Significance
With changes in land use and increased urbanization, the frequency with which pathogens jump species barriers to emerge in new hosts is expected to rise. Knowing which viruses may be more likely to become transmissible among humans, as opposed to only generating dead-end spillover infections, would be of considerable benefit to pandemic planning. Using multivariate modeling and multimodel inference, we sought to both identify and quantify those biological features of viruses that best determine interhuman transmissibility. This analysis revealed that chronic, nonsegmented, non–vector-borne, nonenveloped viruses with low host mortality had the highest likelihood of being transmissible among humans whereas genomic features had little predictive power. Our analysis therefore reveals that multiple virological features determine the likelihood of successful emergence.
Keywords: virus, evolution, transmission, comparative analysis, spill-over
Abstract
The early detection of pathogens with epidemic potential is of major importance to public health. Most emerging infections result in dead-end “spillover” events in which a pathogen is transmitted from an animal reservoir to a human but is unable to achieve the sustained human-to-human transmission necessary for a full-blown epidemic. It is therefore critical to determine why only some virus infections are efficiently transmitted among humans whereas others are not. We sought to determine which biological features best characterized those viruses that have achieved sustained human transmission. Accordingly, we compiled a database of 203 RNA and DNA human viruses and used an information theoretic approach to assess which of a set of key biological variables were the best predictors of human-to-human transmission. The variables analyzed were as follows: taxonomic classification; genome length, type, and segmentation; the presence or absence of an outer envelope; recombination frequency; duration of infection; host mortality; and whether or not a virus exhibits vector-borne transmission. This comparative analysis revealed multiple strong associations. In particular, we determined that viruses with low host mortality, that establish long-term chronic infections, and that are nonsegmented, nonenveloped, and, most importantly, not transmitted by vectors were more likely to be transmissible among humans. In contrast, variables including genome length, genome type, and recombination frequency had little predictive power. In sum, we have identified multiple biological features that seemingly determine the likelihood of interhuman viral transmissibility, in turn enabling general predictions of whether viruses of a particular type will successfully emerge in human populations.
The cross-species transmission of viruses from animals to humans is responsible for the vast majority of emerging infections, including some of the most devastating disease epidemics on record. Important exemplars are the global HIV/AIDS pandemic, the continual appearance of novel subtypes and strains of influenza A virus (1, 2), and the recent outbreak of Ebola in West Africa (3). Despite the widespread mortality and morbidity caused by emerging diseases, it is striking that the majority of such emergence events result only in dead-end “spillover” infections in which the virus is unable to establish stable onward transmission in the novel (human) host. For example, both the H5N1 and H7N9 subtypes of avian influenza virus have repeatedly spilled over from poultry to humans, but there is only limited evidence of human-to-human transmission such that these viruses are not adapted to spread within the human population (4). In contrast, Ebola virus (EBOV), which likely originated in fruit bats, and Middle East respiratory syndrome coronavirus (MERS-CoV), which jumped from camels to humans, have been able to establish transmission networks within human populations (5, 6). Such different outcomes of cross-species transmission highlight the importance of revealing the biological factors that determine why only a subset of viruses are able to establish productive infections in humans (7, 8).
Understanding the drivers and barriers to successful disease emergence has been the subject of increasing research activity. Previous studies have attempted to reveal the links between disease emergence and a variety of socioeconomic factors, including lack of sanitation, limited access to health care, and social and political instability, as well as ecological disruption and climate change (9). More generally, it has been suggested that collating data on the geographic occurrence and distribution of emerging diseases could be used to identify “hot spots” where emergence events are most likely to occur (10). Crucially, however, such models consider all emerging diseases in the same manner, regardless of their transmissibility within human populations, even though only a subset will establish endemic transmission. Other studies have considered the “genetic” barriers to emergence in both hosts and viruses (11), particularly the number and origin of the mutations necessary to allow adaptation to human hosts (12), and the challenges of evolving new tissue tropisms (13). Although of fundamental importance, such characteristics are often highly pathogen-specific such that it is difficult to draw generalities about the likelihood of successful emergence. Herein, we address a more specific question: That is, how might we assess the capability of a particular emerging virus to achieve interhuman transmission using background knowledge of their biology?
Pathogen transmissibility is often quantified by the basic reproductive number, R0, and, to successfully achieve onward transmission in a host population, a virus must satisfy R0 > 1 (14). Given that natural selection will favor human-to-human transmission to increase the number of secondary infections, an accurate database of R0 estimates for individual viruses would undoubtedly assist pandemic prediction. However, such estimates are limited in number because they require sufficient incidence or sequence data and are strongly influenced by epidemiological context, such as whether they are inferred using data from outbreaks or periods of more endemic transmission.
Given these important limitations, we compiled and analyzed a database of 203 human viruses and assessed whether viruses exhibiting particular biological (i.e., virological) features were more often associated with sustained transmission among humans. The biological features considered reflect key aspects of virus life history and ecology and include the following: host mortality rate; genome type (DNA or RNA); genome length (number of nucleotides); the duration of infection (acute or chronic); segmentation of the virus genome (segmented or nonsegmented); frequency of recombination (classified as high or low); the presence or absence of an outer envelope (enveloped or nonenveloped); and the mode of virus transmission (limiting this variable to either vector-borne or non–vector-borne transmission for ease of interpretation). Using an information theoretic approach, we then set out to determine which of these features, singly or in combination, is most often associated with human-to-human transmission, and thus what biological attributes of viruses increase the likelihood of successful emergence.
Methods
Data Collection.
