Significance
Nucleotide excision repair is the sole mechanism for removing bulky adducts from the human genome, including those formed by UV radiation and chemotherapeutic drugs. We used eXcision Repair-sequencing, a genomic assay for measuring DNA repair, to map the kinetics of repair after UV treatment. These genome-wide repair maps, in turn, allowed us to infer how excision repair is influenced by DNA packaging. Active and open chromatin regions were repaired more rapidly than other genomic regions. Repair in repressed and heterochromatic regions is slower and persists for up to 2 d. Furthermore, late-repaired regions are associated with a higher level of cancer-linked somatic mutations, highlighting the importance of efficient DNA repair and linking chromatin organization to cancer mutagenesis.
Keywords: DNA repair, DNA damage, chromatin, transcription, mutation
Abstract
We recently developed a high-resolution genome-wide assay for mapping DNA excision repair named eXcision Repair-sequencing (XR-seq) and have now used XR-seq to determine which regions of the genome are subject to repair very soon after UV exposure and which regions are repaired later. Over a time course, we measured repair of the UV-induced damage of cyclobutane pyrimidine dimers (CPDs) (at 1, 4, 8, 16, 24, and 48 h) and (6-4)pyrimidine-pyrimidone photoproducts [(6-4)PPs] (at 5 and 20 min and 1, 2, and 4 h) in normal human skin fibroblasts. Each type of damage has distinct repair kinetics. The (6-4)PPs are detected as early as 5 min after UV treatment, with the bulk of repair completed by 4 h. Repair of CPDs, which we previously showed is intimately coupled to transcription, is slower and in certain regions persists even 2 d after UV irradiation. We compared our results to the Encyclopedia of DNA Elements data regarding histone modifications, chromatin state, and transcription. For both damage types, and for both transcription-coupled and general excision repair, the earliest repair occurred preferentially in active and open chromatin states. Conversely, repair in regions classified as “heterochromatic” and “repressed” was relatively low at early time points, with repair persisting into the late time points. Damage that remains during DNA replication increases the risk for mutagenesis. Indeed, late-repaired regions are associated with a higher level of cancer-linked mutations. In summary, we show that XR-seq is a powerful approach for studying relationships among chromatin state, DNA repair, genome stability, mutagenesis, and carcinogenesis.
DNA damage blocks transcription and replication and compromises the integrity of the genome. Bulky adducts in DNA, the focus of this work, are caused by a variety of genotoxic agents including UV radiation in sunlight. In human cells, this damage is removed exclusively by the nucleotide excision repair mechanism (1–3). In nucleotide excision repair, a single-stranded oligomer that contains the bulky adduct is excised by dual incisions bracketing the lesion. The resulting gap is filled in by DNA polymerases, and the newly synthesized repair patch is then sealed by DNA ligase. In general excision repair, damage recognition is achieved by the repair factors xeroderma pigmentosum (XP) complementation group C (XPC) together with replication protein A (RPA) and XPA (4–6). In DNA that is being actively transcribed, damage recognition can also be achieved by a stalled RNA polymerase II, which, with the aid of the Cockayne Syndrome B (CSB) protein, accelerates the recruitment of the excision repair factors. This recognition process leads to transcription-coupled repair (7, 8). The subsequent dual-incision reaction is carried out by the core excision repair factors RPA, XPA, transcription factor II H (TFIIH), XPG, and XPF-ERCC1, releasing an excised oligomer that is 24–32 nucleotides (nt) in length (6, 9).
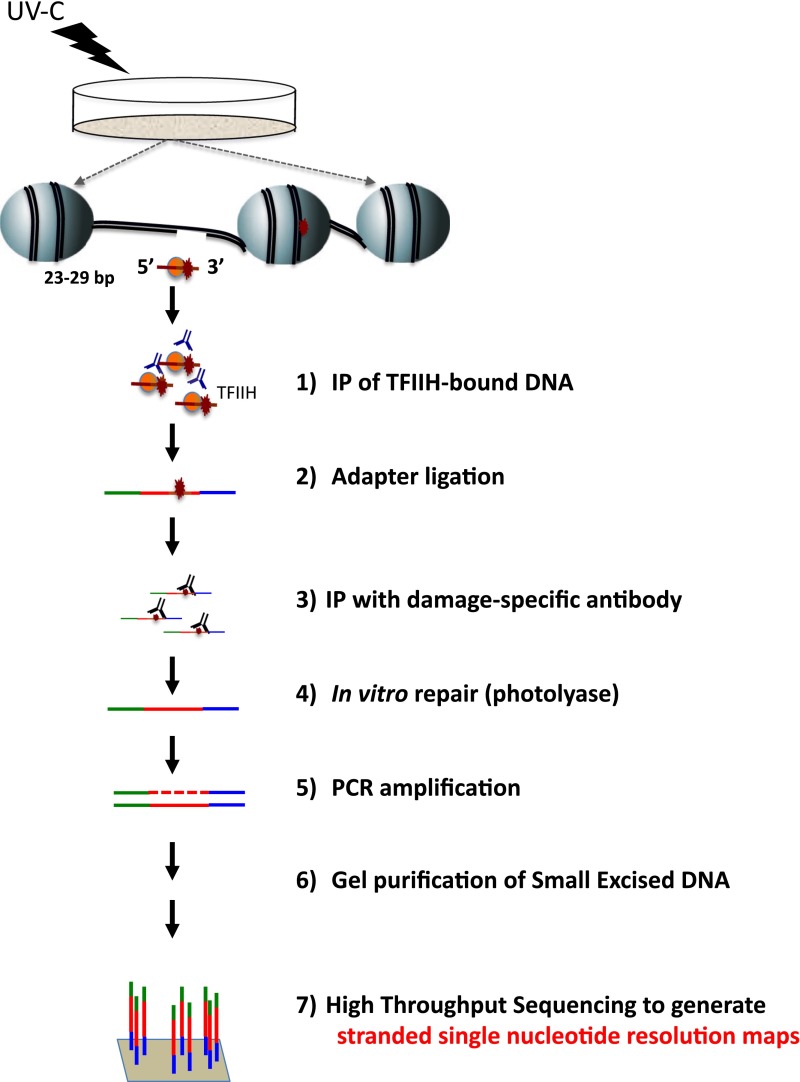
The mechanism of DNA excision repair has been reconstituted in vitro and is well characterized (10). However, a full understanding of repair must take into account how it occurs in live cells, where DNA is packed into chromatin and serves as a template for transcription. A genome-wide comparison of open chromatin status, histone modifications, RNA expression, and DNA damage and repair would allow inference of how these processes are coordinated. We recently developed an assay for mapping DNA excision repair at nucleotide resolution across the human genome called eXcision Repair sequencing (XR-seq) (11). In XR-seq, the excised oligonucleotides, released during repair in vivo, are isolated and sequenced, resulting in a stranded, single-nucleotide resolution map of repair (Fig. S1). In this study, we used XR-seq to measure repair kinetics after UV irradiation of two types of UV-induced damage: cyclobutane pyrimidine dimers (CPDs) and (6-4)pyrimidine-pyrimidone photoproducts [(6-4)PPs], and then related the kinetics of repair to chromatin state and mutagenesis.
Fig. S1.
Schematic of the XR-seq assay. For details, see ref. 11.
Results
Identification of Sites of CPD Damage Repaired Early and Late After UV Exposure in Normal Human Fibroblasts.
