Abstract
Studying the genetic signatures of climate-driven selection can produce insights into local adaptation and the potential impacts of climate change on populations. The honey bee (Apis mellifera) is an interesting species to study local adaptation because it originated in tropical/subtropical climatic regions and subsequently spread into temperate regions. However, little is known about the genetic basis of its adaptation to temperate climates. Here, we resequenced the whole genomes of ten individual bees from a newly discovered population in temperate China and downloaded resequenced data from 35 individuals from other populations. We found that the new population is an undescribed subspecies in the M-lineage of A. mellifera (Apis mellifera sinisxinyuan). Analyses of population history show that long-term global temperature has strongly influenced the demographic history of A. m. sinisxinyuan and its divergence from other subspecies. Further analyses comparing temperate and tropical populations identified several candidate genes related to fat body and the Hippo signaling pathway that are potentially involved in adaptation to temperate climates. Our results provide insights into the demographic history of the newly discovered A. m. sinisxinyuan, as well as the genetic basis of adaptation of A. mellifera to temperate climates at the genomic level. These findings will facilitate the selective breeding of A. mellifera to improve the survival of overwintering colonies.
Keywords: Apis mellifera sinisxinyuan, new subspecies, temperate climates, population history, adaptation
Introduction
Identifying adaptive variation driven by different climate factors is a major focus in evolutionary biology. Both phenotypic and genetic variation among populations can be influenced by climatic factors, as has been demonstrated in a variety of species such as Arabidopsis, Drosophila, sheep, and human (González et al. 2010; Hancock, Brachi, et al. 2011; Hancock, Witonsky, et al. 2011; Lv et al. 2014). Detecting climate-mediated selective signatures is important for understanding not only the genetic basis of adaptation to local environment but also the potential effects of climate change on populations (Balanyà et al. 2009). In addition, identifying adaptive polymorphisms can shed light on functionally important variants (Nielsen et al. 2007).
The western honey bee (Apis mellifera) is an interesting species for studying climate-driven adaptations. Apis mellifera is widely distributed in Africa, western Asia, and Europe, with at least 24 subspecies that can be divided into four lineages based on morphometric and molecular studies: A-lineage in Africa, M-lineage in western and northern Europe, C-lineage in eastern Europe, and O-lineage in the Middle East and western Asia (Ruttner 1988; Garnery et al. 1992; Arias and Sheppard 1996; Whitfield et al. 2006; Han et al. 2012; Wallberg et al. 2014). Apis mellifera originated in tropical/subtropical regions and subsequently radiated into temperate zones, and adaptation to temperate climates is a key step in its evolutionary history (Ruttner 1988; Han et al. 2012). Temperate and tropical populations of A. mellifera show fundamental differences in a variety of traits (Winston 1987), most of which involve coping with long, cold winters (Southwick and Heldmaier 1987; Ruttner 1988; Amdam et al. 2005). However, few molecular studies have been conducted to investigate the genetic responses to temperate climates in A. mellifera.
At present, studies on the adaptation of A. mellifera to temperate climates mainly focus on European honey bees, in particular A. m. mellifera (Maurizio and Hodges 1950; Fluri et al. 1977; Seeley 1985, and references therein; Southwick and Heldmaier 1987; Ruttner 1988; Amdam et al. 2005; Meixner et al. 2007; Seehuus et al. 2013). Apis mellifera mellifera is distributed along the northwest boundary of the range of A. mellifera in Europe, and is well known for its ability to survive harsh winters. However, to enhance our understanding of the species adaptation to temperate climates, it is preferable to also study populations in the northeast region of its range in northwest China, Mongolia, Kazakstan, or southern Russia. We found a new, winter-tolerant population of A. mellifera in Xinyuan prefecture, located in Xinjiang Uygur Autonomous Region, China. The natural environment in Xinyuan is characterized by long, cold winters due to a combination of high latitude and altitude, with a frost season of 185–205 days. However, the genetic diversity and evolutionary history of this group had not been extensively studied.
In this study, we investigated the Xinyuan honey bee population’s demographic history and genome-level adaptation to temperate climates. We collected ten feral colonies at different locations across the Xinyuan prefecture and resequenced the whole genomes of a total of ten individuals, each from a separate colony (supplementary table S1, Supplementary Material online). We also downloaded resequenced data from representative populations in each of the four previously delineated lineages of A. mellifera, including A. m. mellifera, A. m. carnica, A. m. ligustica, A. m. anatoliaca, and A. m. scutellata. Using a population genomic approach, we estimated the relationships and identified genomic polymorphisms in these honey bee populations and reconstructed the demographic history to elucidate how temperate climates shape the divergence of Xinyuan bees and their close relatives. We also identified rapidly evolving genes in honey bees from Xinyuan to gain insight into genetic basis of adaptation to temperate climates.
Results
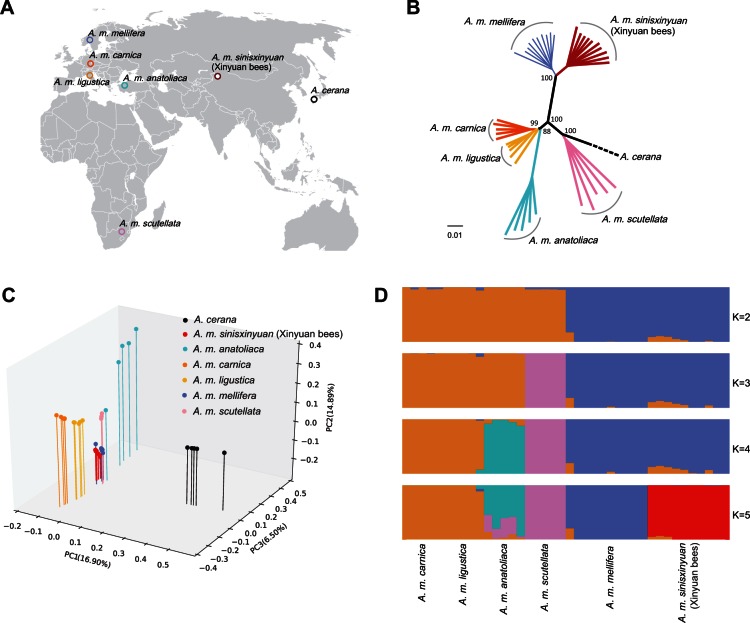
We selected 45 individuals from seven populations (the Xinyuan bees, A. m. mellifera, A. m. carnica, A. m. ligustica, A. m. anatoliaca, A. m. scutellata, and A. cerana as outgroup) (fig. 1A and supplementary table S2, Supplementary Material online). Resequencing of the ten Xinyuan bees generated a total of 178.9 million paired-end reads of 100 bp read length (17.9 Gb of sequence, supplementary table S1, Supplementary Material online). Genome alignment of the Xinyuan bee sequences indicated an average depth of 8.08-fold and coverage of 89.86% relative to the A. mellifera reference genome (234.087 Mb; supplementary table S3, Supplementary Material online) (The Honeybee Genome Sequencing Consortium 2006). After stringent quality filtering, we identified a total of 2,123,243 high-quality population single nucleotide polymorphisms (SNPs) in the 45 individuals (supplementary table S4, Supplementary Material online), and 988,217 small indels (shorter than 50 bp) (supplementary table S5, Supplementary Material online).
Fig. 1.
