Abstract
Although population genetics studies have significantly accelerated the evolutionary and functional interrogations of genes and regulations, limited polymorphism data are available for rhesus macaque, the model animal closely related to human. Here, we report the first genome-wide effort to identify and visualize the population genetics profile in rhesus macaque. On the basis of the whole-genome sequencing of 31 independent macaque animals, we profiled a comprehensive polymorphism map with 46,146,548 sites. The allele frequency for each polymorphism site, the haplotype structure, as well as multiple population genetics parameters were then calculated on a genome-wide scale. We further developed a specific interface, the RhesusBase PopGateway, to facilitate the visualization of these annotations, and highlighted the applications of this highly integrative platform in clarifying the selection signatures of genes and regulations in the context of the primate evolution. Overall, the updated RhesusBase provides a comprehensive monkey population genetics framework for in-depth evolutionary studies of human biology.
Keywords: whole-genome sequencing, population genetics, primate evolution, rhesus macaque, RhesusBase.
Introduction
Recent population genetic studies have significantly accelerated the evolutionary and functional interrogations of genes and regulatory regions in Nematoda (Cutter and Choi 2010), Drosophila (Begun et al. 2007), mouse (Keane et al. 2011), and human (International HapMap 3 Consortium 2010; 1000 Genomes Project Consortium 2012). However, rather limited polymorphism data were reported for rhesus macaque, a unique primate model with genome sequence and composition highly analogous to human. In this regard, candidate gene studies mainly focused on specific genomic regions (Ferguson et al. 2007; Hernandez et al. 2007), whereas several recent resequencing studies with single or limited number of macaque animals did not sufficiently capture a comprehensive macaque polymorphism profile with accurate allele frequency information (Fang et al. 2011; Gokcumen et al. 2013). Such inadequate account of genomic data considerably limits the current use of this unique nonhuman primate model.
Previously, we created “RhesusBase” to address several key unresolved issues regarding this unique model—the limited genomic annotations, error-prone gene structures, and a lack of platform to visualize and assess next-generation sequencing (NGS) data and perform comparative genomics studies (Zhang et al. 2013, 2014). On the basis of the RhesusBase platform and the in-house macaque genomics data, here we report the first genome-wide effort in developing a reference polymorphism map for rhesus macaque. The updated RhesusBase embedded with population genetic data is aimed to enable the primate research community to dissect polymorphism patterns, elucidate selectively constrained genes and regulatory regions in the primate evolution, and accelerate the pace of the human translational study.
Results and Discussion
As rhesus macaque animals are mainly distributed in China and India, with a bigger effective population size in China (Hernandez et al. 2007), we first analyzed the in-house whole-genome sequencing data of 24 independent, Chinese-origin captive animals with high sequencing coverage (median sequencing depth of 40-folds) (Chen et al. 2015; Yang et al. 2015), and further integrated other public genome resequencing data from five Indian-origin and two Chinese-origin animals (median sequencing depth of 20-folds) (Fang et al. 2011; Yan et al. 2011; Gokcumen et al. 2013). Meta-data for these macaque animals, such as their geographic origin, age, gender, as well as the Sequence Read Archive (SRA) accession numbers for the raw sequencing data, are available in supplementary table S1, Supplementary Material online.
