ABSTRACT
Recent studies have demonstrated that DNA demethylation agents can mimic a viral infection by induction of dsRNAs. This viral mimicry leads to an antiviral response mediated by the cytosolic pattern recognition receptor MDA5, followed by MAVS (IPS1) activation, IRF7 nuclear translocation and upregulation of type III Interferon and interferon-stimulated genes.
Keywords: 5-AZA-CdR, colorectal cancer, DNA methylation, decitabine, epigenetic therapy
DNA Demethylating Agents Induce Viral Mimicry
The DNA demethylating agent 5-AZA-CdR is a cytosine analog that can be incorporated into the DNA, trap the DNA methyltransferases resulting in their proteasomal degradation, and global DNA demethylation.1 We and others have recently demonstrated that 5-AZA-CdR (decitabine) treatment can induce the expression of interferon-stimulated genes (ISGs) and antiviral pathways responsive to dsRNAs in several cancer cells.2,3 We found that 5-AZA-CdR treatment increase the levels of cytoplasmic dsRNA from endogenous transcripts (Fig. 1A); including human-endogenous-retrovirus (HERVs).3 These HERVs families are remnant DNA of inactive and ancient retrovirus and represent up to 8% of our genome.4 Using genetic approaches, we identified that the cytoplasmic pattern recognition receptor MDA5 as the main sensor of 5-AZA-CdR-induced endogenous dsRNAs in cancer cells. Moreover, we found that upon 5-AZA-CdR treatment, MDA5 can signal through the mitochondrial adaptor protein MAVS, which forms prion-like aggregates and ultimately induce IRF7 nuclear translocation. The activation of the MDA5/MAVS/IRF7 axis by 5-AZA-CdR treatment induce the expression of ISGs, including type III IFNs (IL28 and IL29) but not type I IFNs in colorectal cancer cells. Interestingly, genetic inactivation of the MDA5/MAVS/IRF7 axis renders cancer cells insensitive to 5-AZA-CdR treatment. Altogether, these results suggests that induction of a viral mimicry state in cancer cells by 5-AZA-CdR is dependent on the activation of the antiviral MDA5/MAVS/IRF7 axis and type III IFNs and suggests a major role of innate immune response to the antitumor effect of DNA demethylating agents3 (Fig. 1A).
Figure 1.
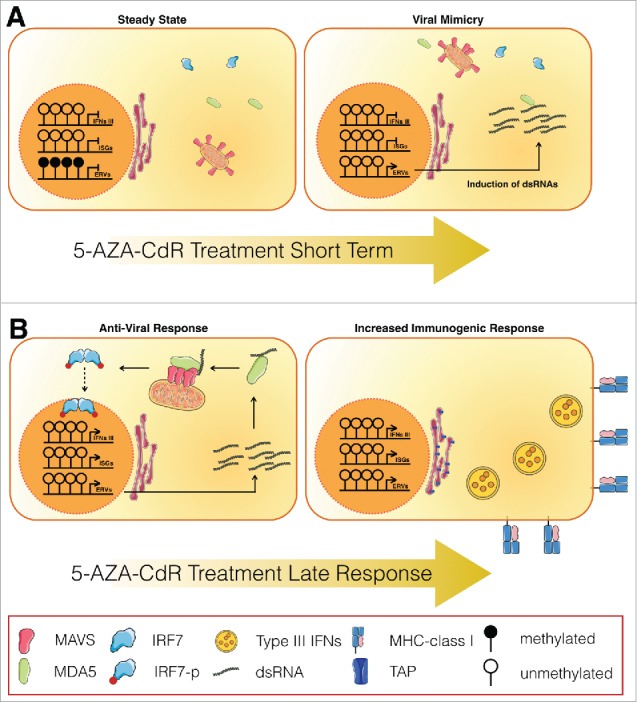
5-AZA-CdR induce viral mimicry is dependent of the MDA5/MAVS/IRF7 axis (A) Colorectal cancer cells in a steady state repress the transcription of endogenous transcript like endogenous retrovirus (ERV) by DNA methylation. Short-term treatment with 5-AZA-CdR induces expression of these dsRNA from endogenous transcript including ERVs, mimicking a viral infection. (B) As a response to the increase of dsRNA, an antiviral response will take place, involving the MDA5/MAVS/IRF7 axis. This antiviral response or ‘Viral Mimicry’ will then mediate the induction of type III IFNs, activation of interferon-stimulated genes (ISGs), increased expression of MHC class I, TAP and neo-antigens; and thus increase cancer cell immunogenicity.