To determine which genomic regions were repaired rapidly after damage and which were repaired more slowly, we conducted a time course of CPD and (6-4)PP repair in normal human skin fibroblasts (NHF1). The bulk repair kinetics of these two types of damage is known to be quite different (4, 12, 13). CPDs, which do not distort the DNA helix as strongly and escape recognition by the general repair machinery, are recognized primarily in a transcription-coupled manner. Their complete repair requires 12–48 h, depending on the UV dose. In contrast, the general repair pathway efficiently removes (6-4)PPs, with most of the damage excised within 4 h of UV treatment.
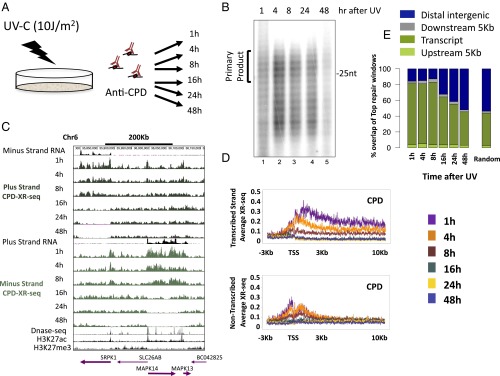
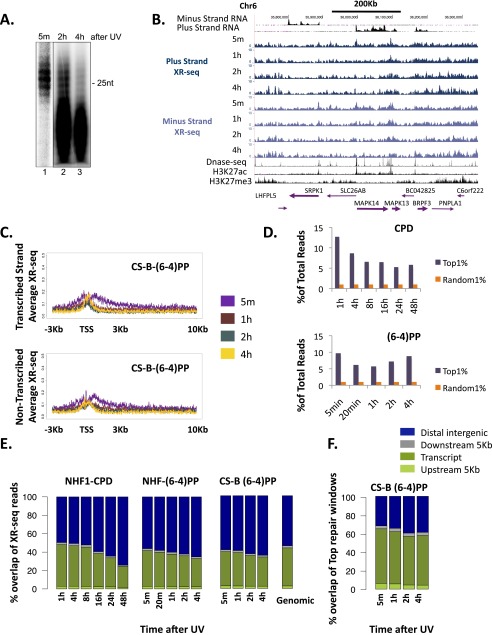
To measure the relatively slow CPD repair, cells were treated with a UV-C dose of 10 J/m2, which results in ∼50% cell survival (14), and time points were collected at 1, 4, 8, 16, 24, and 48 h (Fig. 1A). The time course was performed twice, with each experiment being independent from the other (Materials and Methods). The excised oligomer released in the cells was isolated with anti-CPD antibodies, and analyzed by autoradiography. The primary excision product ranged in size from 24 to 32 nt and rapidly degraded to ∼20 nt (13). Analysis of bulk repair by gel autoradiography revealed that CPD repair peaked at 4 h after UV and continued at substantial levels to 24 h. At 48 h, although at lower levels, CPD repair was still evident (Fig. 1B).
Fig. 1.
Kinetics of the genome-wide repair of CPD in normal human fibroblasts. (A) Schematic of time course of excision repair of the UV-induced CPD. (B) After irradiation with 10 J/m2 UV-C, normal NHF1 fibroblasts were incubated for the indicated time, and the excised oligonucleotides were immunoprecipitated with anti-CPD antibody, radiolabeled at the 3′ end with 32P-cordycepin and analyzed on sequencing gels. (C) Distribution of the XR-seq signal at each time point, separated by strand, for CPD repair along a 400-Kb region of chr6. ENCODE total stranded RNA-seq, DNase-seq, and ChIP-seq signal for H3K27ac and H3K27me3 from NHDF cells are plotted in black. Arrows in the bottom indicate position and directions of annotated genes. Bold arrows indicate expressed genes. (D) Average CPD XR-seq repair signal 3 Kb upstream and 10 Kb downstream of the annotated TSS of genes with the highest quartile of RNA expression in normal human skin fibroblasts (ENCODE, NHDF data). Signal is plotted separately for the transcribed and nontranscribed strands. (E) Percent overlap of the top 1% of repair windows at each of the time points with annotated genes (dark green), 5 Kb upstream of the gene (light green), 5 Kb downstream of the gene (gray), and intergenic regions (blue). Plotted for comparison are the average results of 50 permutations of a random set of 500 mers from the hg19 reference genome.
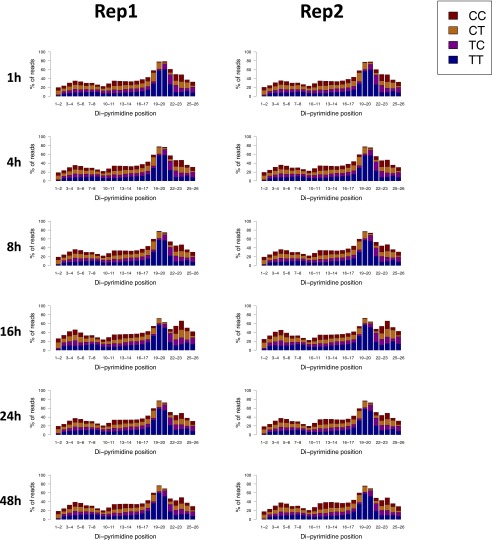
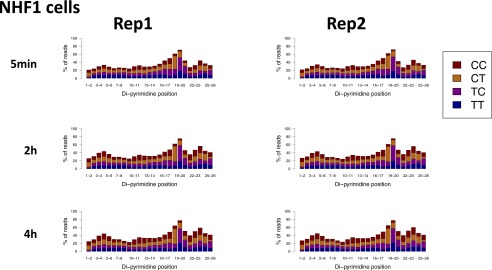
We performed XR-seq (11) to map the repair profile of CPDs genome-wide at each of the time points (Fig. S1). Because the primary excision products (∼30 nt in length) are bound specifically by TFIIH (13), they were isolated by TFIIH immunoprecipitation. This immunoprecipitation was followed by ligation of 5′ and 3′ adapters, compatible with the Illumina small-RNA TruSeq platform, and immunoprecipitation with anti-CPD antibodies. To allow PCR amplification, the photoproducts were removed in vitro by treatment with CPD photolyases. The repaired products were PCR-amplified with primers containing specific Illumina TruSeq small RNA-sequencing (RNA-seq) barcodes (11). As reported previously, XR-seq for CPD identified repair fragments that were 26 nt in length on average and were enriched for TT 6–8 nt from the 3′ end (11). There were no differences in the excised oligomer length or sequence context between the different time points of repair (Dataset S1 and Figs. S2–S4).
Fig. S2.
Similar enrichment of di-pyrimidines 5–7nt from the 3′ end of 26-nt-long reads for CPD XR-seq in normal human fibroblasts at the different time points. Shown are the results of two biological replicates.
Fig. S4.
Same as Fig. S3 except experiments were performed in CS-B cells, defective in transcription-coupled repair.
Fig. S3.
Similar enrichment of di-pyrimidines 6–7 nt from the 3′ end of 26-nt-long reads for (6-4)PP XR-seq in normal human fibroblasts at the different time points. Shown are the results of two biological replicates.
