abstract
The fetal tight junction molecule claudin 6 (CLDN6) is virtually absent from any normal tissue, whereas it is aberrantly and frequently expressed in various cancers of high medical need.
We engineered 6PHU3, a T-cell-engaging bispecific single chain molecule (bi-(scFv)2) with anti-CD3/anti-CLDN6 specificities, and characterized its pharmacodynamic properties.
Our data show that upon engagement by 6PHU3, T cells strongly upregulate cytotoxicity and activation markers, proliferate and acquire an effector phenotype. 6PHU3 exerts potent killing of cancer cells in vitro with EC50 values in the pg/mL range. Subcutaneous xenograft tumors in NSG mice engrafted with human PBMCs are eradicated by 6PHU3 treatment and survival of mice is significantly prolonged. Tumors of 6PHU3-treated mice are strongly infiltrated with activated CD4+, CD8+ T cells and TEM type cells but not Tregs and display a general activation of a mostly inflammatory phenotype.
These effects are only observed upon bispecific but not monospecific engagement of 6PHU3. Together with the exceptionally cancer cell selective expression of the oncofetal tumor marker CLDN6, this provides a safeguard with regard to toxicity.
In summary, our data shows that the concept of T-cell redirection combined with that of highly selective targeting of CLDN6-positive solid tumors is effective. Thus, exploring 6PHU3 for clinical therapy is warranted.
KEYWORDS: Bispecific antibody, ideal target, oncofetal tumor marker, solid tumors, targeted immunotherapy, T cell engagement, tumor-infiltrating lymphocytes, T-cell engager, xenograft mouse model
Abbreviations
- Bi-(scFv)2
bispecific single chain variable fragment antibody
- CLDN6
claudin 6
- E:T
effector to target ratio
- GvH
graft-versus-host reaction
- IgG
immunoglobulin G
- TCM
central memory T cells
- TEM
effector memory T cells
- TEMRA
CD45RA+ effector memory T cells
- TH
T helper cells
- TIL
tumor-infiltrating lymphocytes
- TN
naive T cells
- Treg
regulatory T cells
- VH
variable heavy chain
- VL
variable light chain.
Introduction
Cytotoxic T cells are considered to be the most potent effector cells of the immune system. The idea of using bispecific antibodies to redirect circulating T cells to tumor sites in vivo and engaging them with cancer cells emerged in the 1980s.1-4 Meanwhile, catumaxomab, an anti-EPCAM/anti-CD3 bispecific antibody based on a full IgG-like format, has been approved for the treatment of malignant ascites caused by epithelial cancers.5,6 Single chain variants are engineered by fusing single chain variable fragments (scFv) of different specificities by a flexible linker to obtain bi-(scFv)2, such as the anti-CD19/anti-CD3 bi-(scFv)2 blinatumomab.7 Very recently, the FDA approved blinatumomab (BLINCYTO) for treatment of relapsed/refractory B-cell precursor ALL 8-11 making it the first approved immunotherapy against leukemia.
Cell killing induced by bi-(scFv)2 is not MHC-restricted and does not require costimulatory signals.12 Upon bi-(scFv)2-mediated engagement of a tumor cell with a T cell, an immunological synapse is formed. As a consequence, T cells are activated, proliferate, undergo polyclonal expansion and upregulate various immunomodulatory molecules.13,14 Bi-(scFv)2-mediated effects have been described as highly potent and strictly target dependent.15 Accordingly, the tumor cell selectivity of the target molecule defines the safety profile of a bispecific T-cell engager. One major obstacle for exploitation of this promising platform technology is the scarcity of cancer cell specific surface molecules, in particular for non-hematological cancers of highest medical need. Catumaxomab can, so far, only be administered intraperitoneally for palliative treatment of epithelial cancer derived malignant ascites, as intravenous administration is associated with dose limiting on-target effects on the epithelial organs.16
CLDN6 is an oncofetal tight junction molecule, expression of which in non-cancerous tissue is restricted to early stages of development, as in healthy adult tissues it is transcriptionally silenced,17-23 a fact which has led to consideration of CLDN6 as circulating marker for pregnancy.24 In various cancer types such as ovarian, lung, gastric, breast and pediatric cancers, CLDN6 expression is aberrantly activated.20,21,23,25,30 Thus, CLDN6 is an ideal target for antibody approaches of high potency. In fact, IMAB027, an immune effector mobilizing full IgG1 antibody, has entered clinical development and is being tested in patients with advanced ovarian cancer (NCT02054351).31
This paper describes the preclinical validation of 6PHU3, the first-in-class T-cell-engaging bispecific molecule targeting CLDN6-positive tumor cells. To our knowledge, 6PHU3 is the bispecific T-cell engager with the highest cancer-cell selectivity in non-hematological malignancies and may tap into patient populations, who thus far cannot profit from bispecific antibody treatment.
Results
6PHU3 produced by mammalian cells binds selectively to both CLDN6 and CD3
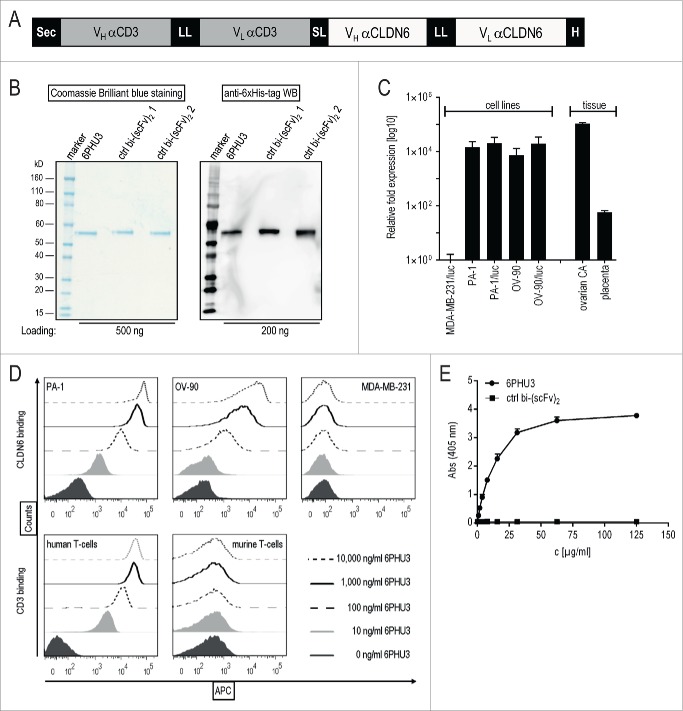
6PHU3 (Fig. 1A) and bi-(scFv)2 control proteins – combining the binding site for human CLDN6 or an irrelevant tumor target with anti-human CD3 – were purified to obtain single bands of 53–55 kDa (Fig. 1B).
Figure 1.
