Abstract
Skin lightening among Eurasians is thought to have been a convergence occurring independently in Europe and East Asia as an adaptation to high latitude environments. Among Europeans, several genes responsible for such lightening have been found, but the information available for East Asians is much more limited. Here, a genome-wide comparison between dark-skinned Africans and Austro-Asiatic speaking aborigines and light-skinned northern Han Chinese identified the pigmentation gene OCA2, showing unusually deep allelic divergence between these groups. An amino acid substitution (His615Arg) of OCA2 prevalent in most East Asian populations—but absent in Africans and Europeans—was significantly associated with skin lightening among northern Han Chinese. Further transgenic and targeted gene modification analyses of zebrafish and mouse both exhibited the phenotypic effect of the OCA2 variant manifesting decreased melanin production. These results indicate that OCA2 plays an important role in the convergent skin lightening of East Asians during recent human evolution.
Keywords: OCA2, East Asians, skin lightening, pigmentation genes, adaptation, natural selection
Introduction
Light skin is thought to be an adaptation gained by modern humans needed to cope with the new environmental and biological factors encountered after exiting Africa about 100,000 years ago (Loomis 1967; Jablonski and Chaplin 2000; Chaplin 2004). Among many environmental factors, ultraviolet (UV) is required for the synthesis of vitamin D, a key nutrient for human bone development (Loomis 1967), and selection on vitamin D synthesis would favor a lighter skin for people living in high latitude regions with relatively weak UV radiation.
Previous investigations into the bases of skin lightening among Europeans found a set of pigmentation genes (e.g., SLC24A5 and SLC45A2) responsible for their light skin (Graf et al. 2005; Lamason et al. 2005; Izagirre et al. 2006; Yuasa et al. 2006; Lao et al. 2007; Norton et al. 2007; Sabeti et al. 2007; Soejima and Koda 2007; Han et al. 2008; Beleza et al. 2013; Wilde et al. 2014). The Ala111Thr variant of SLC24A5 proved the most striking, being nearly fixed in Europeans (>95%) due to local Darwinian positive selection, but extremely rare in Africans and East Asians (Lamason et al. 2005). Curiously, while East Asians share similarly light skin, the identified mutations of the pigmentation genes found in Europeans are rare in East Asians; instead, other pigmentation genes (e.g., OCA2, DCT, and ATRN) have been reported under selection in East Asians. For example, the variants of OCA2 (rs1800414 and rs74653330) were shown to be associated with skin melanin content (Edwards et al. 2010; Eaton et al. 2015). The DCT gene harbored a signature of positive selection in Chinese using multiple neutrality tests (Lao et al. 2007; Myles et al. 2007), and many loci in ATRN showed extended blocks in East Asians, an indication of selection (Norton et al. 2007). These data suggest convergent evolution of light skin in both East Asians and Europeans (Lao et al. 2007; Myles et al. 2007; Norton et al. 2007; Edwards et al. 2010). However, no functional analysis has been conducted to reveal the underlying genetic mechanism of how these pigmentation genes affect melanin production in the skin and cause skin lightening in East Asians.
Compared with Europeans north of the Mediterranean, East Asian populations living between the equator and North Pole manifest a wider spectrum of skin pigmentation (fig. 1A). Previous studies proposed that in the population tree, the dark-skinned Austro-Asiatic speaking populations (abbreviated as A-A) currently residing in southwestern China and Southeast Asia occupy a more basal clade than the light-skinned northern populations who migrated northward some 25,000–30,000 years ago (supplementary fig. S1, Supplementary Material online) (Su et al. 1999; Shi et al. 2005; Consortium et al. 2009). By extension, the A-A group may represent the skin type of early modern human settlers in eastern Asia. Accordingly, a comparative genomic comparison between light-skinned northern populations (e.g., northern Han Chinese, CHN) and dark-skinned A-A as well as African populations may help identify the pigmentation gene(s) responsible for skin lightening in East Asians. Meanwhile, the phenotypic effect of selected mutations in the pigmentation gene(s) can be modeled in laboratory animals to reveal the genetic mechanism.
Fig. 1.
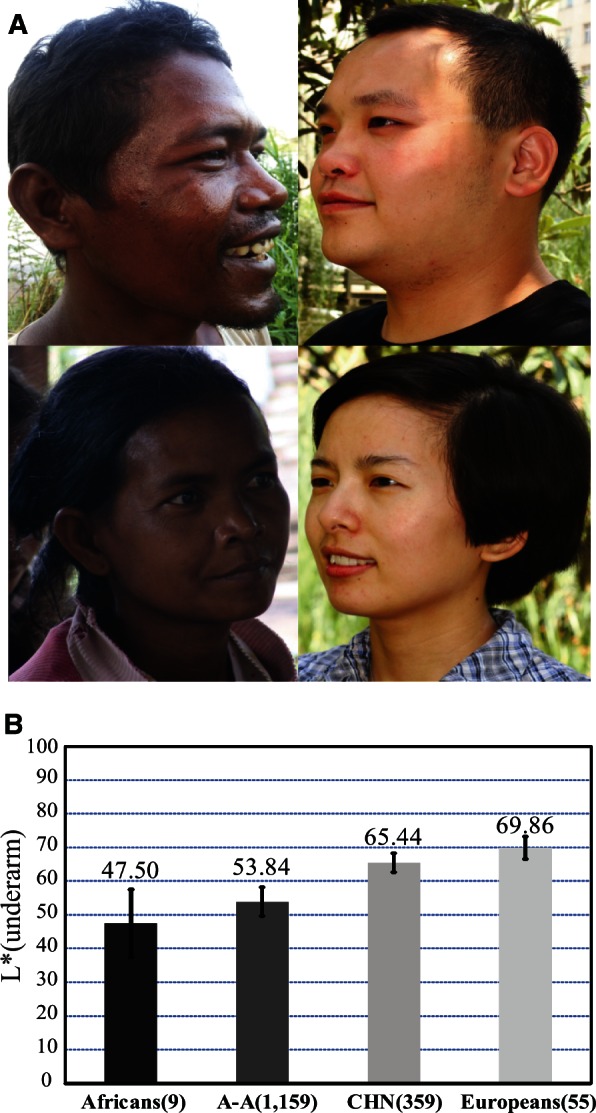
Skin pigmentation variation in world populations. (A) Skin pigmentation comparison between Austro-Asiatic speakers (left) and northern Han Chinese (right); these individuals represent the middle of skin color values of each population. (B) Comparison of skin darkness among world populations. The y axis indicates the L* value of underarm. The average L* value of each population is shown on the histogram, and the error bar indicates standard deviation. The L* values of Africans (African Americans), Europeans (European Americans) were from a previous report (Shriver and Parra 2000). The data of A-A (Austro-Asiatic speakers) and CHN (northern Han Chinese) were collected in this study. Sample size is indicated in the parenthesis.