Population genetics. (A) Geographic locations of the studied honey bees. (B) Neighbor-joining phylogenetic tree of honey bee populations derived from 100 bootstrap replicates. The scale bar represents the evolutionary distances measured by p-distance. (C) Three-way PCA plots of honey bee populations. (D) Genetic structure of the honey bees. Analyses with frappe show the clustering of sample into 2–5 groups. The proportion of the individual’s genome from each ancestral population is shown by the length of each colored segment.
Evolutionary and Demographic History
Population Structure and Divergence
To infer the phylogenetic relationship of the Xinyuan bees and other populations, we constructed a phylogenetic tree using the neighbor-joining algorithm based on the pairwise genetic distances between populations using polymorphic SNPs in whole genome. The tree showed that honey bees from Xinyuan and A. m. mellifera were clustered into a subclade, and that they potentially originated from a common ancestor (fig. 1B). We also used the SNPs in coding and noncoding regions to construct neighbor-joining trees and obtained the same topological structures (supplementary fig. S1, Supplementary Material online). Principle component analysis using EIGENSOFT3.0 (Patterson et al. 2006) and population structure analyses using frappe (Tang et al. 2005) corroborated these patterns. In particular, the first principal component (variance explained = 16.90%, Tracy–Widom P = 1.12E-06) separated the outgroup A. cerana from the other populations. The second principal component (variance explained = 14.89%, Tracy–Widom P = 9.51E-14) indicated that Xinyuan honey bees and A. m. mellifera were separated from the other subspecies (fig. 1C, supplementary fig. S2 and table S6, Supplementary Material online). Clustering analyses with frappe, grouped honey bees from Xinyuan and A. m. mellifera into the same cluster if K was less than 5, but with K equal to 5 they were divided into two groups (fig. 1D). Our resequenced data support the placement of the Xinyuan honey bees in the M-lineage (same as A. m. mellifera), which is consistent with the results of morphometric and mitochondrial analysis (supplementary fig. S3, Supplementary Material online).
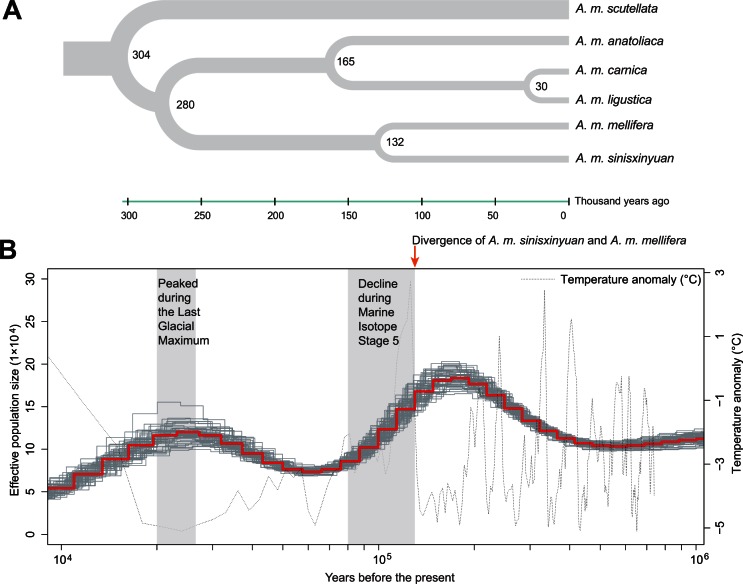
To further characterize the relationship between the Xinyuan bees and A. m. mellifera, we used MCMCtree method in package PAML 4.5 (Yang 2007) to estimate the divergence times between the populations. This analysis revealed that the divergence time between A. m. mellifera and the honey bees from Xinyuan is about 132 ka (fig. 2A). In comparison, divergence time between A. m. carnica and A. m. ligustica, two well-characterized subspecies from the C-lineage, is only about 30 ky before the present, an estimate comparable to that in a previous study (Wallberg et al. 2014). Identity score (IS) analysis also indicated that Xinyuan bees and A. m. mellifera have less similarity than that of A. m. carnica and A. m. ligustica (supplementary fig. S4, Supplementary Material online). Therefore, we propose that the Xinyuan honey bees is a new subspecies of A. mellifera (A. m. sinisxinyuan n. ssp.).
Fig. 2.
Population genetics and demographic history. (A) Time of divergence between populations. Number at each node represents the time of divergence in thousands of years. (B) Demographic history of Xinyuan bees (Apis mellifera sinisxinyuan). The red line indicates the effective population size measured by the scaled mutation rate per generation (0.53 × 10−8), the gray solid lines illustrate the PSMC estimates for 50 rounds of bootstrap. The generation time is 1 year. Shaded areas represent the Last Glacial Maximum (left) and Marine Isotope Stage 5 (right).
Description of A. mellifera sinisxinyuan n. ssp
Holotype Worker bee; China, Xinjiang Uygur Autonomous Region, Xinyuan prefecture, 43 °17′51″N, 83 °42′48″E, elevation 1,709 m. Institute of Apicultural Research honey bee collection, sample number 0999-0096.
Paratypes Several worker bees, same data as holotype.
Etymology The name refers to Xinyuan prefecture in China as the location where this subspecies is first discovered.
Distribution The subspecies is distributed in the Yili River Valley in western China.
Apis mellifera sinisxinyuan is a dark bee with black abdomens and gray hair. Worker bees have very long abdominal hair, longer than the European dark bee, A. m. mellifera, and almost twice as long as A. m. ligustica and A. m. pomonella. The body is slender and small, considerably smaller than its relative A. m. mellifera. The cubital index, sternite 6 index, and tarsus index of A. m. sinisxinyuan are comparable to that of A. m. mellifera. In addition, proboscis of Xinyuan honey bees is short. Mean values and standard deviation of selected morphometric characters are presented in supplementary table S7, Supplementary Material online.
Workers bees of A. m. sinisxinyuan are very aggressive. Preliminary observations found that it has excellent overwintering ability, and the temperature threshold for worker bees to take cleaning flights and to collect honey is low. It is also featured by fast spring build up and high honey production (Liuz et al., unpublished data).
Demographic History
To explore the demographic history of A. m. sinisxinyuan, we first estimated changes in effective population size using a Pairwise Sequentially Markovian Coalescent (PSMC) method (Li and Durbin 2011). The result shows that the effective population size of A. m. sinisxinyuan fluctuates over time with earth climate (fig. 2B), with high effective population size during cold periods. The most dramatic decline in effective population size occurs during Marine Isotope Stage 5 (80–130 ka) (Lisiecki and Raymo 2005; Jouzel et al. 2007), the last major interglacial period in history, after which the effective population size began to increase and peaked during the Last Glacial Maximum (20–26.5 ka) (Clark et al. 2009). Effective population size declined after the Last Glacial Maximum, when the global temperature increased. This pattern strongly suggests a strong effect of global temperature on the effective population size of A. m. sinisxinyuan. Notably, the divergence between A. m. sinisxinyuan and A. m. mellifera (132 ka) occurred at the beginning stage of the effective population size decline, suggesting population decline in the common ancestor of A. m. sinisxinyuan and A. m. mellifera may result in the geographical separation.
