and expected heterozygosity serve as comparison studies to our data if samples from the home gardens and community gardens in Southern California have purported landrace ancestry. Overall, the levels of garden maize genetic diversity and their organization are approximately similar to what has been found in population genetic analyses of landraces in Mesoamerica.
Table 4.
Published Mesoamerican maize diversity statistics used for comparative analysis.
| Authors | Type of Marker | No. SSRs | No. Zea mays L. ssp. mays Accessions | No. of Plants Genotyped | Source | Total No. Alleles | Mean Gene Diversity | Mean Fixation Index |
|---|---|---|---|---|---|---|---|---|
| Doebley et al. (1985) | Isozyme | 23 loci | 94 | 12 per accession | Mexico/Guatemala | 163 (range 3-18) | 0.182 (range 0.01-0.60) | N/A |
| Heraty and Ellstrand, unpubl. Data | SSR | 5.00 | 22 | 10 per accession | Home Gardens and Community Gardens in Southern California | Home Gardens (HG: 25 total) Community Gardens (CG: 19 total) | HG: 0.55±0.10 CG: 0.51±0.16 | HG: 0.05 CG: 0.09 |
| Pressoir and Berthaud (2004) | SSR | 11.00 | 20 | 1 | Mexico | N/A | 0.71 (range 0.65-0.72) | 0.138±0.031 |
| Reif et al. (2006) | SSR | 25.00 | 25 | 497 (total) | Mexico/Guatemala | 196 (range 4-14) | 0.61 | 0.24 (range 0.09-0.47) |
| Sanchez et al. (2000) | Isozyme | 37 loci | 209 | 12 per accession | Mexico | 303 (range 59-120) | N/A | |
| Vigouroux et al. (2008) | SSR | 96 | 945 | 964 | North America, Central America, South America | 3752 Averages within 4 Clusters: Highland Maize (HM: 14.9) Tropical Lowland (TL:14.4) Andean (A: 12.4) Northern US (NUS: 10.6) | HM: 0.81±0.01 TL: 0.80±0.01 A: 0.71±0.02 NUS: 0.72±0.02 | HM: 0.33 TL: 0.331 A: 0.29 NUS: 0.44 |
| Warburton et al. (2011) | SSR | 17 | 59 | 15 per accession | Mexico/Guatemala | N/A | 0.481±0.03 (within landraces from Mexico/Guatemala) | N/A |
Our findings are unique for this area of study. Existing research into maize biodiversity conservation has focused on farmer–based methods of biodiversity conservation of maize in centers of crop diversity and ex situ methods to preserve diversity in controlled environments. It has ignored the importance of human migration and the movement of important cultural food species in establishing new regions of germplasm diversity. Biodiversity research of urban gardens worldwide has been limited in two ways. Geographically, studies have mostly concentrated on tropical home gardens in Asia and South America. Research has also been limited by its focus on species richness and evenness within gardens without addressing the level of genetic diversity within or between species (see Aguilar–Støen et al. 2009; Coomes and Ban 2004; Lamont et al. 1999; Padoch and De Jong 1991; Soemarwoto and Conway 1992; Thompson et al. 2003).
Our study provides compelling evidence that, as compared with available commercial varieties of maize, populations of maize from home gardens and community gardens in Southern California show large variations in genetic diversity both within and between populations. Our comparison with literature–sourced landrace studies provided in Table 4 reveals that urban gardens may serve as underappreciated locations of ex situ diversity managed in situ.
Our study encourages an expansion of research investigating the biogeography of diversity in urban gardens throughout North America and the mechanisms by which migrating farmers select and maintain landraces and crop wild relatives. With the disruption of farming communities and maize diversity in Mexico, immigrants who relocate transnationally have become important stewards in introducing, conserving, and actively managing diverse maize germplasm, ensuring that it finds refuge in new environments where it can continue to evolve and adapt.
Acknowledgements
This research was supported by a National Science Foundation Biocomplexity Grant (DEB–0409984) and a UCMEXUS–CONACYT Collaborative Research Grant awarded to NCE. Cristina Martinez–Canton provided invaluable field support. Laura Smales, Jeffrey Ross–Ibarra, Daniela Soleri, and two anonymous reviewers provided helpful comments to improve earlier drafts of this manuscript.
