Abstract
Qualitative patterns of gene activation and repression are often conserved despite an abundance of quantitative variation in expression levels within and between species. A major challenge to interpreting patterns of expression divergence is knowing which changes in gene expression affect fitness. To characterize the fitness effects of gene expression divergence, we placed orthologous promoters from eight yeast species upstream of malate synthase (MLS1) in Saccharomyces cerevisiae. As expected, we found these promoters varied in their expression level under activated and repressed conditions as well as in their dynamic response following loss of glucose repression. Despite these differences, only a single promoter driving near basal levels of expression caused a detectable loss of fitness. We conclude that the MLS1 promoter lies on a fitness plateau whereby even large changes in gene expression can be tolerated without a substantial loss of fitness.
Keywords: yeast, evolution, expression, MLS1, binding sites.
Introduction
Changes in gene regulation are thought to play an important role in evolution (King and Wilson 1975; Wray 2007; Carroll 2008). Although there are many examples of cis-regulatory changes underlying diverged phenotypes (Wray 2007; Gaunt and Paul 2012), phenotypes more often map to changes in protein-coding sequences (Hoekstra and Coyne 2007; Stern and Orgogozo 2008; Fay 2013; Martin and Orgogozo 2013). One reason for the fewer number of phenotypes attributable to cis-regulatory mutations is the greater difficulty in demonstrating their influence on a phenotype (Stern and Orgogozo 2008). As such, most of our understanding of regulatory evolution is based on gene expression and cis-regulatory sequence divergence irrespective of downstream phenotypes.
One general feature of regulatory evolution that has emerged is conservation of qualitative patterns of gene expression despite divergence in the cis-regulatory sequences driving expression. Quantitative studies of gene expression levels have shown that there is an abundance of quantitative variation within and between species (Whitehead and Crawford 2006; Gordon and Ruvinsky 2012). Yet, qualitative patterns of activation, repression, and tissue-specific expression are generally conserved across distantly related species (Gasch et al. 2004; Chan et al. 2009). In comparison, studies of cis-regulatory sequences have shown that gain and loss of transcription factor binding sites is common (Moses et al. 2006; Doniger and Fay 2007; Kim et al. 2009; Bradley et al. 2010; Schmidt et al. 2010; Yokoyama et al. 2014), and that between distantly related species, cis-regulatory sequences often diverge to the extent that the sequences are unalignable (Wratten et al 2006; Hare et al 2008; Venkataram and Fay 2010; Arnold et al. 2014).
The binding site turnover model explains how gene regulation can be conserved while cis-regulatory sequences diverge (Ludwig et al. 1998 , 2000; Dermitzakis and Clark 2002; Dermitzakis et al. 2003). Under this model, gain and loss of equivalent binding sites within the same regulatory sequence enables high rates of divergence without changes in gene regulation. The binding site turnover model is supported by striking demonstrations that diverged cis-regulatory sequences from distantly related species drive very similar patterns of gene expression when placed in the same genome (Romano and Wray 2003; Ruvinsky and Ruvkun 2003; Markstein et al. 2004; Fisher et al. 2006; Wratten et al. 2006; Hare et al. 2008; Swanson et al. 2011; Arnold et al. 2014). Over long time periods, divergence in cis-regulatory sequences may also be facilitated by transcriptional rewiring, whereby different binding sites can be substituted for one another (Tsong et al. 2006; Tuch et al. 2008). However, the decrease in regulatory conservation as cis-regulatory sequences are placed into more distantly related genomes (Gordon and Ruvinsky 2012; Barrière and Ruvinsky 2014) implies that there are limits to the compatibility of cis-regulatory sequences with distantly related trans-environments.
A major barrier to interpreting patterns of gene expression conservation and divergence is that their influence on outward phenotypes or fitness is unknown. Although in some instances patterns of gene expression divergence themselves are indicative of fitness effects, in most cases expression divergence is assumed to be neutral (Fay and Wittkopp 2008). For example, there is evidence that subtle but consistent changes in the expression of genes in the same pathway or biological process influence fitness (Bullard et al. 2010; Fraser et al. 2010). A further complication is that fitness may depend not only on expression levels. The temporal or developmental patterns of expression may also influence fitness. For example, by comparing the distribution of mutation effects with naturally occurring polymorphism in the gene TDH3, Metzger et al. (2015) inferred that there is abundant purifying selection against mutations that increase cell to cell variation in expression levels. Overall, testing whether cis-regulatory sequences have diverged in their ability to integrate transcription factors, nucleosome positioning, and core transcriptional machinery into proper expression is challenging. However, the consequences of any meaningful regulatory changes should be reflected in fitness.
The direct effects of gene expression on fitness are not often characterized. Ludwig et al. (2005) found complementation of diverged enhancers, although none of the transgenic constructs rescued wild-type fitness levels. Using an inducible promoter, a fitness plateau was found for LCB2 gene expression in yeast (Rest et al. 2013). In this case, fitness increased with gene expression levels, but above a certain level no further changes in fitness were observed (Rest et al. 2013). Although there are also many examples of expression changes that underlie phenotypes likely to influence fitness (Hoekstra and Coyne 2007; Stern and Orgogozo 2008), it is difficult to make generalizations about the nature of these expression changes.