6PHU3 binds selectively to CLDN6 and CD3. (A) Schematic overview of the bi-(scFv)2 6PHU3 sequence cassettes cloned into pcDNA3.1 mammalian expression vector. (B) SDS-PAGE analysis of IMAC-purified 6PHU3 and two different control bi-(scFv)2. (C) CLDN6 expression of human carcinoma cell lines PA-1(/luc), OV-90(/luc) and MDA-MB-231/luc as determined by qPCR. Fold expression of CLDN6 expression has been calculated from two independent experiments. Tissue samples from ovarian carcinoma and placenta served as positive controls, CLDN6– tissues heart and thymus as calibrators. (D) 6PHU3 target binding as measured by flow cytometry. Increasing concentrations of 6PHU3 were incubated with CLDN6+ cells (PA-1, OV-90) or CD3+ human T-cells. MDA-MB-231/luc cells and murine T cells served as negative controls. Bound proteins were detected via their 6xHis-tag. (E) Bispecificity of 6PHU3 as demonstrated by ELISA. 6PHU3 and a control bi-(scFv)2 were captured by a CD3-mimicking peptide. Detection was conducted by an antibody specific for the anti-CLDN6 scFv, and a 2nd detection antibody. ABS indicates absorption; APC, allophycocyanin; bi-(scFv)2, bispecific single chain variable fragment; CA, carcinoma; c, concentration; ctrl, control; H, 6xHis-tag; hu, human; LL, long linker (15–18 amino acids); mu, murine; Sec, secretion signal; SL, short linker (five amino acids); VL, variable light chain; VH, variable heavy chain; WB, western blot.
Before the functional investigation began, the selected human tumor target cell lines PA-1 and OV-90 were confirmed by qPCR to be CLDN6-positive (CLDN6+) and MDA-MB-231/luc to be CLDN6-negative (CLDN6–) (Fig. 1C). It is noteworthy that PA-1 and OV-90 cell lines express lower levels of CLDN6 transcript compared to primary human ovarian cancer specimens.
Specific and robust concentration-dependent binding of 6PHU3 to PA-1 and OV-90 cells but not to the CLDN6– control cells was demonstrated by flow cytometry (Fig. 1D). Analogously, 6PHU3 bound specifically to human but not to murine CD3. It is important to mention that 6PHU3 is cross-reactive with murine CLDN6 (data not shown). Bispecificity of 6PHU3 for human CLDN6 and human CD3 was further confirmed by an ELISA based on simultaneous detection of both binding sites (Fig. 1E). In summary, this data shows that 6PHU3 efficiently engages both T-cells and CLDN6+ target cells.
6PHU3 mediates strong T-cell activation and specific lysis of CLDN6+ target cells
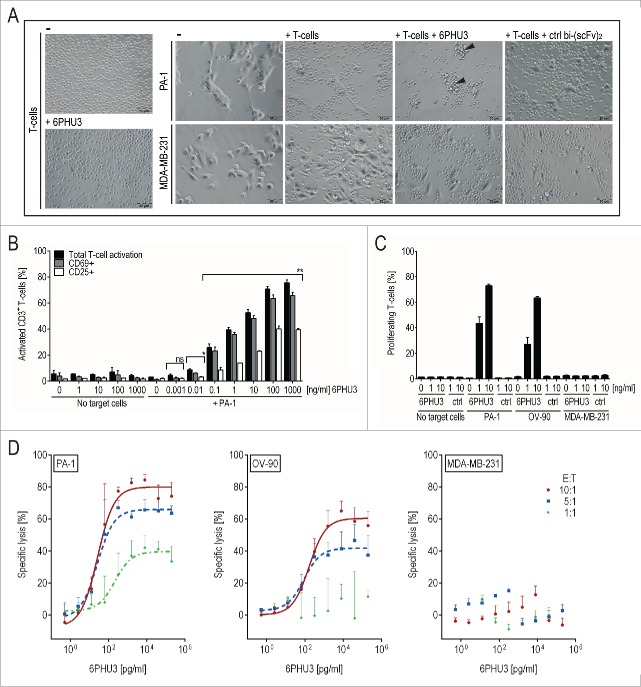
Next, we analyzed the effects of 6PHU3 on effector and on target cells. T-cell clustering as an indicator of T-cell activation, was observed within 24 h of adding 6PHU3 to cultures of T cells and PA-1 target cells (Fig. 2A) but not with CLDN6– targets. Further, 6PHU3 induced a concentration-dependent and profound activation of CD3+ T cells only in the presence of target cells as shown by flow cytometry (Fig. 2B). Upregulation of early (CD69) and late activation markers (CD25) was already significant at 0.01 ng/mL 6PHU3 (p < 0.02). The maximum of about 75% of activated T cells was reached at a concentration of 100 ng/mL 6PHU3 within 48 h of co-incubation.
Figure 2.
6PHU3 mediates strong T-cell-engaging effects only in the presence of target cells. (A) Microscopic image after 24 h of co-culturing CLDN6+ PA-1/luc cells and CLDN6– MDA-MB-231/luc control cells in an E:T ratio of 5:1 +/− 30 ng/mL 6PHU3 or ctrl bi-(scFv)2 as indicated (200× magnification). Arrowheads: clusters of T cells on residual target cells (B) Activation of CD3+ T cells as analyzed by flow cytometry. T cells were incubated with the indicated 6PHU3 concentrations +/− PA-1 cells for 48 h. Percentages of total activated T cells (black columns), CD69+ (gray columns) and CD25+ (white columns) are shown. Ns signifies not significant (p = 0.053); *p < 0.002; **p < 0.0001. (C) Proliferation of CD45+ T-cells as determined by flow cytometry. CFSE-labeled T cells were seeded alone, with PA-1, OV-90 or MDA-MB-231/luc control cells in an E:T ratio of 10:1. Cells were stimulated with bi-(scFv)2 as indicated for 72 h. (D) Specific lysis of 6PHU3 as determined by a luciferase detection cytotoxicity assay. PBMC of three independent donors were co-incubated with CLDN6+ target cells PA-1/luc (left) or OV-90/luc (middle) and with CLDN6– cells MDA-MB-231/luc (right) in E:T ratios of 10:1 (red solid curves), 5:1 (blue dashed curves) and 1:1 (green dotted curve) for 48 h. Curves present the average specific lysis by three donors, error bars show the deviation between donors. A 9-fold serial dilution of 6PHU3 (0.0005–200 ng/mL) was applied.
To analyze proliferation, CFSE-labeled T cells were co-incubated with CLDN6+ or CLDN6– target cells +/− 6PHU3 for 72 h. T-cell proliferation was 6PHU3 concentration-dependent and only occurred in the presence of CLDN6+ cells (Fig. 2C). At a concentration of 10 ng/mL 6PHU3, 60–70% of T cells were found to proliferate.
To assess the capability of 6PHU3 to mediate specific tumor cell lysis, we performed a luciferase-based in vitro cytotoxicity assay using PBMC from three independent donors and determined dose/response relationships over 48 h (Fig. 2D). Specific lysis of PA-1/luc and OV-90/luc cells but not of the CLDN6– control cell line were mediated by 6PHU3. Efficient lysis was obtained and was donor and effector to target ratio dependent (Table 1).
Table 1.
Range of EC50 values and of target cell lysis maxima in dependency of different donors and E:T ratios.
| Target cell line | PA-1/luc |
OV-90/luc |
MDA-MB-231/luc |
|||
|---|---|---|---|---|---|---|
| E:T | EC50 [pg/ml] | Max. lysis [%] | EC50 [pg/ml] | Max. lysis [%] | EC50 [pg/ml] | Max. lysis [%] |
| 10:1 | 12–90 | 76–85 | 130–252 | 56–70 | n.a. | 0–6 |
| 5:1 | 14–90 | 63–71 | 70–145 | 35–53 | n.a. | 0–1.2 |
| 1:1 | 67–473 | 34–48 | n.a. | 0–16 | n.a. | 0 |
At an E:T ratio of 1:1 robust killing of PA-1/luc but not of OV-90/luc was observed. A maximum of 70% PA-1/luc or 55% OV-90/luc cells were killed at an E:T of 5:1 and 85% or 70% at an E:T of 10:1, respectively. Half-maximal lysis concentrations (EC50) were in the pg/mL range at all E:T ratios except for the 1:1 ratio in the case of OV-90/luc cells.