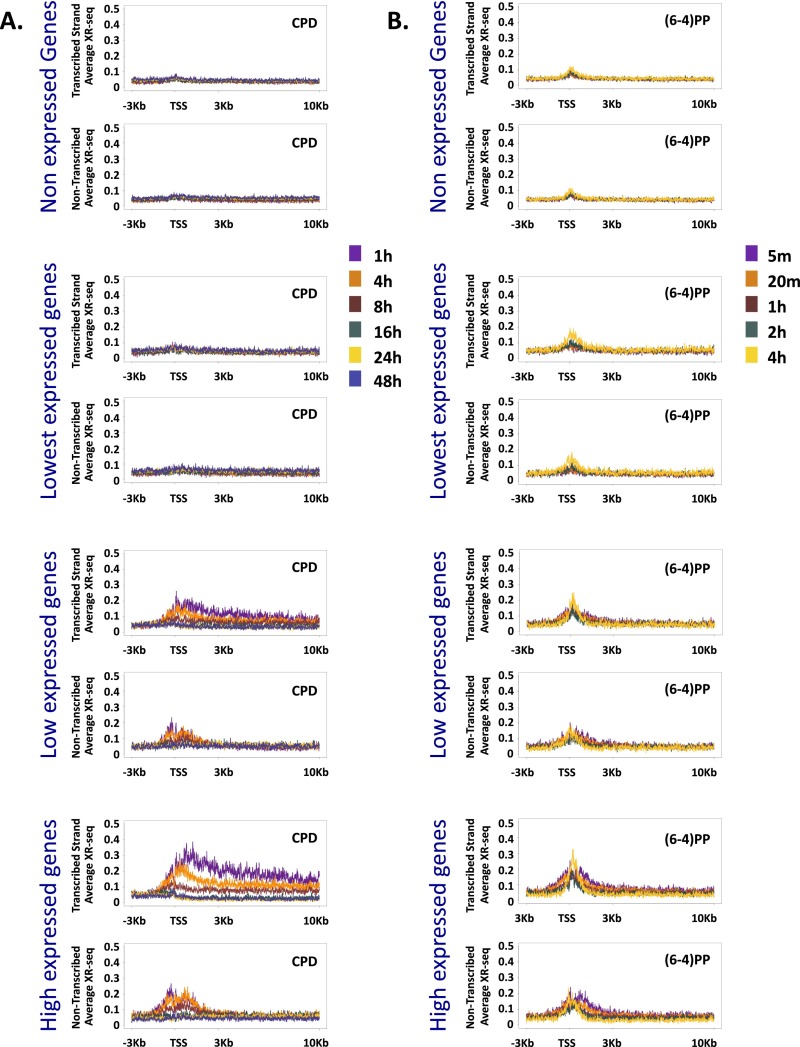
XR-seq reads were mapped to the human genome producing strand-specific, genome-wide profiles of repair at each time point. CPDs are repaired primarily in a transcription-coupled manner (Fig. 1C). This repair was most evident at the earliest time point of 1 h, where repair was enriched specifically over the transcribed strand of expressed genes, starting at the transcription start site (TSS) and extending into the gene body. At later times after UV treatment, repair over genes was completed, with virtually no detectable repair over the template strand at 24 h or 48 h after UV treatment (Fig. 1 C and D). At promoters, repair on the nontemplate strand was slightly higher and extended away from the gene, consistent with repair linked to divergent transcription at promoters. Repair of the nontranscribed strand within gene bodies, and over intergenic regions, persisted at relatively lower levels for up to 48 h after treatment (Fig. 1C). As previously reported (11), there was a positive correlation between the RNA and repair levels, with genes of lower expression levels exhibiting similar but lower-amplitude patterns (Fig. S5).
Fig. S5.
(A) Average CPD XR-seq repair signal 3 Kb upstream and 10 Kb downstream of the annotated TSS of genes separated into groups based on their RNA expression level in normal human skin fibroblasts (ENCODE, NHDF data). Signal is plotted separately for the transcribed and nontranscribed strand. (B) Same as A except plotted is XR-seq signal for (6-4)PP repair.
To define the top repair sites at each time point, XR-seq counts were obtained for 500-nt windows across the human genome. The top 1% of windows (62,743 in total) was identified for each strand. Compared with the whole genome, depending on the sample, repair in these regions was enriched 5- to 12-fold (Fig. S6D). For CPD, >80% of these top repair windows overlapped annotated transcripts up to 8 h after UV. At the later time points, the degree of overlap with transcript regions declined to 48% at 48 h after UV. The overall proportion of top repair windows that overlap transcripts in each of the time points were higher than that calculated for the total of XR-seq reads at each time point or the overlap calculated for a random set of genomic intervals (P < 0.02; Fig. 1E and Fig. S6E).
Fig. S6.
(A) For analysis of the pattern of (6-4)PP repair, CS-B transcription-coupled repair-deficient cells were irradiated with 20 J/m2 UV-C and incubated for the indicated time. The excised oligonucleotides were immunoprecipitated with anti-(6-4)PP antibody, radiolabeled at the 3′ end with 32P-cordycepin, and analyzed on sequencing gels. (B) Distribution of the XR-seq signal at each time point, separated by strand, for (6-4)PP repair in a CS-B cell line, along a 400-Kb region of chr6. ENCODE total stranded RNA-seq, DNase-seq, and ChIP-seq signal for H3K27ac and H3K27me3 from NHDF are plotted in black. Arrows in the bottom indicate positions and directions of annotated genes. Bold arrows indicate expressed genes. (C) Average (6-4)PP XR-seq repair signal from CS-B cells 3 Kb upstream and 10 Kb downstream of the annotated TSS of highly expressed genes in normal human skin fibroblasts (ENCODE, NHDF data). Signal is plotted separately for the transcribed (Upper) and nontranscribed (Lower) strands. (D) The proportion of reads in each sample that fall into the top 1% of repair window. As a control, the same analysis was performed for a random set of 1% of the 500-nt windows in the genome. (E) Genomic distribution of the aligned reads of CPD and (6-4)PP XR-seq experiments at each of the time points between genes (green), 5 Kb upstream of the gene (light green), 5 Kb downstream of the gene (gray), and intergenic regions (blue). Plotted for comparison are the average results of 50 permutations of a random set of 26 mers from the hg19 reference genome. (F) Percent overlap of the top 1% of (6-4)PP repair windows at each of the time points in CS-B cells with the same intervals described in D.
Identification of Sites of (6-4)PP Damage Repaired Relatively Early and Late After UV Exposure in Normal Human Fibroblasts.
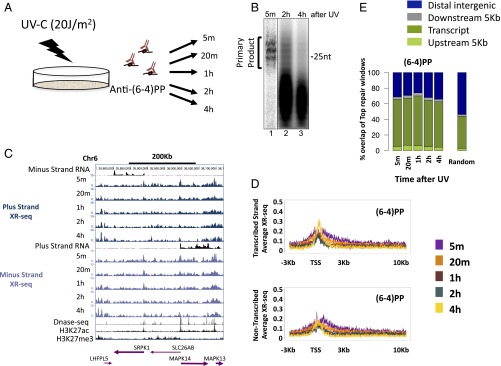
To analyze the repair of (6-4)PPs—which are on average fivefold less abundant than CPDs for a given UV dose (15) and are repaired much more rapidly—cells were irradiated with 20 J/m2 UV-C and harvested at 5 and 20 min and 1, 2, and 4 h after treatment (Fig. 2A). The time course was performed in two biological replicates. The excised oligomer released in the cells was isolated with anti-(6-4)PP antibodies. The general repair machinery efficiently recognizes and repairs (6-4)PPs (4, 15). Autoradiography showed that the primary excision products were detectable 5 min after UV irradiation. By 2 and 4 h after exposure, very low levels of primary products were observed, and the majority of the excised oligomer was degraded (Fig. 2B).
Fig. 2.
