Significance
Interest in male mosquitoes has been motivated by the potential to develop novel vector control strategies, exploiting the fact that males do not feed on blood or transmit diseases, such as malaria. However, genetic studies of male Anopheles mosquitoes have been impeded by the lack of molecular characterization of the Y chromosome. Here we show that the Anopheles gambiae Y chromosome contains a very small repertoire of genes, with massively amplified tandem arrays of a small number of satellites and transposable elements constituting the vast majority of the sequence. These genes and repeats evolve rapidly, bringing about remodeling of the Y, even among closely related species. Our study provides a long-awaited foundation for studying mosquito Y chromosome biology and evolution.
Keywords: Anopheles gambiae, PacBio, RNA-Seq, tandem repetitive DNA, Y-chromosome
Abstract
Y chromosomes control essential male functions in many species, including sex determination and fertility. However, because of obstacles posed by repeat-rich heterochromatin, knowledge of Y chromosome sequences is limited to a handful of model organisms, constraining our understanding of Y biology across the tree of life. Here, we leverage long single-molecule sequencing to determine the content and structure of the nonrecombining Y chromosome of the primary African malaria mosquito, Anopheles gambiae. We find that the An. gambiae Y consists almost entirely of a few massively amplified, tandemly arrayed repeats, some of which can recombine with similar repeats on the X chromosome. Sex-specific genome resequencing in a recent species radiation, the An. gambiae complex, revealed rapid sequence turnover within An. gambiae and among species. Exploiting 52 sex-specific An. gambiae RNA-Seq datasets representing all developmental stages, we identified a small repertoire of Y-linked genes that lack X gametologs and are not Y-linked in any other species except An. gambiae, with the notable exception of YG2, a candidate male-determining gene. YG2 is the only gene conserved and exclusive to the Y in all species examined, yet sequence similarity to YG2 is not detectable in the genome of a more distant mosquito relative, suggesting rapid evolution of Y chromosome genes in this highly dynamic genus of malaria vectors. The extensive characterization of the An. gambiae Y provides a long-awaited foundation for studying male mosquito biology, and will inform novel mosquito control strategies based on the manipulation of Y chromosomes.
Sex chromosomes carry a master switch gene responsible for sex determination (1). They are thought to derive from an ordinary pair of autosomes, and have multiple independent origins across the tree of life (2, 3). In animals with morphologically distinct (heterogametic) sex chromosomes, the Y has ceased crossing over with the X across some or all of its length and the nonrecombining region is transmitted clonally by males (4, 5). The absence of recombination initiates progressive genetic decay—gene loss and accumulation of repetitive sequences—but there is increasing recognition that even relatively old and otherwise highly degenerate Y chromosomes retain functional importance not only for sexual reproduction but for their contributions to global gene regulation, affecting health and survival (6–10). Notwithstanding these critical roles, the Y chromosome remains one of the most recalcitrant and poorly characterized portions of any genome more than a decade into the postgenomic era, with current knowledge resting largely on only two animal groups: mammals and Drosophila (2, 11).
Mosquitoes in the genus Anopheles are the exclusive vectors of human malaria, a disease that claimed nearly 600,000 lives globally in 2013, the majority in sub-Saharan Africa (12). Although 15 y of intensified vector control efforts (mainly insecticide-impregnated bed nets) have successfully averted an estimated 663 million clinical cases of malaria (13), further progress toward elimination in the most malarious regions will depend upon the development of novel methods of vector control complementary to existing approaches (14). One method currently under development entails genetic modification of the mosquito to bias the population sex ratio toward males (which do not bite), with the goal of local population reduction or elimination (15–17). Modeling has shown that the most efficient means toward this end is the engineering of a driving Y chromosome (18). A molecular-level understanding of the Anopheles Y chromosome is important to inform and optimize such a strategy.
Historical cytogenetic studies established that the Anopheles Y chromosome is entirely heterochromatic, but also suggested that, contrary to Drosophila and in common with mammals, it bears partial homology to the X chromosome and plays a male-determining role (19–24). However, efforts to characterize Y chromosome sequences in Anopheles have been thwarted by a lack of directed resources and effective tools. The unsurpassed medical importance of the African malaria mosquito Anopheles gambiae motivated its early selection for whole-genome sequencing (25), making it second only to the model organism Drosophila melanogaster as a fully sequenced insect genome. However, because of formidable obstacles to assembling repeat-dense Y chromosome sequences (26), efforts to assemble or even to assign Y chromosome sequences in the framework of the An. gambiae genome project were largely unsuccessful, despite separate sequencing of males and females (27), leaving its content and organization obscure. Here, we leverage the increased length and reduced bias of single-molecule sequencing (28), along with Illumina-based sex-specific transcriptional profiling and whole-genome sequencing, to identify an extensive dataset of Y chromosome sequences and explore their organization and evolution in the young species radiation known as the An. gambiae complex (29), which contains some of the most important vectors of human malaria. We find massive remodeling of the repeat-dense Y chromosome and remarkably few genes, none of which have counterparts on the X. Only the YG2 gene—the earliest to be expressed in 3-h male embryos—is conserved and exclusive to the Y across species in the complex, and thus is a possible male-determining factor. However, no sequence similarity to YG2 can be detected in the genome of an Asian malaria vector from the same subgenus (30), underscoring the rapid evolution of Y-linked genes in the evolutionarily dynamic genus Anopheles (31). Contrary to the species branching order, the YG2 gene tree from the An. gambiae complex supported the grouping of two major malaria vectors with a history of substantial autosomal introgression (32), consistent with the hypothesis that even the Y chromosome may have crossed species boundaries. Although a Y chromosome assembly awaits further technological developments, the compilation and comparative analysis of Y chromosome sequences in the An. gambiae complex substantially advances our understanding of the composition, organization, and evolution of the Anopheles Y chromosome and lays the groundwork for exploiting the Y chromosome to control disease transmission.
Results
Identification of An. gambiae Y Chromosome Sequences.
Gross cytological estimates suggest that the An. gambiae Y chromosome constitutes ∼10% of the 264-Mb An. gambiae genome (27, 33), yet a mere 0.18 Mb of unordered sequences have previously been assigned to the Y in the PEST reference genome assembly [https://www.vectorbase.org (34)]. Similarly, a recent Anopheles stephensi genome project identified only 57 short unordered Y sequences spanning ∼50 kb from genomic reads, and 11 contigs spanning ∼200 kb from BAC clones that were assigned to the Y chromosome (35). To overcome this impediment, we developed a strategy based on long-read, PacBio single molecule real-time sequencing (36). Template genomic DNA was extracted from male siblings of a single-pair mating that inherited the same paternal An. gambiae Y chromosome and was sequenced to 70× autosomal (35× heterosomal) coverage with PacBio single molecule real-time sequencing (SI Appendix, Text S1.1). Consensus-based error correction with PBcR (37) resulted in 40× autosomal (20× heterosomal) coverage of PacBio-corrected reads with an N50 size of 2,799 bp (SI Appendix, Text S1.2). A whole-genome assembly of the entire PacBio dataset was performed with Celera Assembler (38, 39), resulting in a 294-Mb assembly with an N50 contig size of 101,465 bp (SI Appendix, Text S1.2). However, the moderate coverage and average raw read length of 2,479 bp proved insufficient for de novo reconstruction of the Y chromosome. Initial analysis of this assembly revealed highly fragmented heterosomal contigs and evidence that autosomal and heterosomal sequence had been incorrectly joined. Because of known limitations in assembling heterochromatic sequence and concerns about potential mis-assemblies, we decided to focus exclusively on individual (unassembled) PacBio-corrected reads from genomic DNA for all subsequent Y chromosome analysis.
As a complementary strategy to investigate the organization of large (100 kb) contiguous pieces of the Anopheles Y chromosome, PacBio sequencing of individual An. gambiae BAC clones was performed (SI Appendix, Text S1.3). These BACs were deemed potentially Y-linked based on initial computational analysis of available BAC-end sequences (SI Appendix, Text S1.3). Directed, high-coverage PacBio sequencing, ranging from 300× to 2,000× per BAC, yielded sufficient information to completely assemble each BAC without ambiguity. Successful BAC assembly was a result of localization of the repeat structure as well as the tremendous sequencing depth attained. Because PacBio read lengths are exponentially distributed, such deep sequencing increases the probability of obtaining some very long reads (e.g., >20 kb), which were necessary for the assembly of these highly repetitive sequences. Comprehensive computational analysis supported three of four assembled BACs as originating from the Y chromosome (SI Appendix, Text S1.3, and Figs. S1 and S2).