Literature Cited
- Aguilar–Støen M, Moe SR, Camargo–Ricalde SL. Home gardens sustain crop diversity and improve farm resilience in Candelaria Loxicha, Oaxaca, Mexico. Human Ecology. 2009;37:55–77. doi: 10.1007/s10745-008-9197-y. [DOI] [Google Scholar]
- Altieri MA, Merrick L. In situ conservation of crop genetic resources through maintenance of traditional farming systems. Economic Botany. 1987;41:86–96. doi: 10.1007/BF02859354. [DOI] [Google Scholar]
- Brown AHD, Marshall DR, Frankel OH, Williams JT. The use of plant genetic resources. New York: Cambridge University Press; 1989. [Google Scholar]
- Brush SB. A farmer–based approach to conserving crop germplasm. Economic Botany. 1991;45:153–165. doi: 10.1007/BF02862044. [DOI] [Google Scholar]
- Brush SB. In situ conservation of landraces in centers of crop diversity. Crop Science. 1995;35:346–354. doi: 10.2135/cropsci1995.0011183X003500020009x. [DOI] [Google Scholar]
- Brush SB, Bellon MR, Hijmans RJ, Ramirez QO, Perales HR, van Etten J. Assessing maize genetic erosion. Proceedings of the National Academy of Sciences. 2015;112(1) doi: 10.1073/pnas.1422010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes OT, Ban N. Cultivated plant species diversity in home gardens of an Amazonian peasant village in northeastern Peru. Economic Botany. 2004;58:420–434. doi: 10.1663/0013-0001(2004)058[0420:CPSDIH]2.0.CO;2. [DOI] [Google Scholar]
- Corlett JL, Dean EA, Grivetti LE. Hmong gardens: Botanical diversity in an urban setting. Economic Botany. 2003;57(3):365–379. doi: 10.1663/0013-0001(2003)057[0365:HGBDIA]2.0.CO;2. [DOI] [Google Scholar]
- Crop Wild Relatives and Climate Change. 2013. www.cwrdiversity.org (06 September 2013).
- Doebley JF, Goodman MM, Stuber CW. Isozyme variation in the races of maize from Mexico. American Journal of Botany. 1985;72(5):629–639. doi: 10.2307/2443674. [DOI] [Google Scholar]
- Dyer GA, López–Feldman A, Yúnez–Naude A, Taylor JE. Genetic erosion in maize's center of origin. Proceedings of the National Academy of Sciences. 2014;111(39):14094–14099. doi: 10.1073/pnas.1407033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer GA, López–Feldman A, Yúnez–Naude A, Taylor JE, Ross–Ibarra J. Reply to Brush et al. A wakeup call to crop science. Proceedings of the National Academy of Sciences. 2015;112(1):E2. doi: 10.1073/pnas.1422645112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford–Lloyd BV, Schmidt M, Armstrong SJ, Barazani O, Engels J, Hadas R, Hammer K, Kell SP, Kang D, Khoshbakht K, Li Y, Long C, Lu BK, Ma K, Nguyen VT, Qiu L, Ge S, Wei W, Zhang Z, Maxted N. Crop wild relatives—Undervalued, underutilized, and under threat? Bioscience. 2011;61(7):559–565. doi: 10.1525/bio.2011.61.7.10. [DOI] [Google Scholar]
- Frankel OH. Genetic conservation: Our evolutionary responsibility. Genetics. 1974;78:53–65. doi: 10.1093/genetics/78.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi G, Eyzaguirre PB, Negri V. Home gardens: Neglected hotspots of agro–biodiversity and cultural diversity. Biodiversity and Conservation. 2010;19:3635–3654. doi: 10.1007/s10531-010-9919-5. [DOI] [Google Scholar]
- Hammer, K. 2004. Resolving the challenge posed by agrobiodiversity and plant genetic resources—An attempt. Journal of Agriculture and Rural Development in the Tropics and Subtropics Supplement 76.
- Hancock JF. Plant evolution and the origin of crop species. 2. Oxford, United Kingdom: CABI Publishing; 2004. [Google Scholar]
- Hawkes JG, Maxted N, Ford–Lloyd BV. The ex situ conservation of plant genetic resources. Dordrecht, The Netherlands: Kluwer; 2000. [Google Scholar]
- Heraty, J. 2010. Conservation of maize germplasm as a result of food traditions in Southern California’s immigrant gardens. M.Sc. Thesis, University of California Davis. Ann Arbor: ProQuest/UMI (Publication No. 1485174).
