Summary
There are many transgenic GFP reporter lines that allow visualization of specific populations of cells. Using such lines for functional studies requires a method that transforms GFP into a molecule that enables genetic manipulation. Here we report the creation of a method that exploits GFP for gene manipulation, Cre Recombinase Dependent on GFP (CRE-DOG), a split component system that uses GFP and its derivatives to directly induce Cre/loxP recombination. Using plasmid electroporation and AAV viral vectors, we delivered CRE-DOG to multiple GFP mouse lines, leading to effective recombination selectively in GFP-labeled cells. Further, CRE-DOG enabled optogenetic control of these neurons. Beyond providing a new set of tools for manipulation of gene expression selectively in GFP+ cells, we demonstrate that GFP can be used to reconstitute the activity of a protein not known to have a modular structure, suggesting that this strategy might be applicable to a wide range of proteins.
Introduction
A challenge to the understanding of brain function is the ability to monitor and/or manipulate the activity of the many different cell types comprising the nervous system. To label specific cell types, many transgenic reporter lines using Green Fluorescent Protein (GFP)1, 2 as a marker of gene expression3 have been generated for various model organisms. Notably, over 1,000 transgenic GFP mouse lines are now characterized for labeling specific cell populations in the central nervous system (gensat.org)4–6. Until recently, GFP lines have been used only for labeling purposes. However, studies probing cellular function often require genetic manipulations, e.g. using mouse lines expressing site-specific DNA recombinases in specific cell types to drive the expression of, or deletion of, particular genes. Compared to GFP reporter lines, the availability of mouse lines expressing the widely used Cre recombinase in specific cell types is more limited, and Cre expression patterns have not been as extensively characterized (gensat.org and Allenbrain.org). Manipulation of GFP-labeled cell types previously required the generation of new mouse lines, e.g. using the same cis-regulatory sequences that drove GFP in specific cell types in order to drive Cre expression in the same cell types. For more complex model organisms such as the mouse, such an approach is costly and time-consuming. Furthermore, the expression pattern of Cre in these lines could differ from that of the equivalent GFP reporter line, potentially due to a combination of high sensitivity of Cre and/or position effects exerted by neighboring genomic sequences.
Recently, we developed GFP-dependent transcription factors (T-DDOGs) that enable retrofitting of transgenic GFP lines for functional manipulation studies7. In this system, GFP acts as a scaffold, bringing together modular transcription domains to assemble a hybrid transcription factor for activation of a target gene of interest. GFP recognition is mediated by pairs of GFP binding proteins (GBPs), derived from the antigen recognition portion of camelid antibodies8, 9. These “nanobodies” are easily expressed in living cells and can be used as fusion proteins7–10. T-DDOGs opened the door for quick genetic manipulation of GFP-labeled cells via DNA electroporation, bypassing the need to generate transgenic Cre lines for cell-specific gene manipulations in certain cases7.
Several reasons motivate the continued development of new GFP-dependent systems. First, unlike transcription factors11, 12, many proteins such as enzymes are not composed of simple modular structures. It is unclear whether GFP can be used to directly control the activity of such non-modular proteins. Second, many genetic tools depend on the action of Cre recombinase. Although T-DDOG can induce Cre expression, this effect requires the action of three genetically encodable components. While electroporation can efficiently deliver multiple plasmids encoding T-DOGG to single cells, this approach is restricted to certain tissues and developmental time points. In contrast, the popular recombinant adeno-associated viral (rAAV) vectors can efficiently deliver multiple transgenes to single cells, across diverse tissues and over different developmental and/or mature stages. rAAVs encoding the recombinase-dependent flip-excision (FLEX) switch further enables convenient and selective expression of transgenes in specific cell types when injected into transgenic Cre and/or Flp lines13–15. The integration of GFP-inducible systems with FLEX responder genes in a rAAV delivery scheme would provide a simple protocol for rapid manipulation of many types of genes selectively in GFP-labeled cell types. Furthermore, such a method would increase the number of specific cell types that could be manipulated with intersectional approaches, as transgenic animals with a combination of GFP plus Flp in specific cell types could activate transgenes that are Cre plus Flp-dependent14, 16.
Here, we successfully used GFP to control the activity of Cre, in the form of split Cre-GBP fusion proteins that together make up Cre Recombinase-Dependent On GFP (CRE-DOG). The GBP-Split Cre fusion proteins assemble as a complex on the GFP scaffold, initiating Cre recombination (Fig.1a). This system only requires two components for direct activation of Cre recombination (Supplementary Fig.1). We generated rAAV reagents that enable delivery of CRE-DOG to diverse tissues and brain regions that cannot be electroporated, for rapid and tight GFP-regulated Cre-dependent transgene expression across GFP reporter lines and cell types. To demonstrate the utility of this approach, we showed that CRE-DOG can be used for optogenetic manipulation of GFP-expressing cells in the nervous system.
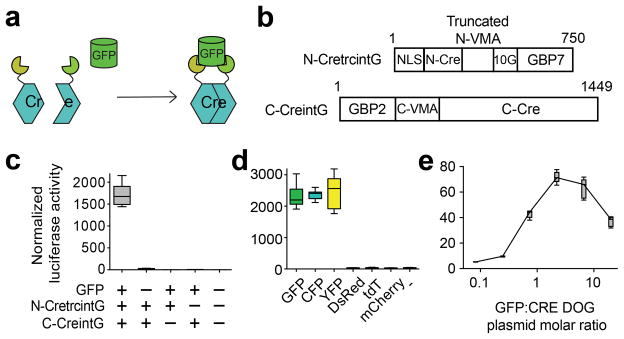
Figure 1. An optimized Cre recombinase dependent on GFP (CRE-DOGOPT).
(a) Schematic of CRE-DOGOPT action. (b) Schematic of chimeric constructs comprising CRE-DOGOPT. (c–e) Luciferase reporter assays in transfected 293T cells assessing CRE-DOGOPT specificity for all components of the system (c), specificity for different fluorescent proteins (d) and dependency on GFP level (e). n=18 per condition for (c and d), n=6 or 9 for (e). (see Online Methods for n definition). Results are representative of at least three independent transfections. Boxplots indicate minimum to maximum range.
Results
CRE-DOG refers to the combined action of two chimeric proteins bearing complementary split Cre fragments17 each fused to a different GBP. We tested the GBP1+GBP6 and GBP2+GBP7 combinations, as they had been previously used to assemble a hybrid transcription factor on the GFP scaffold in living cells7. Using a CAG-loxP-Neo-loxP-luc2 (CALNL-luc2) construct, (or “floxed” luciferase reporter), we screened pairs of the GBP-split Cre fusion proteins for GFP-dependent activity. Almost all GBP-split Cre pairs had either low GFP-induced recombination activity or high background activity (data not shown). We considered whether it is possible to increase efficiency by generating a full-length Cre recombinase via protein splicing18. To this end, we inserted the artificially split S. cerevisiae vacuolar ATPase (VMA1) intein elements19, 20 into the GBP-split Cre constructs and conducted additional reporter screens. None of the constructs based on the GBP1+GBP6 pair yielded any high efficiency recombination. However, a subset of constructs based on the GBP2+GBP7 combination gave the desired GFP-dependent Cre activity with reasonably low background activity. The most desirable pair of fusion constructs showed strong GFP-dependent recombination activity in vitro and in vivo, and was named CRE-DOGOG (Supplementary Fig. 2). CRE-DOGOG refers to the combined action of two fusion protein components, N-CreintG and C-CreintG. In contrast to the T-DDOG system, which carries the caveat of being potentially toxic to cells as a consequence of the squelching phenomenon caused by transcription activation domains21, CRE-DOGOG did not induce any noticeable abnormality in retinal cells.
Although intein elements were intended to create a Cre protein via protein splicing, we failed to find evidence supporting this mechanism in several tests (Supplementary Fig. 3). To promote protein splicing, the split-intein of VMA1 requires a cysteine residue at the boundary between each split-intein and its respective fusion partner22. Double alanine and double serine substitutions at the cysteine positions were expected to disrupt protein splicing22, but were found to have little adverse effect on the GFP-specificity or efficiency of CRE-DOGOG22(Supplementary Fig. 3a). Second, each split VMA element promoted GFP-specificity even in the absence of its protein splicing partner (Supplementary Fig. 3b). Third, under conditions in which GFP could induce the activity of a FLAG-tagged CRE-DOGOG variant, we failed to detect the appearance of spliced, full-length Cre in the presence of GFP (Supplementary Fig. 3c–d). Nonetheless, as these combined fusion proteins provided a more active GFP-dependent Cre, we continued to optimize these constructs.
Although CRE-DOGOG gave GFP-dependent recombination in vitro and in vivo, background activity in the absence of GFP did accumulate noticeably over time in vitro (data not shown). Thus, we conducted additional screens to further optimize CRE-DOGOG activity. We found that the 184 aa N-terminal portion of VMA (N-VMA) could be further truncated without adversely affecting CRE-DOG activity. Serial residue deletion and insertion scans along N-VMA led to the isolation of a 43 aa truncated element that promoted both enhanced GFP-dependent recombination and reduced GFP-independent activity when compared to CRE-DOGOG (Supplementary Fig. 4–5, Supplementary Table 1). This truncated fusion protein, hereafter named N-CretrcintG, was combined with C-CreintG to give the Optimized CRE-DOG (CRE-DOGOPT) (Fig. 1b). CRE-DOGOPT activity depended upon all components of the system and was specific for GFP and its derivatives (Figs. 1c,d). Further, CRE-DOGOPT activity was dependent upon GFP dosage in a manner similar to that observed for T-DDOGs7 (Fig. 1e).