To identify presumptive Y-linked sequences among the unassembled genomic PacBio reads, we implemented two recent computational approaches (SI Appendix, Text S2) that exploit short-read (Illumina) genomic sequencing from sex-specific DNA pools (SI Appendix, Text S1.4, and Table S1). The Y chromosome genome scan (YGS) approach was designed to operate on scaffolds from a genome assembly derived from mixed sexes or males; after identification and masking of identical repeats, scaffolds are classified as Y-linked if they have few or no kmer-length matches to female Illumina sequences (40). As applied to An. gambiae male PacBio-corrected reads, which were treated as “scaffolds,” YGS failed to unambiguously classify Y-linked sequences, apparently because of the extremely small fraction of Y chromosome sequence that is exclusive to the Y in An. gambiae as opposed to highly enriched there (see below and SI Appendix, Table S8). A second approach, the chromosome quotient (CQ) method (30), infers Y-linkage based on the female-to-male ratio of sequence alignments to a reference, in this case An. gambiae female-to-male Illumina sequences aligned to PacBio reads. At a conservative threshold value (CQ ≤ 0.2) imposed across the length of a PacBio read, the CQ method classified 79,475 unassembled reads (246 Mb) as presumptive Y chromosome sequences, which populate a database that we denote Ydb (SI Appendix, Text S2.2, and Tables S4–S7, and Dataset S1). Although the rate of false-positives in Ydb should be low (30) (SI Appendix, Text S2.1), the conservative CQ threshold necessarily means that Ydb is incomplete with respect to possible Y chromosome sequences that share extended sequence identity with other chromosomes, as would be expected for pseudoautosomal regions or sequences recently acquired from elsewhere in the genome. However, it is likely that Ydb represents much of the nonrecombining (male-limited) Y chromosome (NRY) (SI Appendix, Table S4). Greater than 94% of sequence classes comprising Ydb were validated as Y-linked in An. gambiae (Fig. 1 and SI Appendix, Text S3, and Table S9) through genomic PCR and physical mapping by FISH of representative sequences to mitotic chromosomes of male An. gambiae larvae (SI Appendix, Text S4).
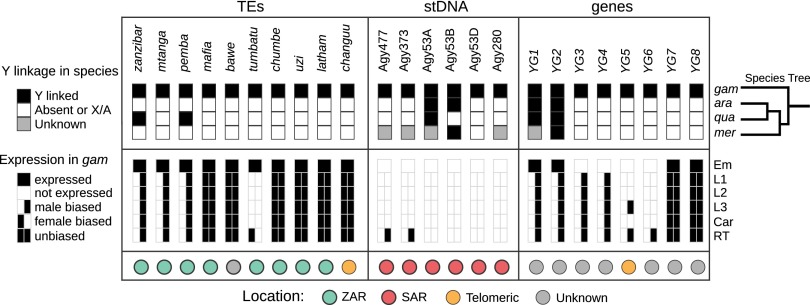
Fig. 1.
Summary of major Y chromosome loci, showing rapid turnover of the Y chromosome content and expression patterns in the An. gambiae species complex. (Top) Black boxes indicate Y-linkage, white boxes indicate either total absence from the species or absence from its Y chromosome, and gray boxes indicate unknown status with regard to Y-linkage. Typically, sequences indicated by gray showed CQ or RCQ values of ∼1, suggesting that they are either on both sex chromosomes or on autosomes. Details are provided in SI Appendix, Tables S6, S7, and S15. At right, the species branching order provides an evolutionary context of the changes in Y chromosome content within the past 2 My. Only YG2 is conserved and exclusively on the Y chromosome in all four species of the An. gambiae complex. (Middle) Sex-specific transcription in An. gambiae was assessed at different developmental stages and tissues, except for embryos (Em). (Bottom) The organization of the Y chromosome loci in An. gambiae, if known. ara, An. arabiensis; Car, adult carcass; Em, embryo; gam, An. gambiae; L1–L3, first- to third-instar larvae; mer, An. merus; RT, adult reproductive tissues; stDNA, satellite DNA; qua, An. quadriannulatus.
The An. gambiae Y Contains Massively Amplified Satellites and Retrotransposons.
We conducted a detailed computational assessment of Y chromosome repeat content based on analysis of Ydb. Our inferences should be minimally affected by redundant and overlapping reads, as they are based on proportional content (relative abundance), and PacBio coverage has limited bias (28, 38). Initially, both PacBio Ydb reads and assembled BAC sequences were screened for interspersed repeats and low-complexity DNA with RepeatMasker 4.0.3 (41), using the An. gambiae PEST RepeatMasker library augmented with previously characterized Y chromosome satellite and retrotransposon sequences (42, 43) (SI Appendix, Text S3). Anticipating that the An. gambiae Y chromosome contains previously unknown repeats, or repeats whose structures differ from those represented in the reference repeat library, we characterized both annotated and unclassified output from RepeatMasker through iterative clustering and consensus building of sequences in Ydb and the Y-linked BACs. This strategy ultimately revealed that ∼98% of bases in Ydb belong to a very few repetitive sequence classes, amplified extensively (Fig. 2A and SI Appendix, Text S3, and Table S9).
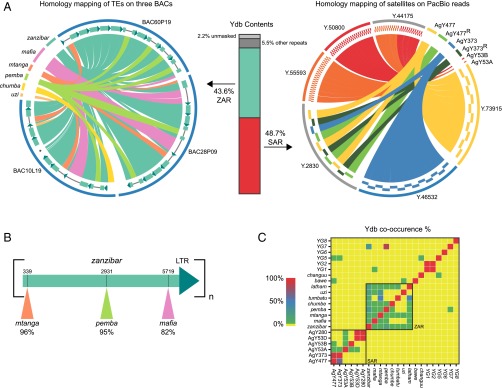
Fig. 2.
The NRY of An. gambiae mainly consists of massively amplified tandem arrays of a small number of satellites and TEs. (A) Two major regions of the An. gambiae Y, the ZAR and SAR, represent 92.3% of the sequences in Ydb (vertical bar plot). Ydb reflects the content of NRY in An. gambiae. Percentages were calculated by masking Ydb using annotated Y chromosome loci. The Left circos plot, created by homology mapping of TEs on three Y chromosome BAC clones, shows the organization of the ZAR in the three BACs. As seen in these BACs, and as independently confirmed in PacBio reads, the ZAR consists of head-to-tail tandem arrays of zanzibar, which sometimes have other transposons inserted. The arrays of zanzibar units inside each BAC are shown schematically directly inside the BAC ideograms (blue semicircular lines) enclosing the circus plot. The dark green arrows of each zanzibar unit (shown enlarged in B) represent the single LTR; lines breaking zanzibar units indicate insertions of other TEs. A few small insertions (∼200 bp) into zanzibar are too small to be visible in this plot. The asterisk in BAC10L19 corresponds to an atypical zanzibar unit that could result from recombination or misassembly. The circos plot at Right, constructed by homology mapping of satellite monomers on PacBio reads from Ydb, shows the organization of the SAR. Shown are representative examples of the occurrence of homo-monomeric tandem arrays (Y73915, Y46532, Y55593), junctions between homo-monomeric tandem arrays (Y44175), and recombinant arrays (Y2830). The recombinant arrays are interspersed with recombinant and nonrecombinant versions of AgY477 and AgY373 satellites (SI Appendix, Fig. S5). (B) Schematic of a single zanzibar unit, consisting of a gag/pol domain and a single LTR; each unit is organized in a head-to-tail tandem array (see Left circus plot in A). Shown by colored triangles are the canonical insertion sites of three other transposons (mtanga, pemba, mafia) into different zanzibar units. Percentages indicate the fraction of Ydb PacBio reads observed to carry TE insertions into zanzibar units at the precise insertion site illustrated (coordinates shown above the gag/pol domain). For example, we observed 243 of 256 (95%) PacBio reads in which pemba was inserted into zanzibar at position 2931. This phenomenon was independently confirmed in whole-genome sequencing Illumina reads. (C) Co-occurrence matrix of Y chromosome loci in PacBio reads from Ydb. These results show that satellite sequences co-occur (in the SAR), as do TEs (in the ZAR), but that the ZAR and SAR regions are largely independent.
Satellite DNA accounts for ∼49% of all bases in Ydb (SI Appendix, Table S9). However, only six different satellite monomers were identified and two—AgY477 and AgY373—predominate, comprising 93% of all satellite DNA sequence in Ydb. Moreover, the satellite sequences are found as long tandem arrays in Ydb, largely devoid of transposable elements (TEs). These data suggest that satellite DNA is an abundant and homogeneous constituent of the NRY, a major Y chromosome sequence feature that we refer to as the satellite amplified region (SAR) (Fig. 2A). The absence of other repetitive sequence classes interspersed within the SAR suggests limited genetic exchange between satellites and other repeats, but we find evidence of recombination and higher-order repeat structures among satellites within the SAR. Different satellite monomers that share extensive sequence similarity (AgY477 and AgY373; AgY280 and AgY53D) frequently co-occur on the same PacBio read, often as interspersed or even chimeric monomers, indicative of sister chromatid or intrachromatid exchange (Fig. 2C, and SI Appendix, Text S3.1, and Figs. S5–S8).