Results
Skin Pigmentation in CHN and A-A Populations
We measured the level of darkness of the constitutive skin areas (underarm and buttock, unexposed to sunlight) and the facultative skin area (hand back, exposed to sunlight) of 1,518 unrelated individuals from A-A (1,159 individuals from 15 geographic regions) and CHN (359 individuals from 8 northern provinces of China) populations (supplementary table S1, Supplementary Material online). The L* value (a measurement of skin pigmentation reflectance) was used to quantify the level of darkness, that is, lower L* values denote darker skin. A-A populations showed markedly darker skin than the CHN population for both the constitutive and facultative areas (fig. 1 and supplementary fig. S2, Supplementary Material online). After adding published data from African Americans and European Americans (underarm) (Shriver and Parra 2000), there was a clear similarity between A-A and African Americans and also between CHN and European Americans (fig. 1B).
Genome-Wide Analysis of Sequence Variations
Using an Affymetrix Genome-wide Human Single Nucleotide Polymorphism (SNP) Array 6.0 (>900,000 SNPs), we genotyped 48 individuals randomly selected from A-A populations of southwestern China. Paired with published Affymetrix 6.0 data of other populations (International HapMap et al. 2010), including Han Chinese from Beijing (CHB, 89 individuals), Han Chinese from Denver (CHD, 90 individuals), Japanese (JPT, 90 individuals), Europeans (CEU, 118 individuals), and Africans (YRI, 120 individuals), principal component analysis (PCA) indicated a close relationship among Asian populations (A-A, CHB, CHD, and JPT) (supplementary fig. S3A, Supplementary Material online), with some local divergence (supplementary fig. S3B, Supplementary Material online).
To identify the genes responsible for skin pigmentation divergence, we looked for genes showing extraordinarily large divergence between CHN (with CHB used to represent CHN) and A-A and Africans (with YRI used to represent Africans) by calculating between-population allele frequency difference of each SNP (measured by FST) (Weir and Hill 2002). Compared with the randomly selected SNPs, SNPs located in pigmentation genes showed larger enrichment scores (P < 0.001, χ2 test) (supplementary fig. S4, Supplementary Material online), suggesting that pigmentation genes may be under selection in CHB.
In total, we identified 1,283 (CHB vs. A-A), 1,640 (CHB vs. YRI), and 1,706 gene regions (YRI vs. A-A) showing large divergences (FST, top 1% regions). Overlapping these gene regions with 171 known pigmentation genes from the human genome (Color Genes database, http://www.espcr.org/micemut/, last accessed September 03, 2015) yielded three gene-sets (15, 21 and 19 genes, respectively), among which three pigmentation genes (OCA2, EDAR, and PAH) showed consistent signals (supplementary table S2, Supplementary Material online). In the genome-wide FSTanalysis, we pooled the samples of three A-A populations from China, and the potential population substructure may cause false negatives. Consequently, the listed gene sets are conservative callings of genes under selection (supplementary table S2, Supplementary Material online). The identified candidate pigmentation genes are mostly different from those in Europeans, supporting the hypothesized convergent evolution of skin pigmentation in East Asia and Europe.
Compared with EDAR and OCA2, the signal of PAH is relatively weak and its functional role in pigmentation remains elusive (Schallreuter et al. 1999). While EDAR has accumulated East Asian specific mutations due to local selection, its functional effect lies in an increased number of active eccrine glands (Kamberov et al. 2013). In contrast, OCA2 showed the largest between-population (light-skinned vs. dark-skinned) divergence and was previously reported to be associated with melanin content (Edwards et al. 2010; Eaton et al. 2015). For further analysis, we focused on OCA2.
OCA2 Accounts for Light Skin in CHN
To search for functional mutation(s) of OCA2, we conducted amplicon sequencing of 40 randomly selected A-A individuals (25 from China and 15 from Cambodia), and obtained sequences of the entire genomic region of OCA2 (344.5 kb) which we compared to data from other populations collected within the 1000 Human Genomes Project (Genomes Project et al. 2012). First, the maps of linkage disequilibrium (LD) (supplementary fig. S5, Supplementary Material online) showed that CHB exhibited the strongest LD, which could be explained either by Darwinian positive selection or population history (e.g., bottleneck effect).
Within the gene region of OCA2, we observed a peak of allele frequency divergence (measured by FST) (Weir and Hill 2002) between CHB and A-A, spanning about 44.2 kb, in which rs1800414 was among a group of SNPs showing large FST values (fig. 2A). Further tests via extended haplotype homozygosity (EHH) (Sabeti et al. 2002) and Fay and Wu’s H statistics (Fay and Wu 2000) (the 44.2 kb region was analyzed) supported the signal of selection in CHB (supplementary fig. S6 and table S3, Supplementary Material online), in line with the previous report using other tests of selection (LSBL statistics, InRH, and Tajima’D test) (Edwards et al. 2010). Conversely, no selective signal was observed in A-A and Europeans (supplementary table S3, Supplementary Material online). rs1800414 is a nonsynonymous SNP located in exon 17 of OCA2, and an A to G mutation causes an amino acid change from histidine to arginine (His615Arg). This missense mutation is located in the melanosomal cytoplasmic topological domain (http://www.uniprot.org/, last accessed September 15, 2015). The geographic distribution of this SNP across world populations (data from HGDP database: http://hgdp.uchicago.edu, last accessed July 10, 2015) showed that the derived G allele of rs1800414 is highly prevalent in most East Asian populations (G allele >50% in Han Chinese), as well as in American Indians, but is in low frequency in A-A populations (G allele = 16.0% [A-A from China]; 21.3% [A-A from Cambodia]), totally absent in Africans, and absent or extremely rare in western Europeans (G allele <0.3%) (fig. 2B). This piece of evidence suggests that rs1800414 is likely an East Asian-specific mutation, consistent with the proposed local selection on this SNP and an independent evolution of skin pigmentation in the area (Edwards et al. 2010).
Fig. 2.
