Significance
A fundamental challenge in materials science is to create synthetic nanoarchitectures that rival the structural complexity found in nature. A promising bioinspired approach is to synthesize sequence-defined polymer chains that fold into precise protein-like structures. Here, we discovered a family of sequence-defined, amphiphilic diblock copolypeptoids that spontaneously form crystalline, hollow nanotubes of uniform diameter, without a central hydrophobic core in aqueous solution. These low–molecular-weight polymers have two chemically distinct domains that are congruent in size and shape, which allows them to behave like molecular tile units that can form brick-like supramolecular lattices. This demonstrates the remarkable structure-directing influence of n-alkane and ethyleneoxy side chains, and that flexible polymers can serve as tile units that can assemble into precision supramolecular architectures.
Keywords: peptoid polymers, sequence-defined polymers, supramolecular assembly, polymer amphiphiles, peptoid nanostructures
Abstract
The folding and assembly of sequence-defined polymers into precisely ordered nanostructures promises a class of well-defined biomimetic architectures with specific function. Amphiphilic diblock copolymers are known to self-assemble in water to form a variety of nanostructured morphologies including spheres, disks, cylinders, and vesicles. In all of these cases, the predominant driving force for assembly is the formation of a hydrophobic core that excludes water, whereas the hydrophilic blocks are solvated and extend into the aqueous phase. However, such polymer systems typically have broad molar mass distributions and lack the purity and sequence-defined structure often associated with biologically derived polymers. Here, we demonstrate that purified, monodisperse amphiphilic diblock copolypeptoids, with chemically distinct domains that are congruent in size and shape, can behave like molecular tile units that spontaneously assemble into hollow, crystalline nanotubes in water. The nanotubes consist of stacked, porous crystalline rings, and are held together primarily by side-chain van der Waals interactions. The peptoid nanotubes form without a central hydrophobic core, chirality, a hydrogen bond network, and electrostatic or π–π interactions. These results demonstrate the remarkable structure-directing influence of n-alkane and ethyleneoxy side chains in polymer self-assembly. More broadly, this work suggests that flexible, low–molecular-weight sequence-defined polymers can serve as molecular tile units that can assemble into precision supramolecular architectures.
Self-assembly of copolymers comprising hydrophilic and hydrophobic blocks in water is a subject of long-standing interest (1, 2). The hydrophilic blocks can be either charged (e.g., polyacrylic acid) or uncharged (e.g., polyethylene oxide) (3, 4), and the hydrophobic blocks are either amorphous (e.g., polybutadiene) or crystalline (e.g., polyethylene) (5, 6). A variety of morphologies such as spheres, disks, cylinders, and vesicles have been identified in these systems (1, 3, 7–10). These morphologies comprise solid cores of the hydrophobic blocks surrounded by solvated hydrophilic chains that extend into the aqueous phase. In all of these cases, the predominant driving force for assembly is the formation of a hydrophobic core that excludes water. In a few cases, it has been shown that the enthalpic gain from the crystallization of polymer blocks can drive micellar self-assembly in organic solvents (11) and can regulate the micellar morphology or stability in aqueous solutions (12).
The recently developed ability to synthesize polymers of precisely controlled monomer sequence (13) opens up the possibility to study the fundamental relationship between polymer sequence and polymer function. Peptoids are one particularly promising class of bioinspired polymer, where the main chain length and monomer sequence can be precisely controlled (14, 15). They are prepared by an automated solid-phase synthesis method where chemically diverse side chains can be efficiently incorporated one at a time from primary amine synthons (16). The synthetic tunability of the peptoid chemical structure, and the availability of newly developed computational tools (17) make them an ideal platform to probe the assembly and crystallization properties of peptoid polymers (18–22).
In this paper, we show that a family of monodisperse amphiphilic diblock copolypeptoids, where both the hydrophilic and hydrophobic blocks are crystalline in the pure copolymer solid, can form uniform supramolecular assemblies in aqueous solution that are structurally distinct from previously reported diblock copolymer structures. Hollow crystalline tubes are formed in abundance, with lengths that exceed 100 nm as determined by electron microscopy, yet have fixed diameters of 5–10 nm. Both blocks remain crystalline in the presence of water as evidenced by differential scanning calorimetry (DSC) and X-ray scattering. Thus, significant portions of the hydrophilic chains do not extend into the aqueous phase, and the hydrophobic block does not collapse to form a central hydrophobic core. Hollow polymer nanotubes have been previously known to form, but have been limited to the domain of biology—polypeptides (23, 24) and DNA (25). In those cases, the assemblies are stabilized by networks of hydrogen bonds. A wide variety of amphiphilic organic small molecules are also known to form supramolecular nanotubes, which typically have membrane-like bilayer structures (26). Alternatively, polymer core–shell rod structures can be cross-linked in their shells and their cores degraded to produce hollow polymer tubes (27). In the present work, supramolecular assembly occurs to directly form a hollow, open structure in the absence of hydrogen-bonding networks, electrostatic or aromatic interactions, without chirality, and without forming a hydrophobic core.