Another 43.5% of bases in Ydb are TE-related, of which only eight distinct TE types, mainly retrotransposons, were identified (SI Appendix, Text 3.2, and Table S9). Remarkably, one particular element alone—a 6.9-kb Ty3/Gypsy LTR retrotransposon that we designate zanzibar—comprises almost 27% of bases in Ydb as a whole, and accounts for more than 61% of the bases classified as TEs (Fig. 2 and SI Appendix, Table S9). Unlike the typical interspersed arrangement of TEs in genomic euchromatin, zanzibar is arranged on the Y in massively amplified head-to-tail tandem arrays, in which one gag/pol region followed by one LTR is repeated in succession like beads on a string: (gag/pol+LTR)n. From its abundance and organization, we infer that this zanzibar amplified region (ZAR) (Fig. 2 A and B)—like the SAR—is another prominent organizational feature of the NRY. Zanzibar monomers (gag/pol+LTR) in the array may carry insertions of a variety of other TEs or TE fragments. Remarkably, every copy of zanzibar carrying a particular TE type (e.g., mtanga) contains precisely the same TE sequence inserted into precisely the same zanzibar sites (Fig. 2 and SI Appendix, Text S3.2), as though replica insertions in different zanzibar copies are not the result of independent transposition events. Taken together, these data—the precise tandem organization of the ZAR and the peculiar clonal nature of insertions—strongly suggest that zanzibar retrotransposons no longer function in the manner expected of autonomous transposable elements. Instead, zanzibar sequences appear to have been the substrate for illegitimate recombination and megabase-spanning tandem amplifications on the Y chromosome, analogous to the process described for the genesis of centromeric repeats in maize (44). Despite its evident origin as an autonomous TE, the present ZAR structure most closely resembles satellite sequence and thus may reflect a general pattern of sequence amplification and evolution on the Anopheles Y chromosome.
Of the remaining ∼7.5% of Ydb bases, we were able to classify ∼5.5% as repetitive, but the last ∼2% could not be clustered (SI Appendix, Table S9). This small unclustered fraction contains a heterogeneous mixture of less abundant types of repetitive sequences, degenerate copies of repeats categorized above, and Y chromosome genes, many of which also appear to be multicopy (see below and SI Appendix, Text S7, and Fig. S11). Only four Ydb reads were classified by a metagenomic analysis as originating from another organism (SI Appendix, Text S2.2), suggesting that nearly all of Ydb is legitimate An. gambiae sequence.
Extensive Structural Dynamism of the Y Chromosome in a Young Species Radiation.
Cytological observations conducted in the 1970s revealed striking differences in sex chromosome heterochromatin among populations and between species in this complex (45, 46). Not only did the staining intensity and pattern vary, but also length of the Y chromosome, ranging from less than half the length of the X in one An. gambiae population to almost the same length as the X in others (45). However, a mechanistic understanding of the phenomenon was lacking. Our finding that ∼98% of the bases in Ydb constitute a highly repetitive sequence organized into tandem arrays suggests that the cytological observations may have their basis in rapid expansion and contraction of tandem repeats on the Y chromosome, through unequal crossover and a variety of other mechanisms (42, 43, 47–49). We applied computational and cytogenetic methods to assess the nature and degree of Y chromosome remodeling within An. gambiae and among sibling species during the relatively brief (2 My) evolutionary history of the species complex.
Intraspecific variation in the SAR and ZAR of An. gambiae was assessed computationally among three laboratory colonies and among 85 male and female mosquitoes sampled from a natural population in Cameroon, available through MalariaGEN’s An. gambiae 1000 Genomes Consortium (Ag1000G) phase 1 AR3 data release (2015) (www.malariagen.net/data/ag1000g-phase1-AR3) (SI Appendix, Text S1.4–S1.5, and Tables S1 and S2). The sample set from Cameroon is one of the few available that contains both sexes. From the An. gambiae Pimperena colony—our reference—and two additional colonies (G3 and Asembo), we generated Illumina sequences from sex-specific genomic DNA pools, and aligned them to the consensus sequences of An. gambiae monomer repeat units compiled from Ydb. Alignments were performed twice, using either a strict read-mapping protocol (for CQ calculations) or a less-stringent mapping protocol used to produce a metric analogous to CQ, termed “relaxed CQ” (RCQ) (SI Appendix, Text S5, and Tables S6 and S7). The presence/absence and relative abundance of each major repeat was estimated from the number of mapped reads; male-bias was estimated from the female-to-male ratio of sequences reflected by CQ and RCQ (SI Appendix, Tables S6 and S7). Using similar strategies, we also interrogated individually sequenced wild-caught An. gambiae mosquitoes of both sexes (40 males, 45 females) from Cameroon (SI Appendix, Figs. S11–S13, and Tables S12–S14).
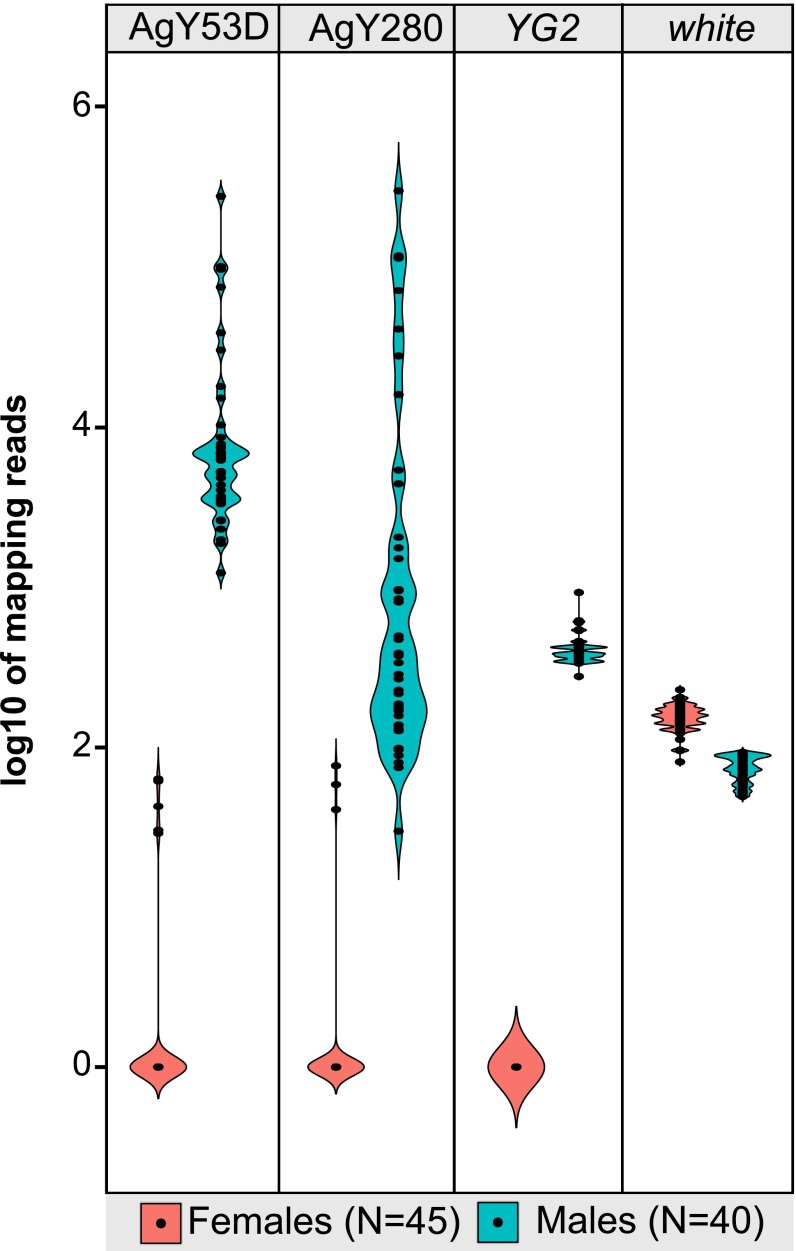
Overall, the pattern of Y-linkage as inferred by CQ and RCQ was qualitatively similar among An. gambiae samples. However, the corresponding copy number of Y-linked sequences was much more labile (Fig. 3 and SI Appendix, Text S5, Figs. S11–S13, and Tables S6, S7, and S13–S15). The most dramatic copy number variation of any SAR or ZAR component was displayed by satellite sequences AgY53D and AgY280 in male samples (Fig. 3). Although numbers of read alignments are not precise reflections of copy number, they can convey a rough approximation of relative abundance if copy number varies across orders of magnitude. Indeed, counts of male alignments to AgY53D and AgY280 spanned four to five orders of magnitude from the Asembo to the Pimperena colony (SI Appendix, Tables S6 and S7), and even within the natural population from Cameroon, normalized alignment counts among individual males spanned three orders of magnitude (SI Appendix, Table S13), suggesting major expansions or contractions in array length. Copy number variation of this magnitude on the Y chromosome would be expected to affect its length. We estimated the combined effect on chromosome length of copy number variation of all SAR and ZAR components among the male An. gambiae sampled from Cameroon. Taking these data together, we find that copy number variation can account for the gain or loss of up to ∼30 Mb of the Y chromosome. Broadly consistent with this in silico estimate, our cytogenetic length estimates also varied by as much as 22 Mb among individual males from the An. gambiae Pimperena colony (from ∼25.9 Mb to ∼47.8 Mb) (SI Appendix, Text S4.2, and Table S11), in conformity with prior studies (50).