Genes under Selective Sweep with Temperate Climates
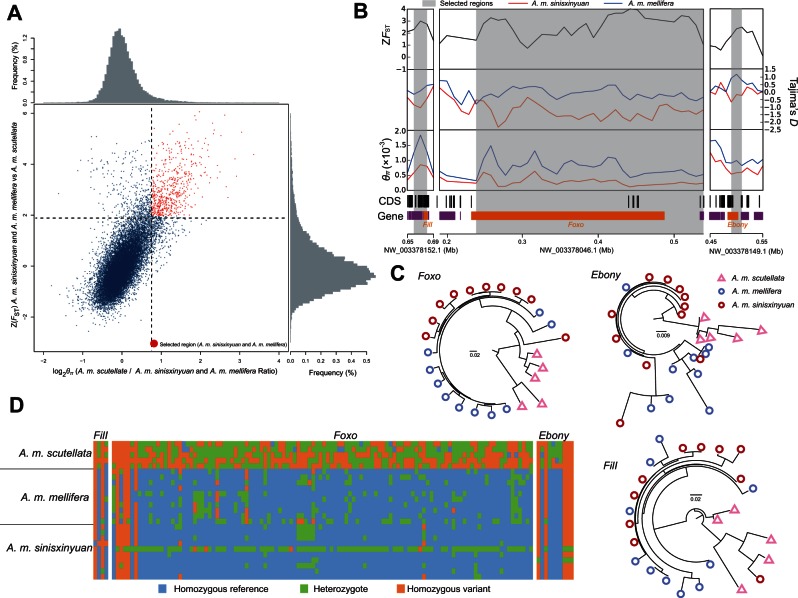
Although A. m. mellifera and A. m. sinisxinyuan are well adapted to temperate climates, the tropical A. m. scutellata do not show the characteristics suitable for a life in temperate regions (Terada et al. 1975; Southwick and Heldmaier 1987). To identify potential mechanisms for the adaptation, we pooled A. m. sinisxinyuan and A. m. mellifera samples and compared with the tropical A. m. scutellata. Genes under selective sweep were classified into Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000) and Gene Ontology (GO) (Ashburner et al. 2000) functional categories based on orthologs in the fruit fly (Drosophila melanogaster). In total, we identified 6.16 Mb of selective sweep regions, encompassing 454 genes with both high FST and high θπ-ratio (fig. 3A). Several highly enriched functional categories were identified, including “membrane depolarization” (four genes, P = 9.64E-05), “dopamine metabolic process” (four genes, P = 1.131E-03), “regulation of lipid storage” (five genes, P = 1.981E-03), “flight behavior” (five genes, P = 2.020E-03) (supplementary table S8, Supplementary Material online), and the Hippo signaling pathway (five genes, P = 3.995289E-02; supplementary table S9, Supplementary Material online).
Fig. 3.
Genomic regions with strong selective sweeps in Apis mellifera sinisxinyuan and A. m. mellifera. (A) Distribution of θπ ratios (θπ, A. m. sinisxinyuan and A, m. mellifera/θπ, A. m. scutellata) and FST values, calculated in 20-kb windows sliding in 10-kb steps. Data points on the right of the vertical dashed line (corresponding to the 5% left tail of the empirical θπ ratio distribution), and above the horizontal dashed line (5% right tail of the empirical FST distribution) were identified as selected regions for A. m. sinisxinyuan and A. m. mellifera (red points). (B) Examples of genes with strong selective sweep signals in A. m. sinisxinyuan and A. m. mellifera. FST, θπ, and Tajima’s D values are plotted using a 10-kb sliding window. Shaded genomic regions were the regions with strong selective signals for A. m. sinisxinyuan and A. m. mellifera. Genome annotations are shown at the bottom (black bar, coding sequences; purple bar, genes). The boundaries of FilI, Foxo, and Ebony are marked in orange. (C) The gene trees for FilI, Foxo, and Ebony for A. m. sinisxinyuan, A. m. mellifera, and A. m. scutellata. (D) Genotypes of SNPs in putative selective sweeps containing FilI, Foxo, and Ebony in A. m. sinisxinyuan, A. m. mellifera, and A. m. scutellata.
Genes Associated with Regulation of Lipid Storage
The enrichment of genes for lipid storage reflects the crucial role of fat body in A. mellifera’s adaptation to temperate climates. We found a selected gene Indy (I’m not dead yet), which is a cotransporter related to adult longevity and is abundantly expressed in fat body (Rogina et al. 2000; Seehuus et al. 2013). Another selected gene Foxo (forkhead box O; fig. 3B–D) is a transcription factor involved in the insulin signaling pathway, and regulates cell number and lipid metabolism (Jünger et al. 2003; Vihervaara and Puig 2008). In A. mellifera, Foxo is hypothesized to play a key role in the longevity of both winter bees and queens, through the regulation of the glycolipoprotein Vitellogenin content in fat body (Corona et al. 2007; Aurori et al. 2013). Other selected genes include δCOP and ζCOP, both are members of the vesicle-mediated Coat Protein Complex I (COPI), a regulator of lipid homeostasis (Beller et al. 2008). Taken together, our results suggest the importance of fat body in overwintering honey bees.
Genes Associated with the Control of Flight Muscle
Two of the overrepresented GO categories were “flight behavior” and “membrane depolarization,” which are related to the control of flight muscle. Gene FilI (flightless I) shows clear signals of a selective sweep in the temperate population, and is involved in the structural organization of indirect flight muscle (fig. 3B–D) (de Couet et al. 1995), the muscle that generates heat (Tautz 2008). We also found that four out of the five genes in the “membrane depolarization” category were selected. Membrane depolarization is a crucial step in the initiation of action potential in muscle fibers, whereas action potential determines muscle contraction. In honey bee, low temperate reduces the frequency of action potential and hence the wing beat frequency (Bastian and Esch 1970), selected genes in membrane depolarization in the temperate populations may enhance their ability to fly under low temperature. Alternatively, they may also be related to efficient generation of heat.
Genes Related to the Neural System
A number of selected genes in top five GO terms in our analysis are related to the neural system. We found that four genes (Tan, Ebony, Manf, and Dysbindin-A-like) were selected in “dopamine metabolic process.” Dopamine is one of the main neurotransmitters in A. mellifera and has profound effects on behaviors (Scheiner et al. 2002; Beggs et al. 2007; Beggs and Mercer 2009; Agarwal et al. 2011). Among these selected genes, Tan and Ebony (fig. 3B–D) code for a pair of N-β-alanyldopamine (NBAD) hydrolase and NBAD-synthase, which function together to regulate the level of dopamine (Borycz et al. 2002; Pérez et al. 2010, 2011). Moreover, three selected genes (Tan, Tbh, and Iav) in “flight behavior” were also related to neurotransmitters, among which Tbh is a tyramine beta-hydroxylase converting tyramine into octopamine (Livingstone and Tempel 1983), and Iav (inactive) is a gene involved in octopamine biosynthesis (Roshina and Pasyukova 2007). One major characteristic setting A. mellifera apart from most other insects is their social behavior, which is closely linked to the neural system (Scheiner et al. 2002; Zayed and Robinson 2012). Selected genes in this category may regulate the behavior under an environment with seasonal climate change.