We first created a catalog of all human viruses, using data available at ViralZone (viralzone.expasy.org/all_by_species/678.html) supplemented with human viruses described in the primary literature. This literature search resulted in a dataset of 203 human virus species from which we determined the following biological properties from the literature: their taxonomic information; genome type, length, and segmentation (i.e., segmented versus nonsegmented); and the presence or absence of an outer envelope (as well as additional features such as duration of infection and host mortality rate) (Dataset S1). To simplify the analysis, we estimated these features assuming average disease progression in nonimmunocompromised human hosts in the absence of medical treatment or intervention. For the purposes of this study, we defined durations of viral infection as either “acute” (i.e., a short duration of infection lasting up to 4 wk) or “chronic” (i.e., an infection of duration longer than 4 wk). Because estimates of recombination rate are often difficult to obtain and sometimes contentious, we used two broad categories of the frequency of intraspecific recombination (or reassortment)—low and high—that reflect the average occurrence of recombination in these viruses as taken from the literature (which also acts to minimize error). Where data on recombination frequency was unavailable, we assumed that the virus in question exhibited the same recombination rate as documented in other members of its family (for example, although reassortment has not been detected in Dhori virus because of small sample size, we assume that the rate of assortment in this case is “high” because it occurs commonly in the Orthomyxoviridae). Finally, we compiled data on the usual mode of transmission (vector-borne, animal bite, direct/indirect contact, bodily fluids, respiratory, fecal–oral, blood-borne, sexual, and unknown), but, due to the high number of categories, we later limited this variable to either “vector-borne” or “non–vector-borne” transmission. A list of the biological features and the justification for their inclusion are given in Table S1.
Table S1.
Table of predictors
| Variable | Levels | Type | Definition | Justification | Fitted in model |
| Human-to-human transmission | Binary (yes = 1; no = 0) | Categorical | Evidence for direct human-to-human transmission, or human-vector-human transmission | The variable of interest | Response |
| Family | Taxonomic family | Categorical | The virus family of the virus species in question as reported in www.ictvonline.org/ | Correction factor (phylogenetic relatedness means that individual data points may not be independent) | Random |
| Envelope status | Enveloped; nonenveloped | Categorical | With or without an outer envelope | Fundamental division in virus structure. Nonenveloped viruses are likely more stable in open-air and can persist longer on surfaces, which may increase the probability of transmission | Fixed |
| Genome type | DNA; RNA | Categorical | DNA (Baltimore classifications I, II, and VII) or RNA (Baltimore classifications III, IV, V, and VI) genomes | These variables represent fundamental differences in genome organization and replication strategy. In addition, viruses in category I consistently evolve more slowly than those viruses in the other categories | Fixed |
| Mode of transmission | Vector-borne; non–vector-borne | Categorical | Viruses that use an arthropod (e.g., mosquito or tick) as a transmission vector are referred to as vector-borne viruses whereas all others are non-vector-borne | These variables are two distinct transmission modes where the vector-borne route allows transmission between two vertebrate species without direct contact | Fixed |
| Duration of infection | Chronic; acute | Categorical | Duration of viral infection in humans: An acute infection is defined here as <1 mo whereas a chronic infection persist >1 mo | The duration of an infection affects the time frame for transmission events and the number of opportunities for interhost transmission | Fixed |
| Recombination frequency | Low; high | Categorical | Broad-scale estimates of recombination frequency within the virus in question | Recombination may allow more genetic flexibility and therefore more rapid host adaptation | Fixed |
| Segmentation | Segmented; nonsegmented | Categorical | The viral genome is divided into different replicating molecules (segmented) or present as a single continuous replicating molecule (nonsegmented) | Fundamental division in virus structure. Segmented viruses are able to generate genetic variation through reassortment | Fixed |
| Mortality level | Percentage (Z-transformed to two SDs) | Numeric | Average case mortality number assuming no treatment, medical intervention, or prevention | Mortality rate affects the opportunity for virus transmission | Continuous |
| Genome length | Number of nucleotides (Z-transformed to two SDs) | Numeric | The number of nucleotides in the complete viral genome | It has previously been shown that genome length is correlated with evolutionary rates in RNA viruses and therefore may play a role in determining human-to-human transmission | Continuous |
The biological variables included in the analysis, how they are coded in the model, and why they are considered important.
The most important variable in our dataset is whether a specific virus is transmissible between humans, such that it can be considered human-“adapted.” We based this classification on the usual mode of transmission for each virus as described above. For example, although there have been documented cases of rabies virus being transmitted among human transplant recipients (15), no cases of bite and nonbite exposures between humans have been reported, such that we regard this virus as not adapted to human transmission. We also assumed that viral characteristics have remained stable over time but noted that some, particularly host mortality rate, may have evolved to their current state in some of the more established human viruses. Overall, we make general classifications and have estimated these variables using the most up-to-date information available in the literature. Importantly, any errors should be random across the dataset, thus having little impact on our results.
It is also important to note that our dataset comprises a wide variety of both RNA and DNA viruses that often do not share homologous genetic regions, preventing sequence alignment and thus phylogenetic inference. In particular, RNA and DNA viruses have no genes in common. Although perhaps unnecessary, this lack of common ancestry prohibited us from explicitly including phylogeny (i.e., evolutionary relatedness) in the model, even though it may in part explain why taxonomically related viruses share common variables. Instead, we explored ancestral associations between viruses by integrating a taxonomic variable at the family level, which is described in detail in Statistical Analyses.
Statistical Analyses.
We used an information theoretic approach to assess predictors of human-to-human transmission in all viruses compiled (Dataset S1). For our purposes, multimodel information theoretic approaches offer many advantages over competing approaches, such as stepwise model selection based on statistical significance or a global model approach with inference restricted to significant terms, both of which overlook model uncertainty (for a discussion, see refs. 16–21). Therefore, generalized linear models (GLMs) were implemented using the “glm” function in the base package within the statistical programming environment R version 3.2.1 (22), which was used for all analyses. A global model was implemented with a binary response variable denoting whether the virus was documented as exhibiting human-to-human transmission, 1, or not, 0, and the model family specified as “binomial” (i.e., a logit-link function). The predictors in the global model were coded like the variables in Table S1 and fitted additively.
We explored taxonomic effects by fitting the family of the virus as a random effect in a generalized linear mixed model (GLMM), along with the aforementioned fixed factors using the “glmer” function in the package lme4 (23). However, the variance component for the family effect in this GLMM was small, and the model had a lower Akaike information criterion corrected for small sample size (AICc) (24) than the global GLM. In addition, the taxonomic GLMM produced fixed effect coefficients identical to the global GLM. Accordingly, we proceeded with GLMs alone (see Tables S2 and S3 for model estimates from the global GLM and GLMM).