Here, we examine the effects of promoter divergence on both gene expression and fitness in yeast using the malate synthase (MLS1) promoter. As part of the glyoxylate cycle, MLS1 is induced in the absence of fermentable carbon and repressed in the presence of glucose (Turcotte et al. 2010). MLS1 converts acetyl-CoA into malate and is necessary for gluconeogenesis and growth on nonfermentable carbon sources (Hartig et al. 1992). The enzymatic function of Mls1 has been demonstrated to be conserved in Kluyveromyces lactis (Georis et al. 2000). Additionally, MLS1 has a well characterized promoter, where the main transcription factor binding sites and regions necessary for activation and repression have previously been identified (Caspary et al. 1997). Activation of MLS1 occurs through two Abf1 binding sites, responsible for basal expression levels, and two Cat8 binding sites, responsible for its large increase in expression following depletion of glucose (Caspary et al. 1997). Cat8 binding sites have also been shown to be bound by Sip4 (Roth et al. 2004). The main transcription factors that control MLS1 expression are conserved across species. Activation of MLS1 by the transcription factor CAT8 is conserved in K. lactis, a species that split before the whole-genome duplication and a shift in metabolism from respiratory to fermentative growth in the presence of oxygen (Georis et al. 2000). Repression of MLS1 occurs through a Mig1 site (Caspary et al. 1997). It has been shown that a MIG1 gene deletion in Saccharomyces cerevisiae can be rescued by MIG1 from Candida utilis (Delfin et al. 2001) and K. lactis (Cassart et al. 1995), indicating that MIG1 has conserved its general function as well.
Assays of orthologous cis-regulatory sequence function in a single species background have previously been valuable in understanding how they evolve. For example, loss of function can be caused by incompatibility between cis-regulatory sequences and trans-acting factors (Barrière et al. 2012) or by gain and loss of binding sites within the same cis-regulatory sequence (Ludwig et al. 2000). Here, we place orthologous MLS1 promoters from eight different yeast species into S. cerevisiae to determine what selective constraints act on this promoter as well as what expression levels and dynamics S. cerevisiae requires for MLS1 function. We expected and found that orthologous promoters caused differences in gene expression levels while maintaining the general pattern of activation and repression. We then used competitive growth assays to show that despite varying expression levels, all but one of the species’ promoters completely rescues competitive fitness in S. cerevisiae. Our results demonstrate that most of the diverse configurations of binding sites within the MLS1 promoter drove expression levels that can be tolerated without substantial fitness effects.
Results
High Sequence Divergence with Conservation of Binding Site Content
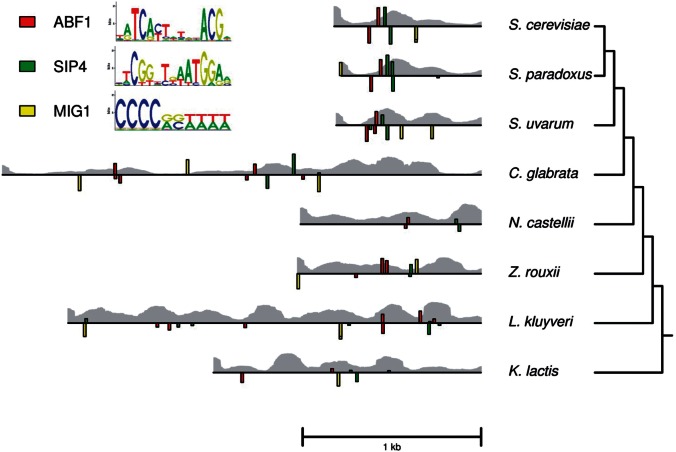
To characterize sequence divergence in the MLS1 promoter, we examined the noncoding sequences between MLS1 and the codon region of the upstream gene in eight yeast species. Similar to genome-wide patterns of promoter evolution in yeast (Venkataram and Fay 2010), the MLS1 promoter exhibits the following: 1) An abundance of conserved sites under purifying selection based on a substitution rate of 0.15 compared with the synonymous substitution rate of 0.21 in the MLS1 coding region (Fay and Benavides 2005); 2) no significant alignment between S. cerevisiae and the more distantly related non-Saccharomyces species compared with alignment of scrambled sequences (Venkataram and Fay 2010); and 3) good matches to known binding sites in most of the species’ promoter sequence (fig. 1 and supplementary fig. S1, Supplementary Material online). Binding sites known to regulate MLS1 expression in S. cerevisiae are two activation sites, which could be bound by either Cat8 or Sip4, a Mig1 repression site, and two Abf1 sites thought to be involved in basal expression (Caspary et al 1997; Roth et al. 2004). Although the number, position, and orientation of matching binding sites are different in all but the Saccharomyces species, they contain good matches to the known binding sites. The one exception is Naumovozyma castellii, which lacks a good TATA and Mig1 site. However, the binding site scores tend to be lower in more distantly related species, as measured by the total binding affinity predicted for each promoter (supplementary table S1, Supplementary Material online).
Fig. 1.
Transcription factor binding and nucleosome occupancy predictions for MLS1 promoters. The noncoding region upstream of the MLS1 start codon from eight yeast species, where the heights of colored bars represent the scores of predicted binding sites for Abf1, Sip4, and Mig1 based on PWMs from MacIsaac et al. (2006). Bars above each line represent sites on the forward strand and those below represent sites on the reverse strand. The probability of nucleosome occupancy at each base pair along the promoters is represented by the height of the gray bars in the background. Phylogenetic relations are based on Scannell et al. (2006).
To examine potential differences in nucleosome occuppancy, we used a sequence-based prediction method (Kaplan et al. 2009), which matches in vivo measurements of nucleosome occuppancy at MLS1 in S. cerevisiae. Similar to other noisy promoters (Blake et al. 2006), MLS1 is characterized by a TATA element with nucleosomes positioned over its other binding sites in the presence of glucose. In ethanol, in vivo nucleosome occupancy goes down but not completely, typical of noisy gene expression found for many condition-specific genes (Kaplan et al. 2009). Occupancy predictions for the other yeast species are similar in that binding sites are often occupied (fig. 1). With the exception of Mig1 sites, the occupancy of binding sites in the non-Saccharomyces species are significantly higher than the median occupancy for each promoter (supplementary table S2, Supplementary Material online).
