Abstract
Myofibroblastic trans-differentiation of hepatic stellate cells (HSCs) is an essential event in the development of liver fibrogenesis. These changes involve modulation of key regulators of the genome and the proteome. Methionine adenosyltransferases (MAT) catalyze the biosynthesis of the methyl donor, S-adenosylmethionine (SAMe) from methionine. We have previously shown that two MAT genes, MAT2A and MAT2B (encoding MATα2 and MATβ proteins respectively), are required for HSC activation and loss of MAT2A transcriptional control favors its up-regulation during trans-differentiation. Hence MAT genes are intrinsically linked to the HSC machinery during activation. In the current study, we have identified for the first time, post-translational modifications in the MATα2 and MATβ proteins that stabilize them and favor human HSC trans-differentiation. Culture-activation of human HSCs induced the MATα2 and MATβ proteins. Using mass spectrometry, we identified phosphorylation sites in MATα2 and MATβ predicted to be phosphorylated by mitogen-activated protein kinase (MAPK) family members [ERK1/2, V-Raf Murine Sarcoma Viral Oncogene Homolog B1 (B-Raf), MEK]. Phosphorylation of both proteins was enhanced during HSC activation. Blocking MEK activation lowered the phosphorylation and stability of MAT proteins without influencing their mRNA levels. Silencing ERK1/2 or B-Raf lowered the phosphorylation and stability of MATβ but not MATα2. Reversal of the activated human HSC cell line, LX2 to quiescence lowered phosphorylation and destabilized MAT proteins. Mutagenesis of MATα2 and MATβ phospho-sites destabilized them and prevented HSC trans-differentiation. The data reveal that phosphorylation of MAT proteins during HSC activation stabilizes them thereby positively regulating trans-differentiation.
Keywords: Hepatic stellate cell, Methionine adenosyltransferases, Phosphorylation
INTRODUCTION
Liver injury leads to the trans-differentiation of hepatic stellate cells (HSCs) from a retinoid-storing to an extracellular matrix (ECM)-producing phenotype known as myofibroblastic state (Miyahara et al, 2000). Deciphering the mechanisms of HSC activation is crucial for our understanding of liver fibrogenesis. HSC activation is associated with induction of protein markers such as alpha-smooth muscle actin (α-SMA), Type I collagen, myocyte enhancer factor 2A (MEF2A) and loss of quiescence- associated factors such as peroxisome-proliferator activated receptor γ (PPARγ) (Miyahara et al, 2000).
Methionine adenosyltransferases (MAT) enzymes catalyze the biosynthesis of the methyl donor, S-adenosylmethionine (SAMe) from methionine and ATP. Mammalian systems express two MAT genes, MAT1A and MAT2A that encode the catalytic subunits, MATα1 and MATα2, of the enzyme (Kotb et al, 1997). MATα2 exists in the native MAT isozyme (MATII) which is associated with a regulatory subunit (MATβ) encoded by the gene, MAT2B (Martínez-Chantar et al, 2003). MATβ regulates the activity of the MATα2 subunit by lowering the inhibition constant (Ki) for SAMe and the Michaelis constant (Km) for methionine (Halim et al, 1999). MAT1A is expressed in adult liver (Gil et al, 1996) whereas MAT2A and MAT2B are widely expressed in extrahepatic tissues (Gil et al, 1996; Horikawa et al, 1992). MAT2A and MAT2B are also expressed in rapidly dividing and de-differentiated liver (Gil et al, 1996). Quiescent HSCs of the liver express MAT2A but not MAT1A (Shimizu-Saito et al, 1992).
We previously demonstrated that rat HSC activation led to induction of MAT2A and MAT2B mRNA and protein (Ramani et al, 2010). This was associated with a drop in MATII enzyme activity and increased global DNA hypomethylation (Ramani et al, 2010). Silencing MAT2A or MAT2B genes in activated HSCs reduces activation and suppresses cell growth (Ramani et al, 2010). MAT2A affects growth and activation by changing intracellular SAMe levels. MAT2B (but not MAT2A) silencing specifically inhibited pro-fibrogenic growth signaling pathways in HSCs such as extracellular signal-regulated kinase (ERK) and phosphatidyl inositol-3-kinase (PI3-K) (Ramani et al, 2010). The MAT2A gene is transcriptionally suppressed by PPARγ in quiescent rat HSCs and induced by PPARβ during HSC trans-differentiation (Ramani and Tomasi, 2012). Hence, there is a strong interplay of MAT genes with the HSC signaling machinery during trans-differentiation.
Phosphorylation is an important post-translational modification that is an integral part of HSC signaling. Pro-fibrogenic growth factors such as platelet-derived growth factor (PDGF) exert their growth promoting effects on HSCs via phosphorylation of ERK or AK strain transforming (Akt) (Borkham-Kamphorst et al, 2004). Phosphorylation of the quiescence factor, PPARγ via a mitogen-activated protein kinase/ERK kinase (MEK)-dependent mechanism may inhibit its transcriptional activity and promote HSC activation (Galli et al, 2004). Phosphorylation of cAMP response element-binding protein (CREB) at S-133 is necessary for blocking the HSC cell cycle and maintaining HSC quiescence (Houglum et al, 1997).
In the current work, we have identified post-translational mechanisms that control the MAT2A and MAT2B-encoded proteins in human HSCs. Our data reveal that phosphorylation stabilizes the MATα2 and MATβ proteins in activated human HSCs and is required for maintenance of the HSC myofibroblastic state.
MATERIALS AND METHODS
Human HSC culture and treatment conditions
Primary, quiescent human HSCs from single donor livers (ScienCell, San Diego, CA) were cultured on plastic dishes for 6 hours (day 0) or further cultured till activation (day 3, 7). Day 7 HSCs were treated with MEK inhibitor, PD98059 (30 μM) (Sigma-Aldrich, St. Louis, MO) or DMSO control for 15 minutes or 60 minutes. The human HSC cell line, LX2 (Xu et al, 2005) were reverted to quiescence with the adipogenic differentiation medium, MDI (0.5mM methylisobutylxanthine, 1μM dexamethasone, and 1μM insulin) (Sigma) for 3 days or 7 days (Zhao et al, 2006).
Mass spectrometry and kinase prediction analysis
Phosphorylated proteins were purified from activated HSCs using phospho-protein columns (Qiagen, Valencia, CA) and enriched phospho-peptides were subjected to mass spectrometry (MALDI-TOF MS/MS) (Applied Biomics Inc., Hayward, CA). Phosphorylated residues were confirmed by MS/MS peak showing the neutral loss of phosphate (Ma et al, 2013). The kinases predicted to phosphorylate MATα2 and MATβ were determined by the NetPhosK 1.0 software (Center for Biological Sequence Analysis, Denmark) (Table 1).
Table 1. MATα2 and MATβ phospho-site mutagenesis.