Results and Discussion
The copolymers used in the study are monodisperse, low–molecular-weight diblock polypeptoids, consisting of two chemically distinct domains: a hydrophobic block of poly-N-decylglycine (pNdc), and a hydrophilic block of poly-N-2-(2-(2-methoxyethoxy)ethoxy)ethylglycine (pNte). This is a unique polymer because the two component monomers are chemically distinct yet congruent in size and shape (28). This results in unusual crystallization behavior in the solid state, where the pNdc block can induce the crystallization of an otherwise amorphous pNte block as previously reported (19). Here, we show that, in aqueous solution, this diblock copolypeptoid acts as a bendable rectangular molecular tile unit that can form planar supramolecular lattices. Typically, molecular tiles are relatively rigid units that either self-assemble in two dimensions on a surface (29), or in solution (30, 31). Recent advances in DNA nanotechnology exploit the ability of DNA-based molecular tile units to assemble into 2D and 3D lattices, as well as nanotubes (32). Molecular tiling with flexible, nonnatural polymer units has not been previously reported.
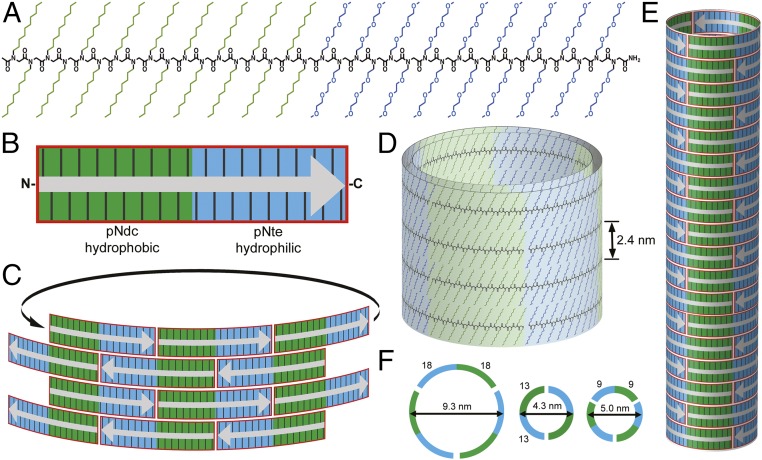
The pNdc-b-pNte block copolypeptoids were synthesized by stepwise solid-phase synthesis (16). This method allows for precision control over chain length and copolymer composition (14, 15). Three polypeptoid tile units of defined length and sequence were investigated in this study, pNdcn-b-pNten, where n = 9, 13, and 18 (Table 1). The copolymers are relatively short, with molar masses between 3.7 and 7.3 kg/mol. These sequences contain an equal number of polar and nonpolar monomers, such that the two blocks are nearly equal in mass. They are devoid of hydrogen bond donors on the side chains, were acetylated at their N termini, and amidated at their C termini such that the constructs are charge neutral. The copolypeptoids were first dissolved in organic solvent, and then diluted into water to induce supramolecular assembly. Although the exact assembly mechanism is not yet understood, the tiles likely align to form half-overlapped brick-like patterns, which can then interlock with themselves to form tubes of uniform diameter (Fig. 1). The polymer chains are thought to be extended, and their backbones aligned orthogonal to the tube axis. Both blocks form crystalline domains that bend significantly along the circumference of the tube, and are stable over a significant range of curvatures. The tube circumference comprises an integer number of polymer molecules (either 2 or 3).
Table 1.
Peptoid block copolymer pNdcn-b-pNten and nanotube characteristics
| n | Weight fraction pNte, % | Molar mass, g/mol, calc/obs | Molecular purity,* % | Nanotube diameter,† nm | Latitudinal stripe periodicity, nm, WAXS/EM | Longitudinal stripe periodicity,† nm |
| 9 | 50.4 | 3,662.7/3,664.9 | 95‡ | 5.0 ± 0.5 | 2.4 ± 0.1/2.4 ± 0.2 | 5.7 ± 0.6 |
| 13 | 50.5 | 5,264.9/5,265.6 | 95‡ | 4.3 ± 0.3 | 2.4 ± 0.1/2.4 ± 0.2 | 7.1 ± 0.5 |
| 18 | 50.6 | 7,267.3/7,263.1 | 90§ | 9.3 ± 0.3 | 2.4 ± 0.1/2.4 ± 0.2 | 11.1 ± 0.9 |
As determined by reverse-phase HPLC and MALDI-MS; the remaining impurities are predominantly full-length polymer missing one monomer.
As determined by analysis of TEM images.
As determined by MALDI (35).
As determined by HPLC.
Fig. 1.