Conserved Regulatory Patterns Despite Changes in Gene Expression Levels
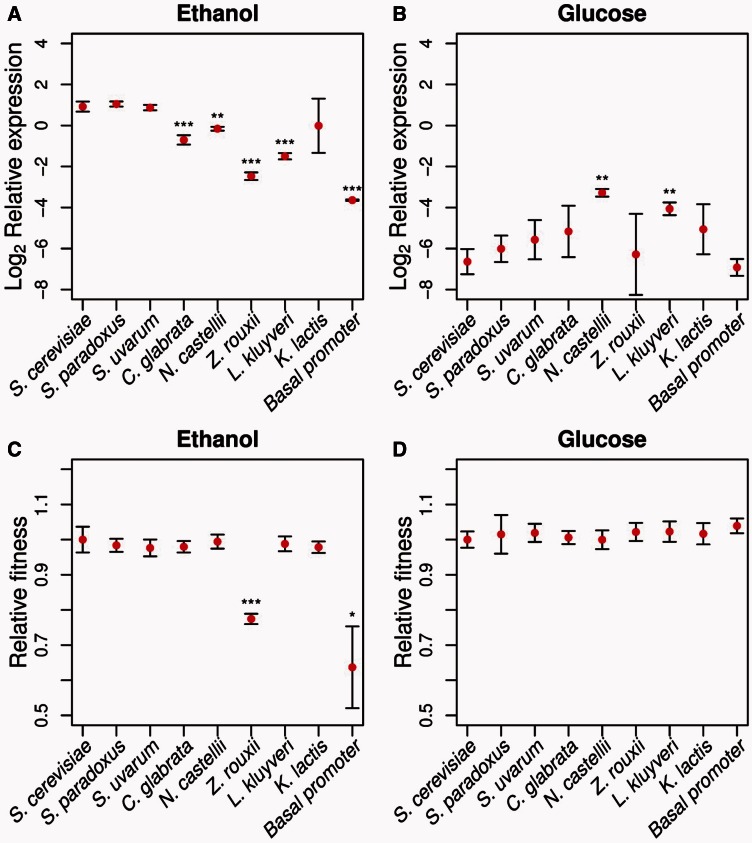
To test whether differences in the position, orientation, and slight changes in binding affinity affect gene expression, we placed each of the eight species’ noncoding regions upstream of the S. cerevisiae MLS1 gene integrated at the URA3 locus to avoid any confounding fitness effects caused by DCP2 which is divergently transcribed from the same intergenic region as MLS1. All the promoters caused significant activation of MLS1 in ethanol compared with glucose, ranging from 5.9- to 188-fold increase in expression (supplementary table S3, Supplementary Material online), demonstrating conservation of the response to carbon source. However, there is a general trend of a loss of both repression and activation in the most distantly related species (fig. 2A and B). Except for K. lactis, all non-Saccharomyces species’ promoters drove significantly lower expression compared with S. cerevisiae (Bonferroni correct P value < 0.05; fig. 2A). In glucose, both N. castellii and Lachancea kluyveri were not as well repressed as S. cerevisiae (Bonferroni corrected P value < 0.05; fig. 2B). Interestingly, the N. castellii promoter does not contain either a Mig1 repressor site (fig. 1) or a proximal TATA element (supplementary fig. S1, Supplementary Material online). The absence of a TATA box is known to correspond with a small dynamic range of expression (Basehoar et al. 2004).
Fig. 2.
Gene expression and fitness of MLS1 promoters in two environments. Relative expression shows expression of each species’ MLS1 promoter and Saccharomyces cerevisiae’s basal promoter relative to the housekeeping gene ACT1 on a log2 scale in (A) 3% ethanol and (B) 2% glucose. Relative fitness represents the growth rate of each strain relative to a reference competitor strain in (C) 3% ethanol and (D) 2% glucose. Error bars indicate one standard deviation, and significant differences in comparison with S. cerevisiae are shown for Bonferroni corrected *P < 0.05, **P < 0.01, and ***P < 0.001.
To gauge the extent to which the activity of the S. cerevisiae MLS1 promoter is influenced by known binding sites, we compared the S. cerevisiae promoter with the following: 1) A promoter lacking the MLS1 proximal Cat8/Sip4 site, previously shown to have a larger effect than the distal site (Caspary et al. 1997), 2) a promoter lacking both Cat8/Sip4 sites, 3) a promoter lacking the Mig1 site, and 4) a basal promoter containing only the proximal 186 bp of the promoter, which includes TATA but lacks the Abf1, Cat8, and Mig1 sites (supplementary fig. S2, Supplementary Material online). Although deletion of either the proximal or both Cat8/Sip4 sites did not affect expression (supplementary fig. S3A, Supplementary Material online), the basal promoter drove expression at much lower levels in ethanol (fig. 2A). However, the basal promoter still caused a 9.7-fold increase in expression in ethanol compared with glucose, similar to the level of activation found for the promoters of N. castellii, L. kluyveri, and Zygosaccharomyces rouxii (fig. 2A and B and supplementary table S3, Supplementary Material online). Similar to a previous study (Caspary et al. 1997), the Mig1 deletion caused a loss of repression (supplementary fig. S3B, Supplementary Material online).