Fig. 3.
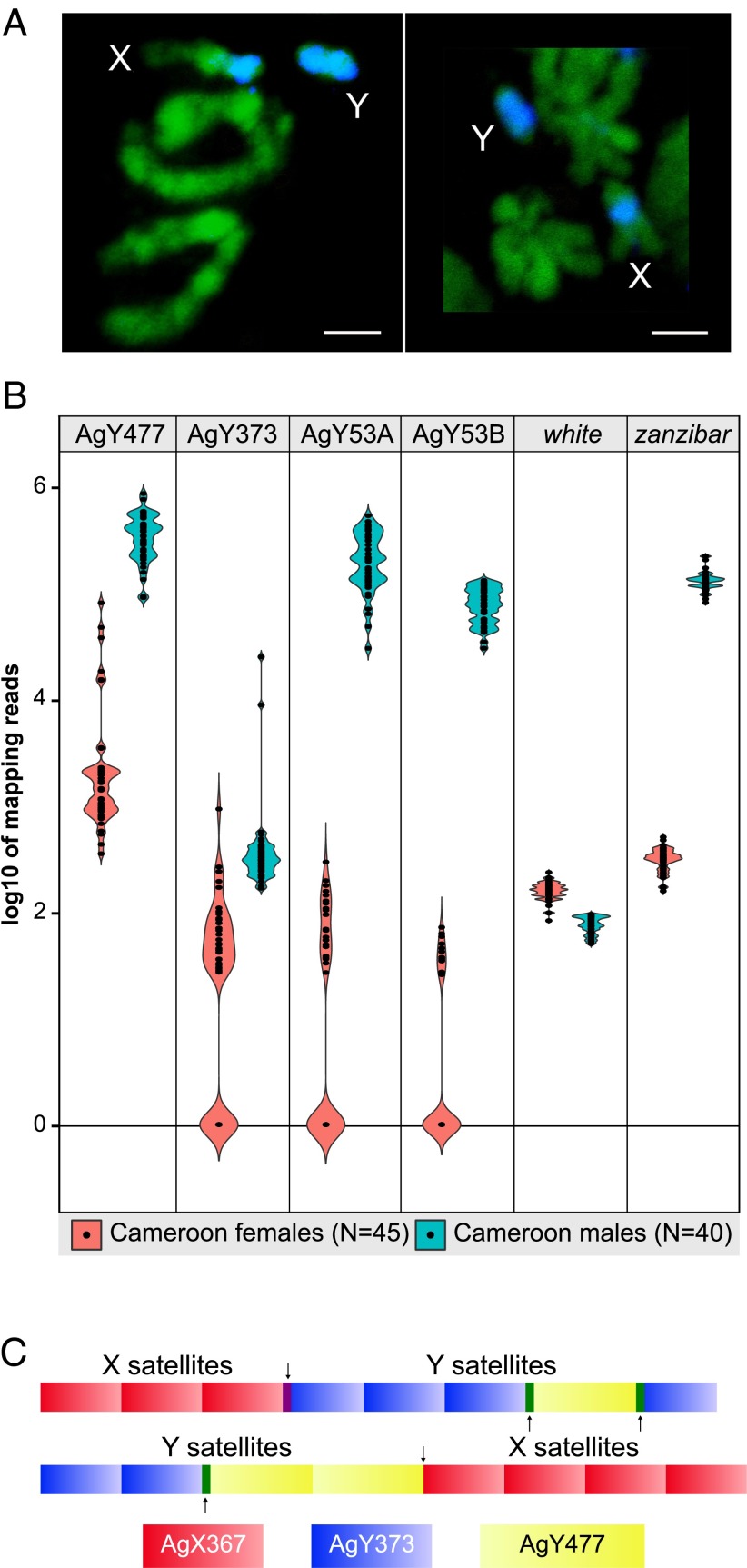
Satellites AgY53D and AgY280 show extensive structural dynamism in males from a natural population of An. gambiae. Shown are violin plots of the log10 numbers of normalized read alignments from Illumina genomic sequence derived from 40 individual male (blue) and 45 individual female (pink) mosquitoes from Cameroon, mapped to satellite monomers of AgY53D and AgY280. For comparison are similar plots of reads mapping to the presumptive male-determining gene, YG2, and to the single-copy X-linked gene, white. Numbers of read alignments to the satellite monomers varies drastically between individuals, in contrast to YG2 and white, suggesting large within-population differences in satellite abundance on the Y. Mapping reads were normalized to library size and locus length.
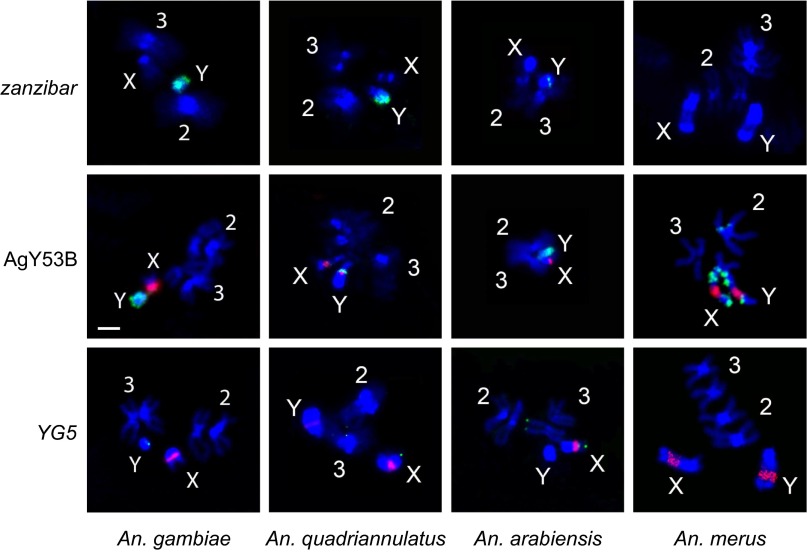
The sibling species complex to which An. gambiae belongs radiated rapidly and recently, within the last 2 My (32). To examine the extent of structural divergence of the Y chromosome between species over this relatively short time frame, we generated Illumina sequences from sex-specific pools of three additional members of the complex (Anopheles arabiensis, Anopheles quadriannulatus, Anopheles merus) (SI Appendix, Table S1), and assessed male bias and relative abundance as described above for An. gambiae (SI Appendix, Text S5.2, and Tables S6, S7, and S15), and by FISH (Fig. 4). Rapid and extensive remodeling of the Y chromosome between species is evidenced by dramatic examples of turnover in both the SAR and ZAR. Satellite AgY477, heavily male-biased and a major component of the Y in An. gambiae, is abundant but not strongly sex-biased in An. merus, and is not detected in An. arabiensis or An. quadriannulatus of either sex (Fig. 1 and SI Appendix, Text S5.2, and Table S15). Similarly, zanzibar is neither strongly sex-biased nor abundant in An. arabiensis and An. merus, yet in An. quadriannulatus (not a sister species of An. gambiae) this retrotransposon has an An. gambiae-like pattern of expanded tandem arrays on the Y chromosome (Figs. 1 and 4, and SI Appendix, Text S5.2, and Table S15).
Fig. 4.
Physical mapping supports structural dynamism of Y chromosome sequences in the An. gambiae complex. FISH of retrotransposon zanzibar, satellite AgY53B, and gene YG5 (green signals) was performed on chromosomes of male An. gambiae Kisumu (zanzibar, YG5), An. gambiae Asembo (AgY53B), An. quadriannnulatus SANGWE, An. arabiensis Dongola, and An. merus MAF. Chromosomes were obtained from imaginal discs except for An. merus chromosomes hybridized to AgY53B, which were obtained from testes. The 18S rDNA probe (red signal) was used in all experiments except with zanzibar. Chromosomes were counterstained with DAPI (blue). (Scale bar, 2 μm; applies to all images.)