Table S2.
GLM model estimates
| Coefficient | Est. | SE | LCI | UCI |
| Intercept | 4.663 | 1.596 | 1.535 | 7.791 |
| Segmentationsegmented | −1.808 | 0.570 | −2.925 | −0.691 |
| Mode of transmissionvector-borne | −3.056 | 0.540 | −4.115 | −1.997 |
| Genome typeRNA | −1.225 | 1.511 | −4.186 | 1.736 |
| Outer envelope statusenveloped | −0.529 | 0.638 | −1.780 | 0.721 |
| Genome length | −0.909 | 0.876 | −2.625 | 0.808 |
| Mortality rate | −0.870 | 0.382 | −1.619 | −0.121 |
| Duration of infectionacute | −1.645 | 1.132 | −3.863 | 0.574 |
| Recombination frequencylow | −0.321 | 0.623 | −1.543 | 0.901 |
Global generalized linear model estimates (Est.) along with their SE and lower to upper 95% confidence interval (LCI to UCI). The Akaike information criterion corrected for small sample size for this model was 174.679.
Table S3.
GLMM model estimates for taxonomic model
| Coefficient | Est. | SE | LCI | UCI |
| Intercept | 4.671 | 1.615 | 1.506 | 7.837 |
| Segmentationsegmented | −1.804 | 0.584 | −2.949 | −0.659 |
| Mode of transmissionvector-borne | −3.061 | 0.555 | −4.149 | −1.972 |
| Genome typeRNA | −1.227 | 1.516 | −4.198 | 1.745 |
| Outer envelope statusenveloped | −0.533 | 0.649 | −1.804 | 0.739 |
| Genome length | −0.909 | 0.879 | −2.632 | 0.813 |
| Mortality rate | −0.870 | 0.384 | −1.622 | −0.117 |
| Duration of infectionacute | −1.645 | 1.133 | −3.867 | 0.576 |
| Recombination frequencylow | −0.323 | 0.631 | −1.559 | 0.913 |
Taxonomic generalized linear mixed model estimates (Est.) along with their SE and lower to upper 95% confidence interval (LCI to UCI). The Akaike information criterion corrected for small sample size for this model was 176.890, and the random factor for the estimated between-family variance was 0.007.
From the global model, a set of candidate models was created using the “dredge” function in the MuMIn package (25). Models were then ranked based on AICc. Rather than restrict our inference to that based on a single “best-fitting” model, which may be subject to model selection uncertainty and model selection bias, we used multimodel inference (20). From the set of candidate models, we obtained a top model set comprising those models with an AICc within two of the top model. Model-averaged GLM coefficients were then obtained using the “model.avg” function in MuMin. For each coefficient, we report the relative importance (RI), adjusted SEs as produced by MuMin (defined in ref. 20), associated 95% confidence intervals (CIs) (1.96 × SE) (26), and the coefficient estimate with shrinkage (sometimes called the “zero method”), which may be less upwardly biased for coefficients with a relative importance less than 1. We note that information theoretic approaches can potentially be misleading when global models have an initially poor fit (20). Therefore, we calculated R2 for the global model following equation 10 in Nakagawa and Schielzeth (27).
Results
A Dataset of Human Viruses.
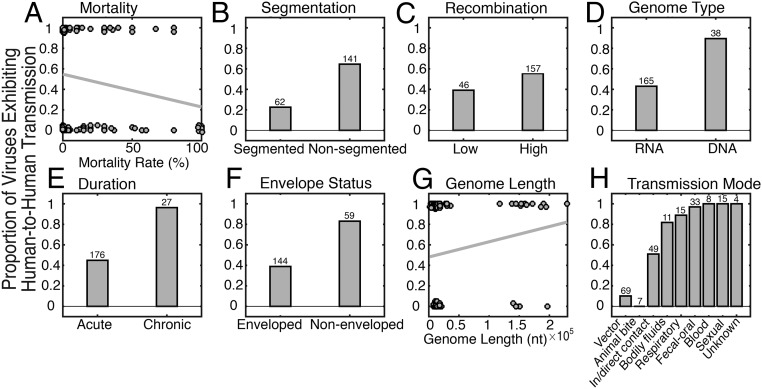
Our final dataset comprised 203 species of human virus, of which 105 (51.72%) exhibited human-to-human transmission, with the remainder associated with only transient spillover infections. These data contained 38 DNA viruses and 165 RNA viruses from 25 different families, of which the Bunyaviridae (negative-sense RNA) was the best represented, containing 37 species. The Flaviviridae and Picornaviridae were also well-represented, containing 23 and 24 species, respectively. The estimated mortality rates in the dataset range from 0% (e.g., some herpes viruses) to 100% (lyssaviruses), and 69 viruses exhibited vector-borne transmission. Strikingly, all viruses transmitted through blood and sexual contact resulted in chronic infections and were transmissible between humans (Fig. 1). In contrast, no viruses transmitted by animal bite were transmissible between humans although we note that Nipah virus has spread human-to-human via saliva after contamination of raw date sap by bats (and subsequent consumption by humans), rather than a bat bite (28).
Fig. 1.
The proportion of human virus species contained in our dataset within each category that have established human-to-human transmission as a function of (A) mortality rate, (B) genome segmentation, (C) recombination frequency, (D) genome type (DNA or RNA), (E) duration of infection (acute versus chronic), (F) envelope status, (G) genome length, and (H) mode of transmission. A line of best-fit is plotted for continuous variables (mortality rate and genome length), along with the raw data, and sample sizes for each categorical variable are shown.