In summary, 6PHU3 is highly potent and strongly mediates efficient T-cell activation, proliferation and cytotoxicity at dose levels in pg/mL range exclusively in the presence of CLDN6-expressing target cells.
T cells engaged by 6PHU3 acquire a pro-inflammatory, cytotoxic type-1 phenotype
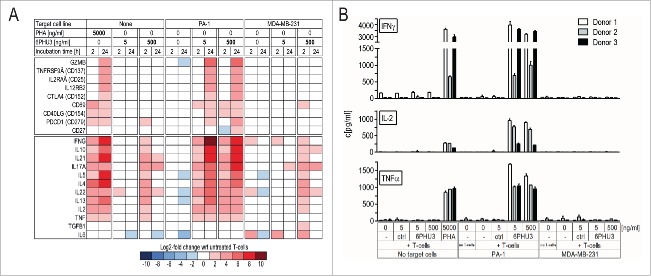
To study 6PHU3-engaged T cells in more detail, we conducted cDNA expression analysis upon co-incubation of T cells, bi-(scFv)2 and target cells (Fig. 3A, the full data set is shown in Fig. S1). 6PHU3 T-cell engagement significantly elevated mRNA transcript levels of various T-cell activation markers and cytokines at a concentration of 5 ng/mL. Upregulation was most prominent for granzyme B (GZMB), CD137 (TNFRSF9A), CD25 (IL2RAA), IL12RB2, IFNγ and IL-17A, which indicate activated, cytotoxic effectors and a general mostly inflammatory phenotype. The transcription of TH2-cell cytokines IL-10, IL-21, IL-5, IL-4, IL-22 and IL-13, associated with stimulated CD4+ T cells, were highly increased. IL-6 and TGFß levels, in contrast, were not altered. Cytokine upregulation was dependent on the presence of both 6PHU3 and target-positive cells. Only at 6PHU3 concentrations above 500 ng/mL – which is far higher than the efficacious concentration range for cell killing – did we detect modest and reversible upregulation of cytokine mRNA in the absence of CLDN6 by 6PHU3 alone.
Figure 3.
T cells activated by 6PHU3 present a cytotoxic, pro-inflammatory phenotype. T cells were incubated with 5 or 500 ng/mL 6PHU3 +/− PA-1 cells or MDA-MB-231/luc control cells. (A) Heatmap showing log2-fold expression with respect to untreated T cells. Gene expression was evaluated using a custom-made panel of commercial primers specific for human T-cell identification-markers as measured by Fluidigm. All values were normalized to CD3. (B) T-cell cytokine release as measured by ELISA. Wrt indicates with respect to.
Cytokine data was further confirmed on a protein level by measuring release of several of the cytokines into the supernatant by ELISA. After 24 h of incubation with 5 or 500 ng/mL 6PHU3, high levels of IFNγ, IL-2, TNFα, (Fig. 3B), but not of IL-6 (data not shown), were measured exclusively in the presence of both 6PHU3 and CLDN6+.
In summary, our data show that soon after engagement to target cells via 6PHU3, T cells are highly activated and acquire a pro-inflammatory effector phenotype.
6PHU3 treatment eradicates advanced tumors in a xenograft mouse model and confers significant survival benefit
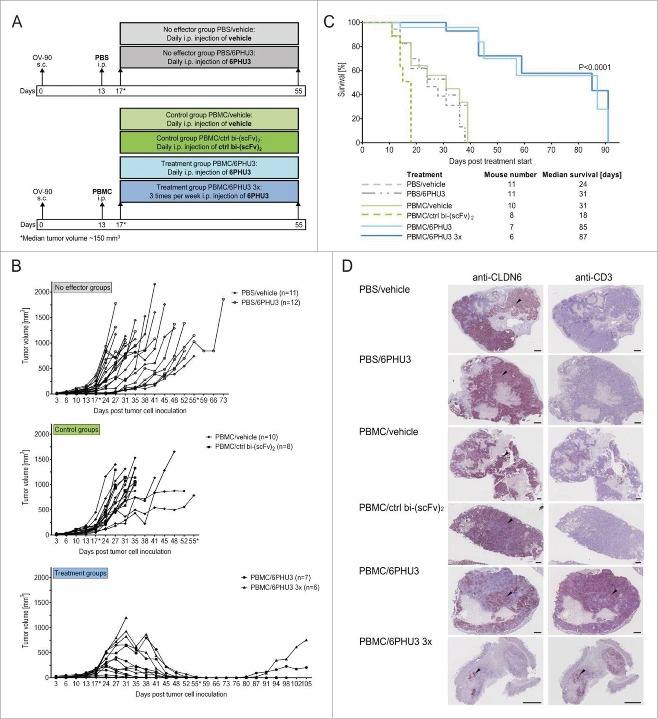
Next, we assessed whether the strong activation of T cells upon engagement to CLDN6+ target cells translates into antitumoral effects in vivo. We made use of a xenograft system with immunodeficient NSG mice permissive for engraftment with human tumor and effector cells. NSG mice were subcutaneously (s.c.) inoculated with CLDN6+ OV-90 cells and stratified at a median tumor volume of ∼50 mm3 to receive either PBS (‘no effector’ groups) or PBMC isolated from humans (‘control’ and ‘treatment’ groups) intraperitoneally (i.p.). Four days later, at a median tumor volume of ∼150 mm3, treatment was started with 200 µg/kg 6PHU3 – due to typical fast clearance of bi-(scFv)2 either daily or three times/week – or with controls (daily vehicle or control bi-(scFv)2) for a total of 38 d (Fig. 4A). In the ‘treatment’ groups that received effectors and 6PHU3, significant tumor growth retardation was observed starting after day 7 of 6PHU3 dosing, leading to nearly complete tumor eradication in all but one of the mice that was treated only three times per week (Fig. 4B). Four mice had to be sacrificed because of graft-vs.-host (GvH) reaction prior to potential relapse incidence. Five mice remained tumor free for another 18–43 d before onset of GvH reaction after treatment was discontinued on day 55. Three mice had a tumor relapse with the earliest being 25 d after the last 6PHU3 injection. Recurrent tumor specimens of relapsed mice were found to express CLDN6 in 5–40% of tumor cells and displayed low numbers of infiltrating human T cells (data not shown).
Figure 4.
6PHU3 treatment eradicates tumors and extends survival in a xenograft mouse model. (A) Injection schedule scheme. Mice with advanced OV-90 xenografts were subjected to the following treatments: ‘No effector’ groups were not engrafted with human PBMC but treated daily with either vehicle or 6PHU3. ‘Control’ groups were engrafted with PBMC and treated daily with either vehicle or the control bi-(scFv)2. ‘Treatment’ groups underwent PBMC grafting and were treated with 6PHU3 according to two different schedules. (B) Tumor growth of all mice and groups. Treatment was applied i.p. between the days marked by an asterisk (*). Each line represents an individual mouse. (C) Kaplan–Meier survival curves of all groups from the day of treatment start. (D) Immunohistochemistry of representative xenograft whole tumor sections. Black arrow heads point to example areas of reddish-brown staining indicating the presence of CLDN6 (left) or CD3 (right) in adjacent sections. Black dashes correlate to 1 mm of tumor size. I.p. indicates intraperitoneal; s.c., subcutaneous.
Median survival of all control groups ranged between 18 and 31 d, whereas the 6PHU3 treatment groups survived significantly longer with a median of 85 and 87 d (p < 0.0001) (Fig. 4C).