Assembly of peptoid polymer tiles into hollow nanotubes. (A) Chemical structure of pNdc18-b-pNte18. The N terminus is acetylated and the C terminus is amidated. (B) Schematic representation of pNdc9-b-pNte9 to indicate its tile shape consisting of hydrophobic (green) and hydrophilic (blue) domains. Gray arrow indicates the chain orientation (from the N to C terminus). (C) Tiles stack in a brick-like pattern to align the polar and nonpolar domains along the tube axis. Nanotubes are composed of stacked horizontal rings, where each ring consists of two to three individual peptoid strands, likely several layers thick, held together through longitudinal side-chain–to–side-chain interactions (D and E). Schematic view (F) showing the approximate chain configuration and dimensions for each cross-sectional ring.
Evidence that both the pNdc and pNte blocks are crystalline was provided by DSC of pNdcn-b-pNten both in the solid state (as a lamellar phase-separated bulk material) and in aqueous solution (as supramolecular assemblies) (Fig. 2). In the solid state, peaks at 66 °C and 138 °C are seen in pNdc18-b-pNte18 due to the melting of pNte and pNdc crystals, respectively. These melting peaks are similar to those obtained from polyethylene oxide (PEO) and polyethylene (PE) homopolymers (33, 34). It is evident that the thermal properties of peptoids are dominated by the properties of the side chains (21). The DSC data obtained from pNdc9-b-pNte9 and pNdc13-b-pNte13 in the solid state are similar, with melting peaks at 43 °C and 93 °C, and at 60 °C and 124 °C, respectively (Fig. 2A). Aqueous solutions of all three pNdcn-b-pNten supramolecular assemblies were analyzed by DSC at a concentration of 0.6 wt % (Fig. 2A). The solutions were not perfectly clear and appeared bluish, typical of solutions containing micelle-like aggregates. The solution samples clearly show melting peaks corresponding to pNte crystals. In fact, in all three cases the melting temperature of the pNte crystals in solution is nearly identical to that of the solid-state block copolymer. This is also true for the pNdc phase in the case of pNdc9-b-pNte9 (however, the pNdc melting peaks for the other two polymers are out of the measurement range). In conventional PEO-containing diblock copolymer aqueous assemblies, the PEO chains are well solvated and random in conformation (1). In contrast, the pNte chains in these block copolypeptoids remain crystalline despite the presence of water.
Fig. 2.
Structural characteristics of the nanotubes. Differential scanning calorimetry (DSC), wide-angle X-ray scattering (WAXS), and small-angle X-ray scattering (SAXS) analysis of peptoid nanotubes. (A) Thermal analysis of pNdcn-b-pNten for (i) n = 9, (ii) n = 13, and (iii) n = 18, in both the solid state (red) and in solution (0.6 wt %) (blue). Tm, Nte is the melting point of pNte and Tm, Ndc is the melting point of pNdc in the block copolypeptoids. In the pNdc18-b-pNte18 and pNdc13-b-pNte13 solution, only Tm, Nte is observed, as Tm, Ndc is outside of the range of measurement. (B) WAXS intensity versus scattering vector, q, for pNdcn-b-pNten in solution. Profiles are vertically offset for clarity. A lamellar structure is observed with q = q*, 2q*, 3q* in pNdc18-b-pNte18, pNdc13-b-pNte13, and pNdc9-b-pNte9. The peak at q = 0.38 nm−1 is due to Kapton window in the sample cell in both cases. (C) Absolute, SAXS intensity I, normalized by polymer concentration, c, versus q for pNdc18-b-pNte18 at 25 °C over a fourfold range of concentration. Solid lines display fit on an absolute scale of a nanotube (hollow cylinder) model of inner radius of 3.2 nm, and a wall thickness of 2.3 nm, yielding an overall diameter of 11 nm. The black line includes a calculation taking into account a finite flexibility of the nanotubes (Kuhn length, 70 nm). See SI Appendix for full description of model that also includes surface roughness (0.7 nm), shape fluctuations (15%), and added crystal scattering.
Insight into the local structure and molecular packing of the nanotubes was provided by solution wide-angle X-ray scattering (WAXS). Sharp scattering peaks at q = q* = 2.6 nm−1 in all three pNdcn-b-pNten polymers are due to Bragg reflections of pNte and pNdc crystals (Fig. 2B). This peak arises from the packing of side chains, which holds the distance between adjacent backbones to 2.4 nm (=2π/q*). The same packing is seen in solid-state pNdc18-b-pNte18 (19). The higher-order scattering peaks at 2q* and 3q* indicate lamellar packing. Qualitatively similar scattering profiles are obtained from pNdc13-b-pNte13 and pNdc9-b-pNte9.