Based on a combination of mass spectrometry peaks showing loss of phosphate and NetPhosK 1.0 kinase prediction analysis, specific phospho-regions in MAT proteins were mutated. The phospho-site mutations in the peptides are marked in bold and underlined. The sequence of the primers used to generate each mutation are provided.
| Amino acid mutation |
Phospho- peptide |
NetPhosK kinase prediction |
Phospho- region mutant label |
Mutagenesis primers |
|---|---|---|---|---|
| MATα2 S283A, Y287F, T288F | AFSGKDYT | ERK1/2 | α2-P1 |
For: 5′-tggggtgctcatggaggaggtgcctttttaggaaaggatgttctaaaggtcgaccgttca-3′ Rev: 5′-tgaacggtcgacctttagaacatcctttcctaaaaaggcacctcctccatgagcacccca-3′ |
| MATα2 Y335F, T337V, S338A | HYGTSQ | ERK1/2 | α2-P2 |
For: 5′-gtttctcatccattatctatctccattttccatttaggtgtcttacagaagagtgagagagagctattagagattgtg-3′ Rev: 5′-cacaatctctaatagctctctctcactcttctgtaagacacctaaatggaaaatggagatagataatggatgagaaac-3′ |
| MATα2 Y371F, T374V | KKPIYQRTA | B-Raf, MAPK | α2-P3 |
For: 5′-gggatctggatctgaagaagccaattttacagagggttgcagcctatggcc-3′ Rev: 5′-ggccataggctgcaaccctctgtaaaattggcttcttcagatccagatccc-3′ |
| MATβ S243A, T247V | MLDPSIKGTF HW | B-Raf | β-P1 |
For: 5′-gaagagaatgctggatccattaattaagggagtctttcactggtctggcaatg-3′ Rev: 5′-cattgccagaccagtgaaagactcccttaattaatggatccagcattctcttc-3′ |
| MATβ T257V, Y259V | QMTKYEMA C | B-Raf | β-P2 |
For: 5′-tttcactggtctggcaatgaacagatgctaaaggttgaaatggcatgtgcaattgcagatgc-3′ Rev: 5′-gcatctgcaattgcacatgccatttcaacctttagcatctgttcattgccagaccagtgaaa-3′ |
Plasmid vectors and siRNA
Plasmids used are: MAT2B-luc (−1319 to +3-MAT2B promoter linked to luciferase)( Yang et al, 2008); MAT2A-DDK (human MAT2A-DDK-tag in pCMV6-entry) (Origene); MAT2B-HA (MAT2B-3X-HA-tag human in pReceiver-M06) (Genecopoeia, Rockville, MD). SiRNA’s for ERK1/2, V-Raf murine sarcoma viral oncogene homolog B1 (B-Raf) and negative control were purchased from Qiagen ((ERK1/2- Cat no. S100300755/SI00605997; B-Raf- Cat no. S100299488; AllStars negative control- Cat No. 1027281).
Transient transfection assays
Plasmid transfections were performed using the Superfect™ reagent (Qiagen) for 48 hours in day 1 or day 7 HSCs. ERK1/ERK2 or B-Raf siRNAs were reverse transfected into day 7 HSCs for 48 hours using RNAiMAX™ (Invitrogen, Carlsbad, CA). For protein stability, cells treated with negative control or ERK1/2 siRNA for 20 hours were pulsed with cycloheximide for different times. For combined over-expression and silencing, day 7 HSCs were reverse-transfected with ERK1/2 siRNA for 72 hours and MAT2B-HA vector was transfected during the last 48 hours of silencing.
Promoter activity assays
Promoter activity in transfected cells described above was assayed using the Dual-Luciferase Reporter Assay system protocol (Promega, Madison, WI) according to the manufacturer’s protocol.
Quantitative RT-PCR
Total RNA reverse transcribed using M-MLV RT (Invitrogen) was subjected to quantitative RT-PCR using TaqMan probes for human MAT2A, MAT2B and the housekeeping gene, GAPDH (ABI, Foster City, CA) (Ramani et al, 2010; Ramani and Tomasi, 2012). The thermal profile consisted of initial denaturation at 95°C for 3 minutes followed by 45 cycles at 95°C for 3 seconds and at 60°C for 30 seconds. The cycle threshold (Ct value) of the target genes was normalized to that of GAPDH to obtain the delta Ct (ΔCt). The ΔCt was used to find the relative expression of target genes according to the formula: relative expression= 2−ΔΔCt, where ΔΔCt= ΔCt of target genes in experimental condition - ΔCt of target gene under control condition (Ramani and Tomasi, 2012).
Western Blotting
Total cellular protein from human HSCs or LX2 cell line prepared using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitor cocktail (Sigma) was separated on SDS-PAGE following standard protocols (Amersham BioSciences, Piscataway, NJ). Antibodies used for Western blotting were: MAT2α2 (Novus Biologicals, CO), MATβ (Sigma) PPARγ (Santa Cruz Biotechology, CA), α-SMA (Novus), MEF2A (Origene), Anti-Phosphorylated Proteins (Pan) antibody (Invitrogen), p-ERK1/2 and B-Raf (Cell Signaling, MA), Type I collagen (Novus), DDK tag antibody (Origene), HA tag antibody (Genecopoeia), α-tubulin (Genetex, Irvine, CA) and actin (Sigma). Detection was done by the chemiluminescence ECL system (Amersham BioSciences). Blots were quantified using the Quantity One™ densitometry program (Bio-Rad laboratories, Hercules, CA) and test protein expression was normalized to α-tubulin control according to previously published reports (Olaso et al, 2001).
Immunocytochemistry
Transfected cells were incubated with a 1:100 dilution of α-SMA antibody according to the manufacturer (Novus) and developed with alexa488 secondary antibody (1:300) (Invitrogen). Nuclei were counterstained with 6-diamidino-2-phenylindole dihydrochloride hydrate. Fluorescence was detected with a high-resolution Zeiss laser-scanning microscope (LSM2, Carl Zeiss, Thornwood, NY).
Immunoprecipitation
Total protein extract was processed for immunoprecipitation as described previously (Tomasi et al, 2012). Briefly, total cellular protein extract was pre-cleared with normal rabbit IgG (Santa Cruz) followed by 30 μl of protein A/G-agarose beads (SantaCruz). 200 Mg of pre-cleared protein was incubated with 2 μg of Pan-phospho antibody (Phos) overnight at 4°C under slow rotation. Immunoprecipitated protein was bound to 40 μl of protein A/G-agarose beads for 1 hour at 4°C under rotation and was washed five times with RIPA buffer containing protease inhibitors. After the final wash beads were heated at 95°C for 8 minutes in loading dye to elute the protein. Samples were processed for Western blotting as described above and developed with Clean-blot™ IP detection reagent (HRP) (Thermo Scientific, Rockford, IL).
in vitro kinase assay
Recombinant MATα2 and MATβ (0.5μM each) (Prospec, East Brunswick, NJ) were mixed with 0.2μg of active ERK1 or ERK2 (Prospec) along with a magnesium/ATP cocktail at 0°C or 37°C for 45 minutes (Millipore, Billerica, MA). Reaction mixture was immunoprecipitated with 0.5μg Pan-phospho antibody (Phos) and immunoblotted with MATα2 or MATβ antibodies.
Protein stability assays
HSCs or LX2 cells treated under specific conditions (described above) were pulsed with 20 μg/ml of cycloheximide (Sigma) and then chased for different times in their respective media. At the experimental end point, total cellular protein was subjected to Western blotting as described above. The relative amount of protein was evaluated by densitometry and normalized to a-tubulin control. Semi-logarithmic plots of log10-densitometric ratios versus time were plotted to determine protein half life as described previously (Rao et al, 2012).