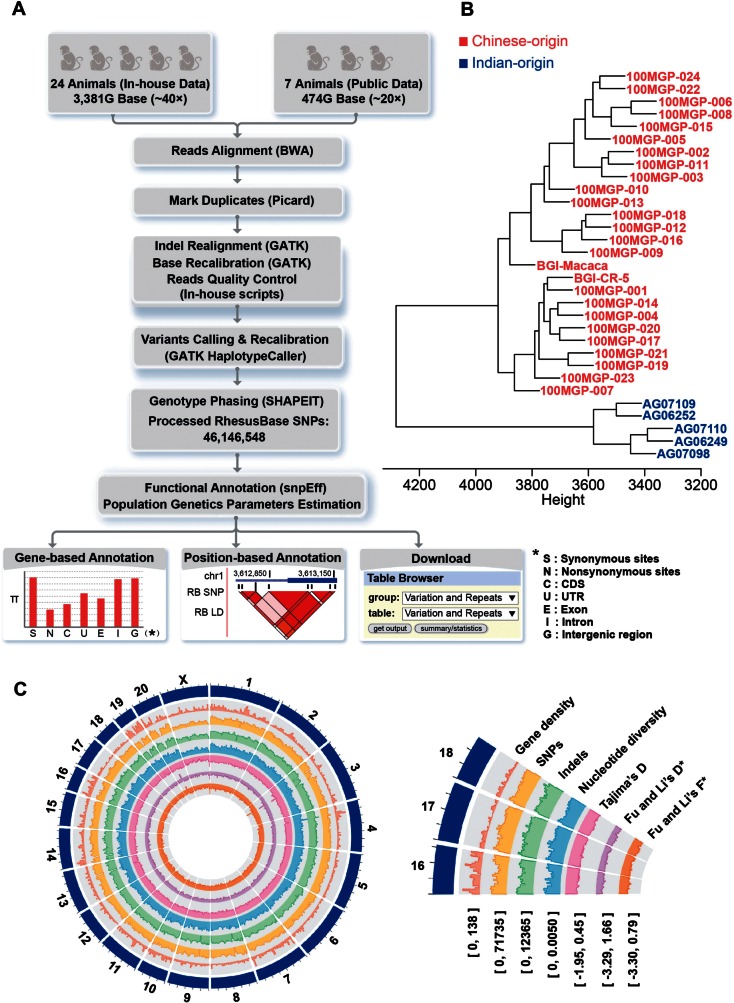
With these whole-genome sequencing data of 31 independent macaque animals (supplementary table S1, Supplementary Material online), we compiled a comprehensive polymorphism map in rhesus macaque, with 46,146,548 polymorphism sites (fig. 1A, supplementary methods, Supplementary Material online). We performed hierarchical clustering analyses on the basis of autosomal polymorphism sites to illustrate the population structure of these independent macaque animals, which clearly reconciled the geological origin of the 31 macaque animals (fig. 1B). For each site, detailed information such as the frequency of alleles, the status of ancestral allele, as well as the haplotype structure were then deduced (supplementary methods, Supplementary Material online).
Fig.1.
Population genetics analyses in rhesus macaque. (A) Overview of the genome-wide study to identify and visualize the population genetics profile in rhesus macaque. (B) Dendrogram generated from autosomal polymorphism sites. IDs of Chinese-origin and Indian-origin macaque animals were marked in red and blue, respectively. Branch height represents dissimilarity. (C) Circos plot illustrates the genome-wide distribution of the polymorphism profiles across the rhesus macaque genome, as well as the population genetics parameters associated with individual profiles. Chromosomes are ordered in clockwise direction, with the sequences binned in 3 Mb. Pie slices below represent selected magnification of the circos subplots. Heights for different parameters shown in the circos plot are relative to the overall distributions of the respective parameters along the genome (numbers at the lower edge of the pie slices indicate the range of values shown in the selected subcircle).
On the basis of these data, multiple population genetics parameters, such as the population mutation rate (θw), nucleotide diversity (π), and Tajima’s D, were further calculated on genome-wide scale to facilitate evolutionary study on single gene or certain genomic regions of interest (fig. 1C and table 1, supplementary tables S3–S5 and S7, Supplementary Material online). In this regard, we separated sex chromosomes from autosomes in the calculations. Since Y chromosome was not constructed in macaque reference genome, we only included the X chromosome. As expected, the parameters on X chromosome differ from that of autosomes (table 1, supplementary tables S3–S5, Supplementary Materials online). Specifically, the polymorphism level of X chromosome is in general lower than that of autosomes, possibly as a consequence of multiple factors such as the smaller population size and efficient selection against slightly deleterious mutations due to its hemizygosity in male. Such a comprehensive population genetics map in rhesus macaque provides a basis for evolutionary interrogations of human biology in the genomic context of rhesus macaque.
Table 1.