The Hippo Signaling Pathway
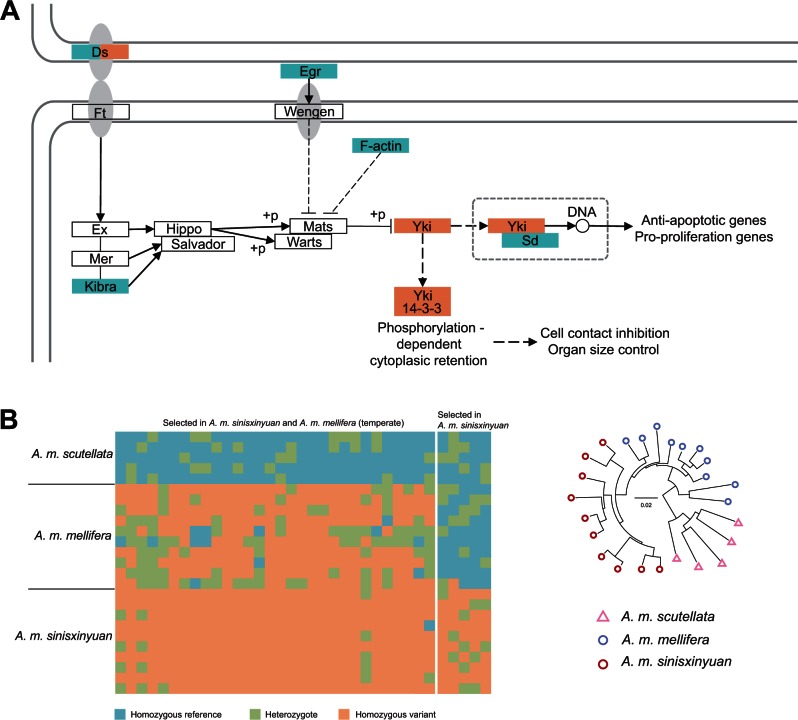
Selected genes in the temperate versus tropical comparison are enriched in the Hippo signaling pathway. Interestingly, we also found enrichment of selected genes in this pathway in an A. m. sinisxinyuan versus A. m. mellifera comparison (three genes, P = 3.8240E-02; supplementary table S10, Supplementary Material online). However, only one of the selected genes is common in both comparisons (fig. 4A). Enrichment of selected genes in this highly conserved pathway indicates its importance in the process of adaptation. One of the selected genes is Ds (Dachsous; fig. 4B), an upstream regulator of the Hippo signaling pathway (Cho and Irvine 2004; Willecke et al. 2008). There were also three selected genes identified in the downstream of the Hippo signaling pathway, namely Yki (Yorkie), Sd (Scalloped), and 14-3-3. Yki is a transcription coactivator which acts as a key regulator of apoptosis and proliferation (Huang et al. 2005); Sd, a partner of Yki, forms a complex with Yki that binds to DNA and regulates the activity of target genes (Goulev et al. 2008); 14-3-3 is an inhibitor of Yki that binds to Yki and prevents it from entering the nucleus to activate corresponding genes (Ren et al. 2010). Yki, Sd, and 14-3-3 together regulate the expression of downstream effector genes in the Hippo signaling pathway (fig. 4A). The Hippo signaling pathway is responsible for the control of organ size during development through cell proliferation and apoptosis (Halder and Johnson 2011; Zhao et al. 2011), and may underlie the differences in size of fat body between winter fat bees and regular bees.
Fig. 4.
(A) Selection in the Hippo signaling pathway. Selected genes in temperate versus tropical comparison and Apis mellifera sinisxinyuan versus A. m. mellifera comparison are in blue and orange, respectively. Annotations, abbreviations, and connections are in accordance with KEGG standards, with solid lines and dashed lines representing direct and indirect relationships, respectively. (B) Genotypes of SNPs and the gene tree of Ds (Dachsous) in A. m. sinisxinyuan, A. m. mellifera, and A. m. scutellata.
Discussion
Our discovery of A. m. sinisxinyuan further extends the eastern boundary of A. mellifera’s distribution into western China. This is the first discovery of an A. mellifera subspecies in China. Apis mellifera sinisxinyuan is also the only subspecies in the M-lineage found outside of Europe. Molecular dating shows that A. m. sinisxinyuan diverged from A. m. mellifera about 132 ka. Analyses of the population history of the cold-tolerant A. m. sinisxinyuan suggest that long-term global temperature has had a substantial influence on its effective population size, with high effective population size under low temperature. A genome-wide scan for signatures of selective sweeps identified a set of candidate genes targeted by natural selection during adaptation to temperate climates. Notably, the Hippo signaling pathway appears to play an important role in the evolution of temperate populations.
Previously, phylogenetic trees constructed from subspecies in the four lineages largely mirrored their geographic range, in that subspecies within the same lineage showed contiguous distributions (Wallberg et al. 2014). Specifically, the ranges of subspecies from the M-lineage are northern and western Europe (Ruttner 1988). However, the discovery of A. m. sinisxinyuan in Asia disrupts this pattern, suggesting a more complex evolutional history. Moreover, although it has been puzzling that the closely related A. mellifera and A. cerana showed allopatric distributions, the area between the distributions of these two species is not well studied (Sheppard and Meixner 2003). Our discovery of A. m. sinisxinyuan in this area further narrows the gap between the distribution of A. mellifera and A. cerana, and further research into this area may provide additional insight into the evolution of both species.
The discovery of A. m. sinisxinyuan raises questions about the origin and colonization of the M-lineage. The existence of A. m. sinisxinyuan in the eastern Tian Shan mountains suggests a northern migration route of the M-lineage from Asia into Europe as proposed by Garnery et al. (1992). Apis mellifera sinisxinyuan may represent an eastern branch during the migration, whereas A. m. mellifera represents the western branch. Alternatively, it is also possible that the origin of the expansion was in the Tien Shan Mountains with A. m. mellifera representing westward expansion of the M-lineage. Similar questions were raised when the O-lineage A. m. pomonella was discovered in the western Tian Shan Mountains, suggesting that the Tian Shan Mountains are also possibly the origin of the O-lineage (Sheppard and Meixner 2003). Further exploration in the Tian Shan Mountains and Xinjiang area may be important in understanding the evolutionary history of A. mellifera.
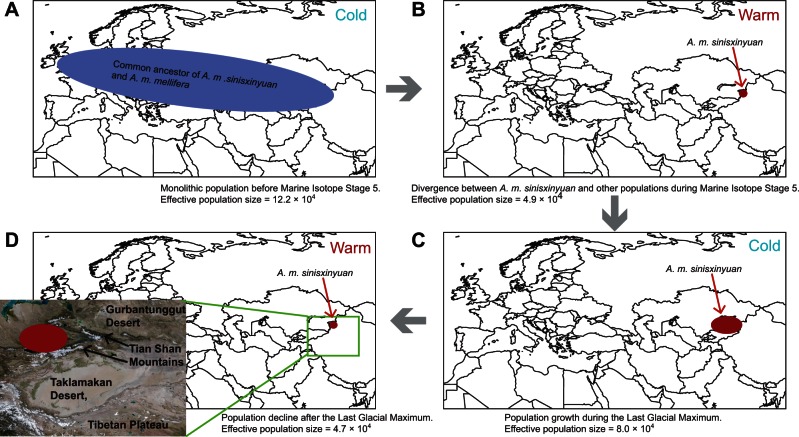
Population genomics of A. m. sinisxinyuan further reveals the history of the newly discovered subspecies. Based on the historical pattern of effective population size of A. m. sinisxinyuan and the divergence time between A. m. sinisxinyuan and A. m. mellifera (fig. 2), we propose a scenario of population history of A. m. sinisxinyuan, in which global temperature plays an important role: 1) Before Marine Isotope Stage 5, when A. m. sinisxinyuan and A. m. mellifera had not separated, the common ancestor of A. m. sinisxinyuan and A. m. mellifera had a monolithic distribution throughout Eurasia (fig. 5A). 2) During Marine Isotope Stage 5, elevated global temperature caused habitat reduction of the common ancestor, resulting in two geographically separated populations: The ancestor of A. m. sinisxinyuan in Asia and the ancestor of A. m. mellifera in Europe (figs. 2B and 5B). 3) After Marine Isotope Stage 5, the ancestor of the A. m. sinisxinyuan expanded its range as the weather became colder, peaking during the Last Glacial Maximum (figs. 2B and 5C). 4) On the one hand, in the post Last Glacial Maximum period, rising global temperature reduced suitable habitat for A. m. sinisxinyuan, whereas on the other hand, migration to new habitat was probably not an option due to the surrounding barriers formed by Tian Shan Mountains, deserts, and the Tibetan Plateau. As a result, A. m. sinisxinyuan experienced another decline in effective population size (fig. 5D).