Tumors of the mice in the ‘no effector’ and ‘control’ groups showed homogeneous CLDN6 expression in non-necrotic tumor areas and low numbers of infiltrating T cells (Fig. 4D). In tumors of 6PHU3-treated mice, CLDN6+ cells were diminished and CLDN6+ areas were surrounded by organized, strong T-cell infiltrates as identified by anti-CD3 IHC staining. Similar results were obtained in a PA-1 xenograft study (Fig. S2) in which the overall CLDN6 expression was lower compared to OV-90 xenografts. Here, the treatment time had to be kept shorter (25 d) due to early GvH reactions in all groups. All except for one tumor showed growth arrest at volumes of 50–300 mm3.
In summary, 6PHU3 treatment proved to be highly efficient against advanced tumors in aggressive xenograft mouse models and conferred a survival benefit.
Treatment with 6PHU3 reshapes the tumor microenvironment and increases immune cell infiltration
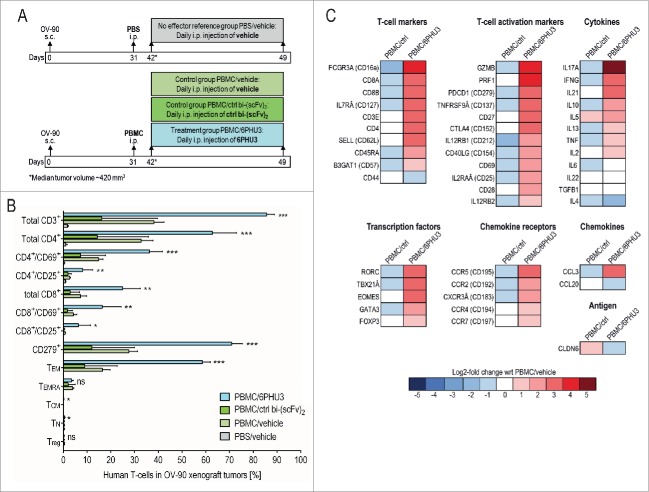
To characterize tumor-infiltrating lymphocytes recruited by 6PHU3, we set up a second treatment experiment (Fig. 5A). NSG mice were s.c. inoculated with OV-90 carcinoma cells and engrafted with human PBMC or PBS as a control (‘no effector’ mice) 31 d later. 11 d post PBMC or PBS injection tumors had a median size of 420 mm3 and treatment was initiated. PBMC engrafted mice were treated i.p. with vehicle, 200 µg/kg bi-(scFv)2 or 6PHU3 for eight consecutive days. The ‘no effector’ mice were treated with vehicle only. Mice were sacrificed and tumors were analyzed using flow cytometry and gene expression analysis.
Figure 5.
T cells infiltrated into subcutaneous xenograft tumors in response to 6PHU3 have an activated and cytotoxic functional status. (A) Injection schedule scheme. (B) Flow cytometric analysis of human T cells in whole tumor tissue. Percentage of TILs from gated live singlet total cells is depicted. Significance was calculated by comparing PBMC/6PHU3 to PBMC/ctrl bi-(scFv)2 and to PBMC/vehicle. (C) Heatmap showing a log2-fold expression of PBMC/ctrl bi-(scFv)2 (‘PBMC/ctrl’) and PBMC/6PHU3 groups with respect to PBMC/vehicle group as reference (mean values of two independent runs). Gene expression was evaluated using a predesigned primer panel specific for 46 human T-cell identification-markers as measured by Fluidigm. Ns indicates not significant; TCM, central memory T cells; TEM, effector memory T cells; TEMRA, CD45RA+ effector memory T cells; TN, naive T cells; Treg, regulatory T cells; *p < 0.03; **p < 0.008; ***p < 0.0009.
Flow cytometric analysis showed a 2–5-fold stronger infiltration of CD3+ T cells into tumors of PBMC-engrafted and 6PHU3-treated mice as compared to controls (Fig. 5B). CD4+ T cells, both early (CD4+/CD69+) and late activated (CD4+/CD25+) were significantly elevated in 6PHU3-treated mice. Total cytotoxic (CD8+) and activated cytotoxic T cells (CD8+/CD69+, CD8+/CD25+) were also enriched significantly by 6PHU3 treatment as was the fraction of PD-1+ T cells (3–6-fold), indicating previous antigen encounter. Effector memory T cells (TEM) were significantly expanded, whereas naive (TN), central memory (TCM), CD45RA+ effector memory (TEMRA) and regulatory (Treg) T cells were not detectable. Other human mononuclear blood cells such as NK cells, B cells or monocytes were not found in any of the tumor specimens.
cDNA expression analysis of the tumors confirmed and further extended the mapping of 6PHU3-specific effects (Fig. 5C). T-cell subset markers CD16a, CD8+, CD127, CD4+ and CD62L (SELL) indicated infiltration by γδ, cytotoxic, mature, TH1/TH2 and TN. T-cell activation markers (CD137 (TNFRSF9A), CD152 (CTLA4), CD212 (IL12RB1), CD69, CD25 (IL2RAα), CD28) were significantly upregulated in response to 6PHU3 as were granzyme B (GZMB) and perforin 1 (PRF1) as markers of high cytotoxicity, CD27 as a marker of TEM as along with CD279 (PDCD1) and CD154 (CD40LG) specifying terminally activated T cells.
The cytokine pattern indicated the presence of TH17 T cells (IL17A), TH1-cells with lytic activity (IFNγ, IL21) and of TH2-cells (IL10, IL5), whereas IL-6 and TNF mRNA levels were not induced by 6PHU3. T-cell specific transcription factors RORC (TH17-cells), TBX21α (TH1-cells), eomesodermin (EOMES, TCM) and GATA3 (TH2-cells) were increased. mRNAs for the chemokine receptors CCR5, CCR2 and CXCR3α involved in T-cell proliferation were augmented, as was chemokine ligand CCL3 mRNA. We did not find significant differences in CCR7 (TN or TCM), TGFB1 and FOXP3 (Treg) encoding mRNA levels.
CLDN6 mRNA level was lowest in the 6PHU3 treatment group, further confirming depletion of target-positive cancer cells.
In summary, 6PHU3 treatment appears to extensively reshape the tumor microenvironment toward an inflammatory response, including infiltration of tumors with TEM.
Discussion
The objective of this study was to characterize the primary pharmacodynamics of 6PHU3, the first recombinant CD3-/CLDN6-bispecifc scFv-antibody. Key findings are as follows:
First, our data shows that 6PHU3 is highly potent in mobilizing resting CD3-expressing T cells. As a consequence, T cells undergo proliferation and acquire an activated, pro-inflammatory, TEM phenotype as demonstrated by transcript profiling and cell surface markers. We found that 6PHU3 mediates cell killing with high efficiency. EC50 values are in the pg/mL range at low E:T ratios of 1:1 to 5:1. This indicates that 6PHU3-engaged T cells execute serial killing as is also claimed for other similar compounds such as blinatumomab.13,32,33
Secondly, we showed that the in vitro pharmacodynamics of 6PHU3, in conjunction with human PBMCs, translates into remarkable antitumor effects in vivo in highly aggressive xenograft tumor models. Treatment of mice bearing CLDN6-expressing tumors results in an almost complete tumor elimination and profoundly prolonged survival, whereas treatment with the control bi-(scFv)2 does not induce reduction of tumor size.