Kinetics of the genome-wide repair of (6-4)PP in normal human fibroblasts. (A) Schematic of time course of excision repair of the UV-induced (6-4)PP. (B) For analysis of the pattern of (6-4)PP repair, normal NHF1 cells were irradiated with 20 J/m2 UV-C and incubated for the indicated time. The excised oligonucleotides were immunoprecipitated with anti-(6-4)PP antibody, radiolabeled at the 3′ end with 32P-cordycepin, and analyzed on sequencing gels. (C) Distribution of the XR-seq signal at each time point, separated by strand, for (6-4)PP repair along a 400-Kb region of chr6. ENCODE total stranded RNA-seq, DNase-seq, and ChIP-seq signal for H3K27ac and H3K27me3 from NHDF cells are plotted in black. Arrows in the bottom indicate positions and directions of annotated genes. Bold arrows indicated expressed genes. (D) Average (6-4)PP XR-seq repair signal 3 Kb upstream and 10 Kb downstream of the annotated TSS of genes with the highest quartile of RNA expression in normal human skin fibroblasts (ENCODE, NHDF data). Signal is plotted separately for the transcribed and nontranscribed strands. (E) Percent overlap of the top 1% of repair windows at each of the time points with annotated genes (dark green), 5 Kb upstream of the gene (light green), 5 Kb downstream of the gene (gray), and intergenic regions (blue). Plotted for comparison are the average results of 50 permutation of a random set of 500 mers from the hg19 reference genome.
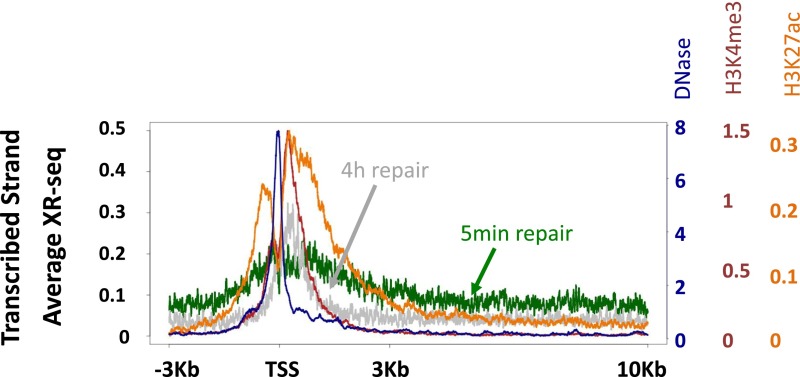
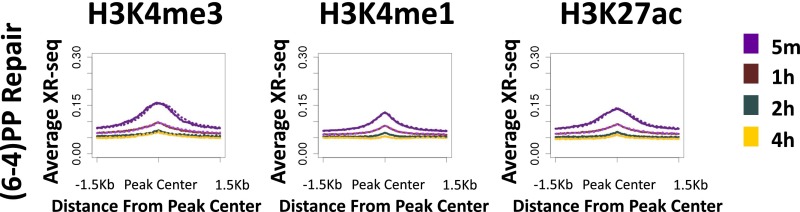
XR-seq for (6-4)PP was performed similarly to CPD, except that immunoprecipitation was performed with anti-(6-4)PP antibodies, and before PCR amplification, the photoproducts were removed in vitro by treatment with (6-4)PP photolyases. At 5 min after UV exposure, repair was evident, particularly at DNase hypersensitivity sites (Fig. 2C). Repair was especially prevalent in the areas surrounding the promoters of highly expressed genes, but unlike CPD, (6-4)PP repair was enriched on both strands, was limited to the regions flanking the TSS, and did not extend into the gene body (Fig. 2 C and D). The profile of the elevated repair of (6-4)PP contains two regions of higher repair flanking the TSS. At 4 h after UV, repair of (6-4)PP is enriched over the TSS, specifically between these initial highly repaired regions (Fig. 2D). This repair appears to be higher on the template strand compared with the nontemplate strand, which could be due to a contribution of transcription-coupled repair of (6-4)PP at these later time points. However, a slight enrichment of transcribed strand repair at late time points is also observed in a CS-B cell line that lacks transcription-coupled recognition (Fig. S6 B and C), suggesting that chromatin states other than transcription affect repair efficiency. It is known that nucleosomes at promoters of active genes are enriched in H3K4me3 and H3K27ac histone modifications (16). We plotted ChIP-sequencing (ChIP-seq) data for these two modifications along with the DNase-seq hypersensitivity signal (17) over these active genes. The early repaired regions followed the pattern of H3K27ac signal. The small “dip” between the two repair peaks contained the DNase hypersensitivity peak and, adjacent to it, the H3K4me3 signal. The late-repaired (6-4)PP region overlapped the sites of H3K4me3 signal (Fig. S7).
Fig. S7.
Similar to Fig. 2D, except that the average profile of DNase-seq (blue), H3K4me3 ChIP-seq (red), and H3K27ac ChIP-seq (orange) signals were plotted in addition to the 5-min (green) and 4-h (gray) repair profiles by using additional axes (color-coded to match the different data).
To identify regions repaired relatively early and relatively late, we defined the top (6-4)PP repair sites as the 1% of genomic 500-nt windows with the highest repair signal. Similarly to CPD repair, ∼70% of the top (6-4)PP repair sites overlap annotated transcripts at all of the measured time points, despite being repaired by the general, and not transcription-coupled, mechanism. At each of the time points, the overlap of the top (6-4)PP repair sites with transcripts was higher than that calculated for all (6-4)PP XR-seq reads or for a random set of genomic intervals (P < 0.02; Fig. 2E and Fig. S6E).
Priority of Repair at Open and Active Chromatin States.
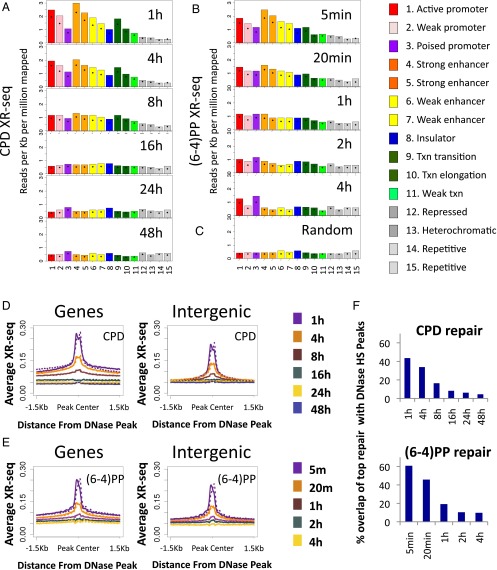
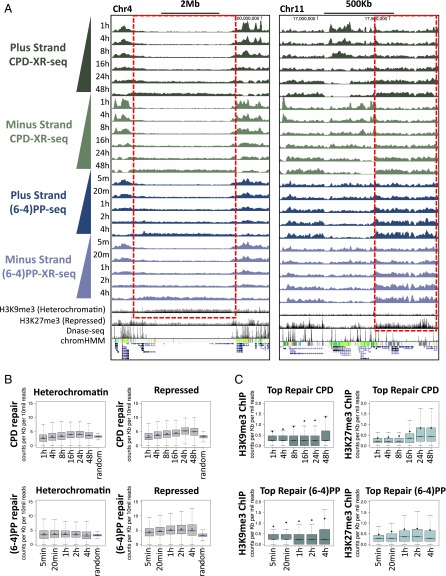
The packaging of DNA into chromatin can hinder the access of repair proteins and affect the efficiency of repair (18). Specific histone modifications are associated with different functional and cytological chromatin states. The ChromHMM algorithm predicts chromatin states based on underlying chromatin modification profiles. We used publicly available chromatin segmentations for an adult human fibroblast cell line [normal human lung fibroblast (NHLF) cells from the Encyclopedia of DNA Elements (ENCODE) (19)]. For each of the 15 predicted states, we calculated the number of XR-seq reads over each state, normalized to the proportion of the genome covered by that state. At the initial time point of 1 h after UV exposure, CPD repair counts were highest over the active promoters or strong enhancers. This enrichment over active states is consistent with CPD repair being primarily transcription-coupled. Less active states (for example, weak or poised promoters and weak enhancers) exhibited lower repair levels. Repressed, heterochromatic, and repetitive states exhibited the lowest XR-seq counts. At later time points, the differences between states diminished, and at the latest time points, they were no longer apparent because repair was largely completed (Fig. 3A).