An initial qualitative exploration of the dataset revealed that all but one of the chronic viruses (25 virus species) exhibited human-to-human transmission, with the single exception being simian foamy virus (although foamy viruses likely codiverge with other primate hosts) (29). In addition, it was notable that, of those viruses that establish a chronic infection, all had nonsegmented genomes and that the vast majority (20 species) had DNA genomes. Interestingly, the only (human-transmissible) chronic, nonsegmented RNA viruses, excluding retroviruses (i.e., HIV-1, HIV-2, and HTLV) and hepatitis D virus (a subviral satellite that requires coinfection with hepatitis B virus), were hepatitis C and human pegiviruses (formally GB viruses), which are both classified within the Flaviviridae. In contrast, only ∼45% of the acute viruses were associated with successful human-to-human transmission, again illustrating the importance of duration of infection in shaping the likelihood of successful viral emergence.
Model Selection and Model Averaging.
Our global GLM that contained all recorded predictors of human-to-human transmission in all viruses as fixed effects had R2 = 0.446, a value that is relatively high for an evolutionary or ecological study (30). A model incorporating duration of infection, outer envelope status, segmentation (i.e., segmented or nonsegmented), mode of transmission, and mortality rate had the lowest AICc (Table 1). However, four other models, including those models that contained genome length and recombination frequency, were within 2 AICc of the favored model (Table 1). The type of genome (DNA or RNA) of the virus was absent from all models in the top model set. Duration of infection, segmentation, mode of transmission, and mortality rate all had a relative importance of 1. In contrast, outer envelope status had a moderate relative importance (0.62) whereas genome length and recombination frequency had a lower relative importance (Table 2).
Table 1.
GLM top model set
| Model form | df | logLik | AICc | ΔAICc | Weight |
| Duration of infection + outer envelope status + segmentation + mode of transmission + mortality rate | 6 | −78.603 | 169.635 | 0.000 | 0.355 |
| Duration of infection + segmentation + mode of transmission + mortality rate | 5 | −80.044 | 170.393 | 0.758 | 0.243 |
| Duration of infection + outer envelope status + segmentation + mode of transmission + genome length + mortality rate | 7 | −78.488 | 171.550 | 1.915 | 0.136 |
| Duration of infection + segmentation + mode of transmission + genome length + mortality rate | 6 | −79.579 | 171.586 | 1.951 | 0.134 |
| Duration of infection + outer envelope status + recombination frequency + segmentation + mode of transmission + mortality rate | 7 | −78.527 | 171.628 | 1.993 | 0.131 |
Top model set based on the Akaike information criterion corrected for small sample size (AICc), the log likelihood of those models (logLik), the difference in AICc between each model and the AICc favored model (ΔAICc), and the model weights.
Table 2.
GLM model-averaged coefficients
| Coefficient | RI | Est. | SE | LCI | UCI | Est. (shrinkage) |
| Intercept | 3.673 | 1.134 | 1.450 | 5.895 | 3.673 | |
| Segmentationsegmented | 1 | −1.742 | 0.477 | −2.677 | −0.807 | −1.742 |
| Mode of transmissionvector-borne | 1 | −3.143 | 0.545 | −4.212 | −2.075 | −3.143 |
| Mortality rate | 1 | −0.992 | 0.386 | −1.748 | −0.237 | −0.992 |
| Duration of infectionacute | 1 | −1.817 | 1.092 | −3.958 | 0.323 | −1.817 |
| Outer envelope statusenveloped | 0.62 | −0.873 | 0.553 | −1.957 | 0.211 | −0.544 |
| Genome length | 0.27 | −0.317 | 0.447 | −1.192 | 0.559 | −0.086 |
| Recombination frequencylow | 0.13 | −0.226 | 0.582 | −1.368 | 0.915 | −0.030 |
Model-averaged generalized linear model estimates (Est.) along with their SE and lower to upper 95% confidence interval (LCI to UCI), and their relative importance (RI). The estimate with shrinkage is also given. Subscripts denote the contrast category for categorical predictors.
Model-averaged coefficients are given in Table 2. Notably, vector-borne viruses were considerably less likely to exhibit human-to-human transmission compared to viruses not transmitted by vectors, and segmented viruses were estimated to be less likely to be associated with human-to-human transmission than nonsegmented viruses. Our models also revealed that increases in host mortality rates were associated with a lower probability of human-to-human transmission. Similarly, viruses with acute durations of infection were estimated to have a lower probability of establishing human-to-human transmission compared to viruses with longer (i.e., chronic) infection although this effect was estimated with poor precision and the 95% CI included zero (Table 2). Three other traits had relative importance less than 0.65, and again the coefficients were estimated with a 95% CI including zero (Table 2). First, enveloped viruses were observed to be less likely to display human-to-human transmission than those viruses that are nonenveloped. Second, viruses with low recombination frequency were less likely to achieve human transmission than viruses that recombine frequently. Finally, increases in genome length, once corrected for genome type, were associated with a marginally decreased probability of human-to-human transmission. Coefficients from the global model and also the single model with the lowest AICc produced the same qualitative conclusions as the multimodel approach, demonstrating that our conclusions are not solely driven by model averaging (Tables S2 and S4).
Table S4.
GLM coefficients from the AICc favored model
| Coefficient | Est. | SE | LCI | UCI |
| Intercept | 3.888 | 1.115 | 1.702 | 6.074 |
| Segmentationsegmented | −1.640 | 0.453 | −2.529 | −0.751 |
| Mode of transmissionvector-borne | −3.006 | 0.519 | −4.022 | −1.989 |
| Mortality rate | −0.903 | 0.368 | −1.624 | −0.182 |
| Duration of infectionacute | −1.842 | 1.079 | −3.957 | 0.273 |
| Outer envelope statusenveloped | −0.903 | 0.536 | −1.955 | 0.148 |
Generalized linear model estimates (Est.) from the top model based on Akaike information criterion corrected for small sample size. For each Est., also given is the SE as well as the lower to upper 95% confidence interval (LCI to UCI).