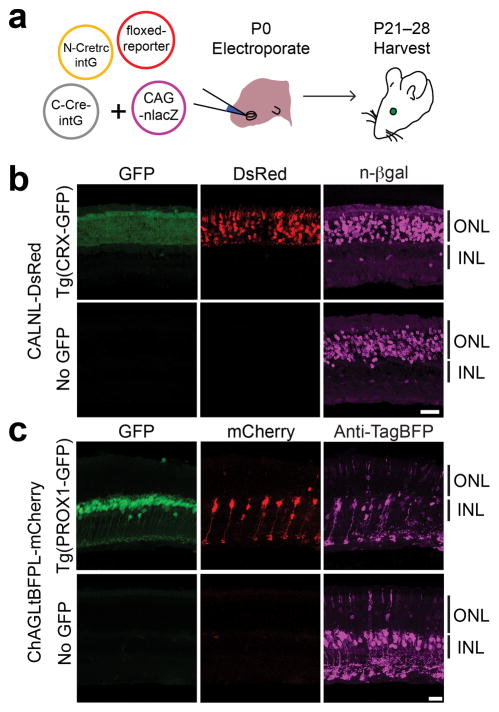
We next tested whether CRE-DOG could retrofit existing transgenic GFP reporter lines for cell type-specific manipulations. The Tg(CRX-GFP) line expresses GFP strongly in photoreceptors and weakly in inner nuclear layer (INL) cells23. Tg(CRX-GFP) retinas electroporated with CRE-DOGOPT and CALNL-DsRed plasmids showed strong DsRed labeling of photoreceptors and occasional labeling of INL cells, whereas electroporated retinas that were GFP-negative showed little to no DsRed expression (Figs. 2a,b, Supplementary Fig. 6a-c). Additionally, 100 +/− 0 % (hereafter mean +/− s.d) of DsRed cells labeled in the outer nuclear layer (ONL) were GFP+ (Supplementary Fig. 6g). CRE-DOGOPT induced DsRed expression in 76 +/− 4% of electroporated cells in the ONL (Supplementary Fig. 6a). This is an estimate of CRE-DOGOPT efficiency in vivo as all electroporated cells in the ONL were GFP+. The value is likely an underestimate of efficiency, as it has not been corrected for the percentage of cells that were co-electroporated with all constructs.
Figure 2. CRE-DOGOPT can be delivered to the mouse retina for retrofitting transgenic GFP lines.
(a–c) Electroporation of Tg(CRX-GFP) (a) and Tg(PROX1-GFP) (b) with plasmids encoding CRE-DOGOPT and the floxed reporter CALNL-DsRed (b) or ChAG-LtBFPL-mCherry (c). CAG-nlacZ, which expresses n-βgal, was added as an electroporation marker in (b). (a) Schematic of electroporation experiment. (b–c) CRE-DOGOPT induced floxed reporter expression in Tg(CRX-GFP) (b) and Tg(PROX1-GFP) (c) retinas, but not in GFP-negative retinas (b–c). Scale bar, 20 μm. Images representative of 3 or 4 retinas per condition. ONL, outer nuclear layer. INL, inner nuclear layer.
As a considerable retinal diversity lies within the bipolar cell class, we tested whether CRE-DOGOPT would be effective in this cell class. We used a bipolar-selective promoter (ChAG) to drive strong expression of CRE-DOGOPT components in various bipolar cell types of the Tg(PROX1-GFP) line, and assayed Cre activity with a ChAG-loxP-TagBFP-loxP-mCherry reporter (ChAG-LtBFPL-mCherry). In contrast to Tg(CRX-GFP), electroporated Tg(PROX1-GFP) retinas showed strong mCherry labeling of bipolar cells, as well as sparse labeling of photoreceptors, and rare labeling of Müller glia and amacrine cells (Fig. 2c). Although only 83 +/− 10% of mCherry+ cells were GFP+ (Supplementary Fig. 6g), analysis of electroporated GFP-negative retinas indicated that the system was absolutely GFP-dependent (Fig 2c and Supplementary Fig. 6d,e). Thus, developmental expression of Tg(PROX1-GFP) in progenitor cells giving rise to multiple cell classes, or weak/transient GFP expression, may contribute to the labeling pattern. These results establish the utility of CRE-DOG for genetic manipulation of specific cell types in transgenic GFP animals.
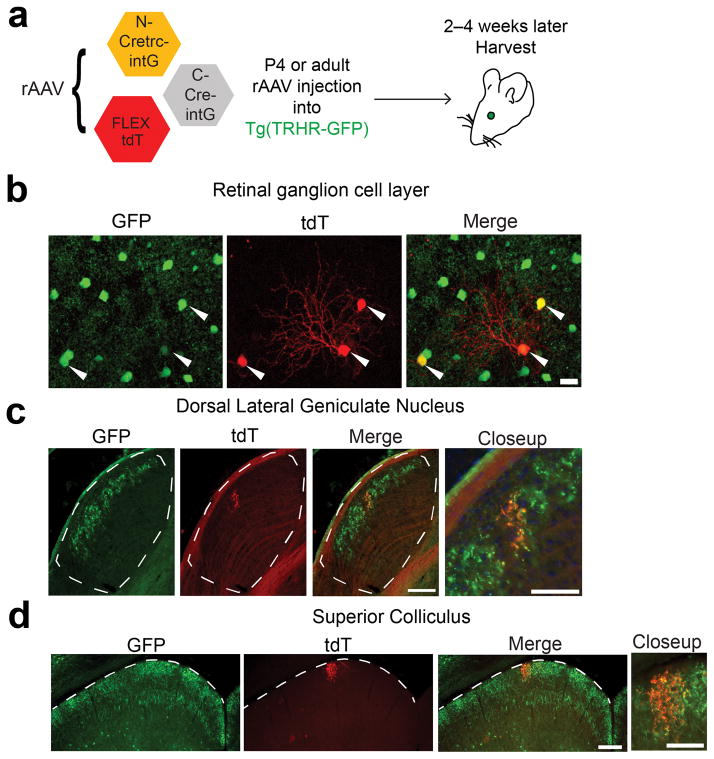
rAAV are widely-used vectors for delivering transgenes to the mammalian nervous system, due to their high infectivity of postmitotic neurons and low toxicity24. The ability to deliver rAAV-encoded CRE-DOGOPT along with rAAV carrying the tight Cre-responsive FLEX switch13 in a single injection mixture would provide a fast and versatile approach for manipulating genes in GFP-labeled cell types across wide areas of the nervous system and across a broad age window. Indeed, rAAV-encoding CRE-DOGOPT could be used to turn on rAAV-FLEX-tdTomato (tdT) in a GFP-dependent manner in vivo (Supplementary Fig. 7a). To test this approach in transgenic GFP lines, we co-injected rAAV-CRE-DOGOPT and rAAV-FLEX-tdT into retinas of Tg(TRHR-GFP)25 and Tg(CDH3-GFP)26 lines, both of which express GFP in specific retinal ganglion cell (RGC) subtypes. Functional studies of RGCs have been hampered by the scarcity of Cre driver lines that are selectively active in the same RGC subtypes seen in multiple GFP reporter lines. Single injection of the aforementioned rAAV mixture into GFP-negative retinas resulted in no detectable tdT expression in Cre immunopositive cells of the ganglion cell layer (GCL) (0 tdT+ cells in 140 anti-Cre+ cells, sampled across 3 retinas; Supplementary Fig. 7b). In contrast, we found highly selective tdT expression in GFP+ RGCs, with 89 +/− 7% of tdT+ cells being TRHR-GFP+ (207 of 243 tdT+ cells, 5 retinas) and 85% of tdT+ cells being CDH3-GFP+ (11 of 13 tdT+ cells, 2 retinas) (Fig. 3a,b) (data not shown for Tg(CDH3-GFP)). The sparser number, as well as nonuniform spatial distribution of CDH3-GFP+ RGCs, may possibly contribute to the low number of cells labeled by tdT27. Importantly, strong tdT expression enabled tracing of labeled RGC projections into various brain targets; the resultant patterns were consistent with the identity of GFP+ RGCs previously characterized for Tg(TRHR-GFP) 25 (Fig. 3c,d). Taking the rAAV and electroporation results together, we have shown that one can use CRE-DOGOPT to selectively target GFP cells residing in all three major cellular layers of a neural tissue, the retina.
Figure 3. rAAV delivery of CRE-DOGOPT to retinas of a GFP line.
(a) Schematic of viral injection into the retina. (b) Expression of tdT in GFP+ retinal ganglion cells (RGCs). Arrows indicate tdT-positive cells. Scale bar, 20 μm. Images representative of 4 retinas. (c–d) Tracing of tdT+ RGC projections into the dorsal lateral geniculate nucleus (c) and superior colliculus (d). White dotted lines delineate boundary of the aforementioned visual pathway region. tdT fluorescence was boosted by immunostaining. Scale bar, 100 μm in whole region view, 50 μm in closeup view. Images representative of 3 brains.