Viral Mimicry Targets Colorectal Cancer-Initiating Cells
One striking consequence of this viral mimicry induction by 5-AZA-CdR was the effect on the frequency of colorectal cancer-initiating cells (CICs). Many studies have described the presence of CICs or cancer stem cells in colorectal cancer and the importance to target them for long-term therapeutic effects.5,6 In our study, we found that transient low dose of 5-AZA-CdR treatment induce a stable decrease of cancer cell proliferation rate maintained up to 20 d. Our data suggests that 5-AZA-CdR treatment is sufficient to significantly decrease the frequency of self-renewing colorectal CICs—as assessed by in vitro and in vivo limiting dilution assays using patient-derived cells. Again, we found that the ability of 5-AZA-CdR treatment in targeting colorectal CICs was dependent on the activation of the MDA5/MAVS/IRF7 virus-sensing pathway, since shRNA directed against MDA5, MAVS or IRF7 abrogate this effect. Moreover, we were able to replicate the effect of 5-AZA-CdR treatment on colorectal CICs by treatment with MDA5 and RIG1 agonists; whereas treatment with ligands of other pattern recognition receptors such as STING (cGAMP) or TLR5 (flagellin) show no effect on the frequency of CICs.3 Altogether, these results suggest that the antitumor effect of DNA demethylating agents are mediated by inducing endogenous transcripts in the form of dsRNA, activation of virus-sensing pathways, leading to the decrease of CIC frequency. Moreover, our results suggest a previously unappreciated link between the type III interferon pathway and the colorectal cancer stem cells properties (Fig. 1B).
Potential of Epigenetic Therapy to Improve Cancer Immunotherapy
Since activation of dsRNA sensor pathways and type III IFNs could potentially lead to an enhanced adaptive immune response, an interesting implication of our findings is that DNA demethylating agents may prime solid tumors to T-cell-mediated immune response and, therefore, may work in synergy with antitumor immunotherapy, such as checkpoint inhibitors. Type III IFNs have been implicated in promoting strong immune responses from type 1 helper CD4+ T cells and CD8+ T cells.7 Type III IFNs induced after viral infection are also known to trigger recruitment and activation of T cells, such as the recruitment of CD4+ T cells by inducing the production of CXCL10.8 Moreover, our gene expression data shows an increase in genes involved in antigen presentation, such as TAP1 and MHC class I, in colorectal cancer cells after 5-AZA-CdR treatment,3 thus enhancing the presentation of tumor antigens to T cells. Altogether, our data suggests a potential to modulate innate and adaptive immune response using DNA demethylating agents. It is worthy to mention that the effect of DNA demethylating agents alone in solid cancer remains poorly effective. An interesting hypothesis is that although DNA demethylating agents may increase immunogenicity of cancer cells and may attract T cells to the tumor site, these T cell are still susceptible to be repressed by immune checkpoint molecules, such as CTLA4, PDL1 and PDL2, allowing immune escape. On the other hand, the tumor then becomes dependent of this immune-repression and, therefore, susceptible to immune checkpoint therapy, such as anti-CTLA4, anti-PD1 or anti-PDL1. Indeed, incidental clinical findings suggest that non-small-cell carcinoma lung cancer patients pre-treated with 5-Azacytidine have better clinical response to subsequent anti-PD1 therapy.9 and mice models of melanoma (B16) do respond better to the combination of 5-Azacytidine plus anti-CTLA4 than 5-Azacytidine alone or anti-CTLA4 alone.2 Several clinical trials are underway or being designed to evaluate this combination of DNA demethylating agents with checkpoint inhibitors in a larger cohort of patients of different cancer types.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work in D.D.C’s laboratory is supported by grants from the Canadian Cancer Society (CCSRI703279 and CCSRI703716), Cancer Research Society (CRS19092 and CRS19091), NSERC (489073), Ontario Institute for Cancer Research (OICR) with funds from the province of Ontario, the Princess Margaret Cancer Foundation, and the University of Toronto McLaughlin Center (MC-2015–02).
References
- 1.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol 2010; 28:1069-78; PMID:20944599; http://dx.doi.org/ 10.1038/nbt.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A et al.. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015; 162:974-86; PMID:26317466; http://dx.doi.org/ 10.1016/j.cell.2015.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roulois D, Yau HL, Singhania R, Wang Y, Danesh A, Shen HY, Han H, Liang G, Jones PA, Pugh TJ et al.. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015; 162:961-973; PMID:26317465; http://dx.doi.org/ 10.1016/j.cell.2015.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths D. Endogenous retroviruses in the human genome sequence. Genome Biol 2001; 2:reviews1017.1-.5; PMID:11423012; http://dx.doi.org/ 10.1186/gb-2001-2-6-reviews1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreso A, O Brien CA, Galen PV, Gan OI, Notta F, Brown AMK, Ng K, Ma J, Wienholds E, Dunant C et al.. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013; 339:543-8; PMID:23239622; http://dx.doi.org/ 10.1126/science.1227670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kreso A, Galen Pv, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, Cao L, Baiazivito R, Du W, Sydorenko N et al.. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med 2014; 20:29-36; PMID:24292392; http://dx.doi.org/ 10.1038/nm.3418 [DOI] [PubMed] [Google Scholar]
- 7.Wack A, Terczyńska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol 2015; 16:802-9; PMID:26194286; http://dx.doi.org/ 10.1038/ni.3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkart C, Arimoto CI, Tang T, Cong X, Xiao N, Liu YC, Kotenko SV, Ellies LG, Zang DE. et al.. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFNλ and elevated secretion of Cxcl10. EMBO Mol Med 2013; 5:967-82; PMID:23681607; http://dx.doi.org/ 10.1002/emmm.201201864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodres K, Hooker CM, Franco N et al.. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011; 1:598-607; PMID:22586682; http://dx.doi.org/; http://dx.doi.org/ 10.1158/2159-8290.CD-11-0214 [DOI] [PMC free article] [PubMed] [Google Scholar]