The Anopheles Y Recombines with the X Chromosome.
Meiotic pairing, chiasma formation, and crossing-over between the sex chromosomes have been reported for three anopheline species in two different subgenera (19, 21, 24), but to our knowledge, similar observations have not been reported in An. gambiae. Indeed, the apparent stability of X- and Y-linked translocations in An. gambiae (51) suggest that legitimate crossing-over between the X and Y chromosomes does not occur. However, our data are difficult to explain without some form of X–Y genetic exchange since the evolution of the sex chromosomes from an ancestral pair of autosomes, an event that must predate anopheline diversification ∼100 Mya. First, there is a very high degree of sequence similarity between Y-associated and X-associated repetitive DNA in An. gambiae. Our initial indication of this was the near-complete failure of the YGS computational method to identify Y-associated sequences in An. gambiae, presumably arising from the fact that such sequences are only rarely exclusive to the Y chromosome, as opposed to highly enriched there. Strongly supporting empirical evidence comes from physical (FISH) mapping of individual components of the SAR (e.g., AgY53B) (Fig. 4 and SI Appendix, Fig. S10), or of fluorescently labeled sequences derived from the entire microdissected Y chromosome (Fig. 5A), which reveals extensive cross-hybridization of Y repeats with X chromosome heterochromatin because of sequence similarity between satellite monomers (SI Appendix, Text 4.1.2). Detailed sequence analysis indicates that satellite monomers normally abundant only on the Y (AgY373, AgY477) share ∼93% pairwise sequence similarity with a satellite monomer from the X chromosome [AgX367 (42)] (SI Appendix, Fig. S6). Moreover, we found the footprints of recombination on individual An. gambiae PacBio reads. These contained AgX367 monomers, together with AgY477 and AgY373 monomers, including AgX367-AgY373 and AgX367-AgY477 recombinants (Fig. 5C), confirming previous evidence of recombination based on PCR amplicon sequencing (42). A second line of evidence for occasional X–Y genetic exchange emerged from individually sequenced male and female An. gambiae from Cameroon (Fig. 5B and SI Appendix, Text S6, and Fig. S11). In a small subset of females, normalized counts of Illumina reads mapping to consensus satellite monomers normally abundant only on the Y chromosome (e.g., AgY53A) were almost two orders of magnitude larger than the median for all females (SI Appendix, Tables S12 and S14). Because this female subset lacked correspondingly high copies of other Y chromosome sequences, contamination by male genomic DNA (whether in the laboratory or via sperm stored in the spermatheca) is an unlikely alternative explanation. Although intrachromosomal genetic exchange on the X may contribute to the strongly bimodal pattern of satellite abundance observed in Cameroon females, this phenomenon must have its basis in periodic X–Y genetic exchange.
Fig. 5.
The An. gambiae X and Y chromosomes are not genetically isolated. (A) Painting of prometaphase (Left) and metaphase (Right) chromosomes from male larval imaginal discs of the An. gambiae Pimperena strain with a probe generated from microdissected Y chromosomes (labeled blue by the WGA3 kit with dNTP-Cy3). Chromosomes are counterstained with YOYO-1 (green). (Scale bars, 2 μm.) (B) Violin plots showing the log10 number of normalized read alignments from 40 individual An. gambiae males (blue) and 45 females (pink) from the Cameroon population, to satellite AgY477, AgY373, AgY53A, and AgY53B monomers, compared with numbers of reads from these sources aligning to the white gene (single-copy and X-linked) and zanzibar (heavily Y-biased). Mapping reads were normalized to library size and locus length. (C) Two examples of single PacBio reads (pacbio_7224704_1 and pacbio_5551309_1) where predominantly X-linked (AgY367; shown in red) and predominantly Y-linked (AgY373, shown in blue; AgY477, shown in yellow) satellites occur in the same PacBio read. Black arrows indicate the junction between the predominantly X-linked and predominantly Y-linked satellites. The purple and orange boxes indicate inferred recombination, between AgX367-AgY373 and AgX367-AgY477, respectively. Green boxes indicate recombination between AgY477-AgY373.
Rapid Turnover of the Small Y Chromosome Genic Repertoire.
Only three Y-linked genes had been identified previously in An. gambiae (30), designated gYG1 to gYG3 (hereafter, YG1 to YG3). Aiming for comprehensive gene discovery on the An. gambiae Y chromosome, we performed extensive transcriptional profiling of developmentally staged An. gambiae embryos (nine time points), sexed larvae (three time points), and adults (whole and dissected males and females) through mRNA sequencing (RNA-Seq)—52 datasets in total (SI Appendix, Text S1.6, and Table S3)—and integrated complementary approaches to gene finding (SI Appendix, Text S7). Gene candidates bearing significant similarity to known TEs or bacterial sequences were discounted. Arising from the combined approaches were eight presumptive genes (YG1–8), including the three previously identified (Fig. 1 and SI Appendix, Text S7).
To be considered valid Y genes, we required that they exhibit male-biased or male-specific expression from RNA-Seq as well as male-specific amplification by genomic PCR, conditions met by YG1–5. With the exception of YG4, this validated set was further confirmed by male-specific RT-PCR. Moreover, we were able to physically localize YG5 to the Y chromosome by FISH (a homolog was also detected on chromosome 3) (Fig. 4 and SI Appendix, Text S7, and Fig. S19). Because we could not identify male-specific SNPs distinguishing YG6–8 from their autosomal homologs, these genes could not be validated despite exclusive expression of YG6 in male accessory glands (SI Appendix, Fig. S20), and elevated numbers of normalized read alignments to YG8 from individually sequenced males versus females in our population sample from Cameroon (SI Appendix, Fig. S11, and Tables S12–S14).
Beyond the strikingly small total number of genes identified as Y-linked in An. gambiae following this intensive search, it is noteworthy that gene number varies even between strains. Both YG3 and YG4 are Y-linked exclusively in the G3 strain of An. gambiae, not in Asembo or Pimperena (SI Appendix, Tables S6 and S7). None of these genes have recognizable gametologs on the X, yet all have partial or complete homologs on the autosomes (SI Appendix, Text S7, and Fig. S14), suggesting that they have been gained on the Y chromosome since its divergence from the X (see below).
To screen for candidate Y-linked genes in three An. gambiae sibling species (An. arabiensis, An. merus, and An. quadriannulatus), we used the male and female Illumina sequences from each species in conjunction with corresponding genome assemblies, mixed-sex transcript sets, and de novo RNA-Seq assemblies (31), as well as An. gambiae genomic resources (SI Appendix, Text S7). After merging the results of these approaches, we made two surprising observations. First, among all of the Y-linked genes identified in An. gambiae, only one—YG2—was computationally detected and confirmed (by male-specific genomic PCR) as Y-linked in each of the other three very closely related species (Fig. 1). None of the other An. gambiae Y genes, with the sole exception of YG1, was Y-linked in any other species examined. For YG1, Y-linkage was validated in An. arabiensis and An. quadriannulatus, but in An. merus the ratio of female-to-male alignments was inconclusive and male-specific genomic PCR amplification was not possible because of highly similar sequence elsewhere in the genome (Fig. 1 and SI Appendix, Text S7, Fig. S14, and Tables S6 and S7). Thus, YG2 is the only gene both conserved on, and exclusive to, the Y chromosome in all four species examined. As it is the earliest to be expressed, at 3 h of male embryonic development (YG1 is not expressed until 4 h), YG2 is a possible male-determining gene. Surprisingly, YG2 is not a single-copy gene. The first suggestion that this might be the case was hinted by the number of read alignments to YG2 from individual males in the Cameroon sample (Fig. 3). However, we have more definitive evidence for multiple, nearly identical copies of YG2 in An. gambiae. Four distinct haplotypes were sampled repeatedly in Ydb PacBio reads (which derived from the same paternal Y chromosome) (SI Appendix, Text S7.1.2, and Fig. S17). Variant positions among YG2 copies were not only validated by sequencing of genomic PCR amplicons from individual male An. gambiae derived from natural populations, but also through RNA-Seq data, which further indicates that multiple YG2 copies are expressed (SI Appendix, Table S17).