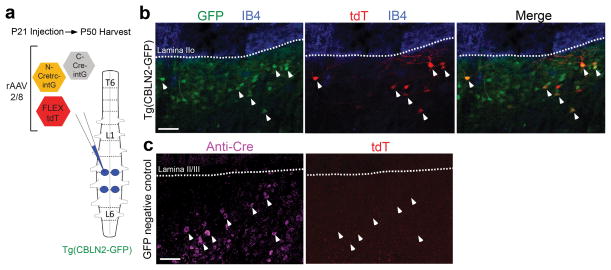
To evaluate whether CRE-DOG is useful outside the retina, we co-injected rAAV-CRE-DOGOPT and rAAV-FLEX-tdT into additional neural tissues. We targeted GFP cells of the spinal cord in the Tg(CBLN2-GFP) line4, which expresses GFP in a subset of interneurons in the dorsal horn (unpublished data, VEA) (Fig. 4a). While rAAV infection of wildtype spinal cords did not result in detectable tdT expression (0 tdT+ out of 108 anti-Cre+ cells, sampled from 2 spinal cords), infection of Tg(CBLN2-GFP) spinal cords led to the appearance of tdT+ cells, some of which expressed tdT at levels strong enough for process visualization (Fig. 4b–c). 75% of tdT+ cells were detected to be GFP+ (n=105 tdT+ cells, sampled from 2 spinal cords). Thus, the labeling pattern may include cells that had transient and/or low levels of GFP, below the detection threshold. To estimate the efficiency of rAAV-CRE-DOGOPT gene manipulation protocol in the spinal cord, we quantified the number of tdT+ cells among the GFP+ cells that also expressed split-Cre fragments, as detected by a polyclonal Cre antibody. Using this approach, we estimate the efficiency to be 51% (n=37 Cre+, GFP+ cells, sampled from 2 spinal cords).
Figure 4. rAAV delivery of CRE-DOGOPT to the lumbar spinal cord of a GFP line labeling dorsal horn interneurons.
(a) Schematic of viral injection into the spinal cord at lumbar level L3–5. Viruses were injected bilaterally into the spinal cord. (b,c) Sagittal lumbar sections of Tg(CBLN2-GFP) (b) and GFP-negative littermate (c) mice. (b) shows GFP antibody-boosted fluorescence (green, left panel), tdT (red, middle panel) and merged image (right panel). IB4 stain (blue) is in all panels and delineates lamina IIo. Arrowheads show obvious examples of GFP and tdT overlap. (c) shows Cre immunostaining (magenta, left panel) and tdT (right panel). DAPI (not shown) was used to delineate lamina II/III. Arrowheads show no tdT induction in Cre+ cells of GFP-negative animals. Scale bar: 100 μm. Images representative of 2 spinal cords per condition.
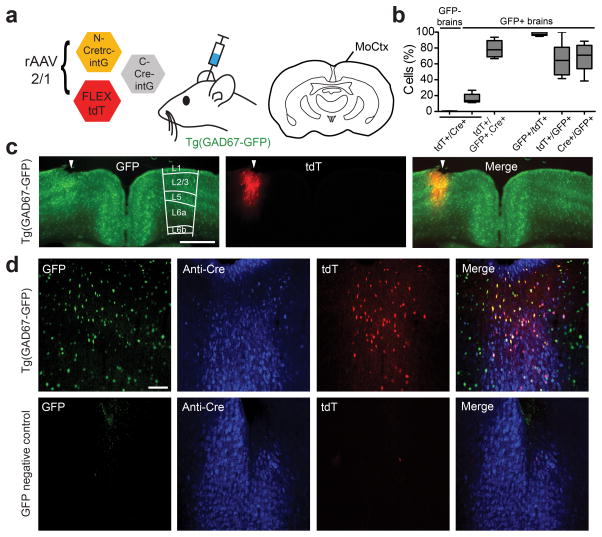
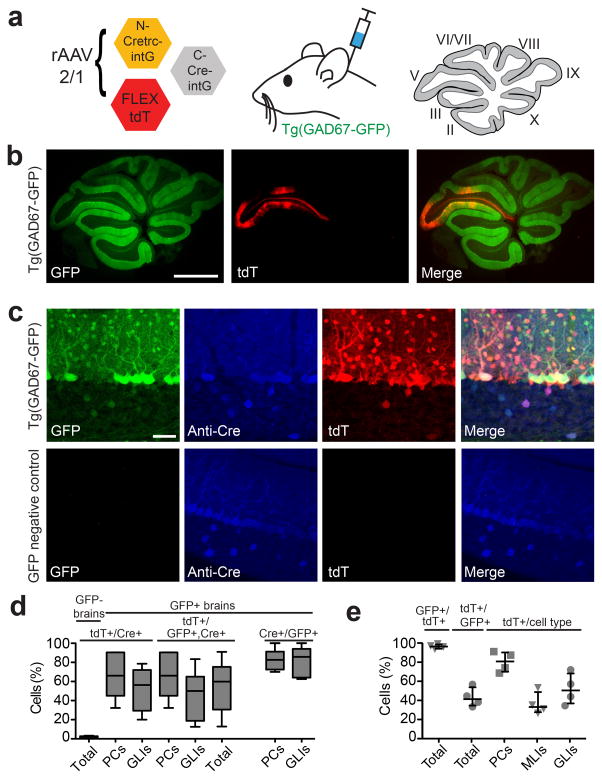
We also tested for CRE-DOGOPT activity in the brain. The Tg(GAD67-GFP) line expresses GFP in GABAergic cell types in multiple brain regions28. We injected the rAAV mixture encoding CRE-DOGOPT and FLEX-tdT into the motor cortex as well as into the cerebellum of Tg(GAD67-GFP) animals (Fig 5 and 6, Supplementary Table 2). Cortical infection resulted in GFP-dependent induction of tdT expression throughout all cortical layers of Tg(GAD67-GFP) brains (Fig. 5b–d). A high percentage of tdT+ cells was GFP+, at 98 +/− 2 % (Fig.5b). In addition, 78 +/− 10% of GFP+ and anti-Cre+ cells were also tdT+, providing an estimate of efficiency of the rAAV-CRE-DOGOPT injection protocol in the brain (Fig 5b). Remarkably, despite efficient infection of cortical pyramidal neurons as evidenced by Cre immunostaining, no pyramidal neurons and rarely any other cell type were tdT+ in GFP-negative mice (8 tdT+ cells in 2560 anti-Cre+ cells, 10 sections, 2 GFP-negative brains; Fig. 5d and Supplementary Table 2). Injection of the rAAV-CRE-DOGOPT and FLEXT-tdT mixture into Tg(GAD67-GFP) cerebella resulted in strong GFP-dependent labeling of multiple GABAergic cerebellar cell types, including Purkinje cells (PCs), molecular layer interneurons (MLIs) and granule layer interneurons (GLIs), as confirmed by morphology and immunostaining for cell type markers (Fig. 6, Supplementary Figs.8 and 9, Supplementary Table 2). Anti-Cre staining demonstrated little to no CRE-DOGOPT activity in GFP-negative cerebella infected with the rAAV mixture (Fig. 6c–d, Supplementary Figs.8 and 9e–g, Supplementary Table 2). In infected Tg(GAD-GFP) cerebella, we found that 96 +/− 2 % of tdT+ cells were GFP+, and in areas of dense infection, 42 +/− 10 % of all GFP+ cells were tdT+ (Fig. 6e). To more accurately evaluate the efficiency of the CRE-DOGOPT system, we quantified the percentage of tdT+ cells that were both anti-Cre+ and GFP+, which was 55 +/− 26 % and ranged between 44–67% depending upon cell type (Fig. 6d, Supplementary Table 2).
Figure 5. rAAV delivery of CRE-DOGOPT to the motor cortex of GFP line.
(a) Left: Schematic of viral injection into mouse motor cortex. rAAV-2/1 viruses were injected into the cortex at 4 weeks of age and brains were harvested 3 weeks post-injection. Right: coronal section of the mouse brain. (b) Quantifications including specificity and efficiency of CRE-DOGOPT labeling in cortical interneurons. (c) Coronal cortical section of Tg(GAD67-GFP) mouse infected with rAAV, showing GFP (green) and tdT (red) expression. Arrowheads indicate injection site. Cortical layers are indicated in GFP+ section. Scale bar, 1 mm. (d) CRE-DOGOPT induces tdT expression in Tg(GAD67-GFP) (top), but not in GFP-negative brains (bottom). Panels show GFP (green), Cre immunostaining (blue), tdT (red) and merged images of all three channels (right panels). Scale bar, 100 μm. Images in (c) and (d) are representative of 3 Tg(GAD67-GFP) and 2 GFP-negative brains. Plots show percentage of cells positive for a marker, given the presence of additional marker(s) (1st marker/2nd, 3rd marker). For GFP+/tdT+ and tdT+/GFP+ parameters, sample size are 735 GFP+ cells, 475 tdT+ cells, 10 sections, 3 Tg(GAD67-GFP) animals), as well as quantification of Cre+ cells that express tdT, GFP, or both. Sample size: 371 GFP+ cells, 203 tdT+ and 1238 Cre+ cells in 9 GFP+ sections, 3 Tg(GAD67-GFP)+ animals). 2560 Cre+ cells and 10 tdT+ cells were counted in 2 GFP-negative animals. Boxplots indicate minimum to maximum range.