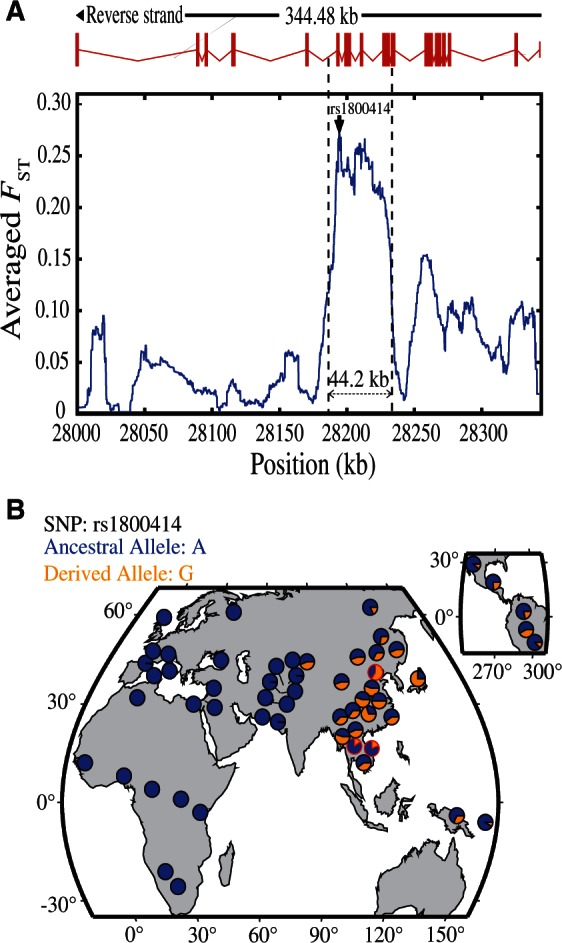
Genetic divergence between A-A and CHN and geographic distribution of rs1800414 in world populations. (A) Genetic divergence between A-A and CHN in the sequenced OCA2 gene region (∼344.5 kb) (measured by the averaged FST). The averaged FST of each position is the simple moving average with a sliding window size of 10 kb. The rs1800414 (showing the largest FST value) is labeled. (B) Geographic distribution of rs1800414 derived allele frequencies in world populations. The rs1800414 genotyping data of one CHN population and two A-A populations (Cambodia and China, marked with “red circles”) were from this study. Data of the other populations were from the HGDP database (http://hgdp.uchicago.edu/, last accessed July 10, 2015).
Assuming rs1800414 being the functional SNP under selection, we estimated the selective intensity and the time of selection using 1000 Human Genomes Project data (Genomes Project et al. 2012). In CHB, the selective coefficient was estimated to be 0.0157 (95% confidence interval [CI]: 0.0127–0.0186). Assuming a generation time of 25 years, the time of selection onset was estimated at 15,224 years ago (95% CI: 12,818–18,750 years ago), which is slightly older than a recently reported age (10,660 years ago with 95% CI of 8,070–15,780 years ago) using a Hidden Markov model (Chen and Slatking, 2015). The estimated age falls into the Upper Paleolithic, following the proposed northward migration of modern humans in East Asia some 25,000–30,000 years ago (Shi et al. 2005).
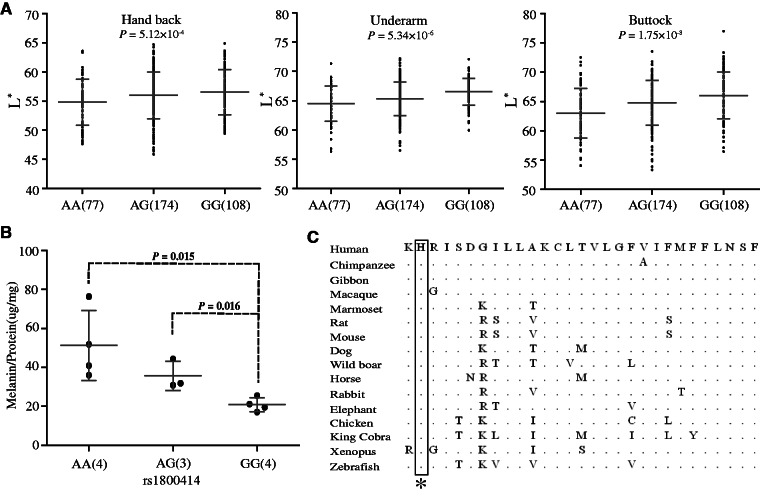
We conducted skin phenotyping of CHN (359 individuals) to determine whether the OCA2 SNPs affect skin pigmentation. We analyzed five SNPs located in the 44.2 kb region and three SNPs outside this region with strong selective signals. Under the additive genetic model, we detected a significant association of rs1800414 with skin pigmentation for both exposed (P = 5.12 × 10−4, hand back) and unexposed (P = 5.34 × 10−6, underarm and P = 1.75 × 10−8, buttock) areas (association remained significant after Bonferroni correction; fig. 3A), consistent with a previous association analysis of rs1800414 (Edwards et al. 2010; Eaton et al. 2015). No significant association was observed for the other seven SNPs (P > 0.05), except for rs12440942, with a marginal significance for skin on the buttock (P = 0.0395) (supplementary table S4, Supplementary Material online). When conditioning for rs1800414, rs12440942 became nonsignificant (P = 0.632).
Fig. 3.
Genetic association of rs1800414 with skin pigmentation in northern Han Chinese. (A) Measurements of skin darkness (by L* value) of three rs1800414 genotypes. The P values (additive genetic model) are shown in the scatted plots. The y axis represents the L* value; the x axis shows three rs1800414 genotypes with sample sizes in the parentheses. The “error bar” indicates standard deviation. (B) The levels of melanin production in human foreskin melanocytes with different rs1800414 genotypes. Two-tailed t-test was used for statistical evaluation. The y axis represents relative abundance of melanin (Melanin/Protein, μg/mg); the x axis shows the three genotypes with sample size in parentheses. The error bar indicates standard deviation. (C) OCA2 protein sequence (human protein position 614–642) alignment among vertebrate species and rs1800414 (labeled with an asterisk) is highly conserved. The protein sequences were from NCBI (http://www.ncbi.nlm.nih.gov/, last accessed July 10, 2015).
Human Skins of Different rs1800414 Genotypes Have Different Melanin Productivity
To test whether rs1800414 affects skin pigmentation, we conducted primary cell culture and purification of melanocytes (human foreskin tissue was used) according to published protocols (Cook et al. 2003), and established 11 melanocyte cell lines covering the three genotypes of rs1800414 (4 AA, 3 AG, and 4 GG). We quantified the relative abundance of melanin in each cell line using the total amount of proteins as a control according to published protocols (Cook et al. 2009). As predicted, homozygotes of the derived G allele (Arg615) had the least amount of melanin (correspondent to a relatively lighter skin), whereas the homozygotes of the ancestral A allele (His615) had the highest amount (correspondent to a relatively darker skin), leaving the heterozygotes between the two types of homozygotes (fig. 3B). Surprisingly, the ancestral allele of rs1800414 (His615) is highly conserved among species from zebrafish to human (fig. 3C). The measurement of GERP’s (Genomic Evolutionary Rate Profiling) score suggested a strong functional constraint on this site during evolution (GERP’s score = 366, P = 2.93 × 10−72) (http://mendel.stanford.edu/SidowLab, last accessed October 20, 2015; Cooper et al. 2005), implying that the Arg615 substitution may change OCA2 function.