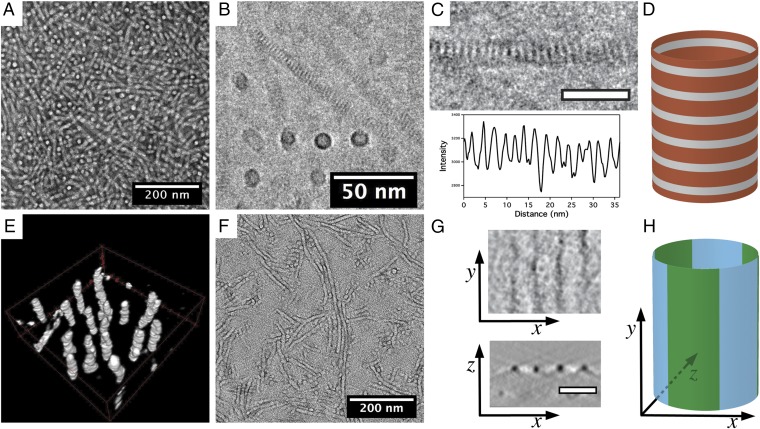
Long cylinders of uniform diameter and hundreds of nanometers in length are produced exclusively as observed by negative-stained transmission electron microscopy (TEM) of the pNdc18-b-pNte18 assemblies (Fig. 3A). Assemblies from the shorter diblock copolypeptoids, pNdc13-b-pNte13 and pNdc9-b-pNte9, also formed nanotubes, with the shortest sequence also forming appreciable amounts of planar nanosheet structures (SI Appendix, Fig. S1). Because the negative-stained samples were adsorbed to a grid surface, then dried and stained, the shorter chain samples were less able to maintain their solution morphology, likely because the tubes they form are less stable. To avoid these sample preparation effects, we next examined the nanostructures in their solution state by cryogenic electron microscopy (cryo-EM). Cryo-EM images obtained from a vitreous (unstained) pNte18-b-pNdc18 thin film are shown in Fig. 3 B and C. Cylinders that lie orthogonal to the plane of the specimen appear circular as expected. Cylinders that lie in the plane of the image contain a remarkably regular periodic pattern of latitudinal stripes (Fig. 3 C and D). The average distance between stripes as measured by cryo-EM is 2.4 ± 0.2 nm (see image analysis in SI Appendix, Fig. S2), which is consistent with the interbackbone spacing determined from WAXS (shown in Fig. 2B). The origin of the contrast observed in the TEM is thought to be due to differences in density between the backbone domain and the crystalline side-chain domains, as discussed below. Analysis of 50 nanotubes in the cryo-EM micrographs reveals that the diameter of the pNdc18-b-pNte18 tubes (9.3 ± 0.3 nm) is highly uniform along the tube length, and from tube to tube (Fig. 3 and SI Appendix, Fig. S2). The two shorter pNdcn-b-pNten variants (n = 9, 13) also formed nanotubes with identical latitudinal striping of 2.4 ± 0.2-nm periodicity, but with smaller diameters as observed by cryo-EM (SI Appendix, Fig. S2, and Table 1). These shorter variants also formed some extended planar nanosheet assemblies that exhibit the same striping pattern (SI Appendix, Fig. S2). This suggests a common packing motif for all of these diblock copolymers, where the chains pack into a planar lattice, and in the tubes are oriented normal to the tube axis.
Fig. 3.
Electron microscopy analysis of pNdc18-b-pNte18 nanotubes. (A) Uranyl acetate negative-stained energy-filtered TEM micrograph of dried nanotubes. (B) Cryo-EM 2D projection image shows nanotubes of uniform diameter in a vitrified thin film. The dark stripe features likely result from the crystallized side chains. (C) Cryo-EM close-up of a single nanotube (scale bar, 20 nm) shows regular latitudinal stripes (Top). A line profile along the longitudinal direction of the nanotube indicates a periodicity of 2.4 ± 0.2 nm (Bottom). (D) Cartoon showing the origin of the latitudinal striping based on the backbone region (gray) and the side-chain regions (orange). (E) Three-dimensional visualized image of reconstructed nanotubes in a vitreous specimen. (F) Energy-filtered TEM micrograph of ruthenium tetroxide-stained dried nanotubes on continuous carbon. The dark region is assumed to be the pNte phase. (G) Slices in tomogram show both the top view and cross-section of ruthenium tetroxide-stained nanotubes, and indicate longitudinal black striping (pNte phase) with a periodicity of 11.1 ± 0.9 nm. The x, y, and z axes are indicated relative to the nanotube in H. (H) Cartoon showing the origin of the longitudinal striping based on the hydrophobic (green) and hydrophilic (blue) domains.
One might have expected the pNdc-b-pNte peptoid chains to pack into cylindrical micelles or bilayers with pNdc hydrophobic cores and pNte coronas in aqueous solution, based on the vast literature on amphiphile self-assembly (1–10). However, the density inside the rings representing the end views of the tubes is comparable to that outside the rings, suggesting that the cylinders are hollow. Cryo-electron tomography data obtained from these samples are consistent with the proposed tubular morphology (Fig. 3E and SI Appendix, Fig. S3, and Movies S1 and S2).