Fig. 3.
Effect of chromatin state on repair. (A) XR-seq read coverage was calculated over genomic intervals assigned to each of the chromatin states predicted for NHLF cells (ENCODE). Shown are results from CPD XR-seq at the different time points. Values were normalized to read depth and interval length. Bars indicate the value of the 75th percentile. Diamonds indicate the mean value. (B) Same as A except calculated was coverage of (6-4)PP XR-seq. (C) Similar to A and B except the coverage was counted for a random set of 26-mer intervals. (D) Average CPD XR-seq profiles for each time point at 1.5-Kb regions flanking the center of DNase hypersensitivity peaks that either overlapped annotated genes (Left) or did not overlap annotated genes (Right). Values were normalized per 10 million mapped reads. The average profile was calculated separately for the plus (solid line) and minus (dashed line) strands. (E) Same as D except plotted is the repair of (6-4)PP. (F) Overlap of the top 1% of repair sites (500-bp windows) with DNase hypersensitivity peaks in each of the time points for CPD repair (Upper) and (6-4)PP repair (Lower).
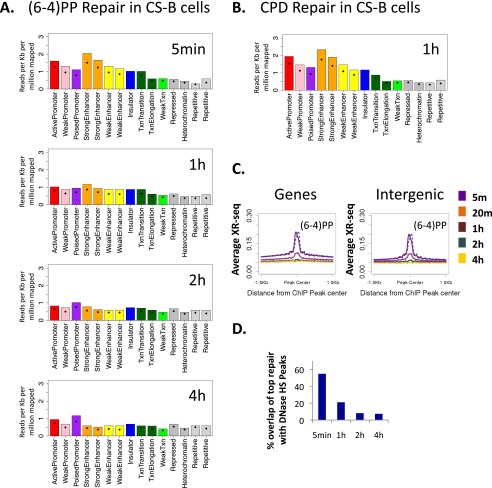
Initial (6-4)PP repair at 5 min after UV was also high in active regions (Fig. 3B). As in CPD repair, the difference between active and inactive states was diminished at later time points; however, repair over active and poised promoters remained higher than the other states, even at the latest measurement 4 h after UV treatment. A similar pattern was observed for both types of damage in a CS-B cell line that lacked transcription-coupled repair, indicating that this enrichment over active regions is a characteristic of general repair and does not require transcription-coupled recognition of (6-4)PP (Fig. S8 A and B).
Fig. S8.
Preferential repair of open and active chromatin states in CS-B cells. (A) XR-seq read coverage was calculated over genomic intervals assigned to each of the chromatin states defined for NHLF cells (ENCODE). Shown are results from (6-4)PP XR-seq at the indicated times after UV. Values were normalized to read depth and interval length. Bars indicate the value of the 75th percentile. Diamonds indicate the mean value. (B) As in A except shown are results of CPD XR-seq at 1 h after UV. (C) Average (6-4)PP XR-seq profiles for each time point at 1.5-Kb regions flanking the center of DNase hypersensitivity peaks that either overlapped genes (Left) or did not overlap genes (Right). Values were normalized per 10 million mapped reads. The average profile was calculated separately for the plus (solid line) and minus (dashed line) strands. (D) Overlap of the top 1% of (6-4)PP repair sites (500-bp windows) with DNase hypersensitivity peaks in each of the time points.
Active promoters and enhancers are characterized by nucleosome loss, which is sometimes referred to as “open” chromatin. We mapped average XR-seq profiles around the center of DNase hypersensitivity peaks in normal human fibroblasts [ENCODE data from NHDF cells (17)]. Knowing that transcription itself enhances repair, we split these peaks into those that overlapped annotated transcripts and peaks that were intergenic. Repair of both CPD and (6-4)PP was significantly enriched at all DNase hypersensitivity peaks at the initial time points, and this enrichment diminished through time (Fig. 3 D and E). A small dip in the repair signal occurred precisely at the center of the peak. At earlier time points, repair of CPD within genes was higher overall in the 3-Kb region surrounding the DNase hypersensitivity peaks. Repair of (6-4)PP surrounding DNase peaks was also higher within genes, although to a lesser extent. Elevated repair of (6-4)PP over genes is not solely due to the effect of transcription-coupled repair of (6-4)PP, as evidenced by an elevated level of repair over transcribed regions even in a CS-B mutant cell line (Fig. S8B).
For both types of damage, there was significant enrichment of repair over intergenic DNase hypersensitivity sites at the initial time points. For (6-4)PP, higher repair over DNAse peaks occurred within 5 min of UV irradiation. For CPD, this enhanced repair required 1 h, because of the slower recognition of CPD by the general repair factors (Fig. 3 D and E).
Analysis of the top repair windows showed that at the initial time points, 60% of top (6-4)PP and 43% of top CPD repair sites overlapped DNase hypersensitivity peaks, which are 14- and 10-fold enriched, respectively, over what is expected by a stochastic repair mechanism (4.3%, P < 0.02; Dataset S2). In both cases, only 10% or fewer of the top repair windows at later time points overlapped the DNase hypersensitivity sites (Fig. 3F).
Early Repair Is Associated with Active Histone Marks.
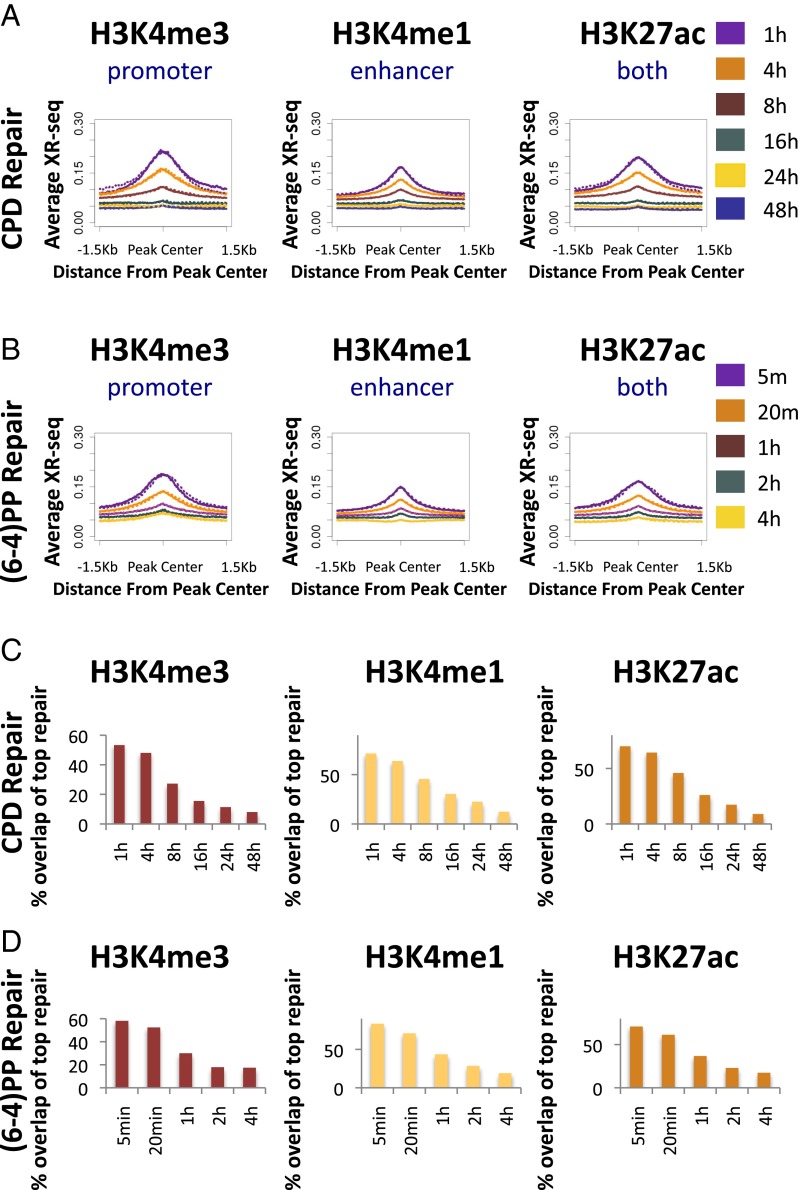
We analyzed repair at sites of histone modifications associated with active chromatin. The modifications we examined were H3K4me3, which is associated with actively transcribed promoters; H3K4me1 which is associated with active enhancers that are also often transcribed; and H3K27ac, which is associated with both (16, 20). It is important to note that, although these marks may be more enriched in promoters or enhancers, they are often found in both.