Equivalent A and G Alleles of rs1800414 in Zebrafish Have Different Functions
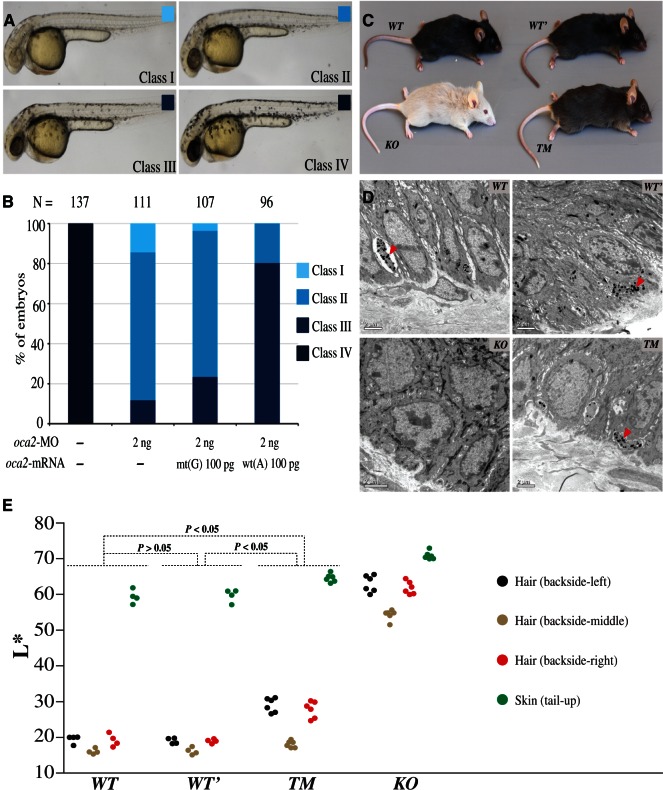
The deficiency of Oca2 leads to hypopigmentation in the skin melanophores and the retinal pigment epithelium, indicating an essential role in melanin synthesis (Beirl et al. 2014). The zebrafish Oca2 protein has His610 residue equivalent to His615 of human OCA2. The underpigmentation of melanophores depleted of oca2 allows us to test functional relevance of the His610Arg mutation genocopying the A to G substitute at rs1800414 in human. The expression of oca2 could be effectively blocked by injection of an oca2-specific antisense morpholino (oca2-MO) (supplementary fig. S7A, Supplementary Material online) and oca2 morphants exhibited progressive loss of melanophore pigmentation in a dose-dependent manner at 36 or 48 h postfertilization (supplementary fig. S7B, Supplementary Material online), which phenocopied oca2 mutants (Beirl et al. 2014). Next, we performed rescue experiments using wild-type zebrafish oca2 mRNA (wt(A) with His610) or mutant oca2 mRNA (mt(G) with His610Arg mutation). Results indicated that coinjection of 100 pg oca2 wt (A) mRNA largely recovered melanophore pigmentation in oca2 morphants whereas coinjection of 100 pg oca2 mt (G) mRNA failed to do so (fig. 4A and B). Hence, His610 of zebrafish Oca2 protein is important for melanin synthesis, supporting the idea that the rs1800414 variant of human OCA2 may be functionally associated with skin coloration.
Fig. 4.
Transgenic and targeted gene modification analyses of zebrafishes and mice. (A and B) Effect of oca2 wt(A) or mt(G) mRNA overexpression in oca2 morphants. (A) The embryos at 36 h postfertilization were classified into four classes based on melanophore pigmentation. Class IV, wild-type pigmentation; Class I–III, different levels of pigmentation reduction. (B) Ratios of different classes of embryos. N, the total number of observed embryos. Note that, unlike wt(A) mRNA, mt(G) mRNA coinjection could not efficiently restore pigmentation in oca2-MO injected embryos. (C) Coloration comparison among different mouse strains, including the wild-type strain (WT), the strain containing two synonymous mutations (WT’), the knockout strain (KO), and the targeted modification strain (TM). (D) The distribution of melanosomes in the tail skin of different mouse strains. The “arrow” indicates the melanosomes in sectioned skin. (E) Quantification of hair and skin pigmentation among the mouse strains (WT, WT’, TM, and KO). The y axis indicates the L* value of the four measured body parts with the error bars indicating standard deviations. The P values are shown in the plot (Wilcoxon rank sum test). More details are presented in supplementary tables S6 and S7, Supplementary Material online.
Mice Homozygous for G Allele-Like Oca2 (His610Arg) Display Light Coloration
To further test the phenotypic effect of rs1800414 on melanin production and skin pigmentation, using the CRISPR/Cas9 approach (Li et al. 2013; Wang et al. 2013; Yang et al. 2013), we generated two gene-modified mouse strains using black hair C57BL/6 mice (WT), including the Oca2 knockout strain (KO) (a frame-shift mutation leading to a truncated Oca2) and the rs1800414 targeted modification strain with His610Arg mutation resembling human His615Arg mutation (#1 mutation, supplementary fig. S8A and B, Supplementary Material online). We also generated a control strain carrying two Oca2 synonymous mutations (WT’) (2 and 3 mutations, supplementary fig. S8A and B, Supplementary Material online) in order to rule out any potential effect of these two synonymous mutations introduced when generating the targeted modification using CRISPR/Cas9. No off-target effect was detected when sequencing the top five predicted potential off-targeted sites in all mouse strains (supplementary table S5, Supplementary Material online).
As expected, the KO mice displayed white body hair, white tail skin, white paws, and pink iris (fig. 4C), in line with the phenotypes reported in previous Oca2 studies (Gardner et al. 1992; Rinchik et al. 1993). These results indicate that Oca2 is indeed a key gene for melanin production. Conversely, mice carrying the two Oca2 synonymous mutations (WT’) showed the same coloration with the wild-type controls, that is, black body hair, dark tail skin, dark paws, and black iris (fig. 4C), implying that the two synonymous mutations do not interfere Oca2 function.
In line with the phenotypic effect observed in the zebrafish, the targeted modification mice (TM), homozygous for the derived allele of rs1800414, showed light coloration, including brown/gray body hair, light tail skin and light paws, but with no obvious color change seen in the iris (fig. 4C). We conducted quantitative measurements of hair and skin pigmentation of the four mouse strains. The TM mice showed a significantly lighter coloration than the WT/WT’ mice (fig. 4E, supplementary fig. S9 and tables S6 and S7, Supplementary Material online). Sequencing of Oca2 RNA transcripts did not detect any change of gene alternative splicing among the mouse strains (supplementary fig. S8C, Supplementary Material online), suggesting that the phenotypic effect of rs1800414 is caused by protein functional alteration due to the observed amino acid change.