Figure 6. rAAV delivery of CRE-DOGOPT to the cerebellum of a GFP line.
(a) Left: Schematic of viral injection into mouse cerebellum. rAAV-2/1 viruses were injected into lobule V and brains were harvested 3 weeks post-injection. Right: sagittal section of the cerebellar vermis with lobules labeled. (b) Sagittal section of Tg(GAD67-GFP)+ cerebellum (green) showing tdT (red) induction in injected lobule. Scale bar, 1 mm. (c) CRE-DOGOPT induces tdT expression in Tg(GAD67-GFP) (top), but not GFP-negative (bottom) cerebellar cortex. Panels show GFP (green), anti-Cre immunostaining (blue) and tdT (red) expression. Scale bar, 50 μm. (d–e) Quantification of infection outcome. Plots show percentage of cells positive for a marker, given the presence of additional marker(s) (1st marker/2nd, 3rd marker). (d) Quantification of tdT+ cells that are also GFP+ and/or Cre+. Sample size: 516 GFP+, 422 Cre+ and 317 tdT+ cells in 7 sections. (e) Additional quantification of CRE-DOG labeling. Sample size: 845 GFP+ and 368 tdT+ cells in 12 GFP+ sections. 643 Cre+ cells and 6 tdT+ cells were counted in 4 Gad– sections. PCs, Purkinje cells. MLIs, molecular layer interneurons. GLIs, granule layer interneurons. Boxplots indicate minimum to maximum range. All data and images reported are gathered from or representative of 4 Tg(GAD-GFP)+ animals and 3 GFP-negative animals.
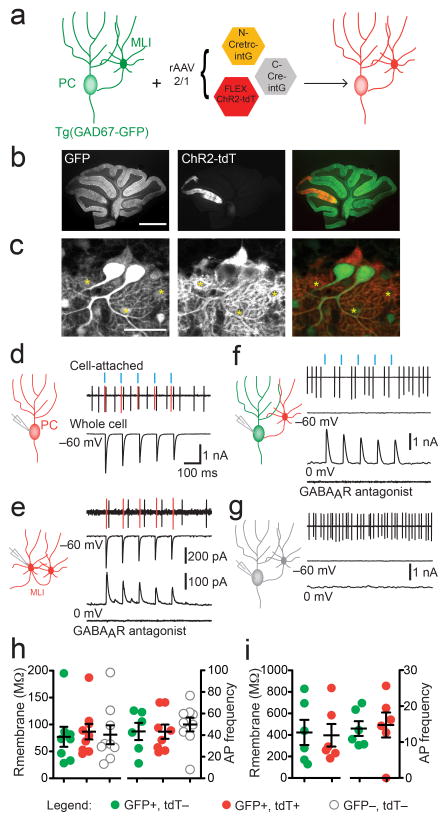
To evaluate the utility of CRE-DOGOPT for functional studies, we co-infected Tg(GAD67-GFP) with rAAV-CRE-DOGOPT and rAAV-FLEX-ChR2-tdT for optogenetic manipulations in GFP+ cells (Fig 7a–c). We were able to directly trigger light-evoked action potentials and photocurrents in ChR2-tdT-labeled PCs (n=9) and MLIs (n=6) (Fig. 7d,e). We also were able to optogenetically inhibit spontaneous activity in ChR2-tdT-negative PCs that received GABAergic synaptic input from ChR2-tdT+ MLIs (n=4) (Fig. 7f). GFP+ PCs in uninfected areas, as well as PCs in infected areas of GFP-negative littermates, did not show any light-induced photocurrents or light-triggered synaptic currents (n=9 for GFP-negative animal, n=8 in uninfected area) (Fig. 7g). Finally, infected PCs and MLIs did not differ from uninfected cells in intrinsic electrical properties, assessed by membrane resistance and spontaneous action potential frequency (p=0.92 and 0.73, respectively, One-way ANOVA for PCs, and p=0.87 and p=0.81, respectively, t-test, for MLIs) (Fig. 7h,i). Together, these results demonstrate the utility of CRE-DOG for cell-specific manipulation of neural circuit activity in the brain.
Figure 7. CRE-DOGOPT allows optogenetic manipulation of GFP-labeled neurons.
(a) Schematic of experiment. PC, Purkinje cell. MLI, molecular layer interneuron. (b–c) Expression of ChR2-tdT in lobule V/VI of parasagittal cerebellar slice(b) and at cellular level (c), in PCs and MLIs (marked by asterisk). Scale bar, 1mm in (b), 50 μm in (c). Note that ChR2-tdT labels membranes. (d–g) Optogenetic control of GFP-labeled cells within the cerebellar circuit. Cell-attached and whole cell recordings are aligned with blue light stimulation (blue vertical bars at top). (d) Light-evoked spikes (marked in red) in addition to spontaneous action potentials (black) in cell-attached recording, and underlying photocurrent in a whole-cell recording of the same cell (bottom trace) in ChR2-tdT+ PC (representative of n=9). Cells were voltage-clamped at the indicated potentials. (e) ChR2-tdT+ MLI shows light-evoked spiking (top trace, red), photocurrent (middle trace) and inhibitory current (bottom trace) from stimulation of a synaptically connected MLI. The GABAA receptor antagonist SR95531 (5 μM) blocked inhibitory current (representative of n=5) (f) Example of GFP+/ tdT– PC (green) receiving inhibitory input from ChR2-tdT+ MLI. Activation of the ChR2-tdT+ MLI briefly inhibits PC spiking in cell-attached recording (top trace), bottom traces: light-evoked inhibitory current blocked by SR95531 (representative of n=4). (g) GFP–, tdT– cells (gray) do not respond to light stimulation (n=9). (h–i) Membrane resistance and spontaneous action potential (AP) frequency of tdT+ (also GFP+; n=8), tdT– (also GFP+; n=6) or GFP– (also tdT–; n=9) PCs are not different across conditions (p= 0.92 p=0.73, respectively, ANOVA) (h) and MLIs (n=6 each, p=0.87 and p=0.81, respectively) (i). PC data were obtained from 4 GFP+ animals injected with CRE-DOG, and 1 injected GFP- animal. MLI data were obtained from 2 injected GFP+ animals. Scatter plots are mean +/− s.e.m.
Discussion
Here, we developed a high signal-to-noise system, CRE-DOGOPT, that provides a reliable and rapid means of genetically manipulating GFP-expressing cells in the many GFP lines available for life science research. We demonstrate the repurposing of GFP reporter lines for cell-specific gene manipulation via electroporation and/or viral infection. Given the widespread use of rAAVs among neuroscientists, rAAV-encoded CRE-DOGOPT should immediately be useful for many neuroscience applications. rAAV allows delivery over a broader time window and into more tissues than electroporation, the method originally used to deliver T-DDOG. It should be straightforward to apply rAAV-encoded CRE-DOGOPT in a single injection mixture with rAAV-encoded Cre-dependent transgenes, or into transgenic GFP animals with Cre-responsive alleles. Further refinement in cell targeting should be possible by intersecting GFP-dependent CRE-DOGOPT activity with other recombinases in intersectional strategies14, 16
The CRE-DOGOPT-encoding plasmids described here also will be immediately useful for targeting GFP cells for gene manipulation via electroporation. The electroporation approach holds an advantage over rAAV delivery in that it is not limited by virus packaging size. This can be exploited in several ways, including the delivery of larger Cre-dependent transgenes and/or larger cis-regulatory elements controlling expression pattern of CRE-DOGOPT and/or Cre-dependent transgenes.
We tested CRE-DOGOPT in multiple neural tissues in vivo and found that its activity was highly dependent on GFP expression. Importantly, the engineered split Cre components retain the ability to induce recombination of responder cassettes in the popular “floxed” and “FLEXed” configurations. In multiple tests described here, CRE-DOGOPT was capable of inducing strong fluorescent reporter protein expression, allowing for visualization of fine neuronal processes for morphological and axonal tracing analysis either with unstained or immunostained tissues (Fig. 2–7 and Supplementary figures 7 and 9). Optogenetic control of specific cell types is a powerful means of dissecting the function of cells within neural circuits29. We showed that CRE-DOGOPT could convert GFP expression into ChR2 expression for cell type-specific and temporally precise manipulation of neural activity (Fig. 7). Beyond light-activated ion channels, many other genetically encodable tools have been developed to uncover the anatomical and physiological features of neurons29–33. We anticipate CRE-DOGOPT to enable activation of any one, or a combination of these readily available tools in any cell type labeled with GFP.