Phos-tag™ analysis
Phos-tag™ analysis. Phos-tag™ acrylamide (Wako Chemicals, Richmond, VA) provides a phosphate affinity SDS-PAGE for mobility shift detection of phosphorylated proteins (Kinoshita et al, 2011). This method was used to separate phosphorylated and non- phosphorylated MATα2 and MATβ proteins as distinct bands of different mobilities. Neutral gels containing zinc nitrate and either 50μM phos-tag™ (MATα2) or 20μM phos- tag™ (MATβ) were prepared according to previously described protocols (Kinoshita et al, 2011) using total cellular extract from HSCs. Western blotting was performed with MATα2 or MATβ antibodies for endogenous expression and anti-tag antibodies for exogenously expressed MATα2 (DDK tag) or MATβ (HA tag).
Site-directed mutagenesis
Single amino acid changes in wild type (WT) MATα2 or MATβ proteins or double/triple mutations to create phospho-region mutants were generated by site-directed mutagenesis of their plasmid vectors using the QuikChange II® site-directed mutagenesis Kit (Stratagene, La Jolla, CA) (Table 1). Mutagenesis primer design was done according to the manufacturer’s instructions (Table 1). Mutant clones were sequenced by the GeneWiz DNA sequencing facility (La Jolla, CA) using gene-specific primers for MAT2A, 5′-TTGTGGACACTTATGGCGGTTG-3′ and MAT2B, 5′-GAAGCAGCTGCTGTTGG-3′.
Statistical analysis
Data are represented as mean ± standard error (mean±S.E.) Statistical analysis was performed using the SPSS 17.0 software tool. Using analysis of variance (One-way ANOVA) followed by Student’s t test, significance was defined as P<0.05.
RESULTS
MAT2A and MAT2B expression and interaction in primary human HSCs
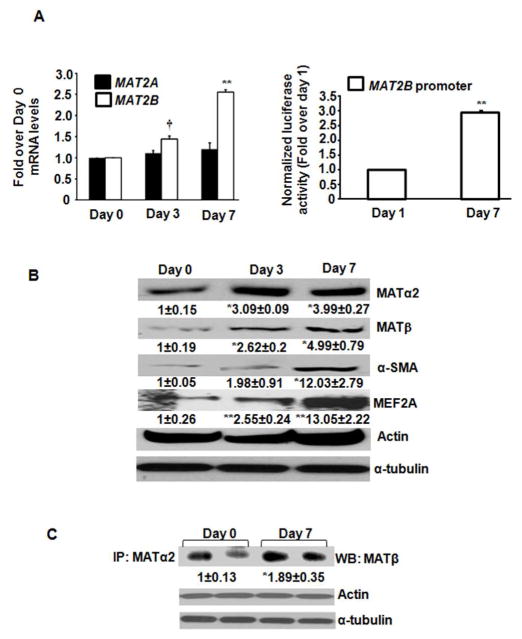
Activation markers, α-SMA and MEF2A were induced during HSC activation (Fig. 1B). MAT2A mRNA remained unchanged from day 0 to day 7 (Fig. 1A, left panel) whereas MAT2B mRNA was enhanced by 2.5-fold at day 7 compared to day 0 (Fig. 1A, left panel). MAT2B-luc promoter activity was increased by 2.8-fold at day 7 compared to day 1 (Fig. 1A, right panel). Despite no change in MAT2A mRNA, a 4-fold induction in MATα2 protein occurred during trans-differentiation (Fig. 1B). The MATβ protein was induced by 5.3-fold during trans-differentiation, significantly higher than the change of its mRNA level (Fig. 1B). Co-immunoprecipitation of MATα2 and MATβ was increased by 1.78-fold at day 7 compared to day 0 (Fig. 1C).
FIGURE 1. MAT2A and MAT2B expression and interaction in primary human HSCs.
A (left panel). Total RNA from day 0, day 3 and day 7 cultured human HSCs was subjected to quantitative RT-PCR for MAT2A and MAT2B as described in Materials and Methods. Results expressed as fold over day 0 are mean ± S.E from three HSC preparations in duplicates. **p<0.005. †<0.001 vs. day 0. A (right panel). Luciferase activity from day 1 or day 7 HSCs transfected with the MAT2B-luc promoter constructs was normalized to that of pGL3-Basic and expressed as fold over day 1 cells. Results represent mean ± S.E. from three experiments. **p<0.005 vs. day 1. B. Total cellular protein from day 0, day 3 and day 7 cells was immunoblotted with MATα2, MATβ, α-SMA, MEF2A, α-tubulin and actin antibodies. Results are densitometric analysis (mean ± S.E.) from four HSC preparations. *p<0.05, **p<0.005 vs. day 0. C. Total protein from day 0 or day 7 HSCs was immunoprecipitated with MATα2 antibody and immunoblotted for MATβ protein. Results are mean ± S.E from four experiments; two representative experiments are shown. *p<0.05 vs. day 0.
MAT protein phospho-site identification and kinase prediction
Mass spectrometry identified phosphorylation sites in MATα2 and MATβ with a protein score CI% threshold of >95% (Supplemental Tables 1 and 2). The NetPhosK 1.0 prediction software that uses neural network predictions of kinase-specific eukaryotic protein phosphorylation sites, predicted mitogen-activated protein kinase (MAPK) family members (B-Raf, ERK1, ERK2, MEK) to be involved in phosphorylating MAT proteins (Table 1). Specific phospho-sites identified by mass spectrometry and predicted to be phosphorylated by specific kinases were chosen for further analysis (shown as bold and underlined phospho-sites in Supplemental tables 1 and 2).
Phosphorylation of MATα2 and MATβ proteins in human HSCs
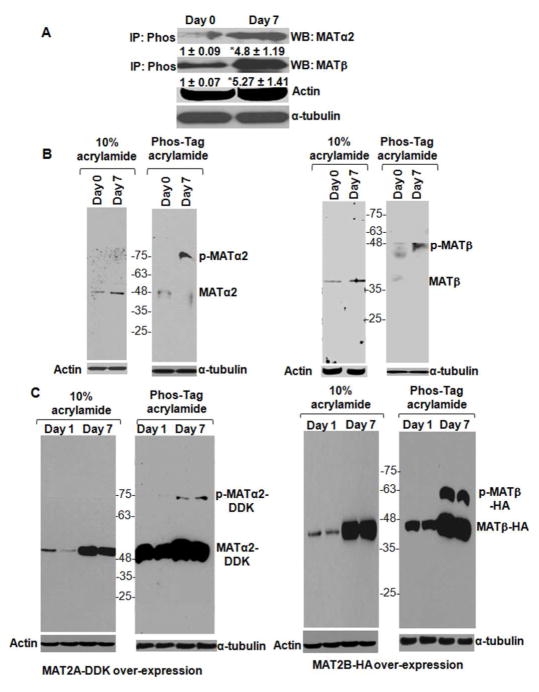
MATα2 or MATβ proteins were identified in the phospho-immunoprecipitates with Pan- phospho antibody (Fig. 2A). The amount of phospho-immunoprecipitated protein was 5 to 6-fold higher at day 7 compared to day 0 (Fig. 2A). Direct phosphorylation of MATα2 and MATβ proteins was confirmed by phos-tag™ analysis. Compared to unphosphorylated MATα2 (47 kDa, day 0), the majority of the protein at day 7 was mobility shifted, phosphorylated MATα2 (~ 75 kDa) (Fig. 2B, left panel). Both unphosphorylated (37 kDa) and phosphorylated (~ 48 kDa MATβ bands were observed at day 0. However, in day 7 HSC extracts, only the phosphorylated MATβ bands were observed at higher levels compared to day 0 (Fig. 2B, right panel). Over-expression level of MATα2-DDK (~ 49 kDa) and MATβ-HA (~ 42 kDa) was significantly higher at day 7 compared to day 1 extracts without phos-tag™ (Fig. 2C, left and right panels). Phosphorylated MATα2-DDK (~ 75 kDa) and MATβ-HA (~ 50 kDa) were detected only on phos-tag™ gels in the day 7 activated HSCs but not in day 1 cells (Fig. 2C, left and right panels), further confirming MAT phosphorylation during HSC activation.