Fitness Is Maintained Despite Changes in Gene Expression Levels
We tested whether any of the differences in expression affect fitness by competing each strain bearing a different MLS1 promoter with a common reference strain in either glucose or ethanol. There were no significant differences in fitness between the S. cerevisiae promoter and that of any other species except for Z. rouxii in ethanol (fig. 2C and D). This indicates that with respect to gene expression levels in ethanol, there is a fitness plateau and a sharp cliff between the expression levels of L. kluyveri and Z. rouxii (fig. 2A and supplementary fig. S4, Supplementary Material online). The expression to fitness relationship for the S. cerevisiae promoter deletions are consistent with this fitness plateau (fig. 2 and supplementary fig. S3, Supplementary Material online). Deletion of all binding sites except the basal promoter had a large impact on fitness, consistent with its low expression level, and deletion of the Sip4/Cat8 and Mig1 sites had little to no impact on fitness in ethanol and a slight increase in fitness in glucose (supplementary fig. S3C, Supplementary Material online). Compared with an S. cerevisiae strain with MLS1 at its endogenous locus, the transgenic S. cerevisiae allele of MLS1 at the URA3 locus exhibited a decrease in expression in ethanol, an increase in fitness in ethanol, and a decrease in fitness in glucose (supplementary fig. S3, Supplementary Material online). The fitness increase of the constructs integrated at the URA3 locus was unexpected, but may be due to the lack of a functional URA3 gene in the strain with MLS1 at its endogenous position.
Dynamic Expression and Fitness in Fluctuating Environments
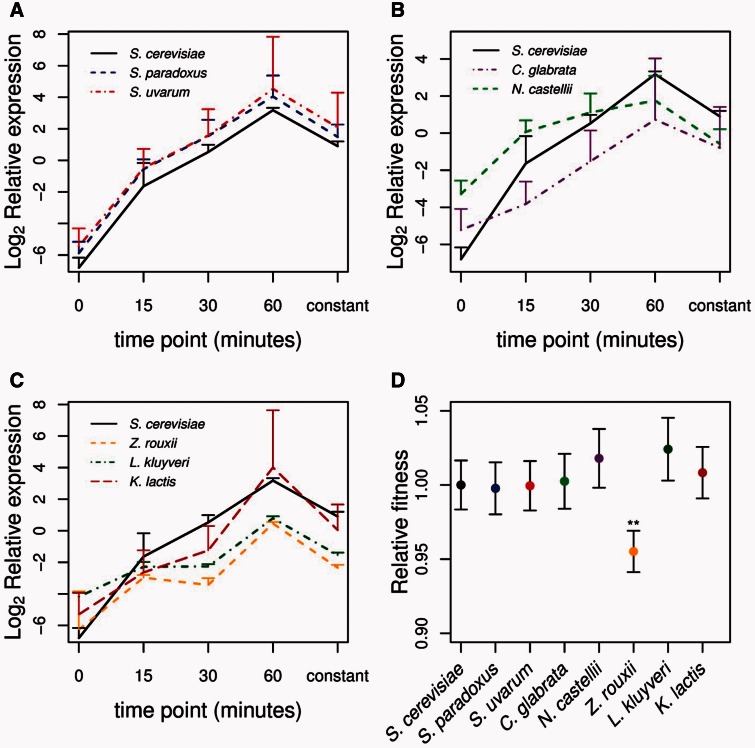
Our previous measurements of gene expression levels and fitness were done during exponential growth after cells were allowed to condition themselves to growth on glucose or ethanol. However, the dynamic response of a promoter to different carbon sources may be as important to fitness as expression levels after adjustment to a single condition. To examine the temporal dynamics of each species’ promoter, we measured expression following a switch from growth on glucose to ethanol. Similar to expression levels after acclimation, expression dynamics after switching from glucose to ethanol are conserved within the Saccharomyces species as is apparent from the consistent response over time (fig. 3A). For all the non-Saccharomyces species, we observed a smaller increase in expression between 0 and 15 min after switching to growth on ethanol (fig. 3B and C and supplementary table S6, Supplementary Material online). Zygosaccharomyces rouxii and L. kluyveri also showed a smaller increase in expression between 15 and 30 min (fig. 3B and C and supplementary table S6, Supplementary Material online). Although the dampened response of non-Saccharomyces species to ethanol is consistent with their lower expression levels after acclimation to ethanol media (fig. 1A), the absolute expression level at 15 min was only different between S. cerevisiae and Candida glabrata (supplementary table S7, Supplementary Material online).
Fig. 3.
Gene expression and fitness of MLS1 promoters in a fluctuating environment. Changes in MLS1 expression for the eight species promoter constructs are shown in A–C and are divided into (A) Saccharomyces species, (B) post–whole-genome duplication species, and (C) pre–whole-genome duplication species. Relative expression levels represent the level of MLS1 relative to the housekeeping gene ACT1 on a log2 scale. Time point 0 is expression in complete media with 2% glucose and subsequent time points are expression levels 15, 30, and 60 min after being placed in complete media with 3% ethanol. The “constant” time point indicates expression after 24 h of exponential growth in ethanol media. Error bars indicate one standard deviation. Relative fitness of each MLS1 species promoter construct (D) measured as the growth rate of each strain relative to a fluorescent competitor strain after 3 days of sequential competition in complete media with 3% ethanol plus 0.2% glucose. Bonferroni corrected P values are indicated for *P < 0.05, **P < 0.01, and ***P < 0.001.
Given the different expression dynamics, we tested whether the MLS1 promoters cause fitness differences in an environment where cells must switch to growth on ethanol once all the glucose has been used. Only the Z. rouxi promoter showed significantly reduced fitness (Bonferroni corrected P value < 0.05; fig. 3D), the only species with lower fitness in a constant ethanol environment (fig. 1C). The higher fitness of the Z. rouxii promoter in a fluctuating carbon source environment, compared with a constant environment, is likely a result of the competition including growth in glucose where no fitness defect was measured. In the fluctuating carbon source environment, there was a slightly higher fitness for the promoter of L. kluyveri (fig. 3D), potentially caused by the weaker MLS1 repression in glucose enabling the strain with the L. kluyveri promoter to start growing earlier (fig. 1B). The other species’ promoter with a significant loss of repression in glucose, N. castellii (fig. 1B), had higher fitness in the fluctuating carbon source environment that is close to significant (fig. 3D).