As with any exogenous construct, the efficiency of CRE-DOGOPT depends upon the effectiveness of gene delivery. We found CRE-DOGOPT to be active in ~44–78% of cells receiving electroporated plasmids or virally delivered CRE-DOGOPT components in vivo. These numbers are likely underestimates of the true efficiency, as we could not completely focus our analysis on cells that received the complete set of CRE-DOGOPT components. For example, rAAV infected cells with only one Cre-fusion protein, or cells with both CRE-DOGOPT components but not FLEX-tdT, would be included in the population of tdT-negative cells. To increase efficiency, it may be beneficial to encode both CRE-DOGOPT components in a single plasmid or rAAV vector, perhaps by linking the two components in a bicistronic cassette. In addition, transgenic lines expressing the two components in a bicistronic cassette would provide activation in a large number of GFP-labeled cells through the use of genetic crosses. Another consideration when using rAAV is the serotype and the promoter used to drive each CRE-DOGOPT component, as different serotypes and promoters may promote differential expression patterns. Such differences are not necessarily a problem. Instead, they may be exploited for intersectional targeting of CRE-DOGOPT activity, for precise cell-specific manipulations16. For example, two different promoters may be used to express each of the CRE-DOGOPT components, resulting in Cre activity in a subset of GFP-expressing cells. Alternatively, it may be possible to intersect the spatial pattern of CRE-DOGOPT activity, as dictated by GFP expression, with the expression pattern of other site-specific recombinases and/or transcription factors, using dual or multiple input-responsive cassettes14, 16, 34.
Both CRE-DOGOPT or T-DDOG may now be used for manipulation of GFP-expressing cells. Several considerations influence the choice of one versus the other. First, over-expression of transcription activation domains may cause undesirable side effects. We found this to be the case for T-DDOG variants using particularly potent activation domains, and overcame the issue by changing the activation domain7. Unlike transcription factors, Cre recombinase does not depend on interactions with endogenous cellular machinery and may be expected to induce fewer complications. Indeed, we did not detect any abnormalities in cells overexpressing CRE-DOGOPT. This is supported by physiology tests on ChR2-expressing cerebellar cells that failed to detect any deleterious effects (Fig. 7). However, we did encounter cytotoxicity when delivering the components to the cerebellum via serotype 8 capsid, but not serotype 1 capsid (data not shown). This is thus a serotype-specific issue and not related to CRE-DOGOPT expression. Serotype-specific cytotoxicity has been reported previously and is important to consider when designing experiments35. Nevertheless, high levels of native Cre have been reported to be toxic in some settings36 Second, T-DDOG may be preferred when temporal control is desired, as T-DDOG can be modified to respond to drug application. It is also possible to modify CRE-DOG to confer drug inducibility, by the use of domains such as ERT2, or destabilizing domains37. However, this would only enable temporal control at the onset of expression, since Cre-mediated recombination is essentially irreversible.
CRE-DOGOPT, as with T-DDOG, is sensitive to high GFP levels in that GFP in excess of the split components can titrate out each component, thereby reducing activity. Nonetheless, empirically we found Cre-dependent recombination to be fairly efficient, even in lines with strong levels of GFP expression (Fig. 2, 5, 6). Also, CRE-DOGOPT was able to detect low GFP levels as demonstrated by our findings with the relatively weakly GFP-expressing RGC lines, which require GFP immunostaining to reveal neuronal processes (Fig. 3). In fact, we failed to detect GFP in some cells with evidence of recombinase activity, presumably because activity was triggered by GFP expression below the threshold of detection, or from transient GFP expression (Fig. 2, 4). This does point out that the proper control for CRE-DOGOPT is a GFP-negative animal; we detected almost no CRE-DOGOPT activity in the GFP-negative tissues examined here.
The irreversibility of Cre recombination can be exploited to trigger stable expression of a transgene, but may also lead to accumulation of background, even if there is only low GFP-independent recombinase activity. We did not find background activity to be an issue for the incubation period of 2–4 weeks used here. However, beyond this incubation period, we did observe increased background in rAAV-CRE-DOGOPT injected GFP-negative animals (data not shown). The accumulation of background may be reduced by optimizing plasmid dose or viral titer, and/or by using tightly drug-regulated CRE-DOGOPT variants that await development. Another contributor to variable background activity may be the differential sensitivity of Cre-responsive gene cassettes; in our experience, FLEX constructs showed less background recombination than loxP-STOP-loxP alleles.
Although split intein elements were found to promote GFP-specificity of CRE-DOG, protein splicing did not seem to be involved. It is unclear why the inclusion of intein elements reduced background activity of CRE-DOG, but one could speculate that the extra elements influenced the orientation or conformation of the split protein fragments such that non-specific CRE-DOG assembly was inhibited in the absence of GFP. Whether split intein elements would also aid in reducing background activity of other ligand-inducible split protein systems is an interesting issue to consider in future studies.
From a broader perspective, our work demonstrates the utility of artificially derived-binding proteins (ex. nanobodies, intrabodies, etc.) for constructing split enzyme systems inducible by intracellular proteins. The ability to rapidly isolate additional high affinity nanobodies should facilitate the extension of this approach to other target proteins38. Our work thus offers a novel approach for engineering protein-inducible systems in addition to providing a practical solution for retrofitting GFP reporter lines for a much broader set of applications.
Online Methods
Animals
All animal experiments performed were approved by the Institutional Animal Care and Use Committee at Harvard University and University of San Diego. All animals studied were Mus musculus. Time pregnant CD1 mice (Charles River Breeding Laboratories, Boston, MA) were used for electroporation experiments. Transgenic GFP mouse lines used were Tg(CRX-GFP)23, Tg(PROX1-GFP) (gensat.org), Tg(TRHR-GFP)25, Tg(CDH3-GFP)26, Tg(CBLN2-GFP) (gensat.org), Tg(GAD67-GFP)28. Strains were kept in C57BL/6 background except Tg(CDH3-GFP), which is in FVB/N and CD1 mixed background.
Molecular Biology Reagents
Ancillary plasmids
pCAG-GFP (Addgene plasmid 11150) 39, pCAG-YFP (Addgene plasmid 11180) 39, pCAG-CFP (Addgene plasmid 11179) 39, pCAG-tdT (Cepko lab, Harvard Medical School), pCAG-mCherry (Cepko lab, Harvard Medical School), pCAG-DsRed (Addgene plasmid 11151) 39, pRL-TK (Promega, #E2241), pRho-GFP-IRES-AP (referred to as Rho-GFP in main text) 40, pCAG-nlacZ (Cepko lab, Harvard Medical School). pAAV-CAG-FLEX-tdT was a gift from Edward Boyden (Addgene plasmid #28306), pCALNL-DsRed (Addgene plasmid #13769) 41.
CRE-DOG chimeric component constructs
Using standard cloning techniques, coding sequences of GBP1, 2, 6 and 7, the sources of which were described in Tang et al 20137, were fused to those of split Cre and split VMA intein fragments in many possible configurations, and the fusion products were inserted into a pCAG vector42. Split Cre fragments were taken from a codon-optimized Cre coding sequence called improved Cre (iCre)43. Two previously described pairs of complementary split Cre fragments were tested; they are 1.) N-split Cre corresponding to residue 19–59 of full-length Cre (iCre19–59), paired with C-split Cre corresponding to residue 60–343 of full-length Cre (iCre60–343). 2.) N-split Cre corresponding to residue 19–104 of full-length Cre, paired with C-split Cre corresponding to residue 106–343 of full-length Cre17. Split VMA intein fragments, termed N-terminal VMA (N-VMA or NVMA) and C-terminal VMA (C-VMA or CVMA) here, were described previously19 and were codon-optimized for mammalian expression by Genewiz (New Jersey).
Notable CRE-DOG fusion plasmids
pCAG-N-CreintG
An AgeI-Kozak-NLS-iCre19–59-NVMA-NheI restriction digested fragment was inserted into the backbone of AgeI/NheI digested pCAG-Gal4DBD-10gly-GBP7 plasmid7, resulting in in-frame fusion to the 10gly-GBP7 coding sequence and replacing Gal4DBD in the original vector.
pCAG-C-CreintG
An NheI/NotI flanked and restriction digested fragment containing CVMA-iCre60–143 (C-CreintG) was inserted into the backbone of NheI/NotI digested pCAG-GBP2-10gly-VPminx3 plasmid7, resulting in in-frame fusion to GBP2 coding sequence and replacing 10gly-VPminx3 in the original vector.
pCAG-N-CretrcintG
An AgeI/NheI flanked and restriction digested fragment containing fusion of NLS, iCre19–59 and the last 129 bp of NVMA (N-CretrcintG) was inserted into the backbone of AgeI/NheI digested pCAG-N-CreintG plasmid, resulting in in-frame fusion to the 10gly-GBP7 coding sequence and replacing NLS-iCre19–59-NVMA in the original vector.
pAAV-EF1a-N-CretrcintG
A BamHI/EcoRI flanked and restriction digested fragment containing Kozak-N-CretrcintG was inserted into the backbone of BamHI/EcoRI digested pAAV-EF1a-FLEX-GTB (Addgene plasmid #26197) 44.
pAAV-EF1a-C-CreintG
A BamHI/EcoRI flanked and restriction digested fragment containing Kozak-C-CreintG was inserted into the backbone of BamHI/EcoRI digested pAAV-EF1a-FLEX-GTB (Addgene plasmid #26197) 44.
pCALNL-luc2
An EcoRI/NotI flanked and restriction digested fragment containing Kozak-luc2 was inserted into the backbone of EcoRI/NotI digested pCALNL-DsRed (Addgene plasmid #13769) 41, replacing DsRed in the original vector.