To probe the detailed nanostructure and morphology of the cylinders, we performed solution small-angle X-ray scattering (SAXS) and detailed theoretical modeling of the pNte18-b-pNdc18 assemblies. The data, shown in Fig. 2C, display a very well-defined scattering pattern with clear minimum and maxima indicating a well-defined structure. The absence of a Guinier regime at low q and the defined minimum at intermediate q, suggests an elongated object with a well-defined cross-section consistent with a tubular structure. At high q, we observe a clear Bragg-like scattering peak at 2.6 nm−1, which can be attributed to the repeating crystalline domains also observed by WAXS (Fig. 2B) and cryo-EM (Fig. 3 B and C). The SAXS data were first analyzed on an absolute scale using a scattering model for a classical core-corona cylindrical scattering model expected for regular amphiphilic block copolymer micelles (35). The analysis did not provide good agreement with the experimental data using reasonable physical parameters (i.e., polymer density and diameter) once the data were constrained to an absolute scale (see SI Appendix for details). Instead, the data were nicely described with a hollow cylindrical model for nanotubes yielding an outer diameter of 11 nm and a wall thickness of 2.3 nm. This suggests that the walls are several atomic layers thick. The hollow structure was also confirmed with a model independent analysis using the indirect Fourier transform method (SI Appendix, Fig. S5).
We propose that the nanotubes are obtained by tiling molecular layers of pNdcn-b-pNten to form a brick-like pattern, which can wrap around to form a series of stacked crystalline rings (Fig. 1). The fully stretched end-to-end chain length of pNdc18-b-pNte18 in the solid state was previously estimated to be 14.3 nm (19). However, in solution assemblies, these peptoids are expected to be shorter because they are not bound in an extended 3D lattice, but rather are solvent exposed and are being pulled together by the aliphatic and ethyleneoxy side-chain packing interactions. In close proximity to one another, these side-chain moieties prefer to adopt straight conformations and pack parallel to each other to maximize neighboring van der Waals interactions (36). It is therefore expected that the latitudinal striping (of 2.4-nm periodicity) is the result of a close-packed crystalline side-chain region surrounded by less dense and less ordered backbone region (gray in Fig. 1D) (37). In this way, the tubes are likely composed of crystalline and less well-ordered domains alternating in the longitudinal direction.
The outer circumference of the pNdc18-b-pNte18 nanotubes is about three times the chain length. We thus propose that, for the pNdc18-b-pNte18 nanotubes, each crystalline ring is made of three polymer chains in a head-to-tail arrangement. Adjacent rings are likely oriented in an antiparallel arrangement to maximize the interfacial contact area of the polar and nonpolar domains with one another. A consequence of this circularly tiled architecture is that it aligns alternating hydrophilic and hydrophobic domains into extended stripes that run parallel to the tube axis. These stripes are chemically distinct, but similar in electron density and degree of crystallinity, and were thus not visible by cryo-EM. To confirm their existence, the nanotubes were dried and stained with ruthenium tetroxide, which preferentially stains the pNte side chains (38). These samples were imaged using energy-filtered TEM (EFTEM) with a 20-eV slit width (Fig. 3 F and G). The EFTEM images revealed tubes with alternating longitudinal bright and dark stripes of equal widths. The periodicity of the stripes (11 ± 0.9 nm) is equal to the peptoid main-chain length as expected. Three stripes would be expected per tube; however, because the staining was performed on dried samples, the tubes are likely collapsed so some stripes may be occluded from view. Importantly, nanotubes formed from the shorter chains, pNdc13-b-pNte13 and pNdc9-b-pNte9, also display this longitudinal striping feature, suggesting the same tiling motif is present (SI Appendix, Fig. S4).
Conclusions
In conclusion, we have shown that monodisperse amphiphilic diblock copolypeptoid tiles can assemble in water to form hollow, multiwalled crystalline nanotubes, with diameters that depend on chain length. Although a full mechanistic understanding of the self-assembly pathway is beyond the scope of this paper, it is clear that crystallization of both the hydrophilic and hydrophobic blocks plays a dominant role. Particularly remarkable is that the intertile packing interactions are strong enough to enable an open tubular structure to form with a significant uncollapsed and solvent-exposed hydrophobic domain. Ethyleneoxy and n-aliphatic packing interactions are surprisingly strong and can crystallize in thin domains that are stable despite significant curvature. The flexibility of the peptoid backbone (39) allows the side chains to coordinate their interactions. The identical size of the hydrophobic and hydrophilic blocks, and the high molecular purity of these low–molecular-weight polymers allow them to behave like molecular tiles that can pack in a brick-like lattice. The sequence-specific submonomer peptoid synthesis method allows these nanotubes to be precisely functionalized and tailored for a variety of potential applications in biomedicine and nanotechnology. More generally, the ability to mimic the precision of biomolecular architecture with sequence-defined synthetic polymers using an ever-growing set of packing interaction motifs will enable the next generation of functional nanomaterials.
Materials and Methods
Synthesis of Peptoid Polymers.