Discussion
Knowing how gene expression affects fitness is important to interpreting patterns of gene expression divergence. Using MLS1 promoters from eight yeast species, we find that large differences in gene expression levels do not generate detectable fitness effects. However, we also find a large drop in fitness below a certain low level of expression, implying that the S. cerevisiae MLS1 promoter resides on a fitness plateau. The high fitness of various configurations of binding sites present in different species provides further experimental support for the flexibility of the cis-regulatory code.
Conservation of Carbon Source Response Combined with Divergence in Expression Levels
Similar to previous promoter studies (Gordon and Ruvinsky 2012), we find that MLS1 promoters are conserved in their ability to respond to glucose and ethanol but also exhibit loss of both activation and repression as divergence between the promoters of these species and S. cerevisiae increases. Also consistent with a previous study of interspecific divergence (Tirosh et al. 2008), we find little correspondence between changes in binding sites and expression levels. First, deletion of one or both Cat8/Sip4 sites did not affect expression and there is no strong correlation between the summed binding affinity of a promoter and its expression level (supplementary table S1, Supplementary Material online). A previous study (Caspary et al. 1997) found that mutation of the proximal Cat8/Sip4 site or both Cat8/Sip4 sites caused a 28% and 80% reduction in expression, respectively. One explanation for why we did not find effects for these sites is that our deletion constructs altered the spacing of other binding sites. For example, Abf1 sites were brought close to TATA. However, the different results could also be a consequence of Caspary et al. (1997) measuring expression from a high-copy episomal plasmid using a reporter assay rather than MLS1 itself, whereas we measured expression of MLS1 integrated into the URA3 locus. A second line of evidence for the importance of sequences besides Cat8/Sip4 sites is that the basal promoter still yielded a 9.7-fold increase in expression in ethanol compared with glucose (supplementary table S3, Supplementary Material online). However, we did find effects associated with the Mig1 binding site: The Mig1 deletion caused a loss of repression in the S. cerevisiae promoter and N. castellii had the highest expression in glucose and also lacked a Mig1 site.
The Fitness-Expression Function in Saccharomyces cerevisiae
Previous work has shown condition-specific fitness costs and benefits of Lac expression in bacteria (Dekel and Alon 2005; Perfeito et al. 2011), and a fitness plateau for LCB2 expression in yeast (Rest et al. 2013). Results for MLS1 expression differ from LCB2 in that endogenous LCB2 expression levels occurred at the edge of the fitness cliff whereas no detectable loss of fitness occurred for up to a 5.4-fold (L. kluyveri) drop below wild-type levels for MLS1. We put forth four explanations for the high level of MLS1 expression in S. cerevisiae. First, low MLS1 expression levels may cause reduced fitness in conditions other than those measured. For example, MLS1 is required for sporulation and low expression could reduce or alter sporulation efficiency. Second, high MLS1 expression could be maintained by small fitness effects that are not detectable by our assays. Because purifying selection can occur on selection coefficients as small as the inverse of the effective population size, the fitness plateau could be covered with undetectable hills. In support of this possibility, there is good evidence for purifying selection on MLS1 binding sites within the Saccharomyces species (supplementary fig. S5, Supplementary Material online; see also Doniger and Fay 2007). Third, the fitness-expression function may only have a small plateau in other genetic backgrounds. Strain differences in LCB2 expression imply that genetic background modulates the fitness-expression function (Rest et al. 2013). Finally, high MLS1 expression may be due to genetic constraints whereby mutations which could lower the expression of MLS1 are constrained by pleiotropic effects on other genes. For instance, mutations may also lower the expression of coregulated genes.
What types of expression changes affect fitness? In addition to expression levels, prior work in yeast showed that noise in TDH3 expression affects fitness (Metzger et al. 2015). MLS1 like other TATA-containing promoters is characterized by large variation in cell-to-cell levels of expression, which may provide a fitness advantage under fluctuating environments through bet hedging (Thattai and Van Oudenaarden 2004; Kaern et al. 2005; Solopova et al. 2014). However, we found fitness effects in a fluctuating carbon source environment (fig. 3) to mirror and be smaller than those under exponential growth on a single carbon source (fig. 2).
One limitation of our approach is that we used heterologous expression and fitness assays. As such, it is possible that MLS1 promoters from the distantly related yeast species do not have reduced activation and repression in their endogenous genome. Both the extensive cis–trans expression interactions found to occur between species (McManus et al. 2010; Swain Lenz et al. 2014) and the dependency of the fitness-expression function on strain background (Rest et al. 2013) indicate that endogenous MLS1 expression in other species may not be the same as that measured in S. cerevisiae. However, a prior study of gene expression following the diauxic shift found that, with the exception of N. castellii, MLS1 is consistently actived between 3.7- and 8.6-fold (Thompson et al. 2013; supplementary table S4, Supplementary Material online). With the exception of SIP4 in Saccharomyces uvarum, CAT8 and SIP4 are also induced following the diauxic shift. Interestingly, comparing the MLS1 fold change from glucose to ethanol from figure 2 to the fold change during the diauxic shift in supplementary table S4, Supplementary Material online, shows a strong correlation between heterologuous and endogenous MLS1 species’ promoters (supplementary table S5, Supplementary Material online). These data support the possibility that the expression divergence observed in figure 2 is representative of the MLS1 expression divergence that has occurred between these species.
Another limitation of the heterologous assays is that we do not know whether the expression fitness function has changed between species. Different species could have different optimal levels of MLS1 expression. For example, the optimal expression of MLS1 in Z. rouxii could be quite low. However, without measurements of endogenous expression in Z. rouxii, it is hard to know whether this is the case. Thus, our use of heterologous measurements limits our interpretations to how different promoters with different outputs affect fitness.