The average repair of both CPD and (6-4)PP was enriched around peak centers of H3K4me3, H3K4me1, and H3K27ac. With time, this enrichment was reduced (Fig. 4 A and B). Once more, a similar enrichment for (6-4)PP repair at sites of active histone marks was observed in CS-B cells, indicating that it is not driven by transcription-coupled recognition of damage (Fig. S9).
Fig. 4.
Initial repair is associated with active histone marks. (A) Average CPD XR-seq profiles for each time point at 1.5-Kb regions flanking the center of ChIP-seq peaks of H3K4me3 (Left), H3K4me1 (Center), and H3K27ac (Right). Values were normalized per 10 million mapped reads. The average profile was calculated separately for the plus (solid line) and minus (dashed line) strands. (B) Same as A except plotted is repair of (6-4)PP. (C) Overlap of the top 1% of CPD repair sites (500-bp windows) with the indicated ChIP-seq peaks. (D) Same as C except plotted is the overlap of the top 1% of (6-4)PP repair sites at each time point.
Fig. S9.
Average (6-4)PP XR-seq profiles in CS-B cells for each time point at 1.5-Kb regions flanking the center of ChIP-seq peaks of H3K4me3, H3K4me1, and H3K27ac.
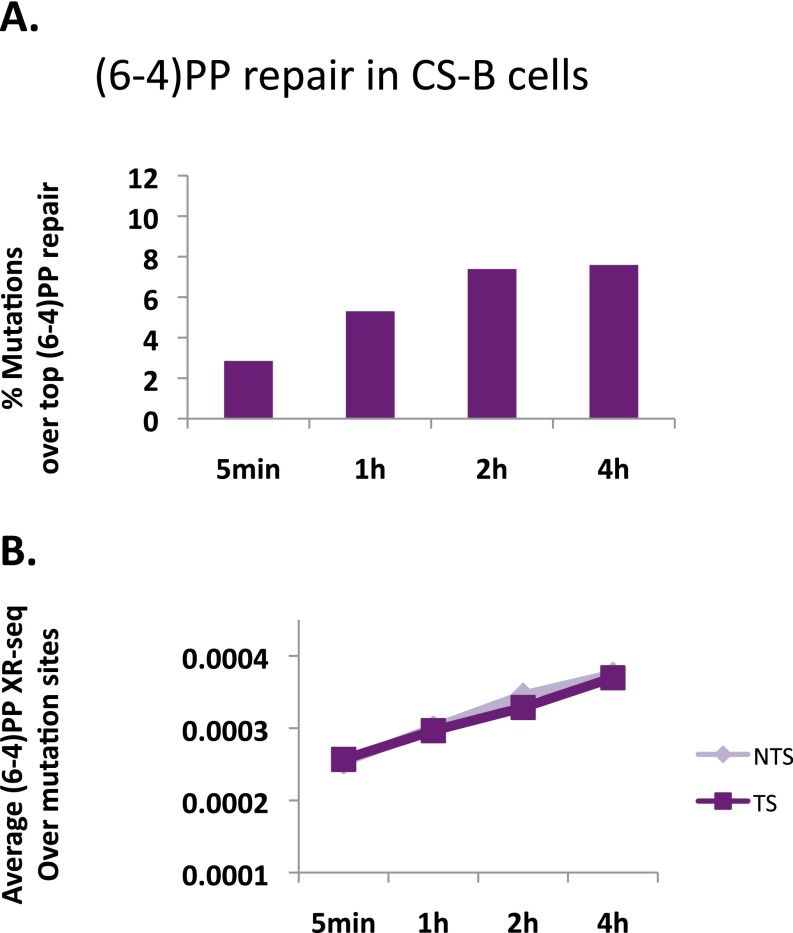
Fig. S10.
(A) The number of melanoma-linked mutations per kilobase that overlap the top 1% of repair windows was calculated for (6-4)PP repair at the different time points in CS-B cells. (B) The average XR-seq count over a 400-bp window centered on the site of melanoma-linked mutations was calculated for (6-4)PP repair in CS-B cells at each of the time points. Repair was calculated separately for template and nontemplate strands.
We analyzed the degree of overlap of the top repair windows at each time point with the peak regions of the active histone marks (Fig. 4 C and D and Dataset S2). More than 50% of the top CPD and (6-4)PP repair windows at the initial time points overlapped H3K4me3, which is associated with actively transcribed promoters (53% and 58% respectively, compared with 7% for a stochastic control; P < 0.02). At later times, the degree of overlap was significantly reduced. An even higher degree of overlap (>70%) was observed for H3K4me1 and H3K27ac peaks (compared with 14% and 12%, respectively, for a uniform stochastic repair control).
Repressed and Heterochromatic States Exhibit Relatively Low Repair that Persists to Late Time Points.
Two chromatin states associated with a lack of gene expression have been called “heterochromatic” and “repressed.” The heterochromatic state is characterized by H3K9me3 enrichment, whereas the repressed state is characterized by H3K27me3 enrichment (19). These marks do not form discrete peaks but, rather, are deposited over broad regions (16, 17). For both types of damage, regions with H3K9me3 or H3K27me3 had low levels of initial repair, which at certain loci appeared higher at the later time points (Fig. 5A).
Fig. 5.
Repair of heterochromatin and repressed regions persist into the late time points. (A) Distribution of the XR-seq signal at each time point, separated by strand, for CPD and (6-4)PP repair in two genomic loci, enriched for H3K9me3 (Left) or H3K27me3 (Right) histone deposition. ChIP-seq signal for H3K9me3 and H3K27me3, DNase-seq, and ChromHMM segmentation tracks (NHDF and NHLF, ENCODE) are plotted in the bottom. Gray in the ChromHMM track depicts repressed or heterochromatic regions; green represents actively transcribed regions. (B) Box plots depicting CPD (Upper) and (6-4)PP (Lower) XR-seq read coverage over heterochromatic (Left) or repressed (Right) genomic intervals as predicted in NHLF cells (ENCODE). Shown are results from each of the time points, in comparison with a random dataset of 26 mers from the hg19 reference genome. Values were normalized to read depth and interval length. Lines indicate median, and diamonds indicate mean values. (C) Box plots depicting the level of histone H3K9me3 (Left) or H3K27me3 (Right) ChIP-seq signal (NHDF, ENCODE) over the top 1% of 500-nt repair windows in each of the time points for CPD (Upper) and (6-4)PP (Lower) XR-seq. Counts were normalized per million mapped reads. Lines indicate median, and diamonds indicate mean values.