Polypeptoids were synthesized by the solid-phase submonomer method (14, 16) on an automated robotic synthesizer or a commercial Aapptec Apex 396 robotic synthesizer on 100 mg of Rink amide polystyrene resin (0.61 mmol/g; Novabiochem). One of the primary amine submonomers, 2-(2-(2-methoxyethoxy)ethoxy)ethyl amine was synthesized as previously described (40). All of the other monomers, solvents, and reagents described here were purchased from commercial sources and used without further purification. The block copolypeptoids pNdc9-b-pNte9, pNdc13-b-pNte13, and pNdc18-b-pNte18 were synthesized and purified as previously described (18, 19). The molecular purities of the isolated polymers were 95%, 95%, and 90%, respectively.
Nanotube Assembly.
Polymer stock solutions were prepared by dissolving the purified peptoids in THF (with gentle heating) to a concentration of 10 mg/mL. To a given amount of peptoid THF solution (0.3–0.6 mL), millipore water was added slowly with gentle stirring to produce a final volumetric ratio of THF:water = 1:2.5. The THF was then slowly removed under house vacuum overnight. The volume of the solution was reduced by 50% to give a final peptoid concentration of 0.6 wt %.
DSC.
DSC experiments of solid-state samples were performed to determine the thermal behavior of the synthesized peptoids using a TA Q200 Differential Scanning Calorimeter. In all tests, a scan rate of 10 K/min was used in the temperature range of −60 °C to 200 °C for three heating and cooling cycles.
Nano-DSC measurements were taken on aqueous solutions of peptoid nanotubes (0.6 wt %) on a CSC Model 6100 Nano Differential Scanning Calorimeter. A scan rate of 1 K/min was used in the temperature range of 15 °C–110 °C.
SAXS.
The SAXS experiments were performed using the automated BM29 bioSAXS beamline (41) at the European Synchrotron Radiation Facility, Grenoble, France. The data were obtained using an energy of 12.5 keV and a detector distance of 2.87 m, covering a Q range [Q = 4π sin (θ/2)/λ, where λ is the wavelength and θ is the scattering angle] of about 0.0047 Å−1 < Q < 0.5 Å−1. The data were calibrated to an absolute intensity scale using water as a primary standard.
WAXS.
The polypeptoid solutions were loaded into the center of Garolite G-10 spacers with 0.125-µm thickness and a central hole with a diameter of 3.86 mm. The samples were then sealed off in a custom-designed sample holder with Kapton windows. Measurements were performed at beamline 7.3.3 at the Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory. Samples were loaded on a custom-built temperature stage and annealed at each temperature for 15–20 min before taking measurements. A silver behenate sample was used as a standard. Full 2D scattering patterns were collected on an ADSC CCD detector. The scattering patterns from the ALS were reduced using the Nika program for Igor Pro available from Jan Ilavsky at Argonne National Laboratory (Argonne, IL) (42).
EM Characterization and Sample Preparation.
The vitrified specimens were prepared using a Vitrobot (FEI, Inc.). Grids with lacey carbon films were glow discharged to produce a hydrophilic surface, and a 3-µL droplet of the aqueous nanotube suspension was deposited on the surface. The droplet was blotted by filter paper and plunged into liquid ethane to obtain a vitrified thin film. The vitrified specimens were examined using a Philips CM200 cryo-EM at −185 °C to obtain 2D projections, and tilt series were collected using Serial EM (43) on JEOL 3100 cryo-EM at 300 kV with a 40-eV slit width. Tilt series included 32 images with a 4° tilting step with accumulated dose about 60 electrons per Å2. Tile series were recorded using the dose fraction mode of a K2 direct detection camera (Gatan) with defocus of 4 µm. The dose fractionated images were aligned using the bandpass filter in DigitalMicrograph (Gatan) and averaged to obtain the micrographs for reconstruction. The pixel size in micrographs was 0.365 nm.
Nanotubes were negative stained using uranyl acetate. A mixture of 1.5 µL of nanotubes aqueous solution and 1.5 µL of uranyl acetate aqueous solution was dropped on the grid with a hydrophilic, continuous carbon film. Nanotubes were also positively stained using ruthenium tetroxide vapor. A grid with dried nanotubes was placed in a well-sealed glass vial that contained 4% (wt/vol) aqueous ruthenium tetroxide solution for 10 min. All stained dried grids were observed using a Tecnai F20 (FEI, Inc.) at 200 kV with a 20-eV slit-width to obtain 2D projections and tilt series. The angular range of tilting was from −70° to 70° with a 1° step. The reconstruction of tilt series and processing and visualization of tomograms were carried out using IMOD (44, 45) and the Avizo visualization package.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Soft Matter Electron Microscopy Program, supported by the Office of Science, Office of Basic Energy Science, US Department of Energy, under Contract DE-AC02-05CH11231. Work at the Molecular Foundry and the Advanced Light Source at Lawrence Berkeley National Laboratory was supported by the Office of Science, Office of Basic Energy Science, US Department of Energy, under Contract DE-AC02-05CH11231. R.L. acknowledges grants from the Norwegian Research Council, under the SYNKNOYT Program (218411 and 228573), and a travel grant from the Peder Sather Center for Advanced Study. Cryo-EM facilities are supported by NIH Grant GM51487. The European Synchrotron Radiation Facility is acknowledged for allocation of beam time at the BM29 bioSAXS instrument.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517169113/-/DCSupplemental.