Fig. 5.
Geography and possible population dynamics of Apis mellifera sinisxinyuan population in history. (A) Monolithic population of the ancestor of A. m. sinisxinyuan and A. m. mellifera spanning Europe and Asia. (B) Divergence of A. m. sinisxinyuan from other populations. (C) Increased population size of A. m. sinisxinyuan during the Last Glacial Maximum. (D) Population decline of A. m. sinisxinyuan after the Last Glacial Maximum.
The drop in effective population size of A. m. sinisxinyuan under elevated global temperature raises concerns for its protection. Apis mellifera sinisxinyuan is currently found only in very limited areas, which have been largely protected from human activities due to mountain barriers. However, global warming, together with increased human activity in this area (including beekeeping), poses a serious threat to the already limited population. In addition, another subspecies from the O-lineage, A. m. pomonella (Sheppard and Meixner 2003), may also exist in this area (Meixner MD, unpublished data). Furthermore, the two subspecies may be separated by a hybrid zone, given that there is no physical barrier between them, and the existence of A. m. pomonella in Xinyuan has the potential to impair the genetic integrity of A. m. sinisxinyuan through hybridization and introgression. Measures should be taken immediately for effective protection of A. m. sinisxinyuan.
Genetic variation in the genome of temperate populations of A. m. sinisxinyuan and A. m. mellifera can be characterized by adaptations to the extended cold winter, particularly rapidly evolving genes associated with the fat body. Several selected genes are related to lipid metabolism, which has been shown to be important in many organisms at high altitudes and/or latitudes (Swanson and Liknes 2006; Cheviron et al. 2012; Qiu et al. 2012; Qu et al. 2013; Liu et al. 2014). In addition, several selected genes related to fat body are also known to influence longevity. Abundant fat body and prolonged life span are characteristics of specialized winter bees, which are observed in temperate populations but not tropical populations (Winston 1980; Winston and Katz 1981). The fat winter bees can live for 6–8 months as opposed to 25–35 days in summer bees (Maurizio and Hodges 1950; Free and Spencer-Booth 1959). Previous studies have shown that a major stored component in fat body, Vitellogenin, is associated with longevity (Amdam et al. 2005). Taken together, fat body may play a central role in promoting longevity in overwintering bees.
The development of fat body may be greatly influenced by the Hippo signaling pathway, another important target of selection. Three lines of evidence support this hypothesis: First, the Hippo signaling pathway has a profound effect on the regulation of liver size in mammals (Halder and Johnson 2011; Hong et al. 2015), and the mammalian liver is analogous to fat body in insects in that it is essential for detoxification, energy storage, and utilization (Liu et al. 2009; Arrese and Soulages 2010). Second, the abundance of the regulatory protein 14-3-3 in the Hippo signaling pathway is influenced by the insulin pathway (Chan et al. 2011), which has a strong effect on fat body (Corona et al. 2007; Nilsen et al. 2011; Ihle et al. 2014). Third, the Hippo signaling is also connected to the insulin signaling pathway (Gotoh et al. 2015).
Another characteristic of overwintering honey bees is the ability to maintain stable nest temperature during winter. Unlike many other insects, the eusocial A. mellifera species do not survive cold winters in a dormant state (Salt 1969; Sinclair 2015). Instead, they have developed a unique socially cooperative mechanism to maintain high nest temperature during winter: Individual bees form a tight winter cluster to minimize heat loss, and within the winter cluster, heater bees generate heat to maintain a temperature of approximately 20 °C. Heat is generated in heater bees by decoupling the flight muscle from wings allowing flight muscles to vibrate without wing movement, generating heat. In addition, foragers warm themselves in the morning by shivering with flight muscles (Tautz 2008). Selection on the selected genes related to flight in temperate populations may involve in heat generation. For example, the FilI gene affect the structure of indirect flight muscle (de Couet et al. 1995), and selection on this gene may result in a muscle structure that can generate heat rapidly and/or efficiently. Selected genes related to flight may also influence the ability of honey bees to fly under low temperature. Cold flight muscles are unable to generate high enough wing beat frequencies for bees to fly (Josephson 1981), probably due to reduced frequency of membrane action potentials under low temperature (Bastian and Esch 1970). Because membrane depolarization is required to generate action potentials, the selected genes in membrane depolarization may enhance the ability of temperate honey bees to fly under cold condition.
Conclusion
Our study benefits from the recent discovery of A. m. sinisxinyuan, and provides insights into the history and genetic basis of adaptation of A. mellifera to temperate climates at the genomic level. Our integrative analyses highlight the role of historical global temperature in the long-term population dynamics of A. m. sinisxinyuan, and shed light on the underlying mechanisms of adaptation to temperate climates in A. mellifera. In addition, our result may facilitate the genetic breeding of A. mellifera to reduce overwintering losses, which have been a severe problem in many main beekeeping countries (van der Zee et al. 2012, 2014; vanEngelsdorp et al. 2012; Spleen et al. 2013; Pirk et al. 2014; Steinhauer et al. 2014; Lee et al. 2015).
Materials and Methods
Sampling
Xinyuan honey bees were collected in Yili River Valley, located in the northwest of Xinjiang Uygur, China, in August, 2011. Workers were collected from 20 colonies either from wild nests or human-made hives populated with wild-caught swarms, in four sites across Xinyuan prefecture. Geographic coordinates for the sample sites are 43 °17′ 39″N, 83 °35′ 23″E, 43 °18′ 7″N, 83 °39′ 48″E, 43 °17′ 51″N, 83 °42′ 48″E, and 43 °18′ 5″N, 83 °39′ 59″E, with elevations of 1,484, 1,566, 1,709, and 1,566 m, respectively. Bees from each colony were preserved in 90% ethanol. Individuals were chosen randomly from each colony and subjected to further analysis.
Sequencing
Ten Xinyuan honey bees, each from a different colony, were sequenced on the Illumina sequencing platform (HiSeq 2500) using standard procedures. The 500-bp paired-end libraries were prepared to generate approximately 8× raw coverage. A total of 17.9 Gb of clean sequence data were generated (supplementary tables S1 and S3, Supplementary Material online).
To analyze the evolutionary history of Xinyuan and other populations, we also downloaded the publicly available sequence data of 35 individuals from other representative populations (Supplementary table S2, Supplementary Material online) (Wallberg et al. 2014). The downloaded data were generated using the AB SOLiD technology with 4–6× coverage per sample.