With the exception of YG2, the near-complete absence of conserved Y-linkage between An. gambiae genes and corresponding genes in the sibling species was reinforced by the converse result, our second surprising observation: all genes identified as Y-linked in any one sibling species could not be assigned to the Y chromosome in any of the other species (SI Appendix, Text S7.2, Tables S6 and S7). In An. quadriannulatus we found three novel Y-linked candidate genes (SI Appendix, Text S7, and Table S20). In An. arabiensis, no genes other than YG1 and YG2 were detected on the Y chromosome. In An. merus, we found evidence supporting the duplication of a multigene segment from chromosome 3R onto the Y since its split from other An. gambiae complex lineages (SI Appendix, Text S7, Table S19). Among seven sequential genes on 3R in this segment (corresponding to AGAP009631 to -37 in An. gambiae), the first three (AGAP009631 to -33) and last two (AGAP009636 and -37) have detectable copies on the Y chromosome in An. merus (based initially on the relative number of normalized read alignments in females and males, later validated by male-specific PCR). Our data are consistent with the two intervening genes (corresponding to AGAP009634 and -35 on 3R) having been lost from the Y, and the five flanking genes becoming amplified, although further experimental evidence will be required to confidently reconstruct these events.
Possible Y Chromosome Introgression Between Hybridizing Malaria Vectors.
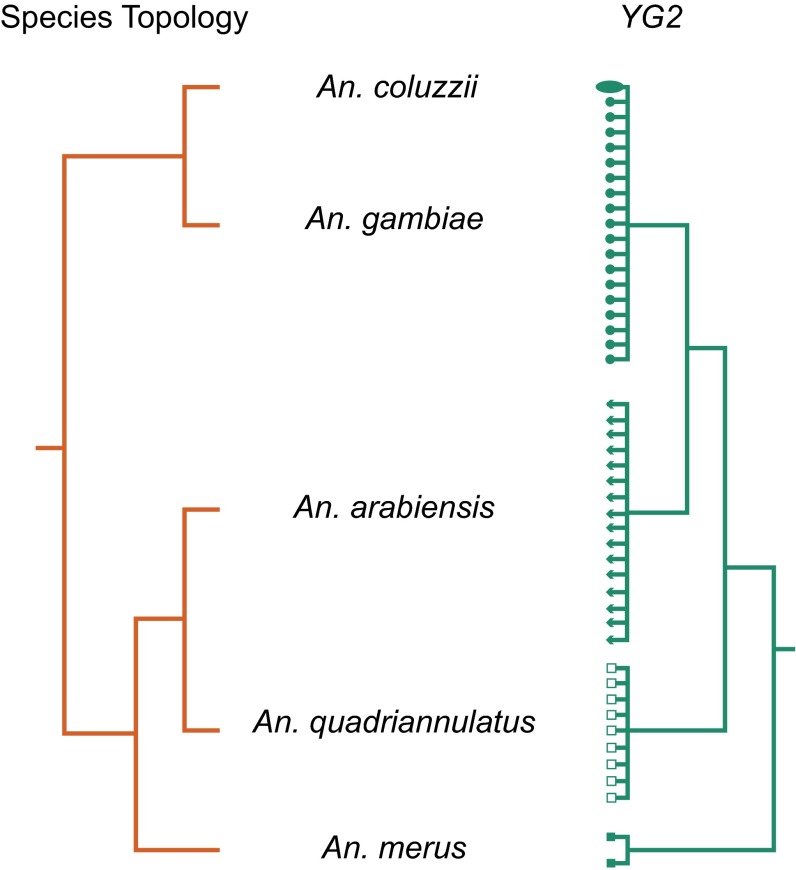
The YG2 gene potentially encodes a short, 56-aa peptide whose possible role in determining maleness is under investigation. In the Asian malaria vector An. stephensi, a Y-linked gene (Guy1) implicated in male determination also encodes a 56-aa sequence whose predicted secondary structure resembles that of the putative YG2 peptide (52) even though primary sequence similarity is not detectable (30) over the relatively short evolutionary span since these lineages separated, ∼30 Mya (53). The fact that YG2 expression is detected in early embryos before any other An. gambiae Y-linked gene, taken together with its uniquely conserved Y chromosome location in all four An. gambiae complex species—in the face of otherwise rampant structural dynamism and genic turnover on the Y—is consistent with a primary role in male determination in this group. For this reason, we predicted that a gene tree reconstructed from YG2 would reflect the known species branching order (32). Although sibling species in the An. gambiae complex are not completely reproductively isolated, contemporary interspecific gene flow is possible only through female F1 hybrids; because their brothers are sterile, the Y chromosome cannot introgress (54). Contrary to this expectation, a YG2 tree built from sequences derived from population samples of the four species considered in this study supported An. gambiae and An. arabiensis as most closely related (Fig. 6 and SI Appendix, Text S8), an arrangement previously shown to be the result of massive historical introgression between these two species that involved most of the autosomes and the proximal ∼10 Mb of the X chromosome (32). The simplest explanation for a gene tree disagreeing with the species tree in a rapid radiation, such as the An. gambiae complex, is incomplete lineage sorting. We performed coalescent simulations to assess the likelihood that the grouping of An. gambiae with An. arabiensis in the YG2 tree is because of incomplete lineage sorting alone. In 62 of 1,000 simulations under the species tree, we recovered An. gambiae and An. arabiensis as sister lineages (i.e., P = 0.062) (SI Appendix, Text S8), indicating that nonintrogressing lineages could produce the observed tree a small fraction of the time. Although we cannot formally reject the null hypothesis at the 0.05 significance level, these results certainly do not rule out Y chromosome introgression. Introgression of the Y chromosome between species is conventionally viewed as unlikely (55), but it is important to consider that the pair of malaria vectors in question have historically exchanged the vast majority of the rest of their genomes, including part of the X chromosome (32). In this context, introgression of the Y chromosome is possible if not likely, as long as the introgression event(s) predated the development of male F1 hybrid sterility barriers between this species pair.
Fig. 6.
Phylogeny inferred from a candidate male-determining gene on the Y chromosome, YG2, differs from the species branching order. Species topology (32) of five members of the An. gambiae complex (red branches) compared with a maximum-likelihood phylogeny inferred from a Y chromosome-specific region of the YG2 gene (green branches) sequenced from male Anopheles coluzzii (ellipse), An. gambiae (filled circles), An. arabiensis (triangles), An. quadriannulatus (open squares), and An. merus (filled squares). Samples were drawn from colonies and natural populations (see text). The YG2 tree was rooted at the midpoint; all nodes are supported by ≥95% bootstrap replicates. The topological disagreement involves An. arabiensis; in the species topology An. arabiensis is sister to An. quadriannulatus, whereas the YG2 topology indicates a sister group relationship of An. arabiensis and An. gambiae + An. coluzzii.
Discussion
From studies of mammals (6, 7, 11, 56, 57) and Drosophila (58–60), it is known that the Y chromosomes in both groups have lost most of their ancestral gene repertoires and have acquired copious amounts of repetitive and ampliconic/palindromic DNA. In the ∼250 My since Drosophila and Anopheles last shared a common Dipteran ancestor, there has been parallel evolution of heteromorphic sex chromosomes from the same ancestral linkage group (61), implying that the Anopheles Y must have undergone a similar fate of massive ancestral gene loss and genomic degradation. In one main characteristic—its male determining role—the Anopheles Y resembles the mammalian Y more than it does Drosophila, in which XO flies are (sterile) males and the scant Y chromosome genes are crucial only for male fertility (2). However, in other respects, the Anopheles and Drosophila Y are much more similar. More than one-third of the human Y chromosome and 99.9% of the mouse Y is euchromatic (56, 62), whereas Drosophila and Anopheles Y chromosomes are entirely heterochromatic. Although relatively few in number, some ancestral X–Y gene pairs have been conserved throughout mammalian evolution because of their vital role as dosage-sensitive regulators of global gene expression (6, 7). Crucially, although cases are known in which a mammalian Y chromosome has acquired autosomal genes (e.g., ref. 63), most extant mammalian Y-linked genes have an X-linked gametolog. In contrast, what little gene content exists on the Y in Drosophila or Anopheles is not only poorly conserved between species, but there are no recognizable ancestral gametologs; all known Y-linked genes in Drosophila seem to have an autosomal origin (58). The recent DNA-based duplication of a gene from chromosome 3R to the Y chromosome in D. melanogaster following its split from Drosophila simulans ∼4 Mya (64) mirrors our finding of a similar event in An. merus since the radiation of the An. gambiae complex, <2 Mya. We conclude that the most salient factor uniting Anopheles and Drosophila Y chromosomes may be the continuous gain of genes and functions from the autosomes (64), in contrast to the conservation of remaining ancestral gametologs seen on the mammalian Y. However, the Anopheles NRY appears to stand apart from both Drosophila and mammalian Y chromosomes in the relative paucity of male-specific content.