According to the genome-wide ChromHMM classifications, repair over heterochromatic and repressed regions was consistently low. As time progressed up to 24 h after UV, there were higher CPD repair levels over these regions (Fig. 5 B, Upper). Repair of (6-4)PP, for which measurements were taken up to 4 h, was relatively constant over heterochromatic regions and appeared slightly elevated at later times over repressed regions (Fig. 5 B, Lower).
Because the H3K9me3 and H3K27me3 marks are broadly distributed, we analyzed the overall levels of the ChIP-seq signal of these marks over the top repair windows at each time point. Levels of both marks were higher over the top repair windows of both types of damage at the later compared with earlier time points, although this change was more significant for H3K27me3, consistent with the elevated repair over repressed regions (Fig. 5C).
Regions Repaired Later Are Associated with Higher Mutagenesis.
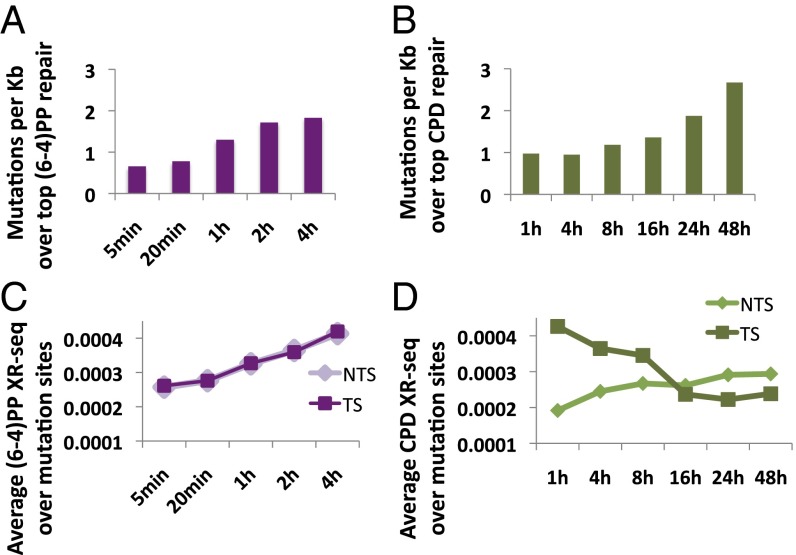
Damage that persists into DNA replication increases the risk of mutagenesis. We hypothesized that regions that are repaired slowly are at a higher risk for mutation. We compared our XR-seq repair data to an existing dataset of cancer-linked mutations in melanoma (21). The set of mutations we analyzed were obtained by exon sequencing and was filtered for somatic mutations and variations present in other cell types or in normal tissue. We calculated the incidence of melanoma-linked mutations per kilobase over the top repair windows at each time point (Fig. 6 A and B). Analysis was limited to those windows that overlapped exons. As a control, we calculated the incidence of these mutations over a random set of size-matched genomic intervals. For both (6-4)PP and CPD, there was a lower incidence of mutations in the top repair sites at the earliest time points compared with the control [0.66 mutations per kilobase for (6-4)PP repair at 5 min, and 0.97 for CPD repair at 1 h, compared with 1.36 mutations per kilobase for control intervals, P < 0.02; Dataset S3)]. Conversely, in the latest time points, there was a higher incidence of mutations [1.83 mutations per kilobase for (6-4)PP repair at 4 h and 2.67 for CPD repair at 48 h compared with 1.36 mutations per kilobase for control intervals, P < 0.02]. The mutations that occurred in the top repaired regions at latest time points also had a higher overlap with heterochromatic and repressed regions (Dataset S4).
Fig. 6.
Repair at late time points is associated with higher somatic-mutation rates. (A) The number of melanoma-linked mutations per kilobase that overlap the top 1% of repair windows was calculated for (6-4)PP repair at the different time points. (B) Same as A except overlaps were counted over the top 1% of CPD repair windows. (C) The average XR-seq count over a 400-bp window centered on the site of melanoma-linked mutations was calculated for (6-4)PP repair at each of the time points. Repair was calculated separately for template and nontemplate strands. Values were normalized for sequencing depth. (D) Same as B except plotted is the average CPD repair at each time point.
We examined the levels of repair over a 400-nt window flanking the mutation sites. We did not ascribe mutations to a specific strand, but because these mutations occur within exons, we can differentiate template strand and nontemplate strand repair (Fig. 6 C and D). For (6-4)PP, there was a higher level of repair over the mutation sites at later time points vs. initial time points, consistent with late-repaired regions having elevated mutagenesis (Fig. 6C). There was no observed difference between the template and nontemplate strands. Average CPD repair over the mutation sites differed for the template and nontemplate strands (Fig. 6D). Nontemplate strand repair over the mutation sites was higher at later time points, similar to (6-4)PP repair. Template strand repair, however, was higher at the early time points, consistent with transcription-coupled repair of this damage.
Discussion
XR-Seq Measures the Kinetics of DNA Repair.
Two characteristics of the XR-seq assay make it amenable to kinetic measurements: (i) The primary, full-length, excised oligomers are isolated by TFIIH immunoprecipitation; and (ii) the excised oligomers are degraded relatively fast compared with the rate at which they are produced (13). As a result, at each measurement, the majority of excised oligomers mapped by XR-seq are specific to that time point. The temporal resolution of repair is evident in later time points, in which repair signal is depleted from those regions in the genomes that were repaired faster and in which repair was completed.
Open and Active Chromatin Regions Are Sites of Priority for Repair.
The packaging of DNA into chromatin has several implications for genome stability. On the one hand, packaging can protect DNA from damaging agents (22, 23). On the other hand, it can hinder damage recognition and the access of repair machinery. Indeed, it has been reported that nucleosomal DNA is less efficiently repaired by the excision repair machinery in vitro (24, 25).
We compared the repair maps at different time points to the chromatin landscape at the time of damage. Our analysis showed that both transcription-coupled and general excision repair are initially enriched over open and active chromatin states. Active regions are characterized with a more open chromatin structure and significant histone acetylation. The association of general repair with these active regions is likely due to the more open chromatin structure rather than transcription per se. Around annotated genes, higher CPD repair is observed, not only at the DNase hypersensitivity peak, but also in the surrounding regions, because of transcription-coupled repair. Conversely, (6-4)PP repair in these same peaks is concentrated at the peak site. There is similar enrichment of CPD and (6-4)PP repair, at the initial measurement, over DNase hypersensitivity sites at intergenic regions. At these regions, although with different efficiency, both types of damage are recognized and repaired by the general excision factors. There is a dip in the pattern of repair specifically over the center of DNase hypersensitivity peaks. The dip in the signal may be explained by a GC-rich sequence or by the binding of a transcription factor within the nucleosome-free region that is blocking the access of repair proteins (26).
We note that our data rule out that the difference in repair levels is driven simply by difference in the underlying damage levels. CPD formation is dictated mostly by sequence, and previous reports of the genome-wide distributions of this type of damage showed a relatively widespread signal that does not follow the enrichment patterns we observed (27, 28). Furthermore, if higher initial repair were solely explained by higher levels of damage, we would expect similar enrichments in all time points. In fact, for the histone modifications, we observed a small spike in repair around the peak center at all time points, which could be explained by higher damage levels.
At the later measurements after UV, we observed reduced enrichment of repair in the open and active regions. The exception was repair of (6-4)PP at the latest measured time point of 4 h. At this time point, we observed enrichment of a region within the promoter of genes that was located between the two peaks of repair in the initial time points (Fig. 2D). We rule out transcription coupled-repair of (6-4)PP because a similar phenomenon is observed in a CS-B cell line deficient in this process. This repair may be at a site in which access to the DNA is initially blocked and only becomes accessible in the later stages, perhaps by an active mechanism. Nucleosomes form a natural block to the repair machinery, and nucleosomes at promoters are enriched for the H3K4me3 modifications (29, 30). The overlap between the elevated repair signal near the TSS at 4 h, and the H3K4me3 ChIP-seq signal, suggests that this late repair occurs at sites initially blocked by well-positioned nucleosomes (16, 20) (Fig. S7).