FIGURE 2. Phosphorylation of MATα2 and MATβ proteins in human HSCs.
A. Total cellular protein from day 0 and day 7 HSCs was immunoprecipitated with Pan-phospho antibody (IP: Phos) and immunoblotted for MATα2 or MATβ. α-tubulin from total protein was used for normalization. Results are densitometric analysis (mean ± S.E.) from four HSC preparations. *p<0.05 vs. day 0. B. Total cellular protein from day 0 and day 7 cells was subjected to SDS-PAGE in the absence or presence of phos-tag™ as described under Materials and Methods and immunoblotted for endogenous MATα2 (left panel) or endogenous MATβ (right panel). A representative image from three HSC preparations is shown. C. Human HSCs were transfected with MAT2A-DDK (left panel) or MAT2B-HA (right panel) constructs as described under Materials and Methods. Extracts from transfected cells at day 1 or day 7 were subjected to SDS-PAGE in the absence or presence of phos-Tag™. Immunoblotting for exogenous MATα2-DDK (left panel) or MATβ-HA (right panel) is shown. Data is representative of four independent experiments. Two representative experiments are shown.
Effect of ERK1, ERK2 or B-Raf on MATα2 and MATβ phosphorylation
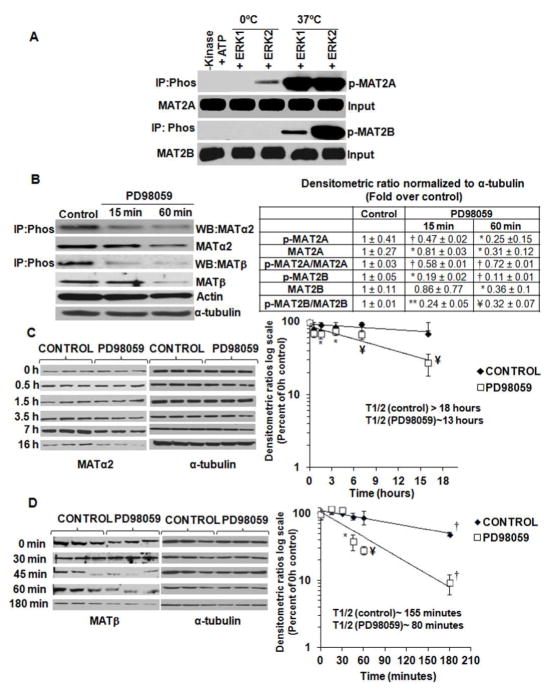
Phosphorylation of recombinant MATα2 and MATβ was significantly enhanced in the presence of active ERK1 or ERK2 enzymes at 37°C compared to 0°C controls or in the absence of active kinases (Fig. 3A). Inhibition of MEK (upstream ERK1/2 activator) in day 7 HSCs with PD98059 till 60 minutes, lowered phospho-immunoprecipitated MATα2 and MATβ by 75 and 80% respectively and total expression by 60% compared to control. The phospho to total MATα2 and MATβ ratios decreased by 40 and 55% respectively, compared to control (Fig. 3B). No change in MAT2A or MAT2B mRNA was observed (data not shown). To examine the stability of MAT proteins during PD98059 treatment, total protein synthesis was blocked by cycloheximide in control and PD98059-treated HSCs. In activated HSCs, MATα2 was highly stable with a half life greater than 18 hours. Treatment with PD98059 decreased the half life of MATα2 to ~ 12 hours (Fig. 3C). The half life of MATβ was decreased by 50% after PD98059 treatment compared to control (Fig. 3D). Blocking MEK activation thereby affected the phosphorylation and stability of MATα2 and MATβ.
FIGURE 3. Role of MAPK family in affecting MATα2 and MATβ phosphorylation.
A Recombinant MATα2 and MATβ proteins were subjected to in vitro kinase assay using active ERK1 and ERK2 enzymes as described under Material and Methods. Phosphorylated proteins were identified by immunoprecipitation-immunoblot using Pan-phospho antibody (Phos). Results are representative of three kinase assays. B. Cells treated with PD98059 were subjected to immunoprecipitation-immunoblotting or regular Western blotting to examine MATα2 or MATβ expression. Results are mean ± S.E. from three independent experiments. *p<0.05, ** p<0.005, †p<0.001, ¥p<0.01 vs. control. C. Cells were treated with PD98059 and chased for different times after cycloheximide treatment as described under Materials and Methods. Results represent the expression of MATα2 from three independent experiments expressed as percent of 0 hour control. Densitometric ratios were normalized to α-tubulin control at each time point and semi-logarithmic graphs were plotted to determine the half life of MATα2. *p<0.05, ¥ p<0.01 vs. control. D. Cells were treated as in ‘C’ and MATβ expression from three independent experiments is expressed as percent of 0 hour control. *p<0.05, ¥ p<0.01, †p<0.001 vs. control.