One of our main findings was rapid turnover in quantity and type of repetitive DNA on the Y chromosome within and between species in the An. gambiae complex. It is known that both satellite and ampliconic DNA regions are prone to rapid divergence in length, structure, and sequence, as a result of unequal sister chromatid exchange between out-of-register repeat units and other mechanisms (49). On the Y chromosome such regions may be subject to accelerated rates of divergence compared with the rest of the genome. Between humans and chimps whose lineages diverged ∼5 Mya, orthologous satellite arrays in the X centromere are collinear and share 93% sequence identity, whereas collinearity declines and sequence conservation drops to 78% between orthologous satellites in Y centromeres (65). Additional evidence of rapid length, structure, and sequence evolution of satellites and ampliconic structures on the Y chromosome has been reported in mice species 1–2 My diverged and mice subspecies separated by only ∼900,000 y (65), between D. melanogaster and D. simulans that split ∼4 Mya (60, 66), and among human males worldwide (67). Despite such pervasive remodeling of the Anopheles Y chromosome over short evolutionary distances, a transgene randomly inserted onto the Y chromosome of an An. gambiae strain in 2014 is transcriptionally active and has been stably integrated ever since, establishing that the Y chromosome is amenable to the molecular manipulation required for Y-linked genetic vector control strategies (68).
The high level of satellite DNA polymorphism within species could have important phenotypic consequences for fitness-related traits (8, 9). Moreover, the dramatic degree of satellite DNA turnover on the Y between closely related species has been implicated in hybrid incompatibility in Drosophila (69–71). Intriguingly, two genes known to cause hybrid incompatibility between D. melanogaster and D. simulans (Hmr and Lhr) function within species to repress transcripts from satellite DNAs and TEs (71). These species differ drastically in satellite DNA content; D. simulans contains fourfold less satellite DNA overall (5% versus 20% of the genome), and is particularly depauperate of the two most abundant satellite types in D. melanogaster (72). In An. gambiae, we found that AgY477 and AgY373 are the most abundant satellites on the NRY, and they are both expressed exclusively in adult male reproductive tissues; these satellite sequences are absent or altered in the other sibling species investigated (Fig. 1). Whether hybridization leads to misregulation of satellite DNA remains to be explored in the An. gambiae complex.
Laborious single-haplotype iterative mapping and sequencing has previously revealed the structure of mammalian Y chromosomes (11). In contrast, single-molecule sequencing now provides individual reads tens of kilobases in length, promising a resource-efficient alternative for characterizing Y chromosomes. Here, we were able to determine the content and structural characteristics of the heterochromatic An. gambiae NRY using this approach. Single-molecule sequencing reads were able to reveal complex repeat structures from whole-genome data and completely assemble heterochromatic BACs without manual finishing. However, the complete reconstruction of heterochromatic Y chromosomes remains a challenging open problem, as a recent PacBio assembly of D. melanogaster failed to completely assemble the Y chromosome (73), but it did successfully resolve the complex regions Mst77Y (74) and FDY (64). The minimum read length, accuracy, and coverage required for the successful assembly of heterochromatin from whole-genome data are currently unknown and will vary by species, but as demonstrated here, An. gambiae Y chromosome BACs can be successfully reconstructed using long sequences collected from deep, directed single-molecule sequencing. These results suggest that continued single-molecule read length and throughput improvements may soon enable the complete reconstruction of Y chromosomes from whole-genome data alone.
Materials and Methods
Please see SI Appendix and Datasets S1–S3 for detailed information about: (i) genomic and transcriptomic datasets; (ii) identification of Y chromosome sequences from PacBio whole-genome sequencing; (iii) repetitive DNA content of the An. gambiae Y; (iv) FISH and size estimation of the Y chromosome; (v) copy number variation on the Y chromosome among individual An. gambiae mosquitoes and across the An. gambiae complex; (vi) Y chromosome recombination; (vii) genic repertoire of the Y chromosome; and (viii) phylogeny reconstruction and coalescent simulations.
Supplementary Material
Acknowledgments
We thank F. Catteruccia and S. N. Mitchell for sharing unpublished data; J. Pease for assistance with simulations; M. Kern, M. Menichelli, M. K. Lawniczak, I. Antoshechkin, T. Persampieri, R. Carballar, and R. D’Amato for technical assistance and discussion; and two anonymous reviewers for helpful suggestions. Genomic sequencing was funded in part by a grant from the Eck Institute for Global Health, University of Notre Dame. RNA-Seq was funded in part from a European Community Seventh Framework Programme (FP7/2007–2013) under Grant 228421 (INFRAVEC). Individual laboratories were funded as follows: National Institutes of Health Grants R01AI076584 (to N.J.B. and M.W.H.), R21AI112734 (to N.J.B. and S.J.E.), R21AI101459 (to N.J.B.), R21AI094289 and R21AI099528 (to I.V.S.), R21AI105575 (to Z.J.T.), and HHSN272200900039C (to S.J.E.); the Foundation of the National Institutes of Health through the VCTR program of the Grand Challenges in Global Health Initiative (to N.J.B., A. Crisanti, P.-A.P., and T.N.); European Commission and Regione Umbria Grant I-MOVE (to R.G., E.F., and P.-A.P.); Rita-Levi Montalcini Career Development Award (to P.-A.P.); Marie Curie Intra-European Fellowship for Career Development PIEFGA-273268 (to T.D.); European Research Council Grant 335724 (to N.W.); National Science Foundation Graduate Research Fellowship Grant DGE-1519168 (to A.B.H.); Department of Education Graduate Assistance in Areas of National Need Fellowship (to A. Steele); and Fralin Life Science Institute of Virginia Tech (to I.V.S. and Z.J.T.). This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (to S.K. and A.M.P.). The contributions of N.H.B., J.H., and D.R. were funded under Agreement HSHQDC-07-C-00020 awarded by the Department of Homeland Security (DHS) Science and Technology Directorate for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a Federally Funded Research and Development Center. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Homeland Security. In no event shall the DHS, NBACC, or Battelle National Biodefense Institute (BNBI) have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. The Department of Homeland Security does not endorse any products or commercial services mentioned in this publication.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequencing data and assemblies have been submitted to NCBI under umbrella BioProject IDs PRJNA254149-152 and SRP045243. BAC assemblies have been deposited in the Sequence Read Archive database as KR610409-411 and KR494253. For a list of individual accession numbers, see SI Appendix, Text S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525164113/-/DCSupplemental.
References
- 1.Bull JJ. The Evolution of Sex Determining Mechanisms. Benjamin/Cummings; Menlo Park, CA: 1983. [Google Scholar]
- 2.Bachtrog D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14(2):113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachtrog D, et al. Tree of Sex Consortium Sex determination: Why so many ways of doing it? PLoS Biol. 2014;12(7):e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355(1403):1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38(4):735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 6.Bellott DW, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508(7497):494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez D, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508(7497):488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 8.Lemos B, Araripe LO, Hartl DL. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science. 2008;319(5859):91–93. doi: 10.1126/science.1148861. [DOI] [PubMed] [Google Scholar]
- 9.Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci USA. 2010;107(36):15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sackton TB, Montenegro H, Hartl DL, Lemos B. Interspecific Y chromosome introgressions disrupt testis-specific gene expression and male reproductive phenotypes in Drosophila. Proc Natl Acad Sci USA. 2011;108(41):17046–17051. doi: 10.1073/pnas.1114690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes JF, Page DC. The biology and evolution of mammalian Y chromosomes. Annu Rev Genet. 2015;49:507–527. doi: 10.1146/annurev-genet-112414-055311. [DOI] [PubMed] [Google Scholar]
- 12. World Health Organization (2014) World Malaria Report: 2014. Available at www.who.int/malaria/publications/world_malaria_report_2014/en/. Accessed March 22, 2016.