Comparison of Population Genetics Features for Protein-Coding Genes in Human and Rhesus Macaque.
| Nucleotide Diversity (π × 103) |
Population Mutation Rate (θw×103) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Autosome |
X Chromosome |
Autosome |
X Chromosome |
||||||
| Median | Mean±SD | Median | Mean±SD | Median | Mean±SD | Median | Mean±SD | ||
| Human | Synonymous Sites | 0.772 | 1.203±1.409 | 0.429 | 0.849±1.176 | 1.649 | 1.895±1.182 | 1.097 | 1.388±1.104 |
| Nonsynonymous Sites | 0.144 | 0.339±0.513 | 0.061 | 0.202±0.405 | 0.552 | 0.693±0.542 | 0.340 | 0.457±0.409 | |
| CDS | 0.331 | 0.481±0.533 | 0.151 | 0.291±0.411 | 0.746 | 0.861±0.547 | 0.439 | 0.551±0.417 | |
| UTR | 0.574 | 0.747±0.720 | 0.277 | 0.434±0.518 | 1.081 | 1.198±0.67 | 0.670 | 0.777±0.500 | |
| Exon | 0.484 | 0.594±0.481 | 0.240 | 0.338±0.351 | 0.915 | 0.99±0.475 | 0.543 | 0.607±0.356 | |
| Intron | 0.758 | 0.818±0.452 | 0.418 | 0.459±0.331 | 1.184 | 1.222±0.387 | 0.736 | 0.775±0.288 | |
| Intergenic Regions | 0.711 | 0.817±0.546 | 0.397 | 0.467±0.361 | 1.186 | 1.229±0.477 | 0.709 | 0.772±0.347 | |
| Macaque | Synonymous Sites | 2.738 | 3.328±2.670 | 1.297 | 1.787±1.785 | 4.446 | 4.989±2.913 | 2.024 | 2.553±1.947 |
| Nonsynonymous Sites | 0.419 | 0.775±1.037 | 0.263 | 0.585±0.837 | 0.939 | 1.329±1.283 | 0.488 | 0.841±0.945 | |
| CDS | 0.977 | 1.263±1.109 | 0.450 | 0.681±0.748 | 1.709 | 2.006±1.343 | 0.706 | 0.969±0.850 | |
| UTR | 1.65 | 2.035±1.709 | 0.758 | 1.071±1.083 | 2.847 | 3.179±1.984 | 1.240 | 1.583±1.302 | |
| Exon | 1.277 | 1.512±1.087 | 0.537 | 0.737±0.703 | 2.146 | 2.368±1.300 | 0.839 | 1.059±0.818 | |
| Intron | 2.274 | 2.334±1.054 | 1.134 | 1.247±0.904 | 3.327 | 3.395±1.165 | 1.515 | 1.625±0.928 | |
| Intergenic Regions | 2.107 | 2.256±1.135 | 1.012 | 1.161±0.717 | 3.3 | 3.355±1.276 | 1.402 | 1.533±0.748 | |
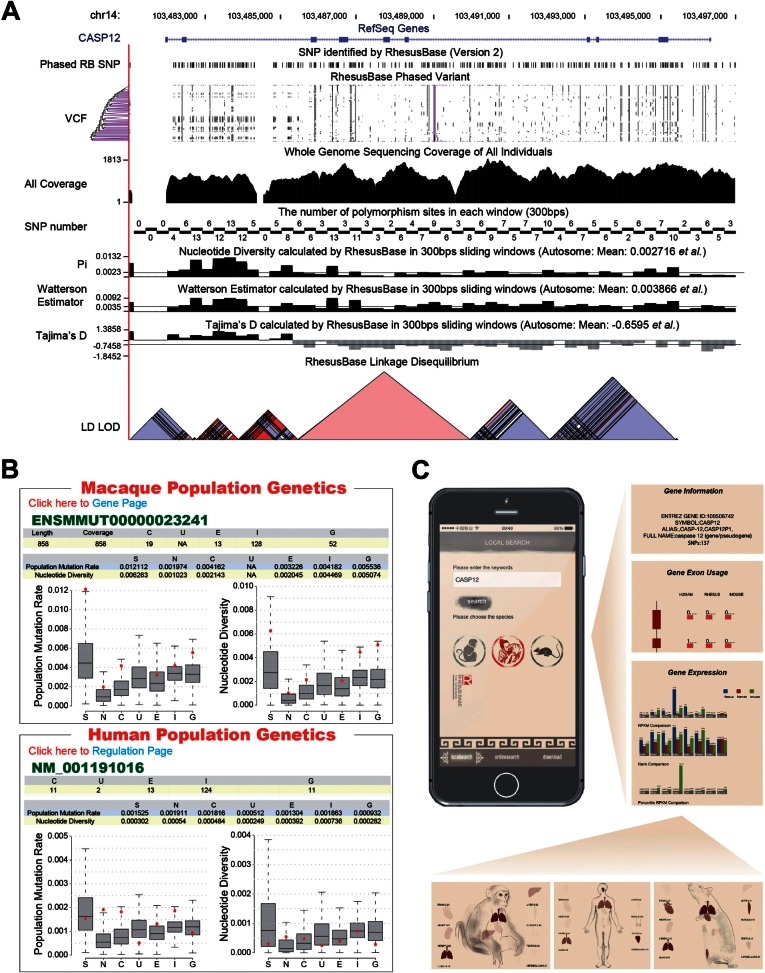
We further developed a user-friendly population genetics gateway, the RhesusBase PopGateway (http://www.rhesusbase.org/popGateway, last accessed February 13, 2016), to expedite the visualization of these macaque population genetics annotations (fig. 2). For each gene in rhesus macaque, we provided on the Population Genetics Page several gene-based genetic parameters, such as θw and π, in a comparative mode of human and rhesus macaque (fig. 2B, supplementary table S2, Supplementary Material