Effect of siRNA-mediated silencing of ERK1/2 and B-Raf on MATα2 and MATβ phosphorylation, expression and stability
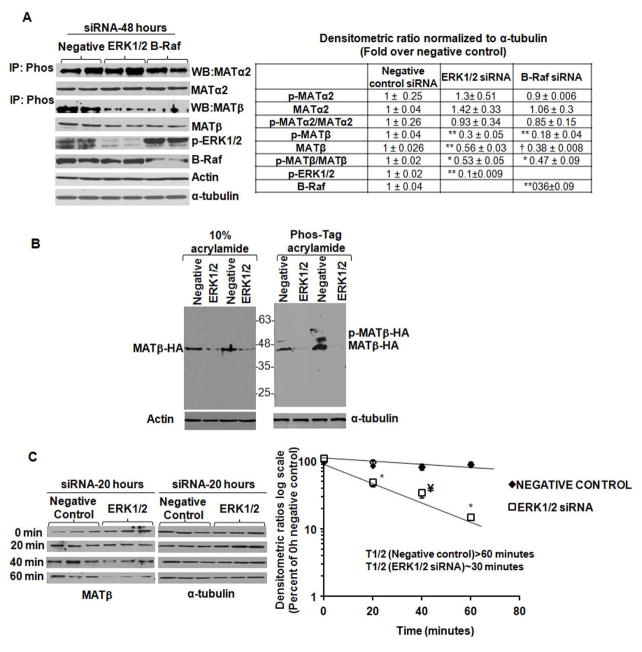
Specific silencing of MAPK family members, ERK1/2 or its upstream activator, B-Raf did not affect phospho-immunoprecipitated or total MATα2 levels (Fig. 4A) However, silencing ERK1/2 or B-Raf lowered phospho-immunoprecipitated MATβ by 70–80% and total MATβ by 50% compared to negative control after 48 hours of knockdown (Fig. 4A). The ratio of phospho to total MATβ dropped by 40–60% compared to negative control (Fig. 4A). No change in MAT2A or MAT2B mRNA was observed after silencing ERK1/2 or B-Raf (data not shown). Silencing ERK1/2 in HSCs blocked phospho-shift of over- expressed MATβ-HA protein on phos-tag™ gels and lowered its expression compared to a negative control siRNA (Fig. 4B), further confirming the results of Fig. 4A. Since MATβ phosphorylation and expression was lowered after silencing ERK1/2, we examined whether this was due to decreased stability. Negative control or ERK1/2 siRNA-transfected HSCs were pulsed with cycloheximide and chased for different times. Silencing ERK1/2 lowered the stability of MATβ by at least 50% compared to negative control (Fig. 4C), thereby indicating that ERK1/2-mediated phosphorylation of MATβ stabilized this protein in human HSCs.
FIGURE 4. Effect of siRNA-mediated knockdown of ERK1/2 and B-Raf on MATα2 and MATβ phosphorylation, expression and stability.
A. Extracts from cells treated with a negative control, ERK1/2 or B-Raf siRNA were subjected to immunoprecipitation-immunoblotting or regular Western blotting to examine MATα2 or MATβ expression. Results are mean ± S.E. from four to six experiments in duplicates. *p<0.05, ** p<0.005; †p<0.001 vs. negative control. B. Total protein from negative control or ERK1/2 siRNA-treated HSCs over-expressing MAT2B-HA vector were subjected to SDS-PAGE in the absence or presence of phos-tag™. Two representative experiments out of four independent analysis are shown. C. Cells were treated with a negative control or ERK1/2 siRNA for 20 hours and chased for different times after cycloheximide treatment as described under Materials and Methods. Results represent the expression of MATβ from three independent experiments expressed as percent of 0 hour negative control. Densitometric ratios were normalized to α-tubulin control at each time point and semi-logarithmic plots were used to determine the half life of MATβ. *p<0.05, ¥ p<0.01 vs. negative control siRNA.
Expression of MAT mRNA and protein and phosphorylation during reversal of HSC activation
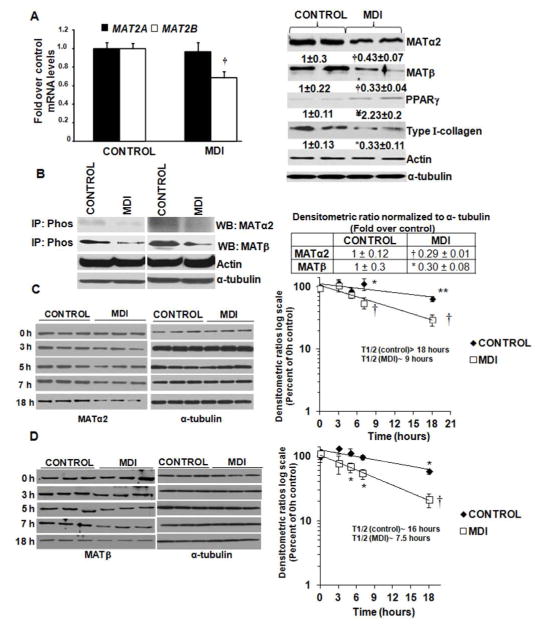
Reversal of LX2 cells to quiescence (MDI) decreased MAT2B mRNA but not MAT2A, by 40% compared to control (Fig. 5A, left panel). MATα2 and MATβ proteins decreased by 50% and 70% respectively, compared to control (Fig. 5A, right panel). Phospho-immunoprecipitated MATα2 and MATβproteins were lowered by 60–70% in MDI-treated cells compared to control (Fig. 5B). Cycloheximide treatment of control and MDI-treated cells showed that MDI treatment lowered the half life of MATα2 and MATβ by at least 50% compared to control (Fig. 5C and D). In LX2 cells, MATβ appears to be more stable at baseline compared to primary HSCs. The reasons for this are unknown but could possibly be due to the fact that LX2 cell line is immortalized spontaneously in the activated state as opposed to primary cells that are activated from a quiescent state in each experiment.
FIGURE 5. Phosphorylation and expression of MAT proteins during reversal of HSC activation.
LX2 cells were reverted to quiescence by MDI treatment as described under Materials and Methods. A (left panel). Expression of MAT2A and MAT2B mRNA was measured by relative quantitative RT-PCR and results are mean ± S.E expressed as fold over control from three experiments in duplicates. †p<0.001 vs. control. A (right panel). Expression of MATα2, MATβ, PPARγ and Type I collagen was determined by Western blotting. Results are mean ± S.E from three experiments in duplicates. †p<0.001, ¥p<0.01, *p<0.05 vs. control B. Control and MDI-treated extracts were immunoprecipitated with Pan-phospho antibody (IP:Phos) and immunoblotted for MATα2 and MATβ, α-tubulin from total cell extract was used as a normalization control. Results are densitometric analysis (mean ± S.E.) from four experiments. Two representative blots are shown, † p<0.001, * p<0.05 vs. control. C. Control and MDI-treated LX2 cells were treated with cycloheximide and assayed for expression of MATα2 at different times. Results represent mean ± S.E. from seven experiments expressed as percent of 0 hour control. Densitometric ratios were normalized to α-tubulin control at each time point and semi-logarithmic plots were used to determine the half life of MATα2. *p<0.05, †p<0.001, ** p<0.005 vs. control. D. Cells treated as in ‘C’ were assayed for MATβ expression. Results represent mean ± S.E. from six experiments expressed as percent of 0 hour control. Densitometric ratios were normalized to α-tubulin control semi-logarithmic plots were used to determine the half life of MATβ. *p<0.05. † p<0.001 vs. control.