Reads Mapping and Quality Control
All reads were preprocessed for quality control and filtered using our in-house script in Perl before mapping. Paired reads were discarded if the number of N’s in either of the paired reads exceeded 10%. Also, the number of low quality (Q ≤ 5) bases in a single read was restricted to less than 50%. 76.6 Gb of high-quality, clean reads were kept and mapped to the latest version of the honey bee apiMel4.5 reference genome using BWA-MEM with default parameters except for the “-t -k 32 -M -R” option (Li and Durbin 2009) (supplementary table S3, Supplementary Material online). Alignment bam files were sorted using samtools and duplicated reads were removed. Sequencing coverage and depth for each sample were calculated (supplementary table S3, Supplementary Material online).
The mapping rates of reads between the 35 individuals in the downloaded data (50–60%) and the 10 Xinyuan individuals (96%–97%) show significant discrepancy due to differences in sequencing platforms. Compared with the mapping technology of LifeScope (Wallberg et al. 2014), BWA set up a set of stricter parameters to judge alignment. To increase the accuracy of SNP calling and population variation analyses, we chose BWA as our read alignment software. We tested the difference of mapping rate with the two methods using data from a sample. The result revealed that the mapping rate of BWA was less than LifeScope by 20%.
SNP Calling and Genotyping
SNPs were detected using SAMtools’ mpileup (Li et al. 2009). The reference genome was subdivided into segments and analyzed in parallel. Population-based genotypes for the 45 individuals were created by SAMtools. About 4.7 × 106 raw SNPs were detected. The output was further filtered meeting the following criteria:
Quality score more than 20.
SNPs within 5 bp around a gap to be filtered.
The sum of read depths across all populations was [4, 1,000].
Ignoring multinucleotide polymorphisms.
The average Phred scaled base quality of the variant allele was ≥20.
The unobserved variant allele constituted ≤50%.
A total of 2.1 × 106 SNPs were retained for downstream analyses after applying the filters above.
Principal Component Analysis
The software EIGENSOFT3.0 (Patterson et al. 2006) was used for principal component analysis with biallelic SNPs of the 45 individuals. We plotted the first three significant components (supplementary fig. S2, Supplementary Material online). To some extent, the discrete points reflect the real structure of population. A Tracy–Widom test was used to determine the significant level of the principal components.
Population Structure
To estimate individual admixture assuming different numbers of clusters, the population structure was investigated using the program frappe (Tang et al. 2005), with a maximum-likelihood method. We increased the coancestry clusters spanning from 2 to 5 and ran analysis with 10,000 iterations.
Phylogenetic Tree
We constructed a neighbor-joining tree using pairwise genetic distances matrix data of all individuals, calculated by TreeBest (http://treesoft.sourceforge.net/treebest.shtml). The bootstrap was set to 1,000 times to assess the branch reliability. With A. cerana as outgroup, the result was in clear separation among all the individual samples. The Xinyuan honey bees and A. m. mellifera derived from the same branch.
Divergence Time Estimation
We used the putative sequence of coding region based on the allele frequency of each population, which was concatenated to one supergene for each species. Divergence times were estimated by MCMCtree (Yang 2007) package in PAML 4.5, using soft fossil constraints under various molecular clock models. The time unit is 100 My. We added three calibrations to edit the tree, one for the most recent common ancestor of A. cerana and A. mellifera, [0.06, 0.09] (Arias and Sheppard 1996), one within A. mellifera, [0.0015, 0.0035], and the last between A. m. ligustica and A. m. carnica [0.0002, 0.00035] (Wallberg et al. 2014). The clock variable was set to 2. The process was ran to sample 10,000 times using HKY85 as the substitution model, with the sample frequency set to 50 after a burn-in of 50,000 iterations.
Linkage-Diseqrilibrium Analysis
Linkage disequilibrium was calculated on the correlation coefficient (r2) statistics of two loci using Haploview software (Barrett et al. 2005), with minor allele frequency (MAF) greater than 0.05. Five individuals were randomly chosen from each population, and SNPs in each population were extracted to perform the analysis. Parameters were set as follows: “-maxdistance 4 -n -dprime -minMAF 0.05.” The longer the physical distance, the lower the value of r2, so the maxdistance was set to 4 K.
Effective Population Size
We used the PSMC model (Li and Durbin 2011) to obtain detailed changes in effective ancestral population sizes of Xinyuan population. The sequencing data of the Xinyuan populations were about 10×. The high sequencing depth was to ensure the quality of consensus sequence. The genotypes of individuals were determined based on the alignment BAM files using SAMTOOLS (Li et al. 2009). Bases of low sequencing depth (a third of the average depth) or high depth (twice of the average) were masked. The utility fq2psmcfa (provided with the PSMC software) was used to convert this diploid consensus sequence to the required input format. The parameters were set as follows: -N30 -t15 -r5 -p 45*2. The generation time (g) was set as an estimate of 1 year. We used a mutation rate (μ) of 5.3 × 10 − 9 per bp per generation.
Gene Annotation
We annotated A. mellifera gene set within functional categories based on the corresponding D. melanogaster orthologs. The genes were annotated by a homology-based method. Apis mellifera coding sequence was blasted to the D. melanogaster peptide sequences downloaded from Ensembl database (Release 74), with the parameter “-p blastx -m8 -e 1e-5 -F F” and the minimum alignment peptides more than 50. The correspondence relationship of A. mellifera gene and ontology category was decided by the best hit score of alignment, and the GO information was obtained from BioMart, with the D. melanogaster reference gene set (BDGP6).
Selection Analyses
The allele frequencies of variable sites were used to identify regions potentially affected by long-term selection using two complementary approaches. We calculated population fixation statistics (FST) and nucleotide diversity (θπ) for each sliding window (in 20-kb windows with 10-kb step size). We Z-transformed the distribution of FST and calculated the log value of θπ ratios. The putative selection targets were extracted based on being in the top 5% of log-odds ratios for both θπ and FST. Our analyses compared A. m. sinisxinyuan (Xinyuan honey bees) and A. m. mellifera with A. m. scutellata, and A. m. sinisxinyuan with A. m. mellifera.
Genomic Similarity
The similarity of the sequenced genomes in pairwise comparisons was calculated by identical score for SNPs. The identity score (IS) (Rubin et al. 2010; Ai et al. 2015) between two samples was measured by the distance score Dsi of allele to the reference allele at a given SNP site i:
| (1) |
The identical scores was the total accumulated score within a 20-kb window divided by the total number of SNPs. The formula of IS was as below:
| (2) |
N is the total number of SNPs within a 20-kb window. The identical scores of a single site were calculated as 1 minus the difference of Ds between the two samples (supplementary fig. S4, Supplementary Material online).
Data Accessibility
All short-read sequence data of this study have been deposited in the NCBI Short Read Archive (SRP065991), under the BioProject PRJNA301648.
Supplementary Material
Supplementary note, figures S1–S4, and tables S1–S10 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Chinese Academy of Agricultural Sciences (The Agricultural Science and Technology Innovation Program, grant No. CAAS-ASTIP-2015-IAR) and the earmarked fund for Modern Agro-industry Technology Research System (grant No. CARDS-45-KXJ1). The authors thank Tianqi Liu and Liqing Zhang for their help in field sampling; Yong Jin and Qian Wang for their assistance in preparing the figures; and Mark Camara for helpful comments and proofreading of the manuscript. They also thank the Oberursel Institute, Germany, for morphological reference data.