- Hodgkin T. Home gardens and the maintenance of genetic diversity. In: Watson JW, Eyzaguirre PB, editors. Proceedings of the second international home gardens workshop, 17–19 July 2001, Witzenhausen, Federal Republic of Germany. Rome: International Plant Genetic Resources Institute; 2002. pp. 14–18. [Google Scholar]
- Hoogendijk M, Williams D. Characterizing the genetic diversity of home garden crops: some examples from the Americas. In: Watson JW, Eyzaguirre PB, editors. Proceedings of the second international home gardens workshop, 17–19 July 2001, Witzenhausen, Federal Republic of Germany. Rome: International Plant Genetic Resources Institute; 2002. pp. 34–40. [Google Scholar]
- International Board for Plant Genetic Resources . The Conservation of Crop Genetic Resources. Rome, Italy: IBPGR; 1985. [Google Scholar]
- Lamont SR, Eshbaugh WH, Greenberg AM. Species composition, diversity, and use of home gardens among three Amazonian villages. Economic Botany. 1999;53(3):312–326. doi: 10.1007/BF02866644. [DOI] [Google Scholar]
- Lopes MS, El–Basyoni I, Baenziger PS, Singh S, Royo C, Ozbek K, Aktas H, Ozer E, Ozdemir F, Manickavelu A, Ban T, Vikram P. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. Journal of Experimental Botany. 2015;66:3477–3486. doi: 10.1093/jxb/erv122. [DOI] [PubMed] [Google Scholar]
- Mano Y, Omori F. Breeding for flooding tolerant maize using teosinte as a germplasm resource. Plant Root. 2007;1:17–21. doi: 10.3117/plantroot.1.17. [DOI] [Google Scholar]
- Maxted N, Dulloo ME, Ford–Lloyd BV, Frese L, Iriondo JM, Pinheiro de Carvalho MAA, editors. Agrobiodiversity conservation: Securing the diversity of crop wild relatives and landraces. Wallingford, Oxfordshire, United Kingdom: CABI; 2012. [Google Scholar]
- Maxted N, Ford–Lloyd BV, Hawkes JG. Complementary conservation strategies. In: Maxted N, Ford–Lloyd BV, Hawkes JG, editors. Plant genetic conservation: The in situ approach. London: Chapman and Hall; 1997. pp. 15–39. [Google Scholar]
- Maxted N, Guarino L, Myer L, Chiwona EA. Towards a methodology for on–farm conservation of plant genetic resources. Genetic Resources and Crop Evolution. 2002;49(1):31–46. doi: 10.1023/A:1013896401710. [DOI] [Google Scholar]
- Maxted N, Kell S, Ford–Lloyd B, Dulloo E, Toledo A. Towards the systematic conservation of global crop wild relative diversity. Crop Science. 2012;52(2):774–785. doi: 10.2135/cropsci2011.08.0415. [DOI] [Google Scholar]
- Nabhan GP. Native crop diversity in Aridoamerica: Conservation of regional gene pools. Economic Botany. 1985;39:387–399. doi: 10.1007/BF02858746. [DOI] [Google Scholar]
- Oldfield ML, Alcorn JB. Conservation of traditional agroecosystems. BioScience. 1987;37:199–208. doi: 10.2307/1310519. [DOI] [Google Scholar]
- Padoch C, De Jong W. The house gardens of Santa Rosa: Diversity and variability in an Amazonian agricultural system. Economic Botany. 1991;45:166–175. doi: 10.1007/BF02862045. [DOI] [Google Scholar]
- Perales H, Brush SB, Qualset CO. Dynamic management of maize landraces in Central Mexico. Economic Botany. 2003;57:21–34. doi: 10.1663/0013-0001(2003)057[0007:LOMICM]2.0.CO;2. [DOI] [Google Scholar]
- Pressoir G, Berthaud J. Patterns of population structure in maize landraces from the central valleys of Oaxaca and Mexico. Heredity. 2004;92:88–94. doi: 10.1038/sj.hdy.6800387. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reif JC, Warburton ML, Xia XCXC, Hoisington DA, Crossa J, Taba S, Muminović J, Bohn M, Frisch M, Melchinger AE. Grouping of accessions of Mexican races of maize revisited with SSR markers. Theoretical and Applied Genetics. 2006;113:177–185. doi: 10.1007/s00122-006-0283-5. [DOI] [PubMed] [Google Scholar]