Influence of MATα2 and MATβphospho-site and whole phospho-region mutations on HSC activation
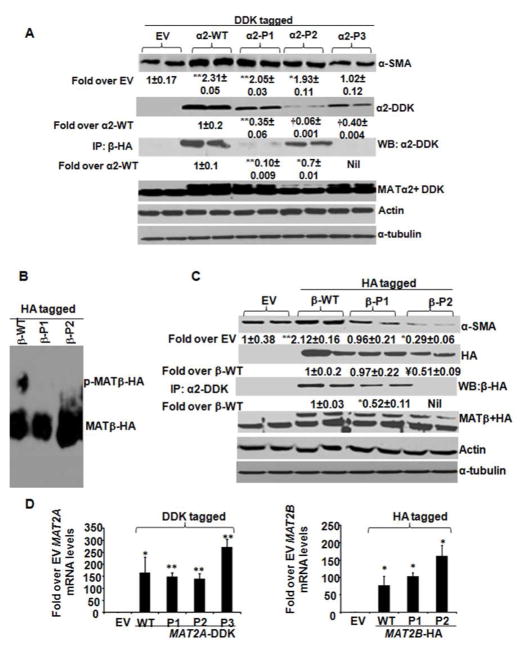
Specific phospho-sites of MAT identified by mass spectrometry were mutated (Table 1). either as a whole phospho-region (Fig. 6, designated as P1, P2 or P3 regions) or individual phospho-sites (Supplemental Fig. 1A and B). The P1, P2 or P3 mutants of MATα2-DDK (Fig. 6A) did not influence their shift on a phos-tag™ gel (data not shown). However, these mutants expressed at lower levels compared to the WT protein (Fig. 6A) without a significant change in their mRNA level (Fig. 6D, left panel), indicating lower stability of the mutants. Over-expression of WT MATα2-DDK in HSCs enhanced HSC activation (α-SMA expression) (Fig. 6A). Over-expression of the P3 mutant of MATα2-DDK (α2-DDK-P3) but not the P1 or P2 mutants (Fig. 6A), prevented the induction of HSC activation observed by the WT protein. Individual phospho-site mutations of MATα2-DDK within the P1, P2 or P3 phospho-regions gave the same effect on HSC activation as the mutation of the whole phospho-region (Supplemental Figure 1A). MATα2-DDK P1 and P2 mutants exhibited 90% and 33% decrease in binding to MATβ-HA respectively, compared to WT whereas the P3 mutant did not bind to MATβ-HA (Fig. 6A). Endogenous MATα2 levels were unchanged throughout the experiments (Fig. 6A, MATα2 + DDK blot). Within MATβ, two phospho-regions were mutated designated as P1 (consisting of S243A and T247V double mutation) and P2 (T257V and T259V double mutation). The P1 and P2 phospho-region mutants of MATβ did not show a phosphorylation-dependent shift on phos-tag™ gels compared to WT (Fig. 6B). The MATβ-HA-P2 mutant was expressed at 50% lower level compared to the WT protein (Fig. 6C). Over-expression of the P1 mutant normalized HSC activation (Fig. 6C) whereas the P2 mutant significantly lowered HSC activation compared to EV (Fig. 6C). Individual phospho-site mutations within the P1 region (S243A, T247V) inhibited HSC activation compared to WT protein and results were similar to the entire P1 mutant region (Supplemental Figure 1B). Within the P2 region, the T257V mutation inhibited HSC activation by 30% compared to EV whereas the T259V mutation normalized activation to EV levels (Supplemental Figure 1B). In combination, the T257V and T259V (P2 region mutant) significantly inhibited HSC activation compared to EV controls (Fig. 6C). The binding of the β-P1 mutant to MATα2-DDK was 50% lower than WT whereas the P2 mutant did not bind to MATα2-DDK (Fig. 6C). Endogenous MATβ levels remained unchanged (Fig. 6C, MATβ+HA blot). The total MAT2B mRNA level remained unchanged. (Fig. 6D, right panel).
FIGURE 6. Influence of MATα2 and MATβ phospho-region mutations on HSC activation.
A. WT and phospho-mutant vectors of MAT2α2 (DDK-tagged α2-P1, P2 and P3) were expressed in HSCs as described under Materials and Methods and total cellular extracts were immunoblotted with MATα2-DDK tag, endogenous MATα2, α-SMA, actin and α-tubulin antibodies. Interaction of MATα2-DDK and its mutants with exogenous MATβ-HA protein were estimated by co-immunoprecipitation (IP: β-HA blots). Densitometric ratios normalized to actin are represented as fold over EV for α-SMA and fold over WT MATα2 for mutant proteins. Results are mean ± S.E. from 6 experiments. *p<0.05, **p<0.005, †p<0.001, vs. EV or α2-DDK over-expression. B. WT and phospho-mutant vectors of MATβ (HA-tagged β-P1 and P2) were over-expressed in HSCs and subjected to phos-tag™ analysis as described in Materials and Methods. Results are representative of three analysis. C. WT and phospho-mutant vectors of MATβ (HA-tagged β-P1 and P2) were expressed in HSCs as described under Materials and Methods and total cellular extracts were immunoblotted with MATβ-HA tag, endogenous MATβ, α-SMA, actin and α-tubulin antibodies. Interaction of MAT β-HA and its mutants with exogenous MATα2-DDK protein were estimated by co-immunoprecipitation (IP: α2-DDK blots). Densitometric ratios normalized to α-tubulin are represented as fold over EV for α-SMA and fold over WT MATβ for mutant proteins. Results are mean±S.E. from 6 experiments. **p<0.005, *p<0.05, ¥p<0.01 vs. EV or β-HA over-expression. D. RNA from cells over-expressing MAT2A-DDK (left panel) or MAT2B-HA (right panel) vectors and their mutants was measured by relative quantitative RT-PCR and expressed as fold over EV. Results are representative of three independent experiments. *p<0.05, **p<0.005 vs. EV.
DISCUSSION
MATα2 and MATβ are strongly induced during rat HSC trans-differentiation and silencing them reduces activation and proliferation (Ramani et al, 2010). Preliminary evidence from our laboratory indicates regulatory cross-talk between MAT proteins and key players associated with HSC activation such as ERK, PI3-K, PPARγ and PPARβ (Ramani et al, 2010; Ramani and Tomasi, 2012). The regulation of MAT2A seems to differ between myofibroblastic human and rat HSCs. Whereas this gene was transcriptionally controlled in rat HSCs (Ramani and Tomasi, 2012), in activated human HSCs we found no changes in MAT2A mRNA levels but a significant increase in the MATα2 protein. This indicates a post-translational level of control of MATα2. MAT2B transcription is enhanced both in human and rat HSCs (Ramani et al, 2010) but the level of MATβ protein is significantly higher in activated human HSCs compared to its mRNA, indicating both transcriptional and post-translational control. The interaction between MATα2 and MATβ subunits is also higher in activated HSCs.
In this work we deciphered that MATα2 and MATβ phosphorylation is significantly induced in trans-differentiated HSCs compared to quiescent cells. Phos-tag™ analysis of quiescent and activated human HSCs indicated that majority of the MATα2 phospho- protein was present in activated cells and the low amount of protein in quiescent cells was essentially unphosphorylated. In the case of MATβ three bands of phospho-form were observed at very low levels in quiescent cells and significant induction in one of those forms was observed in activated cells. Although endogenous MATβ did give a pattern of faint phosphorylation signals in quiescent cells, our results on over-expressed MATβ in quiescent and fully-activated HSCs showed clearly that the phosphorylation of exogenous protein was detected mainly in activated cells.