Supplementary Material
References
- Agarwal M, Guzmán MG, Morales-Matos C, Del Valle Díaz RA, Abramson CI, Giray T. 2011. Dopamine and octopamine influence avoidance learning of honey bees in a place preference assay. PLoS One 6:e25371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H, Fang X, Yang B, Huang Z, Chen H, Mao L, Zhang F, Zhang L, Cui L, He W, et al. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 47:217–225. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Norberg K, Omholt SW, Kryger P, Lourenço AP, Bitondi MMG, Simões ZLP. 2005. Higher vitellogenin concentrations in honey bee workers may be an adaptation to life in temperate climates. Insect Soc. 52:316–319. [Google Scholar]
- Arias MC, Sheppard WS. 1996. Molecular phylogenetics of honey bee subspecies (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol Phylogenet Evol. 5:557–566. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. 2010. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 55:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurori CM, Buttstedt A, Dezmirean DS, Mărghitaş LA, Moritz RF, Erler S. 2013. What is the main driver of ageing in long-lived winter honeybees: antioxidant enzymes, innate immunity, or vitellogenin? J Gerontol A Biol Sci Med Sci. 69:633–639. [DOI] [PubMed] [Google Scholar]
- Balanyà J, Huey RB, Gilchrist GW, Serra L. 2009. The chromosomal polymorphism of Drosophila subobscura: a microevolutionary weapon to monitor global change. Heredity 103:364–367. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- Bastian J, Esch H. 1970. The nervous control of the indirect flight muscles of the honey bee. Z Für Vgl Physiol. 67:307–324. [Google Scholar]
- Beggs KT, Glendining KA, Marechal NM, Vergoz V, Nakamura I, Slessor KN, Mercer AR. 2007. Queen pheromone modulates brain dopamine function in worker honey bees. Proc Natl Acad Sci U S A. 104:2460–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs KT, Mercer AR. 2009. Dopamine receptor activation by honey bee queen pheromone. Curr Biol. 19:1206–1209. [DOI] [PubMed] [Google Scholar]
- Beller M, Sztalryd C, Southall N, Bell M, Jackle H, Auld DS, Oliver B. 2008. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 6:e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Loubani M, Meinertzhagen IA. 2002. tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. J Neurosci. 22:10549–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan QW, Mutti NS, Foster LJ, Kocher SD, Amdam GV, Wolschin F. 2011. The worker honeybee fat body proteome is extensively remodeled preceding a major life-history transition. PLoS One 6:e24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheviron ZA, Bachman GC, Connaty AD, McClelland GB, Storz JF. 2012. Regulatory changes contribute to the adaptive enhancement of thermogenic capacity in high-altitude deer mice. Proc Natl Acad Sci U S A. 109:8635–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Irvine KD. 2004. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development 131:4489–4500. [DOI] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The last glacial maximum. Science 325:710–714. [DOI] [PubMed] [Google Scholar]
- Consortium HGS. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona M, Velarde RA, Remolina S, Moran-Lauter A, Wang Y, Hughes KA, Robinson GE. 2007. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci U S A. 104:7128–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Couet HG, Fong KS, Weeds AG, McLaughlin PJ, Miklos GL. 1995. Molecular and mutational analysis of a gelsolin-family member encoded by the flightless I gene of Drosophila melanogaster. Genetics 141:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluri P, Wille H, Gerig L, Lüscher M. 1977. Juvenile hormone, vitellogenin and haemocyte composition in winter worker honeybees (Apis mellifera). Experientia 33:1240–1241. [Google Scholar]
- Free JB, Spencer-Booth Y. 1959. The longevity of worker honey bees (Apis mellifera). Proc R Entomol Soc Lond. 34A:141–150. [Google Scholar]
- Garnery L, Cornuet J-M, Solignac M. 1992. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol Ecol. 1:145–154. [DOI] [PubMed] [Google Scholar]
- González J, Karasov TL, Messer PW, Petrov DA. 2010. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 6:e1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh H, Hust JA, Miura T, Niimi T, Emlen DJ, Lavine LC. 2015. The Fat/Hippo signaling pathway links within-disc morphogen patterning to whole-animal signals during phenotypically plastic growth in insects. Dev Dyn. 244:1039–1045. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 18:435–441. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. 2011. Hippo signaling: growth control and beyond. Development 138:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J. 2011. Adaptation to climate across the Arabidopsis thaliana genome. Science 334:83–86. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Alkorta-Aranburu G, Beall CM, Gebremedhin A, Sukernik R, Utermann G, Pritchard JK, Coop G, Di Rienzo A. 2011. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 7:e1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Wallberg A, Webster MT. 2012. From where did the Western honeybee (Apis mellifera) originate? Ecol Evol. 2:1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong L, Cai Y, Jiang M, Zhou D, Chen L. 2015. The Hippo signaling pathway in liver regeneration and tumorigenesis. Acta Biochim Biophys Sin. 47:46–52. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122:421–434. [DOI] [PubMed] [Google Scholar]
- Ihle KE, Baker NA, Amdam GV. 2014. Insulin-like peptide response to nutritional input in honey bee workers. J Insect Physiol. 69:49–55. [DOI] [PubMed] [Google Scholar]
- Josephson RK. 1981. Temperature and the mechanical performance of insect muscle In: Heinrich B, editor. Insect thermoregulation. New York: John Wiley & Sons; p. 19–44. [Google Scholar]
- Jouzel J, Masson-Delmotte V, Cattani O, Dreyfus G, Falourd S, Hoffmann G, Minster B, Nouet J, Barnola J-M, Chappellaz J. 2007. Orbital and millennial Antarctic climate variability over the past 800,000 years. Science 317:793–796. [DOI] [PubMed] [Google Scholar]
- Jünger MA, Rintelen F, Stocker H, Wasserman JD, Végh M, Radimerski T, Greenberg ME, Hafen E. 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KV, Steinhauer N, Rennich K, Wilson ME, Tarpy DR, Caron DM, Rose R, Delaplane KS, Baylis K, Lengerich EJ. 2015. A national survey of managed honey bee 2013–2014 annual colony losses in the USA. Apidologie 46:292–305. [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature 475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisiecki LE, Raymo ME. 2005. A Pliocene‐Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 20: [Google Scholar]
- Liu S, Lorenzen ED, Fumagalli M, Li B, Harris K, Xiong Z, Zhou L, Korneliussen TS, Somel M, Babbitt C, et al. 2014. Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157:785–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu H, Liu S, Wang S, Jiang R-J, Li S. 2009. Hormonal and nutritional regulation of insect fat body development and function. Arch Insect Biochem Physiol. 71:16–30. [DOI] [PubMed] [Google Scholar]
- Livingstone MS, Tempel BL. 1983. Genetic dissection of monoamine neurotransmitter synthesis in Drosophila. Nature 303:67–70. [DOI] [PubMed] [Google Scholar]