pAAV-EF1a-GFP
A BamHI/EcoRI flanked and restriction digested fragment containing Kozak-GFP was inserted into the backbone of BamHI/EcoRI digested pAAV-EF1a-FLEX-GTB (Addgene plasmid #26197) 44.
pAAV-EF1a-ZsGreen
A BamHI/EcoRI flanked and restriction digested fragment containing Kozak-ZsGreen was inserted into the backbone of BamHI/EcoRI digested pAAV-EF1a-FLEX-GTB (Addgene plasmid #26197) 44.
pChAG-loxP-TagBFP-loxP-mCherry
First, pChAG was generated. pCAG GFP was digested with SpeI and SnaB1 to excise the CMV enhancer and a ~700 bp Chx10 enhancer element specific for bipolar cells40 was PCR amplified with flanking sequences corresponding to the pCAG backbone. PCR products were cloned in by Gibson assembly. To generate pChAG-loxP-TagBFP-loxP, pChAG was digested with AgeI and NotI. TagBFP was PCR amplified using primers that had overlapping sequence with the digested vector, and Gibson assembly was used. To generate pChAG-loxP-TagBFP-loxP-mCherry, pChAG-loxP-TagBFP-loxP was linearized by NotI digestion. Digested vector and amplified mCherry PCR product were gel purified and assembled by Gibson ligation.
pChAG-N-CretrcintG
An AgeI/NotI flanked and restriction digested N-CretrcintG fragment was inserted into a pChAG vector via AgeI/NotI sites.
pChAG-C-CreintG
An AgeI/NotI flanked and restriction digested C-CreintG fragment was inserted into a pChAG vector via AgeI/NotI sites.
CRE-DOG screens
We tested two combinations of N- and C-terminal split Cre fragments previously reported to be suitable for Rapamycin-induced dimerization17. A split Cre pair comprising of residues 19–59 and residues 60–343 of full-length Cre, were each fused with the split intein portions from yeast VMA19 in specific orientations. These original constructs were found to give strong GFP-dependent recombinase activity coupled with low background activity, giving CRE-DOGOG. This led to additional screens optimizing for CRE-DOG activity, eventually giving CRE-DOGOPT. All screens were assayed with the CALNL-luc2 reporter in 293T cells. CRE-DOG variant components were expressed using the CAG promoter in pCAG. Transfected DNA of the CRE-DOG components were adjusted with filler DNA (pCAGEN) to ensure comparable molar DNA delivery. To assay for GFP-inducibility, pairwise combinations of CRE-DOG component variants were transfected into cells with pCALNL-luc2, pRL-TK, and either pCAG-GFP or an empty CAG vector (pCAGEN). Cells were assayed for luciferase activity at 1 day post-transfection.
In vitro luciferase assays
Plasmids encoding CAG-driven GFP, and N- and C-terminal split Cre chimeric variants were transfected via polyethyleneimine (PEI) into 293T cells along with plasmids encoding CALNL-luc2 and pRL-TK. Between 50–70 ng of total DNA were transfected into single wells of 48 well plates. Cells were ~80–100% confluent at time of transfection. Cells were harvested 24 hours later for Dual Luciferase Assay (Promega). All transfections, except for the dosage curve, were done at a 1:1:1 (GFP:N-CretrcintG:C-CreintG) plasmid molar ratio. Both technical (pipetting of lysate into at least 2–3 different luciferase assay wells) and biological (at least 2–3 different transfection mixes applied to different cell culture wells) replicates were included in each experiment. All experimental results were verified in at least three independent experiments.
Western Blot
293T cells were seeded onto 6 well plates and transfected with CAG-driven N-CreintG and C-CreintG-FLAG, along with CAG-GFP or empty CAG vector. A CALNL-DsRed reporter was included in some cases to confirm GFP-dependent transfection. 24 hours post-transfection, transfected 293T cells were lysed in RIPA buffer (150 mM NaCl, 1.0% NP40, 0.5% Na Deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) for lysate supernatant load or 4x Laemmeli buffer plus β-mercaptoethanol. Samples were run on SDS PAGE and then subjected to Western blot analysis. Anti-FLAG (F3165, Sigma) was used at 1:1000 dilution.
In vivo electroporations
For wildtype, Tg(CRX-GFP) and GFP-negative littermate retina electroporations, postnatal day 0 to 3 (P0–P3) mouse pups were electroporated in vivo as described previously7. DNA solutions (1–1.5 μg/μl) were injected through the sclera and into the subretinal space of the mouse retina. pCAGEN was used as an empty vector substitute for excluded plasmids. P0 CD1 retinas were electroporated with plasmids encoding CAG-driven CRE-DOG components and CAG-driven nuclear β-galactosidase (n-βgal, an electroporation marker expressed from CAG-nlacZ plasmid) along with CALNL-DsRed and different promoter-GFP constructs. Electroporated retinas were harvested at P14, immunostained for n-βgal and then imaged on a Zeiss LSM780 confocal microscope. For Tg(PROX1-GFP) retina electroporations, P2–3 Tg(PROX1-GFP) or GFP-negative littermate retinas were blindly electroporated with plasmids encoding ChAG-driven CRE-DOG components and ChAG-loxP-TagBFP-loxP-mCherry; retinas were harvested between 3–4 weeks of age. Wherever applicable, retinas were immunostained with anti-GFP and anti-β-galactosidase.
rAAV production and injections
rAAV (serotype 2/8 or 2/1) were made from pAAV-EF1a-N-CretrcintG, pAAV-EF1a-C-CreintG, pAAV-FLEX-tdT, pAAV-EF1a-GFP and pAAV-EF1a-ZsGreen. All rAAV, with the exception of rAAV-2/1-CAG-FLEX-ChR2-tdT (UPenn vector core, aliquot generously provided by Bernardo Sabatini lab) were generated by Chen Wang of Zhigang He’s lab (Boston Children’s Hospital). All rAAV were injected in the range between 1014–1015 genome copies/ml, assayed by PCR of rAAV vectors. Primer sequences are GACCTTTGGTCGCCCGGCCT and GAGTTGGCCACTCCCTCTCTGC, which target the ITR region of rAAV vectors.
For Supplementary Figure 9, rAAV-2/8-EF1a-N-CretrcintG, rAAV-2/8-EF1a-C-CreintG and rAAV-2/8-FLEXtdT were co-injected with rAAV-2/8-EF1a-GFP or rAAV2/8-EF1a-ZsGreen into P0 mouse retina. Injected retinas were harvested in adulthood, between P21–30.
For Tg(TRHR-GFP) and Tg(CDH3-GFP) rAAV experiments, retinas were injected with rAAV-2/8 or rAAV-2/1 individually encoding N-CretrcintG, C-CreintG and FLEX-tdT into P4 or 2–3 month old animals. 2–4 weeks post-infection, retina and brain sections were processed as described previously45–47. A brief description is provided in the histology section.
For rAAV infection of the spinal cord, P21 Tg(CBLN2-GFP) mice and GFP-negative littermates of either sex aged were anesthetized with a continuous level of 2% isoflurane. A total volume of 400 nl of the following viral constructs: rAAV2/1-EF1a-N-CretrcintG, rAAV2/1-EF1a-C-CreintG, rAAV2/1-FLEX-tdT was injected over four injection sites between vertebrae L3–L5. 4 weeks later, lumbar spinal cord tissue was collected for immunohistochemistry.
For rAAV infection of cerebellum and the cortex, Tg(GAD67-GFP)+ mice and GFP-negative littermates of either sex aged 3–4 weeks were anesthetized with ketamine/xylazine/acepromazine at 100, 2.5 and 3 mg/kg, respectively, and a continuous level of deep anesthesia was maintained with 5% isoflurane. A total volume of 200 nl of the following viral constructs: rAAV-2/1-EF1a-N-CretrcintG, rAAV-2/1-EF1a-C-CreintG, rAAV-2/1-FLEX-tdT, or rAAV-2/1-CAGGS-FLEX-ChR2-tdT.WPRE.SV40 was injected into cerebellar lobule V or motor cortex using a stereotactic device. 3 weeks later, brain tissue was fixated for immunohistochemistry, or prepared for electrophysiology.
Histology
Retinal immunohistochemistry
Isolated mouse retinas were fixed at room temperature for 30 minutes in 4% paraformaldehyde/PBS solution. Retinas were then transferred to 30% sucrose in PBS, and subsequently into a 50/50 mixture of 30% sucrose/PBS and OCT for sectioning. 20–25 μm retinal cryosections were cut on a Leica CM3050 cryostat (Leica Microsystems), using disposable blades. Retinal cryosections were incubated in blocking solution (5% normal donkey serum, 0.1% Triton-X in PBS) for 1 hour and stained for primary antibody overnight at 4 °C. Immunostained cryosections were washed three times in 0.1% Triton-X in PBS and stained for secondary antibodies in blocking solution for 2 hours at room temperature. Slides were then washed in 0.1% Triton-X in PBS and mounted for imaging in Fluoromount-G (Southern Biotechnology Associates; 0100-01). Retinal sections were taken on a Zeiss LSM780 confocal microscope. Slides were scanned using a 40x or 63x oil immersion objective.