From the prediction analysis, we tested which kinases mediated MATα2 phosphorylation in human HSCs. In the MAPK pathway, signal transduction from Raf kinases (B-Raf, Raf-1or A-Raf) or p21-activated kinase (PAK) to MEK1 or MEK2 causes phosphorylation and activation of these kinases which in turn phosphorylate and activate ERK1/2 (Robinson, 1996). MEK2 activation can also lead to activation of alternative ERKs such as ERK3 (Robinson, 1996). Although recombinant MATα2 protein is phosphorylated by both ERK1 and ERK2, silencing of ERK1/2 or its upstream kinase, B-Raf in activated human HSCs did not affect MATα2 phosphorylation, indicating that this effect was independent of ERK1/2 or B-Raf in cells. However, chemical inhibition of MEK1/2, that is upstream of ERK1/2, lowered MATα2 phosphorylation and also decreased its stability in cycloheximide chase assays. The results indicate a MEK-mediated pathway of MATα2 phosphorylation that is independent of the B-Raf/ERK1/2 loop but may involve alternative upstream activators such as A-Raf or Raf1 or downstream effectors such as ERK3.
Recombinant MATβ protein was phosphorylated by ERK1 or ERK2. In human HSCs, MATβ phosphorylation was lowered by treatment with PD98059 suggesting a MEK1/2-mediated influence on its phosphorylation. Also, specific silencing of ERK1/2 or upstream activator, B-Raf in activated human HSCs also lowered endogenous MATβ phosphorylation. MEK blockage by PD98059 is known to lower ERK1/2 activation (Robinson, 1996). The results suggest that phosphorylation of MATβ is mediated mainly by ERK1/2 activated via the B-Raf/MEK loop. Silencing ERK1/2 in activated cells reduces the stability of MATβ, indicating that phosphorylation via ERK1/2 stabilizes it. In our published work in activated human HSCs, we have shown that silencing the MAT2B gene (but not MAT2A) decreased phosphorylation of ERK1/2 (Ramani et al, 2010). Also, it was recently reported that the MATβ protein forms a scaffold with the G-protein- coupled receptor kinase-interacting protein 1 (GIT1),that recruits and activates MEK/ERK to promote growth of liver and colon cancer cells (Peng et al, 2013). Collectively, these data indicate a novel cross-talk between MATβ and ERK1/2 in different cellular systems. Based on these novel interactions of MATβ, it would be worthwhile to examine how this protein signals to players like ERK1/2 during transition of HSCs to myofibroblastic state and whether the phosphorylation of MATβ is required to transmit these signals.
Studies in culture models suggest that activated HSCs can revert to a quiescent phenotype (Ramani and Tomasi, 2012; Zhao et al, 2006). Here we show that reversal of activation to quiescence in LX2 cells lowered the expression, phosphorylation as well as stability of MAT proteins. These data support our current hypothesis that maintenance of the MAT phosphorylated state favors HSC trans-differentiation.
Mutagenesis studies of MAT phospho-sites confirmed the involvement of certain sites in HSC trans-differentiation. For MATβ, we mutated two phosphorylation regions based on phospho-site analysis by mass spectrometry and prediction assays. Both the P1 and P2 region mutations abolished the shift of the protein on phos-tag™ gels, indicating that these regions were potent sites of protein phosphorylation. The P1 mutant was expressed at similar levels to the WT protein but did not induce HSC activation like the WT. The results suggest that the P1 site does not influence protein stability but does influence trans-differentiation. Individual phospho-site mutations within the P1 region also prevented HSC activation. Interestingly, the P2 mutant was expressed at lower levels compared to WT but drastically reduced HSC activation compared to EV control cells, thereby exhibiting a dominant negative effect. Out of the two phospho-sites in the P2 region, the T257V mutant appeared to negatively regulate HSC activation. The data point towards an important role played by this region in promoting trans-differentiation caused by MATβ, the lack of which can reverse HSC activation.
We studied three predicted phosphorylation regions for MATα2. The MATα2 mutants, P1, P2 and P3 showed the same shift as the WT protein in phos-tag™ gels, indicating that these sites are not potent phosphorylation sites in the protein because their mutation does not abolish the shift of phos-tag™ bound MATα2. This also suggests that other potent phosphorylation sites in MATα2 are present that need to be further studied for their effect on protein stability and HSC trans-differentiation. Although the P1, P2 and P3 regions appear to be less phosphorylated and do not influence the phos- tag™ shift, these mutants are less stable compared to WT protein suggesting that these regions do stabilize the protein. The MATα2 P3 region as well as its individual phospho- site mutations prevented the increased HSC activation caused by the WT protein, indicating that this site might be involved in MATα2’s effect on HSC activation and the mechanism of this effect is yet to be established. The fact that specific mutations in MATβ phospho-sites and certain low potency MATα2 phospho-sites lowered HSC activation support the hypothesis that phosphorylation events at these sites promotes trans-differentiation.
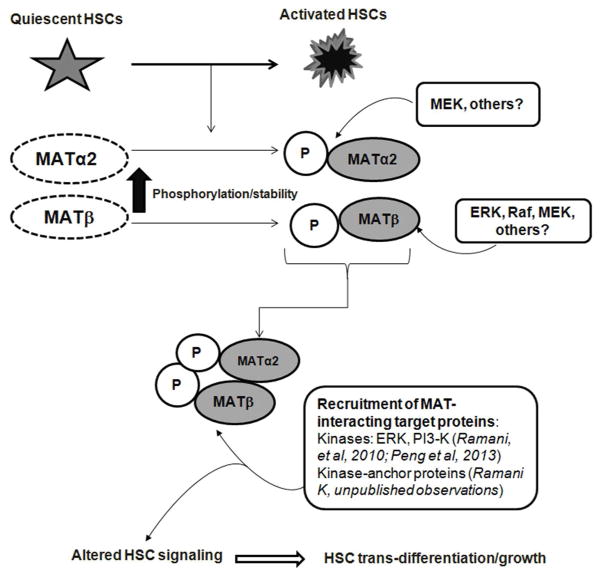
A question raised by this analysis is how do the phospho-sites in MATα2 and MATβ influence human HSC trans-differentiation? We and others have shown that the activity of the MATII enzyme composed of MATα2 and MATβ subunits is decreased during HSC activation despite an induction of MAT genes (Shimizu-Saito et al, 1997; Ramani et al, 2010) leading to a drop in SAMe levels and global DNA hypomethylation (Ramani et al, 2010) promoting HSC activation. Our mutation analysis shows that whereas the MATα2-P1 and P2 mutants (Table 1) do exhibit reduced binding to MATβ, they still promote HSC activation. However, the P3 mutant that does not bind to MATβ does not induce HSC activation. The interplay between phosphorylated subunits at specific protein regions may be important for HSC trans-differentiation. Our results on MATβ phospho-mutants suggests that specific mutations, P1 and P2 (Table 1), lower/block the interaction with MATα2 and also inhibit HSC activation. Hence, the interaction between the subunits at these phospho-sites may play a role in promoting activation. MATβ also exhibits functional interactions with survival signaling proteins (ERK and PI3-K) in HSCs independent of its role in regulating MATα2 (Ramani et al, 2010). Our recent preliminary work has identified interactions of MATα2 and MATβ with kinase-anchor proteins that are involved in HSC proliferation and trans-differentiation and have deciphered that MAT phosphorylation is important for these interactions (Ramani, K unpublished observations). We have also observed that these MAT protein interactions also facilitate phosphorylation of specific targets (Ramani, K unpublished observations). Whether phosphorylation of MAT protein sites recruits these factors for HSC trans-differentiation is yet to be established. The proposed mechanism by which MAT proteins may promote the myofibroblastic phenotype via their phosphorylation and stabilization is summarized in Figure 7. Future crystal structure analysis of MATα2 and MATβ interactions would be required to pinpoint the exact phosphorylation sites that may mediate their binding to each other and to proteins in the HSC signaling machinery required for trans-differentiation.