- Lv F-H, Agha S, Kantanen J, Colli L, Stucki S, Kijas JW, Joost S, Li M-H, Marsan PA. 2014. Adaptations to climate-mediated selective pressures in sheep. Mol Biol Evol. 31(12):3324–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizio A, Hodges FED. 1950. The influence of pollen feeding and brood rearing on the length of life and physiological condition of the honeybee preliminary report. Bee World 31:9–12. [Google Scholar]
- Meixner MD, Worobik M, Wilde J, Fuchs S, Koeniger N. 2007. Apis mellifera mellifera in eastern Europe—morphometric variation and determination of its range limits. Apidologie 38:191–197. [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. 2007. Recent and ongoing selection in the human genome. Nat Rev Genet. 8:857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen K-A, Ihle KE, Frederick K, Fondrk MK, Smedal B, Hartfelder K, Amdam GV. 2011. Insulin-like peptide genes in honey bee fat body respond differently to manipulation of social behavioral physiology. J Exp Biol. 214:1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez MM, Sabio G, Badaracco A, Quesada-Allué LA. 2011. Constitutive expression and enzymatic activity of Tan protein in brain and epidermis of Ceratitis capitata and of Drosophila melanogaster wild-type and tan mutants. Insect Biochem Mol Biol. 41:653–659. [DOI] [PubMed] [Google Scholar]
- Pérez MM, Schachter J, Berni J, Quesada-Allué LA. 2010. The enzyme NBAD-synthase plays diverse roles during the life cycle of Drosophila melanogaster. J Insect Physiol. 56:8–13. [DOI] [PubMed] [Google Scholar]
- Pirk CW, Human H, Crewe RM, vanEngelsdorp D. 2014. A survey of managed honey bee colony losses in the Republic of South Africa–2009 to 2011. J Apic Res. 53:35–42. [Google Scholar]
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nat Genet. 44:946–949. [DOI] [PubMed] [Google Scholar]
- Qu Y, Zhao H, Han N, Zhou G, Song G, Gao B, Tian S, Zhang J, Zhang R, Meng X. 2013. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 4:2071. [DOI] [PubMed] [Google Scholar]
- Ren F, Zhang L, Jiang J. 2010. Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev Biol. 337:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Reenan RA, Nilsen SP, Helfand SL. 2000. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290:2137–2140. [DOI] [PubMed] [Google Scholar]
- Roshina NV, Pasyukova EG. 2007. Genes regulating the development and functioning of the nervous system determine life span in Drosophila melanogaster. Russ J Genet. 43:275–280. [PubMed] [Google Scholar]
- Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464:587–591. [DOI] [PubMed] [Google Scholar]
- Ruttner F. 1988. Biogeography and taxonomy of honeybees. Berlin: Springer-Verlag. [Google Scholar]
- Salt RW. 1969. The survival of insects at low temperatures. Symp Soc Exp Biol. 23:331–350. [PubMed] [Google Scholar]
- Scheiner R, Plückhahn S, Öney B, Blenau W, Erber J. 2002. Behavioural pharmacology of octopamine, tyramine and dopamine in honey bees. Behav Brain Res. 136:545–553. [DOI] [PubMed] [Google Scholar]
- Seehuus S-C, Taylor S, Petersen K, Aamodt RM. 2013. Somatic maintenance resources in the honeybee worker fat body are distributed to withstand the most life-threatening challenges at each life stage. PLoS One 8:e69870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley TD. 1985. Honeybee ecology: a study of adaptation in social life. Princeton (NJ: ): Princeton University Press. [Google Scholar]
- Sheppard WS, Meixner MD. 2003. Apis mellifera pomonella, a new honey bee subspecies from Central Asia. Apidologie 34:367–376. [Google Scholar]
- Sinclair BJ. 2015. Linking energetics and overwintering in temperate insects. J Therm Biol. 54:5–11. [DOI] [PubMed] [Google Scholar]
- Southwick EE, Heldmaier G. 1987. Temperature control in honey bee colonies. BioScience 37:395–399. [Google Scholar]
- Spleen AM, Lengerich EJ, Rennich K, Caron D, Rose R, Pettis JS, Henson M, Wilkes JT, Wilson M, Stitzinger J. 2013. A national survey of managed honey bee 2011–12 winter colony losses in the United States: results from the Bee Informed Partnership. J Apic Res. 52:44–53. [Google Scholar]
- Steinhauer NA, Rennich K, Wilson ME, Caron DM, Lengerich EJ, Pettis JS, Rose R, Skinner JA, Tarpy DR, Wilkes JT. 2014. A national survey of managed honey bee 2012–2013 annual colony losses in the USA: results from the Bee Informed Partnership. J Apic Res. 53:1–18. [Google Scholar]
- Swanson DL, Liknes ET. 2006. A comparative analysis of thermogenic capacity and cold tolerance in small birds. J Exp Biol. 209:466–474. [DOI] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. 2005. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 28:289–301. [DOI] [PubMed] [Google Scholar]
- Tautz J. 2008. The buzz about bees: biology of a superorganism. Berlin: Springer Science & Business Media. [Google Scholar]
- Terada Y, Garofalo CA, Sakagami SF. 1975. Age-survival curves for workers of two eusocial bees (Apis mellifera and Plebeia droryana) in a subtropical climate, with notes on worker polyethism in P. droryana. J Apic Res. 14:161–170. [Google Scholar]
- van der Zee R, Brodschneider R, Brusbardis V, Charrière J-D, Chlebo R, Coffey MF, Dahle B, Drazic MM, Kauko L, Kretavicius J. 2014. Results of international standardised beekeeper surveys of colony losses for winter 2012–2013: analysis of winter loss rates and mixed effects modelling of risk factors for winter loss. J Apic Res. 53:19–34. [Google Scholar]
- van der Zee R, Pisa L, Andonov S, Brodschneider R, Charrière J-D, Chlebo R, Coffey MF, Crailsheim K, Dahle B, Gajda A. 2012. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J Apic Res. 51(10):100–114. [Google Scholar]
- vanEngelsdorp D, Caron D, Hayes J, Underwood R, Henson M, Rennich K, Spleen A, Andree M, Snyder R, Lee K. 2012. A national survey of managed honey bee 2010–11 winter colony losses in the USA: results from the Bee Informed Partnership. J Apic Res. 51:115–124. [Google Scholar]
- Vihervaara T, Puig O. 2008. dFOXO regulates transcription of a Drosophila acid lipase. J Mol Biol. 376:1215–1223. [DOI] [PubMed] [Google Scholar]
- Wallberg A, Han F, Wellhagen G, Dahle B, Kawata M, Haddad N, Simões ZLP, Allsopp MH, Kandemir I, De la Rúa P, Pirk CW, Webster MT. 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat Genet. 46:1081–1088. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Behura SK, Berlocher SH, Clark AG, Johnston JS, Sheppard WS, Smith DR, Suarez AV, Weaver D, Tsutsui ND. 2006. Thrice out of Africa: ancient and recent expansions of the honey bee, Apis mellifera. Science 314:642–645. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. 2008. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci U S A. 105:14897–14902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston ML. 1980. Seasonal patterns of brood rearing and worker longevity in colonies of the Africanized honey bee (Hymenoptera: Apidae) in South America. J Kans Entomol Soc. 53:157–165. [Google Scholar]
- Winston ML. 1987. The biology of the honey bee. Cambridge (MA: ): Harvard University Press. [Google Scholar]
- Winston ML, Katz SJ. 1981. Longevity of cross-fostered honey bee workers (Apis mellifera) of European and Africanized races. Can J Zool. 59:1571–1575. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zayed A, Robinson GE. 2012. Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu Rev Genet. 46:591–615. [DOI] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan K-L. 2011. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 13:877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All short-read sequence data of this study have been deposited in the NCBI Short Read Archive (SRP065991), under the BioProject PRJNA301648.