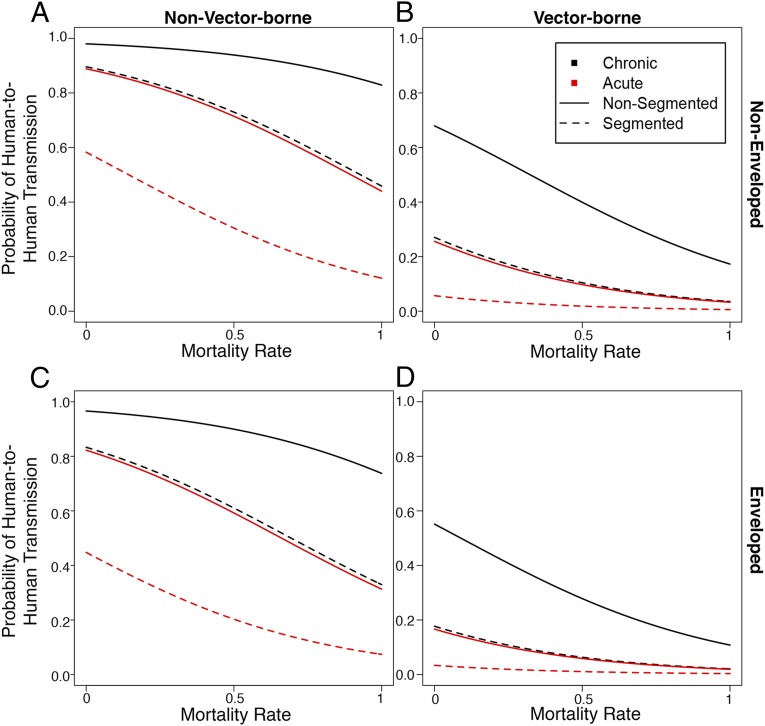
Next, we illustrated the best predictors of human-to-human transmission as a function of mortality rate because the latter is clearly a key determinant of human-to-human transmission (Fig. 2). Specifically, using the model-averaged coefficients, we generated predicted values for a subset of various trait combinations (i.e., those traits estimated to be the strongest predictors of human transmissibility): genome segmentation and duration of infection, for both outer envelope status and mode of transmission. This model averaging demonstrated that the estimated probability of human-to-human transmission decreased as mortality rate increased for all combinations of variables. In addition, the effect of mortality rate differed substantially between vector-borne and non–vector-borne viruses. Chronic, nonsegmented, nonenveloped, non–vector-borne viruses (Fig. 2A, solid, black line) showed the least decline in probability of human-to-human transmission, with a probability of ∼0.8 even with very high mortality (100%). Conversely, acute, enveloped, segmented, vector-borne viruses (Fig. 2D, dashed, red line) showed a very low probability of human-to-human transmission across all mortality rates.
Fig. 2.
Predicted probability of human-to-human transmission based on model-averaged coefficients for duration of infection (black lines for chronic infections; red lines for acute infections) and genome segmentation (solid line for nonsegmented viruses; dashed line for segmented viruses), as a function of the estimated effect of mortality rate. (A and B) Nonenveloped viruses. (C and D) Enveloped viruses. (A and C) Non–vector-borne viruses. (B and D) Vector-borne viruses. All data have been adjusted for frequency of occurrence, and a correction for recombination frequency has been made based on the model-averaged estimate with shrinkage. For coefficients with a relative importance less than 1, the estimate with shrinkage was used.
Finally, it is noteworthy that the full dataset contains a number of viruses (n = 26) that have occurred only rarely in human populations (i.e., fewer than 10 reported human cases) (Dataset S1). For example, only three human cases of Bas-Congo virus have been reported (31), resulting in two deaths, giving a mortality rate of 67%. To assess whether the inclusion of these viruses had biased our analysis, we performed model averaging on a subset of the data containing only those viruses that are more commonly observed: i.e., 10 or more reported cases of human infection (177 virus species) (see Table S5 for model-averaged coefficients based on this data subset). In this reduced dataset, the duration of the infection and mortality rate had very low relative importance, and model-averaged coefficients for these traits were associated with wide 95% CIs. This analysis indicates that many of the uncommon viruses have similar (acute) durations of infection and/or are associated with high human mortality such that they are poor predictors. In contrast, the strong predictive effects of segmentation and mode of transmission remained consistent between the full and reduced datasets.
Table S5.
GLM model estimates for common viruses
| Coefficient | RI | Est. | SE | LCI | UCI | Est. (shrinkage) |
| Intercept | 2.655 | 1.175 | 0.352 | 4.958 | 2.655 | |
| Segmentationsegmented | 1 | −2.345 | 0.536 | −3.396 | −1.294 | −2.345 |
| Mode of transmissionvector-borne | 1 | −3.434 | 0.559 | −4.529 | −2.339 | −3.434 |
| Genome typeRNA | 0.32 | −1.685 | 1.729 | −5.074 | 1.704 | −0.538 |
| Outer envelope statusenveloped | 0.29 | −0.708 | 0.576 | −1.837 | 0.422 | −0.209 |
| Genome length | 0.24 | −1.061 | 1.001 | −3.023 | 0.900 | −0.258 |
| Mortality rate | 0.22 | 0.410 | 0.399 | −0.372 | 1.192 | 0.091 |
| Duration of infectionacute | 0.14 | −0.952 | 1.101 | −3.110 | 1.206 | −0.135 |
| Recombination frequencylow | 0.07 | −0.462 | 0.613 | −1.663 | 0.739 | −0.032 |
Model-averaged generalized linear model estimates (Est.) along with their SE and lower to upper 95% confidence interval (LCI to UCI), and their relative importance (RI), based on only those viruses with greater than 10 recorded cases. The estimate with shrinkage is also given.