online). For studies focusing on regulation regions rather than genic loci, a variety of region-based annotations were intuitively displayed on the Genome Browser of the PopGateway (fig. 2A). Meta-data associated with the calculations of these population genetics parameters, such as the effective sequence coverage and the number of polymorphism sites in the region or the gene of interests, were also included in the RhesusBase PopGateway interfaces (fig. 2A). For each parameter, we also highlighted these values of certain gene/chromosome regions in the context of the genome-wide distribution as the background (fig. 2B). For advanced users, all data in the RhesusBase PopGateway could also be downloaded from our table browser (http://browser.rhesusbase.org/cgi-bin/hgTables, last accessed February 13, 2016) and download page (http://www.rhesusbase.org/download/download.jsp, last accessed February 13, 2016). In particular, the polymorphism profile of the gene, as well as other associated genomic features such as tissue expression and splicing profiles, could also be found in a newly developed mobile APP to enable offline use of RhesusBase annotations in a comparative, cross-species mode (fig. 2C).
Fig. 2.
Evolutionary and functional interrogation of single genes in RhesusBase PopGateway. (A) An expanded view of RhesusBase Genome Browser at the CASP12 locus, shown with comprehensive position-based annotations, such as the gene structure, polymorphisms, nucleotide diversity (π), population mutation rate (θw), Tajima’s D, and linkage disequilibrium. (B) A typical RhesusBase Population Genetics Page showing the comparative population genetics features of CASP12 in human and rhesus macaque. (C) For each gene of interest, three annotation pages—the Gene Information, Gene Exon Usage, and Gene Expression—were developed for visualizing, respectively, the basic gene annotations, the exon splicing efficiency, and gene expression levels in a comparative view of human, rhesus macaque, and mouse. Detailed information could be further accessed by clicking the corresponding links. For example, the transcript expression of the gene of interest is shown in RPKM (Reads Per Kilobase of exon model per Million mapped reads) and marked on the body map, with the color intensity corresponding to its relative expression level in each tissue of the different species.