In conclusion, our finding of a fitness plateau for MLS1 expression provides an explanation for divergence in gene expression levels and configurations of binding sites without an overall change in carbon source response. Current models for the evolution of cis-regulatory sequences hypothesize neutral evolution with a constant transcriptional output. However, when fitness effects are small or absent, many changes in cis-regulatory sequences may evolve under a neutral model despite their effects on gene expression.
Materials and Methods
Binding Site and Nucleosome Predictions
Position weight matrices (PWMs) for Abf1, Sip4, and Mig1 were obtained from MacIsaac et al. (2006). The PWM for Cat8 was obtained from a curated list of motifs (Soontorngun et al. 2007), and the PWM for TATA (NHP6A) was from Zhu et al. (2009). Sequences were searched for binding sites using Patser (Hertz and Stormo 1999). Only binding site scores below a ln(P value) of 7 were considered, where the P value is the expected probability of a random match to the binding site (Hertz and Stormo 1999). Nucleosome occupancy probability was predicted for each MLS1 promoter (Kaplan et al. 2009). The temperature and histone concentration parameters were set to 1 and 0.03, respectively, as in Kaplan et al. (2009).
A single sample Wilcoxon signed-rank test was used to determine if histone occupancy probabilities were higher at binding site positions than the median occupancy of the promoter (supplementary table S2, Supplementary Material online). First, the median occupancy for each promoter was subtracted from the occupancy probability at each binding site. Next, binding sites from the non-Saccharomyces species were pooled and tested for the alternative hypothesis that average occupancy at these sites was greater than the median occupancy. Only non-Saccharomyces species were used because these sequences are more or less phylogenetically independent based on their lack of sequence homology. Each transcription factor and TATA were tested separately.
Species Promoter Constructs
MLS1 promoter regions from eight species were placed into an S. cerevisiae background. First, the pRS306-ScMLS1 plasmid was constructed by inserting MLS1 from S. cerevisiae (S288c) into the integrative plasmid pRS306 (Sikorski and Hieter 1989). The MLS1 region from S288c includes the 893-bp noncoding region upstream of MLS1 as well as the 305-bp region downstream of the MLS1 translation stop site. Second, the promoter of S. cerevisiae MLS1 in pRS306-ScMLS1 was then replaced by the MLS1 promoter in seven other yeast species in the following manner. MLS1 promoter regions were defined as the noncoding region upstream of the MLS1 start codon to the beginning of the next coding region. In the cases of N. castellii and Z. rouxii, the predicted intergenic regions were short (575 bp for Z. rouxii and 250 bp for N. castellii) and therefore the region used for these two promoters was ∼1 kb upstream of the MLS1 start codon. The promoter region of MLS1 from each species (supplementary table S8, Supplementary Material online) was PCR amplified (see supplementary file S1, Supplementary Material online, for primers) and subcloned into the pRS306-ScMLS1 plasmid using the Gibson Assembly method (New England Biolabs, Ipswich, MA). The promoter region as well as the S. cerevisiae MLS1 coding region were sequence confirmed for each construct.
Binding Site Deletions
Binding sites were deleted by removing the region surrounding each binding site from the promoter. Regions deleted are shown in supplementary figure S2, Supplementary Material online. Deletions were generated by amplifying the pRS306-ScMLS1 plasmid with segments of the promoter missing. Primers contained BglII sites on their 5′ end (supplementary file S1, Supplementary Material online). After amplification, the PCR product was digested with BglII and ligated back together to form a circular plasmid. The S. cerevisiae MLS1 promoter deletions and coding region were sequence confirmed.
Plasmid Integrations
The endogenous MLS1 coding region from the strain YJF186 (YPS163 oak isolate, Mat a, HO::dsdAMX4, ura3-140) was deleted by replacement with the KANMX4 cassette to generate the strain YJF604. All pRS306-based plasmids described above were cut in the URA3 coding region with StuI and integrated into YJF604 using lithium acetate transformation (Geitz and Woods 2002) and selected on plates lacking uracil. The competitor strain containing yellow fluorescent protein (YFP) was generated by integrating a YFP-NATMX4 plasmid-containing homology to the HO locus (received from R. Kishony) into YJF186.
Competitive Fitness Assays
Fitness was estimated by competing each strain against a YFP marked reference strain. For each competition, six biological replicates (independent transformants) of each integrated construct were competed against the YFP competitor at 30 °C at 300 rpm in 3 ml media in 18 × 150 mm glass tubes. Ethanol (3%), glucose (2%), and mixed carbon source (3% ethanol and 0.2% glucose) competitions were carried out in complete medium (CM: 0.67% (wt/vol) nitrogen base with ammonium sulfate and amino acids) with the specified carbon sources. All strains were acclimated to each growth medium prior to competition by 3 days of growth, with cells resuspended in fresh medium after each day at an OD600 of 0.07. An OD600 of 1 is ∼107 cells/ml. The YFP competitor strain was mixed with each culture at a 50:50 ratio at a starting cell density of 0.7 × 106 cells/ml. All measurements were taken within the linear range of an OD600 between 1 and 0.1. Competitions in CM with 3% ethanol and CM with 3% ethanol + 0.2% glucose were carried out for 2 days with resuspension in fresh medium after every 23 h of competition. Competitions in CM with 2% glucose were carried out for 1 day with resuspension in fresh medium after every 11 h of competition. There were approximately the same number of generations for the competitions grown in all media (between 8.5 and 9.5 generations). All cultures were switched to new media during exponential growth and never allowed to reach saturation. The proportion of YFP positive strains was determine at the beginning and end of each competition. Cells from each culture were also diluted to an OD600 = 0.2 in sheath fluid and run on a Beckman Coulter FC 500 MPL flow cytometer (Beckman Coulter, Brea, CA). For each sample, 20,000 cells were counted and gated to distinguish between fluorescent and nonfluorescent cells. The false negative rate for YFP cells was 0.003.