Discussion
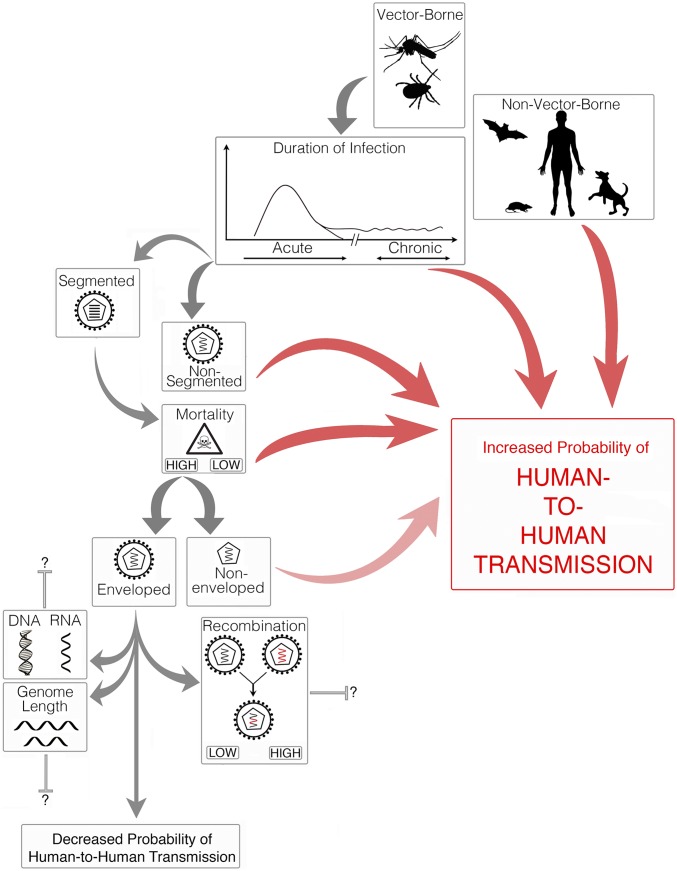
We have revealed those biological features of viruses that show the strongest association with sustained transmission among humans, establishing a framework that can be used to help predict the general types of viruses that may be most likely to successfully emerge in the future. This analysis suggests that the best predictors of transmissibility among humans are the duration of infection, genome segmentation (i.e., segmented or nonsegmented), mode of transmission (i.e., vector-borne or non–vector-borne), mortality rate, and, to a lesser extent, the presence or absence of an outer envelope. In contrast, the frequency of recombination and genome length were less important predictors of transmission success, and, strikingly, genome type (i.e., DNA or RNA) had essentially no predictive power (i.e., appeared in no models in the top model set). Overall, we found that chronic, nonsegmented, non– viruses with low host mortality were most likely to exhibit human-to-human transmission (Fig. 3).
Fig. 3.
Schematic overview of those biological variables associated with an increase (red arrows) and decrease (gray arrows) in the likelihood of a virus establishing human-to-human transmission. The transparency of red arrows indicates the importance of the specific variable in its predictive power (i.e., more important variables are illustrated by a more opaque arrow).
Given that natural selection should always act to increase R0, individual viruses will evidently possess specific biological traits that increase their probability of interhost transmission. On that basis, we aim to offer working hypotheses as to why the traits identified here—particularly low host mortality, chronic infection, non–vector-borne, nonsegmented, and nonenveloped—might facilitate human-to-human transmission. However, we recognize that some of these traits are likely to be confounding, such that they are not independent of each other or additional viral features, and we provide clarifications where such associations might exist.
Our results strongly indicate that human transmissibility decreases as host mortality rate increases. Although the relationship between virulence and transmission is complex (32), the notion that low host mortality will generally allow more time for interhost transmission seems well-founded (33) because, the lower the mortality rate, the fewer the susceptible hosts required to achieve R0 > 1 (34). However, an important caveat is that estimates of host mortality rate rely heavily on precise diagnosis and accurate reporting. Therefore, in the case of rare viruses that are often underreported (e.g., Bas-Congo virus) or those viruses that can establish asymptomatic infections (e.g., enterovirus A71), the mortality rate may be vastly overstated. Indeed, we found that uncommon viruses were often associated with high mortality rates in humans. Despite these shortcomings, our results are clearly in conflict with the theory that vector-borne pathogens have a higher host mortality rate compared with non–vector-borne pathogens (35). In particular, whereas many non–vector-borne viruses exhibited >80% human mortality, the highest human mortality rate in a vector-borne virus was 52% in Chandipura virus (36). Overall, we observed that the average host mortality rate in non–vector-borne viruses was ∼12%, compared with ∼6% for vector-borne viruses, although this difference was not statistically significant.
Our analysis also reveals that the length of time a virus is able to replicate within an individual human host, quantified here as the duration of infection, is an important parameter in determining whether a virus is able to evolve human-to-human transmission. Specifically, chronic viruses were more likely to be transmissible between humans, clearly because extended durations of infection increase the chance of secondary transmission to a new host. Indeed, viruses with long durations of infection, such as retroviruses and some DNA viruses, seem more likely to codiverge with their hosts over evolutionary timescales and thus are often strongly host species-specific (37–39).
Although vector-borne transmission is of equal importance in the model compared to the other predictors discussed here (because they all have a relative importance of 1), it has a much larger overall effect (Table 2). Indeed, of the 69 vector-transmitted viruses in our list, only 6 are transmissible between humans. That vector-borne viruses are less likely to jump to a new host and successfully establish an infection is to be expected, given the complexity of zoonotic transmission cycles that involve invertebrate vectors and vertebrate hosts (8, 39). Remarkably, Zika virus is the only vector-borne virus in our dataset where onward human transmission may not involve the usual zoonotic cycle because sexual transmission has been reported (40). Birds are the most common vertebrate reservoir host for vector-borne viruses in our dataset whereas humans are usually dead-end hosts, presumably because viral loads are insufficient to allow onward transmission through a biting vector (41). In addition, multihost viruses, such as those viruses that are vector-borne, may experience antagonistic pleiotropy (42), which will also act to reduce adaptability in new hosts (43).