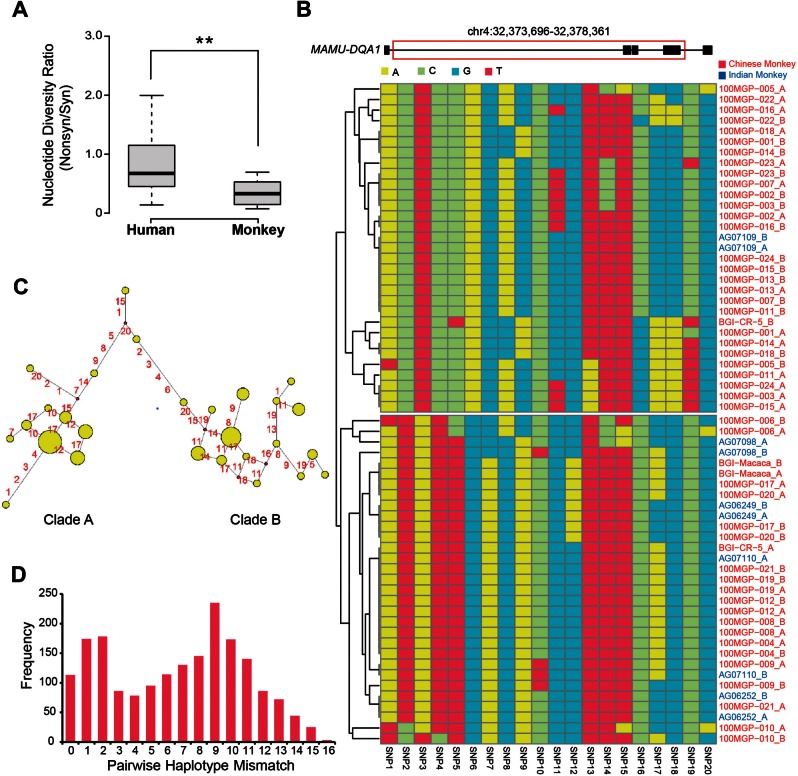
We then highlighted the applications of this well-annotated macaque polymorphism platform in deciphering the selective constraints of specific genes and chromosome regions in primates. First, as the identification of pseudogenes is error-prone due to the poor genome and transcriptome assembly in human outgroup species, we evaluated whether a list of human-specific pseudogenes identified by a recent study are reliable (Wang et al. 2006), from the perspective of population genetics profiles in both human and rhesus macaque. In principle, for a human-specific pseudogene such as CASP12 (Wang et al. 2006), the polymorphism level of nonsynonymous sites and synonymous sites of this gene should be comparable in human. In contrary, the nucleotide diversity for nonsynonymous sites should be significantly lower than that of synonymous sites in rhesus macaque, in that the macaque ortholog of the human-specific pseudogene is still functional on protein level (fig. 2B). According to RhesusBase PopGateway annotations, we found that these candidate pseudogenes show comparable polymorphism level of nonsynonymous sites and synonymous sites in human, but rather significantly lower levels of nonsynonymous sites in rhesus macaque (Wilcoxon one tail test, P-value = 0.0003), indicating that human-specific pseudogenes are indeed enriched in the list, with relaxed selective constrains specifically in human (fig. 3A, supplementary table S6 and methods, Supplementary Material online).
Fig. 3.
Application of the RhesusBase PopGateway in illustrating primate regulations and their functional implications. (A) The ratios of nucleotide diversity (π) for nonsynonymous sites to synonymous sites were summarized in boxplots, for human-specific pseudogenes and their functional orthologs in rhesus macaque, respectively. (B–D) For a candidate genomic region under balancing selection, the haplotype structures in 31 macaque animals (62 haplotypes) were shown in hierarchical clustering chart (B). The evolutionary relationship of these haplotypes were further summarized in (C), with the size of each node proportional to the haplotype frequency, and the numbers on branches representing nucleotide substitution sites. Pairwise mismatch distribution of these haplotypes were further calculated and shown, by counting the number of differences between all pairs of the 62 haplotypes (D). The multimodal distribution recapitulates the existence of two major types of haplotype.
Second, the polymorphism and haplotype annotations in RhesusBase PopGateway also provide clues concerning genomic regions under balancing selection in rhesus macaque. Although it is less likely that the balancing selection has preserved the same haplotype structure throughout evolution, some balancing selection hotspots where environmental pressures independently create multiple adaptive peaks may lead to signals of balancing selection in multiple primate species. Taking one MHC (Major Histocompatibility Complex) gene as an example, which is known to be under balancing selection in human (Solberg et al. 2008), we found that the haplotypes of its macaque ortholog, MAMU-DQA1, could also be clustered into two major types in rhesus macaque (fig. 3B and C), illustrating a multimodal distribution of the pairwise mismatches of candidate haplotypes—a signature of balancing selection (Bamshad et al. 2002; fig. 3D). It is clear that the two divergent haplotypes are seen independently in the Indian-origin and Chinese-origin populations, indicating the divergence more likely resulted from ancient balancing selection, rather than geography-specific selection (fig. 3B). Interestingly, it seems that such cases are not rare across the macaque genome. For another 125 regions reportedly under ancient balancing selection in human and chimpanzee (Leffler et al. 2013), 35 also showed signatures of balancing selection in rhesus macaque according to RhesusBase PopGateway annotations (supplementary table S8, Supplementary Material online).