Thirdly, even though assessment of safety was not the objective of this study, findings on several levels indicate that 6PHU3 is well tolerated and safe. 6PHU3-mediated effects are strictly target dependent and require bispecific binding to both CLDN6 and CD3. Monospecific binding of the control bi-(scFv)2 to CD3-expressing T cells in the absence of tumor target-independent binding does not lead to unspecific activation of the T cells. In our study, the treatment of T cells with 100-fold higher 6PHU3 concentration than the effective dose used in vitro has neither in the presence or absence of CLDN6- cells led to upregulation of genes linked to T-cell activation and has only led to short term upregulation of inflammatory cytokines. An important contribution to the lack of on-target or off-target toxicity of 6PHU3 in mouse experiments is to be attributed to the choice of CLDN6 as target. The vast majority of reports indicate that CLDN6 is exclusively embryofetal and not expressed at all in healthy adolescent human and mouse tissues.17-19,24,25,29,34,38 However, there is also some conflicting data, reporting e.g. transcript detection in human hepatocytes.39 As a matter of fact, a comprehensive and broad profiling of CLDN6 across body maps of both species with valid definitions of positivity has not been published so far. Our own tissue profiling data (manuscript in preparation) conducted with strictly CLDN6-specific primers and antibody reagents, respectively, supports lack of CLDN6 expression in healthy adolescent human cells and tissues. Moreover, even though preliminary, safety monitoring of the ongoing first-in-human testing of the investigational medicinal product IMAB027, from which 6PHU3 has been derived, in more than 40 patients has not indicated on-target hepatic toxicity (manuscript in preparation).31,40 Safety of CLDN6-based targeting approaches remain to be more firmly investigated.
Another important finding is the strong specific effect of 6PHU3 on the tumor microenvironment. Tumors of animals treated with 6PHU3 show a far stronger infiltration with CD3+ T cells compared to control groups. Within these infiltrates activated CD4+ and CD8+ populations and profoundly enriched PD-1-expressing T cells were found in the 6PHU3-treated group only. PD-1 upregulation is frequently linked to initiation of a negative feedback loop to limit inflammation 28,29 or T-cell exhaustion due to persistent antigenic stimulation.30,41 Recent data, however, supports that PD-1+ T cells likely represent a particular differentiation stage or trafficking ability rather than exhaustion.11,42,43 T cells in tumors of 6PHU3-treated mice have the TEM phenotype, as also reported previously for the anti-EpCAM MT110 13,32-34 and anti-CD19 blinatumomab.8 Treg activation, which should be prevented by bispecific antibodies, was not detected in the tumor lesions. In response to 6PHU3, type-1 cytokines IL-2, TNFα and IFNγ that are known to be involved in activation of cell-mediated antitumor responses were expressed at the tumor site, whereas type-2 cytokines IL-4 and TGFβ fostering a pro-tumorigenic state 45-48 were not expressed.
In summary, these data justify further exploration of 6PHU3 to potentially gain a therapeutic that is highly specific and efficient for the treatment of CLDN6+ solid cancers.
Materials and methods
Cells and cell culture
Target cells were the endogenously CLDN6-expressing (CLDN6+) human ovarian carcinoma cell lines PA-1 (teratocarcinoma), OV-90 (adenocarcinoma) (American Type Culture Collection) their firefly luciferase transduced variants (PA-1/luc, OV-90/luc) and the CLDN6-negative (CLDN6–) human breast adenocarcinoma cell line MDA-MB-231/luc (Perkin Elmer). All cell lines have been authenticated by Eurofins in April 2015 (Human Cell Line Authentication Service by STR/DNA Profiling).
As effector cells human peripheral blood mononuclear cells (PBMC) isolated from buffy coats of healthy donors by Ficoll density-gradient centrifugation were used either as bulk or as T cells enriched by magnetic bead separation (Pan T Cell Kit II, human (Miltenyi)).
Construction, affinity purification and analysis of bi-(scFv)2 proteins
Codon optimized scFv sequences were generated by gene synthesis (GeneArt AG). Anti-CLDN6 VH and VL domains directed against the first extracellular loop of CLDN6 (provided by Ganymed Pharmaceuticals AG) 49 were linked by a 15 amino acid encoding linker sequence ((G4S1)3). This anti-CLDN6 scFv was joined by a (G4S1)1 peptide linker coding sequence to anti-CD3ϵ scFv 2,41 and equipped with a secretion signal sequence at the 5′-end and a 6xHis-tag sequence at the 3′-end.
pcDNA3.1/6PHU3 or control bi-(scFv)2 were produced in HEK293 clonal producer cell lines grown in DMEM, 10% FCS supplemented with 0.8 mg/mL G418 (Invitrogen).50 Bi-(scFv)2 culture supernatants harvested from HEK293 producer cell lines in multiple layer Cell Factories (Nunc) were subjected to immobilized metal affinity chromatography (IMAC) (GE Healthcare Life Sciences).50 Eluted proteins were dialyzed against vehicle buffer consisting of 200 mM L-Arginine-monohydrochloride (Roth) in H2O, pH 7.4.
Bi-(scFv)2 quality was tested by Coomassie brilliant blue staining and Western blot analysis 50 carried out using monoclonal anti-6xHis-tag antibody (Dianova) and Fc-specific secondary peroxidase-conjugated goat-anti-mouse IgG antibody (Sigma Aldrich).
qPCR
RNA was extracted and real-time quantitative PCR analysis (qPCR) run on an ABI Prism 7300 Real-Time PCR System (Applied Biosystems) as described elsewhere.51 Relative fold CLDN6 (sense 5′-CTT ATC TCC TTC GCA GTG CAG-3′; antisense 5′-AAG GAG GGC GAT GAC ACA GAG-3′) expression was determined using the ΔΔCt-method by normalization to the housekeeping gene HPRT1 (sense 5′-TGA CAC TGG CAA AAC AAT GCA-3′; antisense 5′-GGT CCT TTT CAC CAG CAA GCT-'3) and calibration to the mean value of CLDN6– human tissue cDNA.
FACS binding assay
OV-90, PA-1, MDA-MB-231/luc (negative control) cells, human T cells and murine T cells (negative control) were incubated with purified 6PHU3. 6PHU3 binding was detected with a primary anti-6xHis-tag antibody (Dianova) followed by incubation with an APC-conjugated goat-anti-mouse secondary antibody (Jackson Immuno Research). Staining controls were either the primary and secondary antibodies or the secondary antibody alone in the absence of 6PHU3. Dead cells were separated by 7-Actinomycin D (7-AAD (Beckman Coulter)) staining. Sample processing was performed on a FACSCantoII (BD Biosciences) and with FlowJo 7 software (Tree Star).
Bispecific ELISA
Streptavidin plates (Nunc) were coated with a biotinylated custom-made CD3-mimicking peptide (JPT Peptide Technologies, patent application submitted). After blocking with 3% BSA, bi-(scFv)2 was applied and an antibody against the CLDN6-recognizing domain of 6PHU3 (Ganymed Pharmaceuticals AG) was added for the detection with an AP-labeled anti-mouse antibody (Jackson Immuno Research) and a pNPP substrate based reaction.
T-cell activation and proliferation assay
Human T cells were incubated with 1 × 105 PA-1/luc cells in an E:T ratio of 5:1 in the presence of 6PHU3 in a six-well plate (Nunc) for 48 h. Effector cell-containing supernatant was collected and re-suspended in FACS-buffer supplemented with fluorescent-conjugated anti-human antibodies anti-CD3-FITC, anti-CD25-PE, anti-CD69-APC (BD Biosciences) and 7-AAD to measure T-cell activation of live cells.