We compared the distributions of melanosomes in the tail skin among different mouse strains by transmission electron microscope. At cellular level, the TM mice showed a clear reduction of mature melanosomes compared with the WT and WT’ mice and the KO mice appeared to lack mature melanosomes (fig. 4D). These results imply that the derived allele of rs1800414 alters the function of Oca2, ultimately affecting the maturation and/or transportation of melanosomes in the melanocytes.
Discussion
Here, we identified a set of pigmentation genes showing large genetic divergence between dark-skinned (A-A and YRI) and light-skinned populations (CHN) (supplementary fig. S4 and table S2, Supplementary Material online), which comprise potential candidates contributing to skin lightening among East Asians. The sustained dark skin of A-A people is likely due to their settling in low latitude tropical areas of Asia, which are similar to those inhabited by our African ancestors. When these groups migrated northward in East Asia about 25,000–30,000 years (Jin and Su 2000; Shi et al. 2005), natural selection would seemingly favor lighter skin, due to the reduced UV radiation at high latitudes. Though multiple pigmentation genes might be involved in this transition, our results strongly indicate that OCA2 served as a major-effect gene conferring skin lightening of East Asians.
OCA2 is one of the key players in the pigmentation pathway, and previous reports associated OCA2 with abnormal skin pigmentation, oculocutaneous albinism type 2 (OCA2) (Gardner et al. 1992; Rinchik et al. 1993; Lee et al. 1994). The protein encoded by OCA2 (a 12-transmembrane protein) is localized on the melanosomal membrane (Rinchik et al. 1993; Rosemblat et al. 1994), and may be involved in the control of l-tyrosine transport (Toyofuku et al. 2002), pH adjustment in melanosome (Puri et al. 2000), glutathione metabolism (Staleva et al. 2002), and tyrosinase processing (Chen et al. 2002). We showed that the nonsynonymous SNP rs1800414 is the most diverged variant between CHN and A-A, and significantly associated with skin pigmentation for the tested skin areas in CHN (fig. 3A). Moreover, the derived G allele homozygotes produced the least amount of melanin in the cultured melanocytes, and showed the lightest skin in northern Han Chinese (fig. 3B). This phenotypic effect was confirmed in the gene-modified zebrafishes and mice (fig. 4), suggesting that the His615Arg mutation at rs1800414 affects the maturation and/or transportation of melanosomes, eventually leading to skin lightening.
Given the demonstrated role of OCA2 for skin lightening of East Asians and previous reports of different pigmentation genes responsible for light skin in Europeans (SLC24A5 and SLC45A2) (Graf et al. 2005; Lamason et al. 2005; Izagirre et al. 2006; Yuasa et al. 2006; Lao et al. 2007; Norton et al. 2007; Sabeti et al. 2007; Soejima and Koda 2007; Han et al. 2008; Beleza et al. 2013; Wilde et al. 2014), our data support a convergent evolution of skin pigmentation in East Asians (Lao et al. 2007; Myles et al. 2007; Norton et al. 2007; Edwards et al. 2010). Although OCA2 also showed a potential signal of selection in Europeans (supplementary table S2, Supplementary Material online; Lao et al. 2007; Norton et al. 2007), the causal SNP rs1800414 enriched in East Asians is nearly absent in Europeans. Intriguingly, we found that the proteins of OCA2, SLC24A5, and SLC45A2 are all trans-membrane proteins located on the melanosomal membrane, implying that similar molecular checkpoints were acted on by natural selection, eventually leading to independent skin-lightening in both East Asians and Europeans, who live in high latitude areas with weak light conditions.
There are also pigmentation genes (DCT, EGFR, DRD2, and MC1R) reportedly under selection in dark-skinned people (e.g., Africans) (Harding et al. 2000; John et al. 2003; Lao et al. 2007), which have been explained by the protection conferred by melanin against the harmful effects of UV radiation, including protection against sunburn and folate destruction. For light-skinned people, because UV is required for the synthesis of vitamin D, a key nutrient for bone development (Loomis 1967), and selection on vitamin D synthesis would favor a lighter skin at these high latitude regions and thereby explain the selective signal of OCA2 in northern Han Chinese. However, we cannot rule out other environmental and biological factors (e.g., diet change and/or sexual selection) that may also create selective pressures on skin lightening. Despite differences in the underlying causes, our evidence for a convergent evolution of skin lightening in Europe and East Asia represents an excellent case of how natural selection has shaped our phenotypic diversity in recent human evolutionary history.
Materials and Methods
Sample Collection and Skin Pigmentation Measurement
We collected blood samples from 1,159 Austro-Asiatic speaking individuals (716 females and 443 males) living in southwestern China (Yunnan province) and northeastern Cambodia as well as 359 blood samples (210 females and 149 males) from northern Han Chinese of China (supplementary table S1, Supplementary Material online). Sample collection was conducted randomly and biologically related individuals were excluded from this study. DNA samples were prepared using the standard phenol–chloroform method.
We used a CR-400 tristimulus colorimeter (Konica Minolta, Tokyo, Japan) to measure skin pigmentation, which generates three numerical readings that can express the color of any object: L* (levels of darkness), a* (amount of green or red), and b* (amount of yellow or blue). Both constitutive skin pigmentation from skin not exposed to sunlight (underarm and buttock) and facultative skin pigmentation that is exposed to sunlight (back of hand) were measured for comparisons. To minimize technical variations, each area was measured three times, and the average used for statistical analysis. Due to the known seasonal variation in skin phenotype, skin pigmentation measurements were conducted in the same season (summer of 2011–2012) for all populations, the same seasons with the published data (Shriver and Parra 2000). GraphPad Prism 5 was used to plot L* distribution.
Written informed consent was obtained from each individual prior to their inclusion in the study. Additional written consent was obtained for use of portrait photos. All protocols of this study were approved by the Institutional Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (approval No: KMDWS-SEYX-20120101010).
Genome-Wide SNP Analysis
We employed an Affymetrix Genome-wide Human SNP Array 6.0, which includes 906,600 SNPs with an average inter-SNP distance of 1.5 kb, to scan 48 randomly selected Austro-Asiatic individuals from southwestern China (20 Wa, 10 Bulang, and 18 De’ang) (supplementary table S1, Supplementary Material online). The published Affymetrix data of Han Chinese from Beijing (CHB, 89 individuals) (International HapMap et al. 2010) was used to represent northern Han Chinese and compared with the A-A population.
PCA was used to show the relationship of the A-A samples with other populations. The SNPs were pruned based on LD for PCA, no particular genomic regions were removed prior to the PCA.