References
- 1.Jain S, Bates FS. On the origins of morphological complexity in block copolymer surfactants. Science. 2003;300(5618):460–464. doi: 10.1126/science.1082193. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Kesselman E, Talmon Y, Hillmyer MA, Lodge TP. Multicompartment micelles from ABC miktoarm stars in water. Science. 2004;306(5693):98–101. doi: 10.1126/science.1103350. [DOI] [PubMed] [Google Scholar]
- 3.Zhang L, Eisenberg A. Multiple morphologies of “crew-cut” aggregates of polystyrene-b-poly(acrylic acid) block copolymers. Science. 1995;268(5218):1728–1731. doi: 10.1126/science.268.5218.1728. [DOI] [PubMed] [Google Scholar]
- 4.Pochan DJ, et al. Toroidal triblock copolymer assemblies. Science. 2004;306(5693):94–97. doi: 10.1126/science.1102866. [DOI] [PubMed] [Google Scholar]
- 5.Won Y-Y, Davis HT, Bates FS. Giant wormlike rubber micelles. Science. 1999;283(5404):960–963. doi: 10.1126/science.283.5404.960. [DOI] [PubMed] [Google Scholar]
- 6.Won Y-Y, Brannan AK, Davis HT, Bates FS. Cryogenic transmission electron microscopy (Cryo-TEM) of micelles and vesicles formed in water by poly(ethylene oxide)-based block copolymers. J Phys Chem B. 2002;106(13):3354–3364. [Google Scholar]
- 7.Percec V, et al. Self-assembly of amphiphilic dendritic dipeptides into helical pores. Nature. 2004;430(7001):764–768. doi: 10.1038/nature02770. [DOI] [PubMed] [Google Scholar]
- 8.Yin L, Hillmyer MA. Disklike micelles in water from polyethylene-containing diblock copolymers. Macromolecules. 2011;44(8):3021–3028. [Google Scholar]
- 9.Discher DE, Eisenberg A. Polymer vesicles. Science. 2002;297(5583):967–973. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- 10.Raez J, Manners I, Winnik MA. Nanotubes from the self-assembly of asymmetric crystalline-coil poly(ferrocenylsilane-siloxane) block copolymers. J Am Chem Soc. 2002;124(35):10381–10395. doi: 10.1021/ja020349h. [DOI] [PubMed] [Google Scholar]
- 11.Gilroy JB, et al. Monodisperse cylindrical micelles by crystallization-driven living self-assembly. Nat Chem. 2010;2(7):566–570. doi: 10.1038/nchem.664. [DOI] [PubMed] [Google Scholar]
- 12.Yin L, Lodge TP, Hillmyer MA. A stepwise “micellization–crystallization” route to oblate ellipsoidal, cylindrical, and bilayer micelles with polyethylene cores in water. Macromolecules. 2012;45(23):9460–9467. [Google Scholar]
- 13.Lutz JF, Ouchi M, Liu DR, Sawamoto M. Sequence-controlled polymers. Science. 2013;341(6146):1238149. doi: 10.1126/science.1238149. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Zuckermann RN. Peptoid polymers: A highly designable bioinspired material. ACS Nano. 2013;7(6):4715–4732. doi: 10.1021/nn4015714. [DOI] [PubMed] [Google Scholar]
- 15.Knight AS, Zhou EY, Francis MB, Zuckermann RN. Sequence programmable peptoid polymers for diverse materials applications. Adv Mater. 2015;27(38):5665–5691. doi: 10.1002/adma.201500275. [DOI] [PubMed] [Google Scholar]
- 16.Zuckermann RN, Kerr JM, Kent SB, Moos WH. Efficient method for the preparation of peptoids [oligo (N-substituted glycines)] by submonomer solid-phase synthesis. J Am Chem Soc. 1992;114(26):10646–10647. [Google Scholar]
- 17.Mannige RV, et al. Peptoid nanosheets exhibit a new secondary-structure motif. Nature. 2015;526(7573):415–420. doi: 10.1038/nature15363. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Teran AA, Liao X, Balsara NP, Zuckermann RN. Nanoscale phase separation in sequence-defined peptoid diblock copolymers. J Am Chem Soc. 2013;135(38):14119–14124. doi: 10.1021/ja404233d. [DOI] [PubMed] [Google Scholar]
- 19.Sun J, Teran AA, Liao X, Balsara NP, Zuckermann RN. Crystallization in sequence-defined peptoid diblock copolymers induced by microphase separation. J Am Chem Soc. 2014;136(5):2070–2077. doi: 10.1021/ja412123y. [DOI] [PubMed] [Google Scholar]
- 20.Nam KT, et al. Free-floating ultrathin two-dimensional crystals from sequence-specific peptoid polymers. Nat Mater. 2010;9(5):454–460. doi: 10.1038/nmat2742. [DOI] [PubMed] [Google Scholar]
- 21.Rosales AM, Murnen HK, Zuckermann RN, Segalman RA. Control of crystallization and melting behavior in sequence specific polypeptoids. Macromolecules. 2010;43(13):5627–5636. [Google Scholar]