A more puzzling observation is that nonsegmented viruses seem more able to be transmitted among humans compared to viruses with segmented genomes. In this context it is important to note that none of the positive-sense single-stranded RNA (+ssRNA) viruses in our dataset possess segmented genomes. Accordingly, the true cause of the predictive power of nonsegmented viruses may reflect the preponderance of +ssRNA compared with negative-sense (–ssRNA) viruses among the human-transmitted set. Indeed, the replication cycle of +ssRNA viruses can be considered simpler than that of –ssRNA viruses, with the positive-sense RNA acting as an mRNA from which translation can proceed immediately after infection whereas –ssRNA viruses are required to go through an additional transcription step before translation. It is therefore possible that this simpler, and presumably quicker, replication process may benefit host adaptation. However, our analysis also revealed that the distinction between DNA and RNA genomes is only a weak predictor of the likelihood of establishing human-to-human transmission. A similar confounding association is that all of the segmented viruses in our dataset develop acute infections, which is itself associated with a decreased probability of human-to-human transmission. In addition, many DNA viruses establish a chronic infection and are never segmented. Elucidating the apparently increased ability for nonsegmented viruses to generate sustained infections in humans is clearly an important area for future study.
Finally, we observed that nonenveloped viruses were more likely to establish human-to-human transmission than enveloped viruses; only ∼39% of the enveloped viruses in our dataset were transmissible between humans, compared with 83% of the nonenveloped viruses. It is possible that nonenveloped viruses are more environmentally stable than their enveloped counterparts (44) because the glycoproteins and lipids that comprise the envelope are easily degradable, which in turn increases the probability of interhost transmission through contact with exposed surfaces. Indeed, nonenveloped viruses are resistant to common ethanol disinfectant, and this resistance is associated with epidemics in areas with abundant human interaction, especially institutional settings such as schools or hospitals (45) [for example, outbreaks of human norovirus (46)]. The frequency with which nonenveloped viruses are found in “extreme” environments, such as oceans (47), and preserved intact in ice cores (48) is further evidence for their stability. However, the majority of viruses in our dataset are enveloped (144, compared with 59 nonenveloped), indicating that viruses of this type possess additional beneficial characteristics, such as an enhanced ability to evade the host immune response. Indeed, the ability to evade the adaptive immune response may have been a key selection pressure for the origin of the viral envelope (49).
In marked contrast, our analysis reveals that the frequency with which viruses recombine has little predictive power for interhuman transmissibility. Recombination rates vary extensively among RNA viruses, from seemingly clonal in nonsegmented negative-sense RNA viruses (i.e., the order Mononegavirales) to per site rates that are greater than that of mutation in the case of HIV-1 and that undoubtedly have a major impact on their evolution and epidemiology (39). Although recombination has the potential to facilitate transmissibility by accelerating the rate at which advantageous genetic combinations are produced compared with mutation alone, frequent recombination will also break up beneficial genetic configurations, and clonal viruses like those species of the Mononegavirales are readily able to emerge in new hosts (for example, Ebola virus) (50). Indeed, there are few cases in which recombination has been shown to underpin successful cross-species transmission and emergence (39).
Until recently, the focus of much research on new emerging diseases was to reveal the processes that lead to pathogen emergence, both the ecological factors that precipitate emergence and the genetic factors that enable host adaptation (or the host barriers to this process), rather than the subsequent transmissibility of pathogens in the new host species (11). Herein, we have revealed factors that may explain why some viruses are more readily transmitted among the human population than others. More generally, our work offers a framework for predicting the transmissibility of emerging pathogens among humans. By identifying the major biological features of successfully emerging viruses, our analysis can be used to generate broad-scale predictions of the likelihood that a virus of a specific family will achieve human-to-human transmission and thus epidemic spread.
Supplementary Material
Acknowledgments
J.L.G. and A.M.S. are supported by the Judith and David Coffey fellowship from the Charles Perkins Centre, University of Sydney. F.D.G. is supported by Swiss National Science Foundation Grant P2ZHP3_151594. E.C.H. is funded by National Health and Medical Research Council Australia Fellowship AF30 and NIH Grant R01 GM080533.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521582113/-/DCSupplemental.
References
- 1.Garske T, et al. Assessing the severity of the novel influenza A/H1N1 pandemic. BMJ. 2009;339:b2840. doi: 10.1136/bmj.b2840. [DOI] [PubMed] [Google Scholar]
- 2.Horimoto T, Kawaoka Y. Influenza: Lessons from past pandemics, warnings from current incidents. Nat Rev Microbiol. 2005;3(8):591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 3.Chertow DS, et al. Ebola virus disease in West Africa: Clinical manifestations and management. N Engl J Med. 2014;371(22):2054–2057. doi: 10.1056/NEJMp1413084. [DOI] [PubMed] [Google Scholar]
- 4.Lam TT, et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522(7554):102–105. doi: 10.1038/nature14348. [DOI] [PubMed] [Google Scholar]
- 5.Azhar EI, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 6.Gire SK, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345(6202):1369–1372. doi: 10.1126/science.1259657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes EC. Evolution in health and medicine Sackler colloquium: The comparative genomics of viral emergence. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1742–1746. doi: 10.1073/pnas.0906193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolhouse MEJ, Haydon DT, Antia R. Emerging pathogens: The epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20(5):238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morse SS. Factors in the emergence of infectious diseases. Emerg Infect Dis. 1995;1(1):7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrish CR, et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72(3):457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell CA, et al. Improving pandemic influenza risk assessment. eLife. 2014;3:e03883. doi: 10.7554/eLife.03883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taber SW, Pease CM. Paramyxovirus phylogeny: Tissue tropism evolves slower than host specificity. Evolution. 1990;44(2):435–438. doi: 10.1111/j.1558-5646.1990.tb05210.x. [DOI] [PubMed] [Google Scholar]
- 14.May RM, Gupta S, McLean AR. Infectious disease dynamics: What characterizes a successful invader? Philos Trans R Soc Lond B Biol Sci. 2001;356(1410):901–910. doi: 10.1098/rstb.2001.0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasan A, et al. Rabies in Transplant Recipients Investigation Team Transmission of rabies virus from an organ donor to four transplant recipients. N Engl J Med. 2005;352(11):1103–1111. doi: 10.1056/NEJMoa043018. [DOI] [PubMed] [Google Scholar]
- 16.Burnham K, Anderson D, Huyvaert K. AIC model selection and multimodel inference in behavioral ecology: Some background, observations, and comparisons. Behav Ecol Sociobiol. 2011;65(1):23–35. [Google Scholar]
- 17.Hegyi G, Garamszegi L. Using information theory as a substitute for stepwise regression in ecology and behavior. Behav Ecol Sociobiol. 2011;65(1):69–76. [Google Scholar]
- 18.Grueber CE, Nakagawa S, Laws RJ, Jamieson IG. Multimodel inference in ecology and evolution: Challenges and solutions. J Evol Biol. 2011;24(4):699–711. doi: 10.1111/j.1420-9101.2010.02210.x. [DOI] [PubMed] [Google Scholar]
- 19.Whittingham MJ, Stephens PA, Bradbury RB, Freckleton RP. Why do we still use stepwise modelling in ecology and behaviour? J Anim Ecol. 2006;75(5):1182–1189. doi: 10.1111/j.1365-2656.2006.01141.x. [DOI] [PubMed] [Google Scholar]
- 20.Burnham KP, Anderson DR. Model Selection and Mulitmodel Inference: A Practical Information-Theoretic Approach. 2nd Ed Springer; New York: 2002. [Google Scholar]
- 21.Mundry R. Issues in information theory-based statistical inference: A commentary from a frequentist’s perspective. Behav Ecol Sociobiol. 2011;65(1):57–68. [Google Scholar]
- 22.R-Development-Core-Team 2015 R: A language and environment for statistical computing, Version 3.2.1. Available at www.r-project.org.