We also calculated the enrichment score for each FST bin when comparing A-A and CHB among genes involved in the pigmentation pathways and those not involved. We divided the FST values into five bins with the value interval of 0.05 in each bin. Simply, the FST values assigned to each bin were (0–0.05), (0.05–0.1), (0.1–0.15), (0.15–0.2), and (≥0.2), as shown in supplementary fig. S4, Supplementary Material online. The genes spanning the SNP were assigned to the relevant FST bin based on the FST value of that SNP. The enrichment score in each bin was calculated based on 1,000 bootstrapping data sets from random SNPs or pigmentation-associated SNPs, the score was the fraction of SNPs in each bin, divided by the fraction of all SNPs from known genes except for the pigmentation associated genes.
To identify genomic regions showing deep divergence between dark-skinned (A-A and YRI) and light-skinned populations (CHN), we used the moments estimate of FST described by Weir and Hill to measure between-population genetic divergence (Weir and Hill 2002). Overlapping with 171 known pigmentation genes from the human genome (Color Genes database, http://www.espcr.org/micemut/, last accessed September 20, 2015) yielded 15 (CHB vs. A-A), 21 (CHB vs. YRI), and 19 (YRI vs. A-A) pigmentation gene regions with large divergence (FST, top 1% gene regions).
Resequencing of OCA2 and Molecular Evolution Analysis
For sequencing, we randomly selected 40 A-A individuals (25 from China and 15 from Cambodia) and designed a series of primers to cover the entire 344.483 kb genomic region of OCA2. Using data from the 1000 Human Genomes Project (Genomes Project et al. 2012), we extracted all OCA2 SNPs of individuals from populations of Han Chinese (CHB 97), western Europeans (CEU 85), and Africans (YRI 88).
Haploview software (D’ algorithm) (Bandelt et al. 1999) was used to analyze LD pattern of OCA2 in different populations. Genetic divergence between A-A and CHN in the sequenced OCA2 genomic region (∼344.5 kb) was measured by the averaged FST. The averaged FST of each position is the simple moving average with a sliding window size of 10 kb. We also used the EHH test described by Sabeti et al. (2002) for detecting recent positive selection displaying incomplete selective sweep, that is, the selected allele has not reached fixation. We carried out tests of neutrality following the method developed by Fay and Wu (2000), using the program DnaSPv5 (Librado and Rozas 2009). A significant negative Fay and Wu’s H indicates an excess of high-frequency derived alleles due to Darwinian positive selection.
Inferring Allele Age and Intensity of Selection of rs1800414
The SNP data covering a region of 942.7 kb for 97 CHB individuals was obtained from the 1000 Genomes Project (Genomes Project et al. 2012). We used fastPHASE (Scheet and Stephens 2006) to infer the haplotype phase from SNP genotypes. The inferred haplotypes were then used in further analysis. Starting from the putatively selected mutant position, we ran a hidden Markov model to identify ancestral haplotypes retained around the vicinity of the selected mutant. The extent of ancestral haplotypes was recorded and the likelihood function was built based on the haplotype extent. We adopted the importance sampling approach by Chen and Slatkin (2013) to infer the parameters, including selection intensity and allele age (see the original paper for the details of the Materials and Methods). A generation time of 25 years was used.
Genetic Association Analysis
According to the result of molecular evolution analysis of OCA2, we chose eight weak linkage SNPs as possible for association analysis, including five SNPs located in the 44.2 kb region with strong selective signal and other three SNPs outside this region. A total of 359 blood samples (210 females and 149 males) from Han Chinese individuals (CHN, light-skinned) from northern China (supplementary table S1, Supplementary Material online) were used for skin pigmentation association analysis. Genotyping was conducted by the SnapShots method on an ABI 3130 sequencer (Applied Biosystems, Forster city, CA, USA). Skin pigmentation association of single SNP with L* (hand back, underarm, and buttock) were tested by utilizing PLINK v1.07 (Purcell et al. 2007), using the linear regression option, with age and sex as covariates.
Melanocyte Culture and In Vitro Analysis of Melanin Synthesis
We collected foreskin tissue samples of male individuals (8–20 years old) who had circumcisions at the local hospitals. Written informed consent was obtained from each donor or donor’s parents prior to sample collection. The published method by Cook et al. (2003) was used to culture and purify melanocytes. We used the melanin analytical method by Cook et al. (2009) to calculate the total amount of melanin and protein of melanocytes in each cell culture dish. Total melanin and protein were isolated from cells with different rs1800414 genotypes (4 AA, 3 AG, and 4 GG). The protocol of this study was approved by the Institutional Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (approval No: KMDWS-SEYX-20120101010).
Functional Study of oca2 in Zebrafish Embryos
Wild-type embryos of Tuebingen strain were used. Embryos were incubated in Holtfreter’s solution at 28.5°C and staged as previously described (Kimmel et al. 1995). Ethical approval was obtained from the Animal Care and Use Committee of Tsinghua University.
The coding sequence of oca2 was amplified and cloned into the pXT7 vector using enzymatic assembly method (Gibson et al. 2009), with Kozak sequence added to enhance the translation efficiency. The A>G nucleotide substitution at + 1829, which resulted in His610Arg mutation, was made with site direct mutation.
The oca2-MO, synthesized by Gene Tools, LLC, targets 5′-CTGCCAAATAAGTGAATGAAATGAT-3′ in the 5′-untranslated region (UTR) of oca2. The reporter construct poca2-EGFP was made by fusing 5′-UTR and adjacent coding sequence to EGFP cds in frame and used to test the effectiveness of oca2-MO. Capped mRNAs were synthesized using T7 mMessage mMachine Kit (Ambion) and purified using RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. MO was injected alone or in combination with mRNA into the yolk of zebrafish embryos at the one-cell stage. Pigment phenotypes were observed and imaged with stereomicroscope (Nikon SMZ 1500). Because higher doses (>150 pg) of oca2 mRNA impaired early embryonic development, we chose to combine a lower dose (100 pg) of mRNA with a lower dose of oca2-MO.