Proliferation was measured with the Cell Trace CFSE reagent (Molecular Probes). In a 96-well plate, 2 × 104 target cells were seeded together with 2 × 105 CFSE-labeled human T cells and bi-(scFv)2 protein. After 72 h of incubation at 37°C, 5% CO2, T cells were harvested and stained with anti-CD45-APC (BD Biosciences) and the viability dye eFluor 506 (eBioscience) to gate live CD45+ T cells. Sample processing was performed on a FACSCantoII and with FlowJo 7 software.
T-cell phenotyping assays
1 × 106 T cells were co-incubated with medium or with 2 × 105 tumor cells and 6PHU3. Treatment of T cells with 5 µg/mL Phytohemagglutinin (PHA) served as a positive control. Cytokine release into the supernatant was measured after 24 h by ELISA (eBioscience). For gene expression analysis, RNA was extracted and cDNA was synthesized using PrimeScriptTM RT Reagent Kit with gDNA Eraser (Takara Bio Inc.). Samples were processed according to the Fluidigm® Gene Expression Advanced Development Protocol 28 – Fast Gene Expression Analysis using human TaqMan® Gene Expression Assays for 46 T-cell-specific genes and TaqMan® PreAmp MasterMix (LifeTechnologies). Chip arrays were analyzed via a Fluidigm BioMarkTM HD system (Fluidigm). Data sets were analyzed according to the ΔΔCt-method by normalization to CD3 and calibration to the mean value of untreated T cells of all time points to evaluate T-cell signals only.
Cytotoxicity assay
1 × 104 luciferin transduced target cells were plated together with effector cells into white 96-well flat bottom plates (Nunc). 6PHU3 was added to the experimental lysis samples (Lexp), protein buffer to the minimum (Lmin) and maximum lysis (Lmax) control wells, all in triplicates. Plates were incubated for 48 h before lysis of Lmax-cells with Triton X-100 and addition of a buffered luciferin solution (BD Monolight luciferin, BD Biosciences). Luminescence arising from oxidation of luciferin by luciferase-expressing viable cells was measured in a microplate-reader Infinite M200 (Tecan). Percentage of specific target cell lysis was calculated by the formula: Specific lysis [%] = [1 – (Lexp– Lmax)/(Lmin– Lmax)] × 100.
Data was analyzed by a sigmoidal dose-response (variable slope) algorithm integrated into PRISM 6 software (GraphPad).
For microscopic analysis, the assay was performed in transparent 48-well plates with enriched T cells. Photographs were recorded at a magnification of 200x with an Olympus IX53 inverted microscope (Olympus) after 24 h of incubation.
Xenograft mouse model for the assessment of therapeutic efficacy
All mice were used in accordance with the guidelines from the Institutional Animal Care Committee of the Johannes Gutenberg University, Mainz, Germany.
Immunodeficient male and female NOD.Cg-Prkdscid IL2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory) at 8–12 weeks of age and with a body weight of 20–32 g were used for the study. CLDN6+ OV-90 or PA-1 cells served as tumor cells and PBMC as effector cells.
1 × 107 tumor cells were inoculated subcutaneously (s.c.), 1 × 107 PBMC were intraperitoneally (i.p.) administered, ‘no effector’ groups received 1x PBS. Mice were treated daily with control bi-(scFv)2 or vehicle by i.p. injections and with 6PHU3 either daily or three times per week. Tumor volumes were calculated by the formula: Tumor volume [mm3] = length [mm] × (width [mm])2/2. Termination criteria were tumor volumina of 1500 mm3 or weight loss of 20% e.g., in case of GvH reaction. For generation of Kaplan–Meier survival curves, the day of treatment start was considered day 0.
Immunohistochemistry (IHC)
IHC analysis was described previously.20 Briefly, 3 µm tissue sections of xenograft tumors were incubated with a primary rabbit anti-CLDN6 antibody (IBL-America) or a polyclonal anti-CD3 antibody (Abcam) followed by incubation with PowerVision polymer-HRP conjugated anti-Rabbit secondary antibody (Immunologic). Binding reactions were visualized by the Vector NovaRED kit (Vector Laboratories Ltd.) and hematoxylin counterstaining (Carl Roth). Stained tumor sections were scanned with the Leica SCN 400 (Leica Biosystems).
Flow cytometric analysis of TIL
Single tumor cell suspensions were generated by manual disruption. Two subsets of anti-human antibodies were used. Subset 1: CD45, CD3, CD25, CD69, CD62L, CD45RA, CD14, CD16, CD19 (all BD Biosciences), CD8+ (eBioscience), CD4+ and CD197 (both BioLegend). Subset 2: CD45, CD3, CD25, CD45RA, CD103, CD127, CD279, FoxP3 (all BD Bioscience), CD8+ (eBioscience) and CD4+ (BioLegend). Dead cells were stained with LIVE/DEAD Fixable Blue Dead Cell Stain Kit (Invitrogen). Samples were processed on an LSRFortessa flow cytometer (BD Biosciences) and data was analyzed as whole percentage of live singlet cells using FlowJo software 7.
Gene expression analysis of xenograft tumors
RNA from tumor slices was isolated by QIAzol Lysis Reagent and the TissueLyser II, followed by the RNeasy® Lipid Tissue Mini Kit procedure including the RNeasy® Mini Kit spin purification method (Qiagen). cDNA synthesis and gene expression analysis via Fluidigm BioMarkTM HD system was performed as described above. Data sets were evaluated according to the ΔΔCt-method by normalization to the housekeeping gene HPRT1 and calibration to the mean values of the ‘no effector’ reference tumor samples.
Statistical analysis
Kaplan–Meier survival curves were compared by using log-rank (Mantel-Cox) test, T-cell activation by One-way ANOVA and TIL flow cytometry data by one-tailed Students t-test (unpaired). Differences between groups were considered significant in cases where p < 0.05. All analyses were integrated in GraphPad PRISM 6.
Disclosure of potential conflicts of interest
C Stadler is Head of Bispecifc Antibodies at BioNTech AG and has ownership interest in a patent application. H Baehr-Mahmud is Deputy Head of Bispecifc Antibodies at BioNTech AG and has ownership interest in a patent application. L Plum has ownership interest in a patent application. O Tuereci is a cofounder of Ganymed Pharmaceuticals AG and has ownership interest in a patent application. U Sahin is a cofounder of BioNTech AG and has ownership interest (including patents) in TRON gGmbH and BioNTech AG.
Supplementary Material
Acknowledgments
We particularly thank René Roth, Alexandra Roth, Bernhard Hebich and Silvia Weßel for their excellent and dedicated technical assistance. We also highly appreciate the technical assistance of Martin Suchan, Larissa Ralla and Irina Eichelbrönner. Moreover, we owe special thanks to Dr Mustafa Diken for his support in Fluidigm data interpretation, Dr Tim Beißert for critical reading and discussions and Richard Rae for native speaker reviewing.