For rAAV infected transgenic GFP retinas, mice were transcardially perfused with 1xPBS followed by 4% PFA. Eyes were fixed in 4% PFA for 3–4 hours and retinas dissected and rinsed in 1xPBS. Retinas were incubated for 2 hours in blocking solution (10% goat serum, 0.25% Triton-X, 1x PBS) at room temperature followed by incubation with block solution containing primary antibodies at 4 °C overnight. Tissue was washed in 1x PBS (3×, 30 min). Retinas were incubated in block solution containing secondary antibody for 2 hours at room temperature, and washed in PBS (3×, 30 min). Then, retinas were mounted and cover slipped with Vectashield containing DAPI (Vector Laboratories) and then imaged using either an epifluorescent Zeiss Axio Imager 2 or a Zeiss LSM 780 confocal microscope47.
Spinal cord immunohistochemistry
Mice were transcardially perfused with 4% PFA in PBS (pH=7.4) and the spinal cord was post-fixed overnight at 4 °C in the same solution. Sagittal slices of the lumbar spinal cord were cut at 50 μm thickness on a Leica VT1000S vibratome. Free-floating sections were immunostained using standard methods for spinal cord histology (for a detailed protocol see reference48). Stained slices were then mounted on Superfrost plus slides (VWR) with Fluoromount-G (SouthernBiotech). Images were acquired with an LSM700 confocal microscope.
Cerebellar immunohistochemistry
Mice were transcardially perfused with 4% PFA in PBS (pH=7.4) and the brain was post-fixed overnight at 4 °C in the same solution. Parasagittal vermal slices of the cerebellum were cut at 40 μm thickness on a Leica VT1000S vibratome. Free-floating sections were immunostained using standard methods. Slices were then mounted on Superfrost slides (VWR) using Prolong Diamond mounting medium (Invitrogen). Images were acquired with an Olympus FV1000 or FV1200 confocal microscope. Whole brain sections were imaged on an Olympus MVX10 MacroView dissecting scope.
Antibodies and stains
For immunohistochemistry of retinas: rabbit anti-GFP (1:500 dilution) (A-6455, Invitrogen), chicken anti-β-galactosidase (1:1000 dilution) (ab9361, Abcam), rabbit anti-TagRFP (1:1000) (AB233, Evrogen). Note that anti-TagRFP also recognizes TagBFP and shows cross-reactivity to mCherry. The mCherry cross-reactivity did not cause problems for our analysis. In fact, it was advantageous in our identification of electroporated cells in the Tg(PROX1-GFP) experiment. Secondary antibodies raised against the relevant species were obtained from Jackson ImmunoResearch or Invitrogen.
For immunohistochemistry of rAAV-infected transgenic GFP retinas: rabbit anti-DsRed (1:2000 dilution) (632292, Clontech), chicken anti-GFP (1:2000 dilution) (1020, Aves Labs Inc).
For spinal cord immunohistochemistry: chicken anti-GFP (1:1000 dilution) (1020, Aves Labs Inc), rabbit anti-DsRed (1:1000 dilution) (632292, Clontech). rabbit anti-Cre (1:200 dilution) (69050-3, Novagen) Isolectin GS-IB4 (AlexaFlour conjugated, Molecular Probes, 1:500)
For cerebellar and cortical immunohistochemistry: chicken anti-GFP (1:200 dilution) (ab13970, Abcam), rabbit anti-Calbindin (1:500 dilution) (AB1779, Millipore), rabbit anti-Parvalbumin (1:1000 dilution) (PV25, Swant), rabbit anti-Cre (1:200 dilution) (69050-3, Novagen). Secondary antibodies: goat anti-rabbit and chicken anti-rabbit coupled to Alexa 647 (A-21443 and A21245, Invitrogen).
Slice preparation for electrophysiology
Mice were anaesthetized with isoflurane and decapitated. The brain was quickly removed and immersed in oxygenated ice-cold cutting solution containing (in mM) 110 choline chloride, 25 glucose, 25 NaHCO3, 11.5 sodium ascorbate, 7 MgCl2, 3 sodium pyruvate, 2.5 KCl, 1.25 NaH2PO4, 0.5 CaCl2. Parasagittal cerebellar slices were cut at 270 μm thickness on a Leica VT1200S and subsequently allowed to recover for 30 min at 32 °C in ACSF equilibrated with 95% O2 and 5% CO2, containing (in mM) 125 NaCl, 26 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 1, MgCl2, 2 CaCl2, and 25 glucose. Slices were then kept at room temperature for up to 5 hours.
Electrophysiological recordings
Slices were superfused with 31–32 °C warm ACSF at a flow rate of ~3 ml/min in a recording chamber heated by an inline heater (Warner instruments). PCs and MLIs were visualized using an Olympus BX51WI microscope equipped with differential interference contrast (DIC). GFP+ and ChR2-tdT+ were imaged using a custom two-photon laser-scanning microscope with 750 nm illumination49. Visually guided recordings were performed with ~2 MΩ (PCs) and 3.5 MΩ (MLIs) borosilicate glass pipettes (Sutter Instrument). The internal solution for voltage-clamp recordings contained the following (in mM): 140 Cs-methanesulfonate, 15 HEPES, 0.5 EGTA, 2 TEA-Cl, 2 MgATP, 0.3 NaGTP, 10 phosphocreatine-tris2, and 2 QX 314-Cl (pH adjusted to 7.2 with CsOH). For current-clamp recordings the internal solution contained 150 K-gluconate, 3 KCl, 10 HEPES, 0.5 EGTA, 3 MgATP, 0.5 GTP, 5 phosphocreatine-tris2, and 5 phosphocreatine-Na2 (pH adjusted to 7.2 with KOH). Recordings were performed with a 700B Axoclamp amplifier (Molecular Devices) and were controlled with custom software written in Matlab (generously provided by Bernardo Sabatini, Harvard Medical School). ChR2-tdT+ cells were excited using a 473 nm wavelength blue laser (OptoEngine) coupled through the excitation pathway of the microscope. Laser light was focused onto slices through a 40× water-immersion objective. Brief light pulses (0.5–1 ms) at an intensity of 3–5 mW/mm2 evoked ChR2-mediated photocurrent and stimulus-locked action potentials.
Data analysis
Imaging data was analyzed and processed with ImageJ (National Institutes of Health, Bethesda, MD), Imaris (Bitplane, Zurich, Switzerland), ZEN (Carl Zeiss Microscopy, Göttingen, Germany) and/or Photoshop (Adobe Systems, Mountain View, CA) software. Prism (GraphPad, La Jolla, CA) was used to generate graphs. Electrophysiological data was analyzed with IgorPro (Wavemetrics, Lake Oswego, OR), AxographX (Axograph Scientific, Sydney, Australia) and Prism (GraphPad, La Jolla, CA). For luciferase assays, two-tailed Student’s t test assuming unequal variance was used for all comparisons. p > 0.05 is judged as statistically significant. For cerebellar data, statistical significance was assessed with two-tailed Student’s t test (Fig. 7i) assuming unequal variance or One-way ANOVA (Fig. 7h). Statistical significance was assumed when p<0.05.
Quantifications
Quantification of cells was performed on the imaging software ZEN (Zeiss) or ImageJ (National Institutes of Health, Bethesda, MD). The ImageJ plugin, Cell Counter, was used to aid counting.
Quantification of electroporated retinas
Confocal Z-stack images were taken of retina sections with dense expression of electroporation markers. Electroporated cells were marked by the presence of electroporated GFP, n-βgal (from CAG-nlacZ) or the combination of TagBFP and mCherry (driven from ChAG-loxP-tBFPL-loxP-mCherry cassette; combination is detected by a polyclonal TagRFP antibody that detects both TagBFP and mCherry). Cells expressing a given marker (ex. Y) were selected using ImageJ Cell Counter plug-in, and then evaluated for co-expression of a second marker of interest (ex. X). Counted number of the second marker were divided by selected number of cells expressing the given marker, and multiplied by 100 to get % of X+ cell given Y+ cell (ex. % X+/Y+). This general quantification scheme was also used for rAAV-infected tissues. To avoid counting green fluorescent DsRed molecules in GFP and DsRed colocalization analysis, we imaged for Anti-GFP expression in the blue or far red channel, and occasionally used the native GFP fluorescence as aid for determining GFP expression.
Electroporated retina cells labeled by either n-βgal or TagBFP immunostaining were quantified for DsRed or mCherry fluorescence, respectively. Using ImageJ, a mask for the chosen cell body was created on the Anti-βgal or Anti-TagBFP channel and transferred to the DsRed or mCherry fluorescence channel, respectively. Measurements of the mean pixel intensity on single confocal slices were taken for the chosen cell bodies. Each measurement was paired with a background fluorescence intensity measurement from the same slice, chosen at a location in the slice where no cells were present. Each measurement was then subtracted from the background to obtain the net fluorescence, in arbitrary units (a.u.). We found mCherry to show slight aggregation in retinal cells, especially INL cell types. Thus, we avoided quantifying cellular regions with strong mCherry aggregation. To quantify fluorescence, we drew a shape circling the soma region with minimal aggregation in the confocal slice corresponding to the strongest mCherry fluorescence.