Targeted Gene Modification Experiment in Mouse
Using a CRISPR design tool (http://crispr.mit.edu/, last accessed July 10, 2015), we selected the sgRNA target sequence (TAAAGCACAGGATTTCAGAC) for murine Oca2 to direct Cas9 cleavage. The murine Oca2 sgRNA sequence was cloned into pDR274 (a sgRNA cloning vector from Addgene, Cambridge MA) to form a T7 promoter-mediated sgRNA expression vector (Hwang et al. 2013). With DraI digestion, the linearized expression vector was purified using a QIAquick Gel Purification Kit (QIAGEN) and used as a DNA template to produce sgRNA by a MAXIscript T7 kit (Life Technologies). For Cas9 mRNA production, T7 promoter-contained px330 vector (Cong et al. 2013) was digested with NotI and then purified using a QIAquick Gel Purification Kit (QIAGEN). Subsequently, the linearized and purified vector was used as a DNA template to synthesize Cas9 mRNA using a mMESSAGE mMACHINE T7 Ultra Kit (Life Technologies). Cas9 mRNA and sgRNA were purified using RNA Clean & Concentrator-25 (ZYMO Research) and dissolved in RNase-free water. RNA concentrations were measured using a NanoDrop ND1000. Finally, 2 µl each of sgRNA and Cas9 mRNA were mixed with an equal volume of formamide, respectively, and the denatured mixtures were run on a DNA agarose gel to evaluate RNA quality.
We synthesized 180-bp single-stranded donor oligonucleotides (ssODN) to introduce the designed mutations by homologous recombination mediated repair. As shown in the supplementary figure S8, Supplementary Material online, mutation #1 is the expected mutation to cause His610Arg substitution (by mutating Oca2 A→G); mutation #2 to delete protospacer adjacent motif sequence of NGG to avoid repeated Cas9 cleavage of target DNA; mutation #3 to delete PshAI cut site (GACNNNNGTC) for convenience of subsequent genotyping. Mutations #2 and #3 do not cause amino acid changes. According to the literature(Wang et al. 2013; Yang et al. 2013; Singh et al. 2015) and our previous injection experience (Zhong et al. 2015), sgRNA and Cas9 mRNA mixture (containing 50 ng/µl sgRNA, 100 ng/µl Cas9 mRNA, and 100 ng/µl ssODN) were prepared using RNase-free water. RNA was microinjected into the cytoplasm of C57BL/6 inbred zygotes. After injection, surviving zygotes were immediately transferred into oviducts of ICR albino pseudopregnant females. All mice were maintained under cycles of 12-h light and 12-h dark. The Oca2 genotypes of the generated mouse strains were confirmed by sequencing (supplementary fig. S8B, Supplementary Material online). All animal operations were approved by the Institutional Animal Care and the Institutional Review Board of Baylor College of Medicine and Kunming Institute of Zoology, Chinese Academy of Sciences (approval No: KMDWS-SEYX-20120101010).
Quantitative measurements of hair and skin pigmentation were conducted for the four mouse strains. Adult mice (∼5 months) were measured (4 WT, 4 WT’, 6 TM, and 6 KO) with a CR-400 tristimulus colorimeter. Hair pigmentation was measured in three different regions of the back (left, middle, and right), and we also obtained skin pigmentation measures of the tail (supplementary fig. S9, Supplementary Material online).
Transmission electron microscopic analysis was performed. Mouse skin tissues were prepared by conventional methods: fixations, staining, and dehydration, embedding in resin and sectioned to obtain thin slices and viewed under a JEOL JEM-1101 transmission electron microscope at room temperature. Refer to the published method of Varani et al. (2001) for details.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to all the voluntary donors in this study. Special thanks to persons who agreed to provide their portrait photos for publication in this paper. The authors thank Prof. Yongtang Zheng for his help in collecting blood samples. They also thank Xiaofeng Ma, Shuangjuan Yang, Wenting Wan and Xiaolu Li for their technical assistances. This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB13010000); the National Natural Science Foundation of China (91131001, 91231203, 31321002, 31123005, 31371268, and 31371269); the Natural Science Foundation of Yunnan Province (2010CI044); State Key Laboratory of Genetic Resources and Evolution grant (GREKF15-06); and the Personnel Training Project of Yunnan Province (KKSY201526061).
References
- Bandelt HJ, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 16:37–48. [DOI] [PubMed] [Google Scholar]
- Beirl AJ, Linbo TH, Cobb MJ, Cooper CD. 2014. oca2 regulation of chromatophore differentiation and number is cell type specific in zebrafish. Pigment Cell Melanoma Res. 27:178–189. [DOI] [PubMed] [Google Scholar]
- Beleza S, Johnson NA, Candille SI, Absher DM, Coram MA, Lopes J, Campos J, Araujo II, Anderson TM, Vilhjalmsson BJ, et al. 2013. Genetic architecture of skin and eye color in an African-European admixed population. PLoS Genet. 9:e1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin G. 2004. Geographic distribution of environmental factors influencing human skin coloration. Am J Phys Anthropol. 125:292–302. [DOI] [PubMed] [Google Scholar]
- Chen H, Hey J, Slatkin M. 2015. A hidden Markov model for investigating recent positive selection through haplotype structure. Theor Popul Biol. 99:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Slatkin M. 2013. Inferring selection intensity and allele age from multilocus haplotype structure. G3 3:1429–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Manga P, Orlow SJ. 2002. Pink-eyed dilution protein controls the processing of tyrosinase. Mol Biol Cell. 13:1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AL, Chen W, Thurber AE, Smit DJ, Smith AG, Bladen TG, Brown DL, Duffy DL, Pastorino L, Bianchi-Scarra G, et al. 2009. Analysis of cultured human melanocytes based on polymorphisms within the SLC45A2/MATP, SLC24A5/NCKX5, and OCA2/P loci. J Invest Dermatol. 129:392–405. [DOI] [PubMed] [Google Scholar]
- Cook AL, Donatien PD, Smith AG, Murphy M, Jones MK, Herlyn M, Bennett DC, Leonard JH, Sturm RA. 2003. Human melanoblasts in culture: expression of BRN2 and synergistic regulation by fibroblast growth factor-2, stem cell factor, and endothelin-3. J Invest Dermatol. 121:1150–1159. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. 2005. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton K, Edwards M, Krithika S, Cook G, Norton H, Parra EJ. 2015. Association study confirms the role of two OCA2 polymorphisms in normal skin pigmentation variation in East Asian populations. Am J Hum Biol. 27:520–525. [DOI] [PubMed] [Google Scholar]
- Edwards M, Bigham A, Tan J, Li S, Gozdzik A, Ross K, Jin L, Parra EJ. 2010. Association of the OCA2 polymorphism His615Arg with melanin content in East Asian populations: further evidence of convergent evolution of skin pigmentation. PLoS Genet. 6:e1000867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wu CI. 2000. Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JM, Nakatsu Y, Gondo Y, Lee S, Lyon MF, King RA, Brilliant MH. 1992. The mouse pink-eyed dilution gene: association with human Prader-Willi and Angelman syndromes. Science 257:1121–1124. [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6:343–345. [DOI] [PubMed] [Google Scholar]