References
- 1.Clark MR, Waldmann H. T-cell killing of target cells induced by hybrid antibodies: comparison of two bispecific monoclonal antibodies. J Natl Cancer Inst 1987; 79:1393-401; PMID:3121901 [PubMed] [Google Scholar]
- 2.Lanzavecchia A, Scheidegger D. The use of hybrid hybridomas to target human cytotoxic T lymphocytes. Eur J Immunol 1987; 17:105-11; PMID:3102250; http://dx.doi.org/ 10.1002/eji.1830170118 [DOI] [PubMed] [Google Scholar]
- 3.Sahin U, Hartmann F, Senter P, Pohl C, Engert A, Diehl V, Pfreundschuh M. Specific activation of the prodrug mitomycin phosphate by a bispecific anti-CD30/anti-alkaline phosphatase monoclonal antibody. Cancer Res 1990; 50:6944-48; PMID:2170012 [PubMed] [Google Scholar]
- 4.Staerz UD, Kanagawa O, Bevan MJ. Hybrid antibodies can target sites for attack by T cells. Nature 1985; 314:628-31; PMID:2859527; http://dx.doi.org/ 10.1038/314628a0 [DOI] [PubMed] [Google Scholar]
- 5.Ruf P, Gires O, Jäger M, Fellinger K, Atz J, Lindhofer H. Characterisation of the new EpCAM-specific antibody HO-3: implications for trifunctional antibody immunotherapy of cancer. Br J Cancer 2007; 97:315-21; PMID:17622246; http://dx.doi.org/ 10.1038/sj.bjc.6603881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seimetz D. Novel monoclonal antibodies for cancer treatment: the trifunctional antibody catumaxomab (removab). J Cancer 2011; 2:309-16; PMID:21716847; http://dx.doi.org/ 10.7150/jca.2.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanford M. Blinatumomab: first global approval. Drugs 2015; 75:321-27; PMID:25637301; http://dx.doi.org/ 10.1007/s40265-015-0356-3 [DOI] [PubMed] [Google Scholar]
- 8.Bargou R, Leo E, Zugmaier G, Klinger M, Goebeler M, Knop S, Noppeney R, Viardot A, Hess G, Schuler M et al.. Tumor regression in cancer patients by very low doses of a T cell-engaging antibody. Science 2008; 321:974-77; PMID:18703743; http://dx.doi.org/ 10.1126/science.1158545 [DOI] [PubMed] [Google Scholar]
- 9.Garber K. Bispecific antibodies rise again. Nat Rev Drug Discov 2014; 13:799-801; PMID:25359367; http://dx.doi.org/ 10.1038/nrd4478 [DOI] [PubMed] [Google Scholar]
- 10.Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol 2015; 67(2 Pt A):95-106, EPub ahead of print; PMID:25637431; http://dx.doi.org/ 10.1016/j.molimm.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 11.Topp MS, Kufer P, Gökbuget N, Goebeler M, Klinger M, Neumann S, Horst H, Raff T, Viardot A, Schmid M et al.. Targeted therapy with the T-cell-engaging antibody blinatumomab of chemotherapy-refractory minimal residual disease in B-lineage acute lymphoblastic leukemia patients results in high response rate and prolonged leukemia-free survival. J Clin Oncol 2011; 29:2493-98; PMID:21576633; http://dx.doi.org/ 10.1200/JCO.2010.32.7270 [DOI] [PubMed] [Google Scholar]
- 12.Offner S, Hofmeister R, Romaniuk A, Kufer P, Baeuerle PA. Induction of regular cytolytic T cell synapses by bispecific single-chain antibody constructs on MHC class I-negative tumor cells. Mol Immunol 2006; 43:763-71; PMID:16360021; http://dx.doi.org/ 10.1016/j.molimm.2005.03.007 [DOI] [PubMed] [Google Scholar]
- 13.Haas C, Krinner E, Brischwein K, Hoffmann P, Lutterbüse R, Schlereth B, Kufer P, Baeuerle PA. Mode of cytotoxic action of T cell-engaging BiTE antibody MT110. Immunobiology 2009; 214:441-53; PMID:19157637; http://dx.doi.org/ 10.1016/j.imbio.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann P, Hofmeister R, Brischwein K, Brandl C, Crommer S, Bargou R, Itin C, Prang N, Baeuerle PA. Serial killing of tumor cells by cytotoxic T cells redirected with a CD19-/CD3-bispecific single-chain antibody construct. Int J Cancer 2005; 115:98-104; PMID:15688411; http://dx.doi.org/ 10.1002/ijc.20908 [DOI] [PubMed] [Google Scholar]
- 15.Brischwein K, Schlereth B, Guller B, Steiger C, Wolf A, Lutterbuese R, Offner S, Locher M, Urbig T, Raum T et al.. MT110: a novel bispecific single-chain antibody construct with high efficacy in eradicating established tumors. Mol Immunol 2006; 43:1129-43; PMID:16139892; http://dx.doi.org/ 10.1016/j.molimm.2005.07.034 [DOI] [PubMed] [Google Scholar]
- 16.Mau-Sørensen M, Dittrich C, Dienstmann R, Lassen U, Büchler W, Martinius H, Tabernero J. A phase I trial of intravenous catumaxomab: a bispecific monoclonal antibody targeting EpCAM and the T cell coreceptor CD3. Cancer Chemother Pharmacol 2015:1065-73; PMID:25814216; http://dx.doi.org/ 10.1007/s00280-015-2728-5 [DOI] [PubMed] [Google Scholar]
- 17.Abuazza G, Becker A, Williams SS, Chakravarty S, Truong H, Lin F, Baum M. Claudins 6, 9, and 13 are developmentally expressed renal tight junction proteins. Am J Physiol Renal Physiol 2006; 291:F1132-41; PMID:16774906; http://dx.doi.org/ 10.1152/ajprenal.00063.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza T, Sherman-Baust CA, Poosala S, Mullin JM, Morin PJ. Age-related changes of claudin expression in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med Sci 2009; 64:1146-53; PMID:19692671; http://dx.doi.org/ 10.1093/gerona/glp118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashizume A, Ueno T, Furuse M, Tsukita S, Nakanishi Y, Hieda Y. Expression patterns of claudin family of tight junction membrane proteins in developing mouse submandibular gland. Dev Dyn 2004; 231:425-31; PMID:15366020; http://dx.doi.org/ 10.1002/dvdy.20142 [DOI] [PubMed] [Google Scholar]
- 20.Micke P, Mattsson JSM, Edlund K, Lohr M, Jirström K, Berglund A, Botling J, Rahnenfuehrer J, Marincevic M, Pontén F et al.. Aberrantly activated claudin 6 and 18.2 as potential therapy targets in non-small-cell lung cancer. Int J Cancer 2014:2206-14; PMID:24710653; http://dx.doi.org/ 10.1002/ijc.28857 [DOI] [PubMed] [Google Scholar]
- 21.Rendón-Huerta E, Teresa F, Teresa GM, Xochitl G, Georgina A, Veronica Z, Montaño LF. Distribution and expression pattern of claudins 6, 7, and 9 in diffuse- and intestinal-type gastric adenocarcinomas. J Gastrointest Cancer 2010; 41:52-59; PMID:19960275; http://dx.doi.org/ 10.1007/s12029-009-9110-y [DOI] [PubMed] [Google Scholar]
- 22.Turksen K, Troy T. Permeability barrier dysfunction in transgenic mice overexpressing claudin 6. Development 2002; 129:1775-84; PMID:11923212 [DOI] [PubMed] [Google Scholar]
- 23.Ushiku T, Shinozaki-Ushiku A, Maeda D, Morita S, Fukayama M. Distinct expression pattern of claudin-6, a primitive phenotypic tight junction molecule, in germ cell tumours and visceral carcinomas. Histopathology 2012; 61:1043-56; PMID:22803571; http://dx.doi.org/ 10.1111/j.1365-2559.2012.04314.x [DOI] [PubMed] [Google Scholar]
- 24.Brinch M, Hatt L, Singh R, Møller K, Sommer S, Uldbjerg N, Christensen B, Kølvraa S. Identification of circulating fetal cell markers by microarray analysis. Prenat Diagn 2012; 32:742-51; PMID:22570279; http://dx.doi.org/ 10.1002/pd.3894 [DOI] [PubMed] [Google Scholar]