FIGURE 7. Proposed mechanism of action of MATα2 and MATβ during human HSC trans-differentiation.
Transition of quiescent HSCs to activated state leads to increased phosphorylation and stabilization of MATα2 and MATβ proteins. Phosphorylation of MATα2 involves a MEK-mediated mechanism whereas MATβ is phosphorylated via an ERK/MEK-Raf loop. Increased phosphorylation at certain sites facilitates enhanced interaction between MATα2 and MATβ subunits. The enhanced phospho-stabilization of MAT proteins may promote human HSCs trans-differentiation by causing altered recruitment of MAT-interacting target proteins previously identified such as ERK and PI3-K kinases in human HSCs (Ramani et al, 2010; Peng et al, 2013) or MAT-interacting kinase-anchor proteins ( Ramani. unpublished observations). These interactions might lead to de-regulated HSC signaling thereby promoting HSC activation and cell growth.
In conclusion, we provide for the first time, evidence of phosphorylation-mediated stabilization of MATα2 and MATβ proteins during myofibroblastic trans-differentiation of quiescent HSCs. The findings suggest a novel role of MATα2 and MATβ phosphorylation in mediating HSC activation and open new areas of investigation in understanding the cross-talk of these proteins in HSCs during liver fibrogenesis.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Institute of Health, Contract grant number: 5R00AA017774-05 (Komal Ramani)
Footnotes
Conflict of Interest: None
References
- Borkham-Kamphorst E, Herrmann J, Stoll D, Treptau J, Gressner AM, Weiskirchen R. Dominant-negative soluble PDGF-beta receptor inhibits hepatic stellate cell activation and attenuates liver fibrosis. Lab Invest. 2004;84:766–777. doi: 10.1038/labinvest.3700094. [DOI] [PubMed] [Google Scholar]
- Galli A, Crabb D, Price D, Ceni E, Salzano R, Surrenti C, et al. Peroxisome proliferator-activated receptor gamma transcriptional regulation is involved in platelet-derived growth factor-induced proliferation of human hepatic stellate cells. Hepatology. 2000;31:101–108. doi: 10.1002/hep.510310117. [DOI] [PubMed] [Google Scholar]
- Gil B, Casado M, Pajares MA, Bosca L, Mato JM, Martin-Sanz P, et al. Differential expression pattern of S-adenosylmethionine synthetase isoenzymes during rat liver development. Hepatology. 1996;24:876–881. doi: 10.1002/hep.510240420. [DOI] [PubMed] [Google Scholar]
- Halim AB, LeGros L, Geller A, Kotb M. Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem. 1999;274:29720–29725. doi: 10.1074/jbc.274.42.29720. [DOI] [PubMed] [Google Scholar]
- Horikawa S, Tsukada K. Molecular cloning and developmental expression of a human kidney S-adenosylmethionine synthetase. FEBS Lett. 1992;312:37–41. doi: 10.1016/0014-5793(92)81405-b. [DOI] [PubMed] [Google Scholar]
- Houglum K, Lee KS, Chojkier M. Proliferation of hepatic stellate cells is inhibited by phosphorylation of CREB on serine 133. J Clin Invest. 1997;99:1322–1328. doi: 10.1172/JCI119291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics. 2011;11:319–323. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- Kotb M, Mudd SH, Mato JM, Geller AM, Kredich NM, Chou JY, et al. Consensus nomenclature for the mammalian methionine adenosyltransferase genes and gene products. Trends Genet. 1997;13:51–52. doi: 10.1016/s0168-9525(97)01013-5. [DOI] [PubMed] [Google Scholar]
- Ma H, Qazi S, Ozer Z, Zhang J, Ishkhanian R, Uckun FM. Regulatory phosphorylation of Ikaros by Bruton’s tyrosine kinase. PLoS One. 2013;8:e71302. doi: 10.1371/journal.pone.0071302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Chantar ML, García-Trevijano ER, Latasa MU, Martín-Duce A, Fortes P, Caballería J, et al. Methionine adenosyltransferase IIB subunit gene expression provides a proliferative advantage in human hepatoma. Gastroenterology. 2003;124:940–948. doi: 10.1053/gast.2003.50151. [DOI] [PubMed] [Google Scholar]
- Miyahara T, Schrum L, Rippe R, Xiong S, Yee HF, Jr, Motomura K, et al. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J Biol Chem. 2000;275:35715–35722. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- Olaso E, Ikeda K, Eng FJ, Xu L, Wang LH, Lin HC, et al. DDR2 receptor promotes MMP-2-mediated proliferation and invasion by hepatic stellate cells. J Clin Invest. 2001;108:1369–1378. doi: 10.1172/JCI12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Dara L, Li TW, Zheng Y, Yang H, Tomasi LM, et al. Methionine adenosyltransferase 2B-GIT1 interplay activates MEK1-ERK1/2 to induce growth in human liver and colon cancer. Hepatology. 2013;57:2299–2313. doi: 10.1002/hep.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani K, Yang H, Kuhlenkamp J, Tomasi L, Tsukamoto H, Mato JM, et al. Changes in the Expression of Methionine Adenosyltransferase Genes and S-adenosylmethionine Homeostasis during Hepatic Stellate Cell Activation. Hepatology. 2010;51:986–995. doi: 10.1002/hep.23411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani K, Tomasi ML. Transcriptional regulation of methionine adenosyltransferase 2A by peroxisome proliferator-activated receptors in rat hepatic stellate cells. Hepatology. 2012;55:1942–1953. doi: 10.1002/hep.25594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao V, Guan B, Mutton LN, Bieberich CJ. Proline-mediated proteasomal degradation of the prostate-specific tumor suppressor NKX3.1. J Biol Chem. 2012;287:36331–40. doi: 10.1074/jbc.M112.352823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MJ, Cheng M, Khokhlatchev A, Ebert D, Ahn N, Guan KL, et al. Contributions of the mitogen-activated protein (MAP) kinase backbone and phosphorylation loop to MEK specificity. J Biol Chem. 1996;271:29734–29739. doi: 10.1074/jbc.271.47.29734. [DOI] [PubMed] [Google Scholar]
- Shimizu-Saito K, Horikawa S, Kojima N, Shiga J, Senoo H, Tsukada K. Differential expression of S-adenosylmethionine synthetase isozymes in different cell types of rat liver. Hepatology. 1997;26:424–443. doi: 10.1002/hep.510260224. [DOI] [PubMed] [Google Scholar]
- Tomasi ML, Tomasi I, Ramani K, Pascale RM, Xu J, Giordano P, et al. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology. 2012;56:982–93. doi: 10.1002/hep.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ara AI, Magilnick N, Xia M, Ramani K, Chen H, et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Chen W, Yang L, Chen L, Stimpson SA, Diehl AM. PPARgamma agonists prevent TGFbeta1/Smad3-signaling in human hepatic stellate cells. Biochem Biophys Res Commun. 2006;350:385–391. doi: 10.1016/j.bbrc.2006.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.