Quantification of rAAV-infected tissues
Confocal Z-stack images of rAAV-infected retinal, spinal cord, or brain samples were manually analyzed on ZEN or ImageJ for marker expression. For GFP-positive retinas, entire retinas were analyzed for tdT expression. For GFP-negative samples, tissue areas showing Cre-immunostaining were imaged and analyzed for tdT expression amongst Cre-immunopositive cells.
To estimate the rough efficiency of labeling in brain infections, GFP+ cells in densely infected areas were counted, and the number of overlapping tdT+ cells was determined. The GFP-overlapping tdT+ cell count was divided by the number of counted GFP+ cells, and multiplied by 100 to obtain % tdT+/GFP+ parameter. For cerebellum, this analysis was performed in slices immunostained against GFP to enhance GFP brightness. To refine the efficiency measure in infected spinal cords and brains, we counted GFP cells that were immunostained with polyclonal Cre antibody, and determined the number of tdT+ cells overlapping with this GFP+,Cre+ cell population. The number of tdT+ cells overlapping with GFP+, Cre+ cells were divided by the total number of counted GFP+, Cre+ cells, and multiplied by 100 to obtain % tdT+/GFP+,Cre+ parameter. The efficiency in MLIs could not be quantified in this manner because the dense anti-Cre-staining of PC dendrites obscured MLIs in the cerebellar molecular layer. To estimate leak expression of tdT+ in GFP-negative animals, all or randomly selected cells immunopositive for Cre were counted, and then the number of overlapping tdT+ cells was determined. This measure provides an estimate of the percentage of Cre-expressing cells showing background CRE-DOG activity. The same calculation in GFP+ animals allows an estimate of induction.
Additional Quantifications
To determine GFP specificity CRE-DOG in electroporated and rAAV infected tissues, cells expressing the Cre-dependent reporter (tdT, DsRed or mCherry) were either randomly counted or counted in their entirety within an image. The number of overlapping GFP+ cells was then determined. This value was divided by the number of counted tdT+, DsRed+ or mCherry+ cells, and then multiplied by 100 to obtain % GFP+/tdT, GFP+/DsRed+ or GFP+/mCherry+ parameter.
A supplementary methods checklist is available.
Supplementary Material
Acknowledgments
We thank Chen Wang of the Zhigang He lab for rAAV production (core service supported by grant NEI 5P30EY012196-17), Didem Goz for help with immunohistochemistry, Ben Huang, Dimphna Meijer and the Cepko/Tabin/Dymecki lab members for inputs on the manuscript, the Neurobiology Imaging Facility (supported by NINDS P30 Core Center Grant NS072030) for consultation and instrument availability. We are grateful to Ulrich Rothbauer and Heinrich Leonhardt for providing GBPs. This work was funded by the Howard Hughes Medical Institute (C.L.C.), the Nancy Lurie Marks Foundation (W.G.R.), the Lefler Foundation (W.G.R.), the Knights Templar Eye Foundation (O.S.D.), the McKnight Foundation (A.D.H.), The Pew Charitable Trusts (A.D.H.), the Glaucoma Research Foundation (A.D.H.), NIH RO1 EY022157-01 (A.D.H.) and R01 NS32405 (W.G.R.). S.R. is funded by an Alice and Joseph Brooks fellowship and a F32 NS087708 training grant and E.D. is funded by a F31 AG041582 (NIA) training grant.
Footnotes
J.C.Y.T. and C.L.C. initiated and coordinated the entire project. J.C.Y.T. originated the idea and carried out all in vitro molecular biology, electroporation and proof-of-principle rAAV experiments to validate the concept. S.R. coordinated brain injections, performed electrophysiological recordings of cerebellar slices, immunohistochemistry, imaging and analysis of cerebellar and cortical data. O.D. and A.D.H. performed viral injections into retinas of GFP lines and subsequent tissue processing, imaging and analysis. V.E.A. and S.C. performed viral injections into spinal cords and subsequent tissue processing, imaging and analysis. E.D. and S.L. constructed the ChAG-loxP-TagBFP-loxP-mCherry construct. S.L. performed electroporation into the Tg(PROX1-GFP) line and subsequent tissue processing. I.R.D. performed viral injections into the cortex and cerebellum and provided feedback on experiments. A.D.H. and W.G.R. supervised aspects of the project involving rAAV injection into transgenic GFP retinas and brains, respectively. C.L.C. supervised the entire project. J.C.Y.T and C.L.C wrote the paper, with contributions from all co-authors.
Competing Financial Interests
The authors declare no financial interests
References
- 1.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 2.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 5.Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 6.Siegert S, et al. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- 7.Tang JC, et al. A nanobody-based system using fluorescent proteins as scaffolds for cell-specific gene manipulation. Cell. 2013;154:928–939. doi: 10.1016/j.cell.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 9.Kirchhofer A, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- 10.Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol. 2012;19:117–121. doi: 10.1038/nsmb.2180. [DOI] [PubMed] [Google Scholar]
- 11.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 12.Ho SN, Biggar SR, Spencer DM, Schreiber SL, Crabtree GR. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 13.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenno LE, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pivetta C, Esposito MS, Sigrist M, Arber S. Motor-circuit communication matrix from spinal cord to brainstem neurons revealed by developmental origin. Cell. 2014;156:537–548. doi: 10.1016/j.cell.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- 17.Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vila-Perello M, Muir TW. Biological applications of protein splicing. Cell. 2010;143:191–200. doi: 10.1016/j.cell.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nat Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 20.Mootz HD, Muir TW. Protein splicing triggered by a small molecule. J Am Chem Soc. 2002;124:9044–9045. doi: 10.1021/ja026769o. [DOI] [PubMed] [Google Scholar]
- 21.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 22.Anraku Y, Mizutani R, Satow Y. Protein splicing: its discovery and structural insight into novel chemical mechanisms. IUBMB Life. 2005;57:563–574. doi: 10.1080/15216540500215499. [DOI] [PubMed] [Google Scholar]
- 23.Samson M, Emerson MM, Cepko CL. Robust marking of photoreceptor cells and pinealocytes with several reporters under control of the Crx gene. Dev Dyn. 2009;238:3218–3225. doi: 10.1002/dvdy.22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Betley JN, Sternson SM. Adeno-associated viral vectors for mapping, monitoring, and manipulating neural circuits. Hum Gene Ther. 2011;22:669–677. doi: 10.1089/hum.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivlin-Etzion M, et al. Transgenic mice reveal unexpected diversity of on-off direction-selective retinal ganglion cell subtypes and brain structures involved in motion processing. J Neurosci. 2011;31:8760–8769. doi: 10.1523/JNEUROSCI.0564-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osterhout JA, et al. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron. 2011;71:632–639. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osterhout JA, El-Danaf RN, Nguyen PL, Huberman AD. Birthdate and outgrowth timing predict cellular mechanisms of axon target matching in the developing visual pathway. Cell Rep. 2014;8:1006–1017. doi: 10.1016/j.celrep.2014.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamamaki N, et al. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- 29.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Nagai T, Horikawa K, Saito K, Matsuda T. Genetically encoded Ca(2+) indicators; expanded affinity range, color hue and compatibility with optogenetics. Front Mol Neurosci. 2014;7:90. doi: 10.3389/fnmol.2014.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sternson SM, Roth BL. Chemogenetic tools to interrogate brain functions. Annu Rev Neurosci. 2014;37:387–407. doi: 10.1146/annurev-neuro-071013-014048. [DOI] [PubMed] [Google Scholar]
- 32.Wickersham IR, Feinberg EH. New technologies for imaging synaptic partners. Curr Opin Neurobiol. 2012;22:121–127. doi: 10.1016/j.conb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venken KJ, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howard DB, Powers K, Wang Y, Harvey BK. Tropism and toxicity of adeno-associated viral vector serotypes 1, 2, 5, 6, 7, 8, and 9 in rat neurons and glia in vitro. Virology. 2008;372:24–34. doi: 10.1016/j.virol.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semprini S, et al. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques. Nucleic Acids Res. 2007;35:1402–1410. doi: 10.1093/nar/gkl1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emerson MM, Cepko CL. Identification of a retina-specific Otx2 enhancer element active in immature developing photoreceptors. Dev Biol. 2011;360:241–255. doi: 10.1016/j.ydbio.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 43.Shimshek DR, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- 44.Haubensak W, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhande OS, et al. Genetic dissection of retinal inputs to brainstem nuclei controlling image stabilization. J Neurosci. 2013;33:17797–17813. doi: 10.1523/JNEUROSCI.2778-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huberman AD, et al. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cruz-Martin A, et al. A dedicated circuit links direction-selective retinal ganglion cells to the primary visual cortex. Nature. 2014;507:358–361. doi: 10.1038/nature12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes DI, et al. Morphological, neurochemical and electrophysiological features of parvalbumin-expressing cells: a likely source of axo-axonic inputs in the mouse spinal dorsal horn. J Physiol. 2012;590:3927–3951. doi: 10.1113/jphysiol.2012.235655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drobizhev M, Makarov NS, Tillo SE, Hughes TE, Rebane A. Two-photon absorption properties of fluorescent proteins. Nat Methods. 2011;8:393–399. doi: 10.1038/nmeth.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.