- 25.Ben-David U, Nudel N, Benvenisty N. Immunologic and chemical targeting of the tight-junction protein Claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat Commun 2013; 4:1992; PMID:23778593; http://dx.doi.org/ 10.1038/ncomms2992 [DOI] [PubMed] [Google Scholar]
- 26.Birks DK, Kleinschmidt-DeMasters BK, Donson AM, Barton VN, McNatt SA, Foreman NK, Handler MH. Claudin 6 Is a Positive Marker for Atypical Teratoid/Rhabdoid Tumors. BRAIN PATHOL 2010; 20:140-50; PMID:19220299; http://dx.doi.org/ 10.1111/j.1750-3639.2008.00255.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan LM, Yankovich T, Le P, Martinez D, Santi M, Biegel JA, Pawel BR, Judkins AR. Claudin-6 is a nonspecific marker for malignant rhabdoid and other pediatric tumors. Am J Surg Pathol 2012; 36:73-80; PMID:21989342; http://dx.doi.org/ 10.1097/PAS.0b013e31822cfa7e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Z, Zhang X, Liu Z, Liu Q, Wang L, Lu Y, Liu Y, Wang M, Yang M, Jin X et al.. The distinct expression patterns of claudin-2, -6, and -11 between human gastric neoplasms and adjacent non-neoplastic tissues. Diagn Pathol 2013; 8:133; PMID:23919729; http://dx.doi.org/ 10.1186/1746-1596-8-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Jin X, Lin D, Liu Z, Zhang X, Lu Y, Liu Y, Wang M, Yang M, Li J et al.. Clinicopathologic significance of claudin-6, occludin, and matrix metalloproteinases −2 expression in ovarian carcinoma. Diagn Pathol 2013; 8:190; PMID:24245968; http://dx.doi.org/ 10.1186/1746-1596-8-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Väre P, Soini Y. Twist is inversely associated with claudins in germ cell tumors of the testis. APMIS 2010; 118:640-47; PMID:20718715; http://dx.doi.org/ 10.1111/j.1600-0463.2010.02638.x [DOI] [PubMed] [Google Scholar]
- 31.Sahin U, Jaeger D, Marme F, Mavratzas A, Krauss J, de Greve J, Vergote I, Türeci Oezlem. First-in-human phase I/II dose-escalation study of IMAB027 in patients with recurrent advanced ovarian cancer (OVAR): Preliminary data of phase I part. J Clin Oncol, 2015. ASCO Meeting Abstracts; 33:15_suppl 2015: 5537); http://meeting.ascopubs.org/cgi/content/abstract/33/15_suppl/5537 [Google Scholar]
- 32.Baeuerle PA, Reinhardt C. Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res 2009; 69:4941-44; PMID:19509221; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0547 [DOI] [PubMed] [Google Scholar]
- 33.Brischwein K, Parr L, Pflanz S, Volkland J, Lumsden J, Klinger M, Locher M, Hammond SA, Kiener P, Kufer P et al.. Strictly target cell-dependent activation of T cells by bispecific single-chain antibody constructs of the BiTE class. J Immunother 2007; 30:798-807; PMID:18049331; http://dx.doi.org/ 10.1097/CJI.0b013e318156750c [DOI] [PubMed] [Google Scholar]
- 34.Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI, Guseh JS, Rajagopal J, Melton DA. Genetic targeting of the endoderm with Claudin-6(CreER). Dev Dynam 2008; 237:504-12; PMID:18213590; http://dx.doi.org/ 10.1002/dvdy.21437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assou S, Le Carrour T, Tondeur S, Ström S, Gabelle A, Marty S, Nadal L, Pantesco V, Réme T, Hugnot J et al.. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells 2007; 25:961-73; PMID:17204602; http://dx.doi.org/ 10.1634/stemcells.2006-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turksen K, Troy TC. Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Dev Dyn 2001; 222:292-300; PMID:11668606; http://dx.doi.org/ 10.1002/dvdy.1174 [DOI] [PubMed] [Google Scholar]
- 37.Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol 2007; 312:509-22; PMID:17980358; http://dx.doi.org/ 10.1016/j.ydbio.2007.09.049 [DOI] [PubMed] [Google Scholar]
- 38.Sahin U, Türeci Ö, Koslowski M, Helftenbein G, Korden W, Wöll S, Oprea GE Methods and compositions for diagnosis and treatment of cancer: Google Patents; 2010. http://www.google.ga/patents/WO2010094499A1?cl=en [Google Scholar]
- 39.Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol 2007; 81:12465-71; http://dx.doi.org/ 10.1128/JVI.01457-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaeger D, Sahin U, Türeci O. A first-in-human dose escalation and dose-finding phase I/II trial of IMAB027 in patients with recurrent advanced ovarian cancer (GM-IMAB-002-01). J Clin Oncol 2014; ASCO Meeting Abstracts; 32:15_suppl, 2014: TPS5623; http://meeting.ascopubs.org/cgi/content/abstract/32/15_suppl/TPS5623?sid=86b60114-f2ab-4a71-9649-e785cb232e0d [Google Scholar]
- 41.Traunecker A, Lanzavecchia A, Karjalainen K. Bispecific single chain molecules (Janusins) target cytotoxic lymphocytes on HIV infected cells. EMBO J 1991; 10:3655-59; PMID:1834458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong JJ, Amancha PK, Rogers K, Ansari AA, Villinger F. Re-evaluation of PD-1 expression by T cells as a marker for immune exhaustion during SIV infection. PLoS ONE 2013; 8:e60186; PMID:23555918; http://dx.doi.org/ 10.1371/journal.pone.0060186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol 2012; 24:207-12; PMID:22236695; http://dx.doi.org/ 10.1016/j.coi.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kischel R, Hausmann S, Klinger M, Baeuerle PA, Kufer P. Effector memory T cells make a major contribution to redirected target cell lysis by T cell-engaging BiTE antibody MT110. AACR poster presentation 2009 [Google Scholar]
- 45.DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev 2010; 29:309-16; PMID:20405169; http://dx.doi.org/ 10.1007/s10555-010-9223-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruffell B, DeNardo DG, Affara NI, Coussens LM. Lymphocytes in cancer development: polarization towards pro-tumor immunity. Cytokine Growth Factor Rev 2010; 21:3-10; PMID:20005150; http://dx.doi.org/ 10.1016/j.cytogfr.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiao SL, Ganesan AP, Rugo HS, Coussens LM. Immune microenvironments in solid tumors: new targets for therapy. Genes Dev 2011; 25:2559-72; PMID:22190457; http://dx.doi.org/ 10.1101/gad.169029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC et al.. Abrogation of TGF beta signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 2008; 13:23-35; PMID:18167337; http://dx.doi.org/ 10.1016/j.ccr.2007.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erdeljan M, Hubner B, Koslowski M, Kreuzberg M, Sahin U, Türeci Ö, Walter K, Weichel M, Wöll S Antibodies for treatment of cancer expressing claudin 6: Google Patents; 2012. https://www.google.ga/patents/WO2012156018A1?cl=en [Google Scholar]
- 50.Green MR, Sambrook J. Molecular cloning: A laboratory manual 4th ed Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Pr; 2012 [Google Scholar]
- 51.Wagner M, Koslowski M, Paret C, Schmidt M, Türeci O, Sahin U. NCOA3 is a selective co-activator of estrogen receptor α-mediated transactivation of PLAC1 in MCF-7 breast cancer cells. BMC Cancer 2013; 13:570; PMID:24304549; http://dx.doi.org/ 10.1186/1471-2407-13-570 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.