- Sanchez JJ, Goodman MM, Stuber CW. Isozymatic and morphological diversity in the races of maize of Mexico. Economic Botany. 2000;54(1):43–59. doi: 10.1007/BF02866599. [DOI] [Google Scholar]
- Senior ML, Murphy JP, Goodman MM, Stuber CW. Utility of SSRs for determining genetic similarities and relationship in maize using an agarose gel system. Crop Science. 1998;38:1088–1098. doi: 10.2135/cropsci1998.0011183X003800040034x. [DOI] [Google Scholar]
- Simmonds N. Variability in crop plants, its use and conservation. Biological Reviews. 1962;37:422–465. doi: 10.1111/j.1469-185X.1962.tb01620.x. [DOI] [Google Scholar]
- Soemarwoto O, Conway G. The javanese home garden. Journal for Farming Systems Research–Extension. 1992;2:95–118. [Google Scholar]
- Soleri D, Cleveland DA, Aragón CF, Fuentes LMR, Rios LH, Sweeney SH. Understanding the potential impact of transgenic crops in traditional agriculture: maize farmers’ perspectives in Cuba, Guatemala and Mexico. Environmental Biosafety Research. 2005;4:141–166. doi: 10.1051/ebr:2005019. [DOI] [PubMed] [Google Scholar]
- Thompson K, Austin KC, Smith RM, Warren PH, Angold PG, Gaston KJ. Urban domestic gardens (I): Putting small–scale plant diversity in context. Journal of Vegetation Science. 2003;14:71–78. doi: 10.1111/j.1654-1103.2003.tb02129.x. [DOI] [Google Scholar]
- van Heerwaarden J, Hellin J, Visser R, Van Eeuwijk F. Estimating maize genetic erosion in modernized smallholder agriculture. Theoretical and Applied Genetics. 2009;119:875–888. doi: 10.1007/s00122-009-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heerwaarden J, Ortega del Vecchyo D, Alvarez–Buylla E, Bellon MR. New genes in traditional seed systems: Diffusion, detectability, and persistence of transgenes in a maize population. PLoS ONE. 2009;7(10) doi: 10.1371/journal.pone.0046123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux Y, Glaubitz JC, Matsouka Y, Goodman MM, Sanchez JG, Doebley J. Population structure and genetic diversity of new world maize races assessed by DNA microsatellites. American Journal of Botany. 2008;95(10):1240–1253. doi: 10.3732/ajb.0800097. [DOI] [PubMed] [Google Scholar]
- Warburton ML, Wilkes G, Taba S, Charcosset A, Mir C, Dumas F, Madur D, Dreisigacker S, Bedoya C, Prasanna BM, Xie CX, Hearne S, Franco J. Gene flow among different teosinte taxa and into the domesticated maize gene pool. Genetic Resources and Crop Evolution. 2011;58(8):1243–1261. doi: 10.1007/s10722-010-9658-1. [DOI] [Google Scholar]
- Watson JW, Eyzaguirre PB, editors. Home gardens and in situ conservation of plant genetic resources in farming systems—Proceedings of the second international home gardens workshop, 17–19 July 2001, Witzenhausen, Federal Republic of Germany. Rome: International Plant Genetic Resources Institute; 2002. [Google Scholar]
- Yeh, F. C., R. C. Yang, T. Boyle, Z. H. Ye, and J. X. Mao. 1997. POPGENE. The user–friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Canada.
- Zaldivar ME, Rocha OJ, Aguilar G, Castro L, Castro E, Barantes R. Genetic variation of cassava (Manihot esculenta Crantz) cultivated by Chibchan Amerindians of Costa Rica. Economic Botany. 2004;58:204–213. doi: 10.1663/0013-0001(2004)058[0204:GVOCME]2.0.CO;2. [DOI] [Google Scholar]