- Graf J, Hodgson R, van Daal A. 2005. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 25:278–284. [DOI] [PubMed] [Google Scholar]
- Han J, Kraft P, Nan H, Guo Q, Chen C, Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, et al. 2008. A genome-wide association study identifies novel alleles associated with air color and skin pigmentation. PLoS Genet. 4:e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJ, Ellis NS, Flanagan N, Todd C, Dixon C, Sajantila A, Jackson IJ, Birch-Machin MA, et al. 2000. Evidence for variable selective pressures at MC1R. Am J Hum Genet. 66:1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUGO Pan-Asian SNP Consortium Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, Chaurasia A, Chen CH, Chen J, et al. 2009. Mapping human genetic diversity in Asia. Science 326:1541–1545. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 31:227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, et al. 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izagirre N, Garcia I, Junquera C, de la Rua C, Alonso S. 2006. A scan for signatures of positive selection in candidate loci for skin pigmentation in humans. Mol Biol Evol. 23:1697–1706. [DOI] [PubMed] [Google Scholar]
- Jablonski NG, Chaplin G. 2000. The evolution of human skin coloration. J Hum Evol. 39:57–106. [DOI] [PubMed] [Google Scholar]
- Jin L, Su B. 2000. Natives or immigrants: modern human origin in East Asia. Nat Rev Genet. 1:126–133. [DOI] [PubMed] [Google Scholar]
- John PR, Makova K, Li WH, Jenkins T, Ramsay M. 2003. DNA polymorphism and selection at the melanocortin-1 receptor gene in normally pigmented southern African individuals. Ann N Y Acad Sci. 994:299–306. [DOI] [PubMed] [Google Scholar]
- Kamberov YG, Wang S, Tan J, Gerbault P, Wark A, Tan L, Yang Y, Li S, Tang K, Chen H, et al. 2013. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell 152:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn. 203:253–310. [DOI] [PubMed] [Google Scholar]
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. 2005. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310:1782–1786. [DOI] [PubMed] [Google Scholar]
- Lao O, de Gruijter JM, van Duijn K, Navarro A, Kayser M. 2007. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Ann Hum Genet. 71:354–369. [DOI] [PubMed] [Google Scholar]
- Lee ST, Nicholls RD, Bundey S, Laxova R, Musarella M, Spritz RA. 1994. Mutations of the P gene in oculocutaneous albinism, ocular albinism, and Prader-Willi syndrome plus albinism. N Engl J Med. 330:529–534. [DOI] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, et al. 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 31:681–683. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. [DOI] [PubMed] [Google Scholar]
- Loomis WF. 1967. Skin-pigment regulation of vitamin-D biosynthesis in man. Science 157:501–506. [DOI] [PubMed] [Google Scholar]
- Myles S, Somel M, Tang K, Kelso J, Stoneking M. 2007. Identifying genes underlying skin pigmentation differences among human populations. Hum Genet. 120:613–621. [DOI] [PubMed] [Google Scholar]
- Norton HL, Kittles RA, Parra E, McKeigue P, Mao X, Cheng K, Canfield VA, Bradley DG, McEvoy B, Shriver MD. 2007. Genetic evidence for the convergent evolution of light skin in Europeans and East Asians. Mol Biol Evol. 24:710–722. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Gardner JM, Brilliant MH. 2000. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J Invest Dermatol. 115:607–613. [DOI] [PubMed] [Google Scholar]
- Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, Avidano KM, Jong MT, Nicholls RD. 1993. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 361:72–76. [DOI] [PubMed] [Google Scholar]
- Rosemblat S, Durham-Pierre D, Gardner JM, Nakatsu Y, Brilliant MH, Orlow SJ. 1994. Identification of a melanosomal membrane protein encoded by the pink-eyed dilution (type II oculocutaneous albinism) gene. Proc Natl Acad Sci U S A. 91:12071–12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallreuter KU, Moore J, Tobin DJ, Gibbons NJ, Marshall HS, Jenner T, Beazley WD, Wood JM. 1999. alpha-MSH can control the essential cofactor 6-tetrahydrobiopterin in melanogenesis. Ann N Y Acad Sci. 885:329–341. [DOI] [PubMed] [Google Scholar]
- Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 78:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, Chakraborty R, Jin L, Su B. 2005. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet. 77:408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ. 2000. Comparison of narrow-band reflectance spectroscopy and tristimulus colorimetry for measurements of skin and hair color in persons of different biological ancestry. Am J Phys Anthropol. 112:17–27. [DOI] [PubMed] [Google Scholar]
- Singh P, Schimenti JC, Bolcun-Filas E. 2015. A mouse geneticist's practical guide to CRISPR applications. Genetics 199:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima M, Koda Y. 2007. Population differences of two coding SNPs in pigmentation-related genes SLC24A5 and SLC45A2. Int J Legal Med. 121:36–39. [DOI] [PubMed] [Google Scholar]
- Staleva L, Manga P, Orlow SJ. 2002. Pink-eyed dilution protein modulates arsenic sensitivity and intracellular glutathione metabolism. Mol Biol Cell. 13:4206–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, et al. 1999. Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet. 65:1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku K, Valencia JC, Kushimoto T, Costin GE, Virador VM, Vieira WD, Ferrans VJ, Hearing VJ. 2002. The etiology of oculocutaneous albinism (OCA) type II: the pink protein modulates the processing and transport of tyrosinase. Pigment Cell Res. 15:217–224. [DOI] [PubMed] [Google Scholar]
- Varani J, Spearman D, Perone P, Fligiel SE, Datta SC, Wang ZQ, Shao Y, Kang S, Fisher GJ, Voorhees JJ. 2001. Inhibition of type I procollagen synthesis by damaged collagen in photoaged skin and by collagenase-degraded collagen in vitro. Am J Pathol. 158:931–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Hill WG. 2002. Estimating F-statistics. Annu Rev Genet. 36:721–750. [DOI] [PubMed] [Google Scholar]
- Wilde S, Timpson A, Kirsanow K, Kaiser E, Kayser M, Unterlander M, Hollfelder N, Potekhina ID, Schier W, Thomas MG, et al. 2014. Direct evidence for positive selection of skin, hair, and eye pigmentation in Europeans during the last 5,000 y. Proc Natl Acad Sci U S A. 111:4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa I, Umetsu K, Harihara S, Kido A, Miyoshi A, Saitou N, Dashnyam B, Jin F, Lucotte G, Chattopadhyay PK, et al. 2006. Distribution of the F374 allele of the SLC45A2 (MATP) gene and founder-haplotype analysis. Ann Hum Genet. 70:802–811. [DOI] [PubMed] [Google Scholar]
- Zhong H, Chen Y, Li Y, Chen R, Mardon G. 2015. CRISPR-engineered mosaicism rapidly reveals that loss of Kcnj13 function in mice mimics human disease phenotypes. Sci Rep. 5:8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.