Fitness Calculations
Fitness measurements were calculated using as in Hartl and Clark (1997), where and are the starting frequencies of the YFP strain and the nonfluorescent competitor strain, respectively. Here,. Similarly, and represent these frequencies at the end of the competition. Relative fitness of a given strain is equal to , where is the average fitness of the S. cerevisiae strains.
MLS1 mRNA Expression Analysis
MLS1 measurements during exponential growth were measured as follows. Using four of the same replicates for each promoter from the competition, each strain was acclimated and cells were sampled 4 h after being resuspended in fresh CM with 3% ethanol or CM with 2% glucose medium at an OD600 of 1 on the third day. The equivalent of 1 ml cells at an OD600 of 0.3 were sampled. Cells were centrifuged, supernatant was removed, pellets were frozen in liquid nitrogen, and stored at −80 °C.
MLS1 mRNA expression during the switch from glucose to ethanol was obtained from 4 time points. After 3 days acclimation cells were placed in 3 ml CM with 2% glucose at an OD600 = 1 and grown for 4 h. Cells were centrifuged for 30 s at 3,000 rpm, supernatant was removed and cells were washed with 1 ml CM with 3% ethanol and centrifuged again. Supernatant was removed and cells were resuspended in 3 ml of CM with 3% ethanol and cultures were placed in the incubator. Cells were then sampled 15, 30, and 60 min after cells were initially placed into CM with 3% ethanol, centrifuged, and pellets were frozen in liquid nitrogen and stored at −80 °C.
MLS1 expression was measured using QuantiGene (Affymetrix, Santa Clara, CA) following manufacturer’s instructions. In total, 200 μl of homogenization buffer (Affymetrix) was added to each pellet, resuspended, centrigued, and supernatant was removed. Pellels were resuspended in 100 μl of ZYM buffer (Clontech, Mountain View, CA) and 10 μl of zymolase (Clontech) and allowed to digest for 1 h at 30 °C at 300 rpm. After digestion, 150 μl of homogeniztion buffer was added to each well. The content of each well was then diluted 1:100 in homogenization buffer. Next, 40 μl of these 1:100 diluted samples were added to 60 μl of “working bead mix” described in Steps 4–6 of the “Purified RNA or in vitro Transcribed RNA” protocol in the QuantiGene 2.0 Plex Assay User Manual (Panomics Solutions P/N 16659 Rev.C 020912). The Purified RNA or in vitro Transcribed RNA protocol was then followed exactly from Step 7 onwards. Probes were designed to the MLS1 and ACT1 coding regions of S. cerevisiae. A total of 40 μl of the 1:100 diluted samples was added to 60 μl of mastermix. Measurements were obtained on a Bio-Plex 200 System (Life technologies, Carlsbad, CA) and analyzed using the Bio-Plex Manager 6.1 software. Standard curves for each analyte were generated by a 4-fold serial dilution of one of the S. cerevisiae MLS1 promoter strains sampled in ethanol media.
Statistical Analysis of Fitness and Expression
Six biological replicates (independent integrations at the URA3 locus) of each promoter construct were measured for fitness. Four biological replicates of each promoter were measured for expression in exponential growth after acclimation to either glucose or ethanol. Outliers from each group were removed using the Grubbs’ test (P < 0.05). Significant differences were measured by t-tests with unequal variance. Bonferonni correction was used for the seven hypotheses that another species promoter was different than the S. cerevisiae promoter. For measurements of the dynamics of gene expression from glucose to ethanol, three biological replicates were used and no outliers were removed. A nested analysis of variance was used to measure the differences between each species’ promoter at each time point as well as the rate of change (slope) between each time point. This was done in R where level∼(species/time).
Supplementary Material
Supplementary tables S1–S8, figures S1–S5, and file S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors would like to thank Xueying Li, Ping Liu, Kim Lorenz, Linda Riles, Ching-Hua Shih, and Katie Williams with help on competition assays; Holly Brown with help on the QuantiGene assay; as well as three anonymous reviewers for helpful comments and suggestions. The research was supported by the National Institutes of Health training grant GM007067 to A.C.B. and GM08669 to J.C.F.