- 23.Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. arXiv:1406.5823v1.
- 24.Hurvich CM, Tsai C-L. Regression and time series model selection in small samples. Biometrika. 1989;76(2):297–307. [Google Scholar]
- 25.Bartoń K. 2015 MuMIn: Multi-model inference. R package version 1.15.1. Available at https://cran.r-project.org/web/packages/MuMIn/index.html.
- 26.Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: A practical guide for biologists. Biol Rev Camb Philos Soc. 2007;82(4):591–605. doi: 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa S, Schielzeth H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol. 2013;4(2):133–142. [Google Scholar]
- 28.Gurley ES, et al. Person-to-person transmission of Nipah virus in a Bangladeshi community. Emerg Infect Dis. 2007;13(7):1031–1037. doi: 10.3201/eid1307.061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Switzer WM, et al. Ancient co-speciation of simian foamy viruses and primates. Nature. 2005;434(7031):376–380. doi: 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]
- 30.Møller AP, Jennions MD. How much variance can be explained by ecologists and evolutionary biologists? Oecologia. 2002;132(4):492–500. doi: 10.1007/s00442-002-0952-2. [DOI] [PubMed] [Google Scholar]
- 31.Grard G, et al. A novel rhabdovirus associated with acute hemorrhagic fever in central Africa. PLoS Pathog. 2012;8(9):e1002924. doi: 10.1371/journal.ppat.1002924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bull JJ, Lauring AS. Theory and empiricism in virulence evolution. PLoS Pathog. 2014;10(10):e1004387. doi: 10.1371/journal.ppat.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: History, current state of affairs and the future. J Evol Biol. 2009;22(2):245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 34.Anderson RM, May RM. The population dynamics of microparasites and their invertebrate hosts. Philos T Roy Soc B. 1981;291(1054):451–524. [Google Scholar]
- 35.Ewald PW. Evolution of Infectious Disease. Oxford Univ Press; Oxford: 1994. [Google Scholar]
- 36.Ghosh S, Dutta K, Basu A. Chandipura virus induces neuronal death through Fas-mediated extrinsic apoptotic pathway. J Virol. 2013;87(22):12398–12406. doi: 10.1128/JVI.01864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villarreal LP. Viruses and the Evolution of Life. ASM Press; Washington, DC: 2005. [Google Scholar]
- 38.Villarreal LP, Defilippis VR, Gottlieb KA. Acute and persistent viral life strategies and their relationship to emerging diseases. Virology. 2000;272(1):1–6. doi: 10.1006/viro.2000.0381. [DOI] [PubMed] [Google Scholar]
- 39.Holmes EC. The Evolution and Emergence of RNA viruses. Oxford Univ Press; Oxford: 2009. [Google Scholar]
- 40.Foy BD, et al. Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis. 2011;17(5):880–882. doi: 10.3201/eid1705.101939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2(10):789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elena SF, Agudelo-Romero P, Lalić J. The evolution of viruses in multi-host fitness landscapes. Open Virol J. 2009;3:1–6. doi: 10.2174/1874357900903010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woelk CH, Holmes EC. Reduced positive selection in vector-borne RNA viruses. Mol Biol Evol. 2002;19(12):2333–2336. doi: 10.1093/oxfordjournals.molbev.a004059. [DOI] [PubMed] [Google Scholar]
- 44.Howie R, Alfa MJ, Coombs K. Survival of enveloped and non-enveloped viruses on surfaces compared with other micro-organisms and impact of suboptimal disinfectant exposure. J Hosp Infect. 2008;69(4):368–376. doi: 10.1016/j.jhin.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Eterpi M, McDonnell G, Thomas V. Disinfection efficacy against parvoviruses compared with reference viruses. J Hosp Infect. 2009;73(1):64–70. doi: 10.1016/j.jhin.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Greig JD, Lee MB. A review of nosocomial norovirus outbreaks: Infection control interventions found effective. Epidemiol Infect. 2012;140(7):1151–1160. doi: 10.1017/S0950268811002731. [DOI] [PubMed] [Google Scholar]
- 47.Culley AI, Lang AS, Suttle CA. High diversity of unknown picorna-like viruses in the sea. Nature. 2003;424(6952):1054–1057. doi: 10.1038/nature01886. [DOI] [PubMed] [Google Scholar]
- 48.Ng TF, et al. Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc Natl Acad Sci USA. 2014;111(47):16842–16847. doi: 10.1073/pnas.1410429111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buchmann JP, Holmes EC. Cell walls and the convergent evolution of the viral envelope. Microbiol Mol Biol Rev. 2015;79(4):403–418. doi: 10.1128/MMBR.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon-Loriere E, Holmes EC. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9(8):617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.