Taken together, on the basis of the whole-genome sequencing of 31 independent macaque animals, we profiled the most comprehensive polymorphism map in rhesus macaque to date. We also developed a user-friendly interface, the RhesusBase PopGateway, to facilitate the in-depth visualization of these data and information. We further incorporated RhesusBase annual update on transcriptome and regulatory annotations, with the current version of the database containing 172 million functional annotation records derived from 1,761 NGS data sets. This updated RhesusBase, equipped with the population genetics gateway, would thus help the primate research community to address the fundamental questions in human biology and translational study, by highlighting functional genes and regulatory regions in the context of the primate evolution.
Supplementary Material
Supplementary methods and tables S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors acknowledge Dr Bin Z. He at Harvard University and Yong E. Zhang at Institute of Zoology, Chinese Academy of Science for insightful suggestions, and Dr Jia Yu and Ms Xin Wen at Peking Union Medical College Hospital for assistance in the study. The authors acknowledge the anonymous reviewers for valuable suggestions on this work. This work was supported by grants from the National Key Basic Research Program of China [2013CB531202, 2012CB518004], the National Natural Science Foundation of China [31522032, 31471240, 31171269, 31221002], and the National Young Top-Notch Talent Support Program of China.
References
- 1000 Genomes Project 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Watkins WS, Wooding S, Stone AC, Jorde LB, Weiss RB, et al. 2002. A strong signature of balancing selection in the 5' cis-regulatory region of CCR5. Proc Natl Acad Sci U S A. 99:10539–10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5:e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Shen QS, Zhou WZ, Peng J, He BZ, Li Y, Liu CJ, Luan X, Ding W, Li S, et al. 2015. Emergence, retention and selection: a trilogy of origination for functional de novo proteins from ancestral LncRNAs in primates. PLoS Genet. 11:e1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter AD, Choi JY. 2010. Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae. Genome Res. 20:1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Zhang Y, Zhang R, Yang L, Li M, Ye K, Guo X, Wang J, Su B. 2011. Genome sequence and global sequence variation map with 5.5 million SNPs in Chinese rhesus macaque. Genome Biol. 12:R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B, Street SL, Wright H, Pearson C, Jia Y, Thompson SL, Allibone P, Dubay CJ, Spindel E, Norgren RB., Jr. 2007. Single nucleotide polymorphisms (SNPs) distinguish Indian-origin and Chinese-origin rhesus macaques (Macaca mulatta). BMC Genomics 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokcumen O, Tischler V, Tica J, Zhu Q, Iskow RC, Lee E, Fritz MH, Langdon A, Stutz AM, Pavlidis P, et al. 2013. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci U S A. 110:15764–15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, et al. 2007. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science 316:240–243. [DOI] [PubMed] [Google Scholar]
- International HapMap 3 Consortium 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, et al. 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, Segurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, et al. 2013. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 339:1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, Thomson G. 2008. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 69:443–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J. 2006. Gene losses during human origins. PLoS Biol. 4:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, et al. 2011. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol. 29:1019–1023. [DOI] [PubMed] [Google Scholar]
- Yang XZ, Chen JY, Liu CJ, Peng J, Wee YR, Han X, Wang C, Zhong X, Shen QS, Liu H, et al. 2015. Selectively constrained RNA editing regulation crosstalks with piRNA biogenesis in primates. Mol Biol Evol. 32:3143–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Shi M, Kong L, Chen JY, Zhou WZ, Zhu X, Yu P, Wang J, Yang X, et al. 2013. RhesusBase: a knowledgebase for the monkey research community. Nucleic Acids Res. 41:D892–D905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Yu P, Zhong X, Chen JY, Yang X, Peng J, Yan S, Wang C, Zhu X, et al. 2014. Evolutionary interrogation of human biology in well-annotated genomic framework of rhesus macaque. Mol Biol Evol. 31:1309–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.