- 13.Bhatt S, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526(7572):207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. malERA Consultative Group on Vector Control (2011) A research agenda for malaria eradication: Vector control. PLoS Med 8(1):e1000401. [DOI] [PMC free article] [PubMed]
- 15.Galizi R, et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat Commun. 2014;5:3977. doi: 10.1038/ncomms4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Windbichler N, Papathanos PA, Crisanti A. Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. PLoS Genet. 2008;4(12):e1000291. doi: 10.1371/journal.pgen.1000291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burt A. Heritable strategies for controlling insect vectors of disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1645):20130432. doi: 10.1098/rstb.2013.0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deredec A, Godfray HC, Burt A. Requirements for effective malaria control with homing endonuclease genes. Proc Natl Acad Sci USA. 2011;108(43):E874–E880. doi: 10.1073/pnas.1110717108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakai RK, Baker RH, Raana K, Hassan M. Crossing-over in the long arm of the X and Y chromosomes in Anopheles culicifacies. Chromosoma. 1979;74(2):209–218. [Google Scholar]
- 20.Redfern CP. Satellite DNA of Anopheles stephensi Liston (Diptera: Culicidae). Chromosomal location and under-replication in polytene nuclei. Chromosoma. 1981;82(4):561–581. doi: 10.1007/BF00295013. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell SE, Seawright JA. Recombination between the X and Y chromosomes in Anopheles quadrimaculatus species A. J Hered. 1989;80(6):496–499. doi: 10.1093/oxfordjournals.jhered.a110790. [DOI] [PubMed] [Google Scholar]
- 22.Marchi A, Mezzanotte R. Inter- and intraspecific heterochromatin variation detected by restriction endonuclease digestion in two sibling species of the Anopheles maculipennis complex. Heredity (Edinb) 1990;65(Pt 1):135–142. doi: 10.1038/hdy.1990.80. [DOI] [PubMed] [Google Scholar]
- 23.White GB. Academic and applied aspects of mosquito cytogenetics. In: Blackman RL, Hewitt GM, Ashburner M, editors. Insect Cytogenetics. Blackwell Scientific Publications; Oxford: 1980. pp. 245–274. [Google Scholar]
- 24.Fraccaro M, Laudani U, Marchi A, Tiepolo L. Karotype, DNA replication and origin of sex chromosomes in Anopheles atroparvus. Chromosoma. 1976;55(1):27–36. doi: 10.1007/BF00288324. [DOI] [PubMed] [Google Scholar]
- 25.Holt RA, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298(5591):129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 26.Carvalho AB, et al. Y chromosome and other heterochromatic sequences of the Drosophila melanogaster genome: How far can we go? Genetica. 2003;117(2-3):227–237. doi: 10.1023/a:1022900313650. [DOI] [PubMed] [Google Scholar]
- 27.Krzywinski J, Nusskern DR, Kern MK, Besansky NJ. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics. 2004;166(3):1291–1302. doi: 10.1534/genetics.166.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross MG, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14(5):R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White BJ, Collins FH, Besansky NJ. Evolution of Anopheles gambiae in relation to humans and malaria. Annu Rev Ecol Evol Syst. 2011;42:111–132. [Google Scholar]
- 30.Hall AB, et al. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genomics. 2013;14:273. doi: 10.1186/1471-2164-14-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neafsey DE, et al. Mosquito genomics. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science. 2015;347(6217):1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontaine MC, et al. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 2015;347(6217):1258524. doi: 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharakhova MV, et al. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007;8(1):R5. doi: 10.1186/gb-2007-8-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giraldo-Calderón GI, et al. VectorBase Consortium VectorBase: An updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43(Database issue):D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X, et al. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol. 2014;15(9):459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 37.Koren S, et al. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14(9):R101. doi: 10.1186/gb-2013-14-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koren S, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30(7):693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers EW, et al. A whole-genome assembly of Drosophila. Science. 2000;287(5461):2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 40.Carvalho AB, Clark AG. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 2013;23(11):1894–1907. doi: 10.1101/gr.156034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smit AFA, Hubley R, Green P. 2013 RepeatMasker Open 4-0. Available at www.repeatmasker.org. Accessed March 22, 2016.
- 42.Krzywinski J, Sangaré D, Besansky NJ. Satellite DNA from the Y chromosome of the malaria vector Anopheles gambiae. Genetics. 2005;169(1):185–196. doi: 10.1534/genetics.104.034264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohr CJ, Ranson H, Wang X, Besansky NJ. Structure and evolution of mtanga, a retrotransposon actively expressed on the Y chromosome of the African malaria vector Anopheles gambiae. Mol Biol Evol. 2002;19(2):149–162. doi: 10.1093/oxfordjournals.molbev.a004067. [DOI] [PubMed] [Google Scholar]
- 44.Sharma A, Wolfgruber TK, Presting GG. Tandem repeats derived from centromeric retrotransposons. BMC Genomics. 2013;14:142. doi: 10.1186/1471-2164-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonaccorsi S, Santini G, Gatti M, Pimpinelli S, Colluzzi M. Intraspecific polymorphism of sex chromosome heterochromatin in two species of the Anopheles gambiae complex. Chromosoma. 1980;76(1):57–64. doi: 10.1007/BF00292226. [DOI] [PubMed] [Google Scholar]
- 46.Gatti M, Santini G, Pimpinelli S, Coluzz M. Fluorescence banding techniques in the identification of sibling species of the Anopheles gambiae complex. Heredity (Edinb) 1977;38(1):105–108. doi: 10.1038/hdy.1977.11. [DOI] [PubMed] [Google Scholar]
- 47.Cohen S, Agmon N, Sobol O, Segal D. Extrachromosomal circles of satellite repeats and 5S ribosomal DNA in human cells. Mob DNA. 2010;1(1):11. doi: 10.1186/1759-8753-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J, Jackson SA. Retrotransposon accumulation and satellite amplification mediated by segmental duplication facilitate centromere expansion in rice. Genome Res. 2006;16(2):251–259. doi: 10.1101/gr.4583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 50.Gatti M, Bonaccorsi S, Pimpinelli S, Coluzzi M. Polymorphism of sex chromosome heterochromatin in the Anopheles gambiae complex. In: Steiner WWM, Tabachnick WJ, Rai KS, Narang S, editors. Recent Developments in the Genetics of Insect Disease Vectors. Stipes Publishing; Champaign, IL: 1982. pp. 32–48. [Google Scholar]
- 51.Curtis CF, Akiyama J, Davidson G. Genetic sexing system in Anopheles gambiae species A. Mosq News. 1976;36(4):492–498. [Google Scholar]
- 52.Criscione F, Qi Y, Saunders R, Hall B, Tu Z. A unique Y gene in the Asian malaria mosquito Anopheles stephensi encodes a small lysine-rich protein and is transcribed at the onset of embryonic development. Insect Mol Biol. 2013;22(4):433–441. doi: 10.1111/imb.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamali M, et al. Multigene phylogenetics reveals temporal diversification of major African malaria vectors. PLoS One. 2014;9(4):e93580. doi: 10.1371/journal.pone.0093580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davidson G, Paterson HE, Coluzzi M, Mason GF, Micks DW. The Anopheles gambiae complex. In: Wright JW, Pal R, editors. Genetics of Insect Vectors of Disease. Elsevier; Amsterdam: 1967. [Google Scholar]
- 55.Payseur BA. Y not introgress? Insights into the genetics of speciation in European rabbits. Mol Ecol. 2009;18(1):23–24. doi: 10.1111/j.1365-294X.2008.04011.x. [DOI] [PubMed] [Google Scholar]
- 56.Skaletsky H, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423(6942):825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 57.Hughes JF, et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483(7387):82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koerich LB, Wang X, Clark AG, Carvalho AB. Low conservation of gene content in the Drosophila Y chromosome. Nature. 2008;456(7224):949–951. doi: 10.1038/nature07463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bachtrog D, Hom E, Wong KM, Maside X, de Jong P. Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol. 2008;9(2):R30. doi: 10.1186/gb-2008-9-2-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Méndez-Lago M, et al. A large palindrome with interchromosomal gene duplications in the pericentromeric region of the D. melanogaster Y chromosome. Mol Biol Evol. 2011;28(7):1967–1971. doi: 10.1093/molbev/msr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toups MA, Hahn MW. Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics. 2010;186(2):763–766. doi: 10.1534/genetics.110.118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soh YQ, et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159(4):800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saxena R, et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat Genet. 1996;14(3):292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- 64.Carvalho AB, Vicoso B, Russo CA, Swenor B, Clark AG. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA. 2015;112(40):12450–12455. doi: 10.1073/pnas.1516543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pertile MD, Graham AN, Choo KH, Kalitsis P. Rapid evolution of mouse Y centromere repeat DNA belies recent sequence stability. Genome Res. 2009;19(12):2202–2213. doi: 10.1101/gr.092080.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wei KH, Grenier JK, Barbash DA, Clark AG. Correlated variation and population differentiation in satellite DNA abundance among lines of Drosophila melanogaster. Proc Natl Acad Sci USA. 2014;111(52):18793–18798. doi: 10.1073/pnas.1421951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Repping S, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38(4):463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 68.Bernardini F, et al. Site-specific genetic engineering of the Anopheles gambiae Y chromosome. Proc Natl Acad Sci USA. 2014;111(21):7600–7605. doi: 10.1073/pnas.1404996111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferree PM, Barbash DA. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009;7(10):e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326(5959):1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Satyaki PR, et al. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 2014;10(3):e1004240. doi: 10.1371/journal.pgen.1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lohe AR, Brutlag DL. Identical satellite DNA sequences in sibling species of Drosophila. J Mol Biol. 1987;194(2):161–170. doi: 10.1016/0022-2836(87)90365-2. [DOI] [PubMed] [Google Scholar]
- 73.Berlin K, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33(6):623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- 74.Krsticevic FJ, Schrago CG, Carvalho AB. Long-read single molecule sequencing to resolve tandem gene copies: The Mst77Y region on the Drosophila melanogaster Y chromosome. G3 (Bethesda) 2015;5(6):1145–1150. doi: 10.1534/g3.115.017277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.