References
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet. 46:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Gordon KL, Ruvinsky I. 2012. Coevolution within and between regulatory loci can preserve promoter function despite evolutionary rate acceleration. PLoS Genet. 8:e1002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière A, Ruvinsky I. 2014. Pervasive divergence of transcriptional gene regulation in Caenorhabditis nematodes. PLoS Genet. 10:e1004435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699–709. [DOI] [PubMed] [Google Scholar]
- Blake WJ, Balázsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. 2006. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 24:853–865. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC. 2010. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 8:e1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Mostovoy Y, Dudoit S, Brem RB. 2010. Polygenic and directional regulatory evolution across pathways in Saccharomyces. Proc Natl Acad Sci U S A. 107:5058–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134:25–36. [DOI] [PubMed] [Google Scholar]
- Caspary F, Hartig A, Schüller HJ. 1997. Constitutive and carbon source-responsive promoter elements are involved in the regulated expression of the Saccharomyces cerevisiae malate synthase gene MLS1. Mol Gen Genet. 255:619–627. [DOI] [PubMed] [Google Scholar]
- Cassart JP, Georis I, Östling J, Ronne H, Vandenhaute J. 1995. The MIG1 repressor from Kluyveromyces lactis: cloning, sequencing and functional analysis in Saccharomyces cerevisiae. FEBS Lett. 371:191–194. [DOI] [PubMed] [Google Scholar]
- Chan ET, Quon GT, Chua G, Babak T, Trochesset M, Zirngibl RA, Aubin J, Ratcliffe MJ, Wilde A, Brudno M, et al. 2009. Conservation of core gene expression in vertebrate tissues. J Biol. 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekel E, Alon U. 2005. Optimality and evolutionary tuning of the expression level of a protein. Nature 436:588–592. [DOI] [PubMed] [Google Scholar]
- Delfin J, Perdomo W, García B, Menendez J. 2001. Isolation and sequence of the MIG1 homologue from the yeast Candida utilis. Yeast 18:597–603. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Bergman CM, Clark AG. 2003. Tracing the evolutionary history of Drosophila regulatory regions with models that identify transcription factor binding sites. Mol Biol Evol. 20:703–714. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG. 2002. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol. 19:1114–1121. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Fay JC. 2007. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput Biol. 3:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC. 2013. The molecular basis of phenotypic variation in yeast. Curr Opin Genet Dev. 23:672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. 2005. Hypervariable noncoding sequences in Saccharomyces cerevisiae. Genetics 170:1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Wittkopp PJ. 2008. Evaluating the role of natural selection in the evolution of gene regulation. Heredity 100:191–199. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. 2006. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312:276–279. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Moses AM, Schadt EE. 2010. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci U S A. 107:2977–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. 2004. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2:e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt SJ, Paul YL. 2012. Changes in cis-regulatory elements during morphological evolution. Biology 1:557–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitz RD, Woods RA. 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350:87–96. [DOI] [PubMed] [Google Scholar]
- Georis I, Krijger JJ, Breunig KD, Vandenhaute J. 2000. Differences in regulation of yeast gluconeogenesis revealed by Cat8p-independent activation of PCK1 and FBP1 genes in Kluyveromyces lactis. Mol Gen Genet. 264:193–203. [DOI] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I. 2012. Tempo and mode in evolution of transcriptional regulation. PLoS Genet. 8:e1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. 2008. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 4:e1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig A, Simon MM, Schuster T, Daugherty JR, Yoo HS, Cooper TG. 1992. Differentially regulated malate synthase genes participate in carbon and nitrogen metabolism of S. cerevisiae. Nucleic Acids Res. 20:5677–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. 1997. Principles of population genetics. Sunderland (MA: ): Sinauer Associates. [Google Scholar]
- Hertz GZ, Stormo GD. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563–577. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61:995–1016. [DOI] [PubMed] [Google Scholar]
- Kaern M, Elston TC, Blake WJ, Collins JJ. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 6:451–464. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, He X, Sinha S. 2009. Evolution of regulatory sequences in 12 Drosophila species. PLoS Genet. 5:e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–116. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. 2000. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403:564–567. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. 2005. Functional evolution of a cis-regulatory module. PLoS Biol. 3:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, Kreitman M. 1998. Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125:949–958. [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. 2004. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131:2387–2394. [DOI] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67:1235–1250. [DOI] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 20:816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger BP, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ. 2015. Selection on noise constrains variation in a eukaryotic promoter. Nature 521:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Pollard DA, Nix DA, Iyer VN, Li XY, Biggin MD, Eisen MB. 2006. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput Biol. 2:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L, Ghozzi S, Berg J, Schnetz K, Lässig M. 2011. Nonlinear fitness landscape of a molecular pathway. PLoS Genet. 7:e1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rest JS, Morales CM, Waldron JB, Opulente DA, Fisher J, Moon S, Bullaughey K, Carey LB, Dedousis D. 2013. Nonlinear fitness consequences of variation in expression level of a eukaryotic gene. Mol Biol Evol. 30:448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano LA, Wray GA. 2003. Conservation of Endo16 expression in sea urchins despite evolutionary divergence in both cis and trans-acting components of transcriptional regulation. Development 130:4187–4199. [DOI] [PubMed] [Google Scholar]
- Roth S, Kumme J, Schüller HJ. 2004. Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr Genet. 45:121–128. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Ruvkun G. 2003. Functional tests of enhancer conservation between distantly related species. Development 130:5133–5142. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440:341–345. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, Kuipers OP. 2014. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci U S A. 111:7427–7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soontorngun N, Larochelle M, Drouin S, Robert F, Turcotte B. 2007. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol Cell Biol. 27:7895–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62:2155–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain Lenz D, Riles L, Fay JC. 2014. Heterochronic meiotic misexpression in an interspecific yeast hybrid. Mol Biol Evol. 31:1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Schwimmer DB, Barolo S. 2011. Rapid evolutionary rewiring of a structurally constrained eye enhancer. Curr Biol. 21:1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, Van Oudenaarden A. 2004. Stochastic gene expression in fluctuating environments. Genetics 167:523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Roy S, Chan M, Styczynsky MP, Pfiffner J, French C, Socha A, Thielke A, Napolitano S, Muller P, et al. 2013. Evolutionary principles of modular gene regulation in yeasts. Elife 2:e00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Bezalel D, Kaganovich M, Barkai N. 2008. On the relation between promoter divergence and gene expression evolution. Mol Syst Biol 4:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443:415–420. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte B, Liang XB, Robert F, Soontorngun N. 2010. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 10:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataram S, Fay JC. 2010. Is transcription factor binding site turnover a sufficient explanation for cis-regulatory sequence divergence? Genome Biol Evol. 2:851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. 2006. Variation within and among species in gene expression: raw material for evolution. Mol Ecol. 15:1197–1211. [DOI] [PubMed] [Google Scholar]
- Wratten NS, McGregor AP, Shaw PJ, Dover GA. 2006. Evolutionary and functional analysis of the tailless enhancer in Musca domestica and Drosophila melanogaster. Evol Dev. 8:6–15. [DOI] [PubMed] [Google Scholar]
- Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 8:206–216. [DOI] [PubMed] [Google Scholar]
- Yokoyama KD, Zhang Y, Ma J. 2014. Tracing the evolution of lineage-specific transcription factor binding sites in a birth-death framework. PLoS Comput Biol. 10:e1003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. 2009. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.