- 22.Murnen HK, Rosales AM, Jaworski JN, Segalman RA, Zuckermann RN. Hierarchical self-assembly of a biomimetic diblock copolypeptoid into homochiral superhelices. J Am Chem Soc. 2010;132(45):16112–16119. doi: 10.1021/ja106340f. [DOI] [PubMed] [Google Scholar]
- 23.Hamley IW. Peptide nanotubes. Angew Chem Int Ed Engl. 2014;53(27):6866–6881. doi: 10.1002/anie.201310006. [DOI] [PubMed] [Google Scholar]
- 24.Valéry C, Artzner F, Paternostre M. Peptide nanotubes: Molecular organisations, self-assembly mechanisms and applications. Soft Matter. 2011;7(20):9583–9594. [Google Scholar]
- 25.Yin P, et al. Programming DNA tube circumferences. Science. 2008;321(5890):824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu T, Masuda M, Minamikawa H. Supramolecular nanotube architectures based on amphiphilic molecules. Chem Rev. 2005;105(4):1401–1443. doi: 10.1021/cr030072j. [DOI] [PubMed] [Google Scholar]
- 27.Huang K, Rzayev J. Well-defined organic nanotubes from multicomponent bottlebrush copolymers. J Am Chem Soc. 2009;131(19):6880–6885. doi: 10.1021/ja901936g. [DOI] [PubMed] [Google Scholar]
- 28.Hill JP, et al. Self-assembled hexa-peri-hexabenzocoronene graphitic nanotube. Science. 2004;304(5676):1481–1483. doi: 10.1126/science.1097789. [DOI] [PubMed] [Google Scholar]
- 29.Whitelam S. Examples of molecular self-assembly at surfaces. Adv Mater. 2015;27(38):5720–5725. doi: 10.1002/adma.201405573. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Hu C, Comotti A, Ward MD. Supramolecular Archimedean cages assembled with 72 hydrogen bonds. Science. 2011;333(6041):436–440. doi: 10.1126/science.1204369. [DOI] [PubMed] [Google Scholar]
- 31.Xu L, et al. Self-assembly of ultralong polyion nanoladders facilitated by ionic recognition and molecular stiffness. J Am Chem Soc. 2014;136(5):1942–1947. doi: 10.1021/ja410443n. [DOI] [PubMed] [Google Scholar]
- 32.Lin C, Liu Y, Rinker S, Yan H. DNA tile based self-assembly: Building complex nanoarchitectures. ChemPhysChem. 2006;7(8):1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 33.Romankevich O, Frenkel SY. Equilibrium melting point of polyethylene oxide. Polym Sci. USSR. 1980;22(11):2647–2654. [Google Scholar]
- 34.Włochowicz A, Eder M. Distribution of lamella thicknesses in isothermally crystallized polypropylene and polyethylene by differential scanning calorimetry. Polymer (Guildf) 1984;25(9):1268–1270. [Google Scholar]
- 35.Lund R, et al. Equilibrium chain exchange kinetics of diblock copolymer micelles: Effect of morphology. Macromolecules. 2011;44(15):6145–6154. [Google Scholar]
- 36.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev. 2005;105(4):1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 37.Wirix MJ, Bomans PH, Friedrich H, Sommerdijk NA, de With G. Three-dimensional structure of P3HT assemblies in organic solvents revealed by cryo-TEM. Nano Lett. 2014;14(4):2033–2038. doi: 10.1021/nl5001967. [DOI] [PubMed] [Google Scholar]
- 38.Li B, Li L, Wang B, Li CY. Alternating patterns on single-walled carbon nanotubes. Nat Nanotechnol. 2009;4(6):358–362. doi: 10.1038/nnano.2009.91. [DOI] [PubMed] [Google Scholar]
- 39.Rosales AM, Murnen HK, Kline SR, Zuckermann RN, Segalman RA. Determination of the persistence length of helical and non-helical polypeptoids in solution. Soft Matter. 2012;8(13):3673–3680. [Google Scholar]
- 40.Sun J, Stone GM, Balsara NP, Zuckermann RN. Structure-conductivity relationship for peptoid-based PEO-mimetic polymer electrolytes. Macromolecules. 2012;45(12):5151–5156. [Google Scholar]
- 41.Pernot P, et al. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J Synchrotron Rad. 2013;20:660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilavsky J. Nika: Software for two-dimensional data reduction. J Appl Cryst. 2012;45:324–328. [Google Scholar]
- 43.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 44.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 45.Mastronarde DN. Dual-axis tomography: An approach with alignment methods that preserve resolution. J Struct Biol. 1997;120(3):343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.