Repair of Heterochromatic and Repressed Chromatin Persists into Late Time Points.
We examined the repair of the heterochromatic, repressed, and repetitive states in the genome. Because low read counts over repetitive states could be attributed to the low mappability of these regions, we did not address them. Initially, repair of heterochromatic and repressed regions was substantially lower than repair of active regions. These chromatin regions are characterized by the histone modifications H3K9me3 and H3K27me3, respectively, and we report higher levels of these marks at later-repaired regions. The current “access–repair–restore” model for excision repair in chromatin suggests active removal of nucleosomes to allow repair (31). Given that the sequencing data is normalized to read depth, we cannot distinguish whether elevated repair levels at later time points are a result of completion of repair in other regions or the result of an active process, including histone modification, chromatin remodeling, or gene activation in response to UV, that makes these regions more accessible (32, 33). However, our assay will be a powerful tool to test for candidates involved in facilitating repair of tightly packed chromatin.
Slow Repair Is Associated with Higher Somatic Mutagenesis in Cancer Cells.
UV in sunlight is a known mutagen and causative agent of skin cancer (34, 35). Mutations in excision repair proteins cause severe genetic disorders such as xeroderma pigmentosum and elevate the risk of cancer (36, 37). A recent study reported an association between lower somatic mutation frequency and open chromatin regions that was significantly lower in cells that were excision repair deficient (38). Our data adds mechanistic insight, showing a negative correlation between repair timing and mutagenesis. This association is clear for general excision repair of (6-4)PP. In the skin fibroblasts used in our study, the top repair regions at the latest time points are associated with the highest incidence of melanoma-linked mutations. Conversely, sites of melanoma-linked mutations exhibited higher average repair at the later time points. For CPDs, there is also a higher incidence of mutations in the top repair regions at the latest time points. However, the pattern of average repair over mutations sites differs for the template and nontemplate strand. Average nontemplate strand repair over mutation sites is higher at the later time points. Conversely, template-strand repair is higher at the earlier time points, consistent with transcription-coupled repair, preferentially removing damage from transcribed exons. We did not assign mutations to a specific strand. The pattern of repair, however, suggests that mutations originated from the nontemplate strand. Higher mutation levels on the nontranscribed strand would be consistent with reports of transcription-based strand-asymmetries in mutagenesis (21, 39).
In general, mutation rates associated with CPD repair are lower if compared with the mutation rate associated with (6-4)PP repair at the parallel time points (1–4 h). These lower mutation rates could be because mutations associated with CPD repair sites are derived only from the nontranscribed strand or perhaps because of the ability of mammalian cells to bypass this damage in an error-free mechanism (40).
DNA replication may also affect repair efficiencies and, in addition, has been implicated in mutagenesis strand asymmetries (39). Future work in synchronized cells will be able to address the effect of replication timing on DNA repair efficiencies.
Our work here investigates how the initial chromatin state at the time of damage affects the efficiency and priority of DNA repair. However, the chromatin landscape is not static, and as a result of genotoxic stress, it is altered. DNA damage induces a transcriptional response and is associated with changes in histone modifications. Histone modifications play an active role in the response to DNA damage and in the recruitment of repair proteins (18). Future studies will incorporate measurements of DNA damage, DNA repair, RNA levels, and histone modifications in response to damage and will further improve our understanding for how DNA repair is orchestrated within chromatin.
Nucleotide excision repair is the sole mechanism for removing bulky adducts from the human genome, including those formed by chemotherapeutic drugs such as cisplatin and oxaliplatin. Improving our understanding of DNA repair is beneficial, not only for understanding carcinogenesis, but also for understanding the processes cancer cells use to cope with chemotherapy. Such information is expected to aid in improving currently used chemotherapy regimens.
Materials and Methods
Cell Culture and UV Irradiation.
Telomerase-immortalized normal human fibroblast NHF1 was obtained from W. K. Kaufmann, University of North Carolina at Chapel Hill, Chapel Hill, NC (41). CS-B (CS1ANps3g2, GM16095) mutant human skin fibroblasts were purchased from the National Institute of General Medical Sciences Human Genetic Cell Repository (Coriell Institute). CS-B cells were cultured in DMEM supplemented with 10% (vol/vol) FBS at 37 °C in a 5% atmosphere CO2 humidified chamber. NHF1 cells were maintained under the same conditions with the addition of 2 mM glutamine.
UV irradiation was performed as described with the indicated dose (12). After incubation at 37 °C for the indicated time, cells were washed and collected in cold PBS.
Detection of Excision Products.
Excision products from UV-irradiated cells were purified, radiolabeled, and separated by electrophoresis as described (13). Short DNA fragments were extracted by modified Hirt’s method and subjected to immunoprecipitation against anti-(6-4)PP or -CPD antibodies. Purified excised oligonucleotides were 3′ radiolabeled by terminal deoxynucleotidyl transferase (NEB) and [α-32P]-3′-deoxyadenosine 5′-triphosphate (cordycepin 5′-triphosphate) (Perkin-Elmer) and resolved in 10% denaturing sequencing gels.
XR-Seq Library Preparation.
XR-seq libraries were prepared as described (11). Briefly, primary excision products pulled down by TFIIH coimmunoprecipitation were ligated to both 5′ and 3′ adaptors. Ligation products containing (6-4)PP or CPD were purified by immunoprecipitation with corresponding antibodies and repaired by specific photolyases. Repaired DNA were PCR-amplified with Index primers and purified by 10% native polyacrylamide gels.
Sequencing and Genome Alignment.
Libraries were sequenced on a Hiseq 2000 platform by the University of North Carolina High-Throughput Sequencing Facility. Flanking adapter sequences were removed from the reads by using trimmomatic (42). Reads were aligned to the hg19 human genome by using bowtie (43) with the command options -q–nomaqround–phred33-quals -m 4 -n 2 -e 70 -l 20–best –S. Uniquely aligned reads were obtained by using samtools. We obtained a total of at least 19 million uniquely mapped reads in each of the time points (Dataset S1). For comparison of the DNA-repair signal, we normalized all of the count data by the sequencing depth; data are available for viewing as a track hub on the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgGateway) by pasting the link: http://trackhubs.its.unc.edu/sancarlb/XRseqTimeCourse/hub.txt. Sequencing data for (6-4)PP repair at 1 h in NHF1 and CS-B cells and CPD repair at 1 h in CS-B cells were taken from the published dataset (11). The raw data and bigwig tracks are available under the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/, accession no. GSE76391).
ENCODE Data.
NHDF long total stranded RNA-seq [ENCODE Data Coordination Center (DCC) accession no. ENCSR00CUH], H3K4me1 (accession no. ENCSR000ARV), H3K4me3 (accession no. ENCSR000DPR), H3K27ac (accession no. ENCSR000APN), H3K27me3 (accession no. ENCSR000APO), H3K9me3 (accession no. ENCSR000ARX), and DNase-seq (accession no. ENCSR000EMP) fastq, aligned reads .bam files, and peak files, as well as the NHLF chromHMM chromatin state segmentation (UCSC accession no. wgEncodeEH000792), were downloaded from the ENCODE portal (genome.ucsc.edu/ENCODE/) or viewed on the UCSC browser.
Analysis of Top Repair Windows.
The hg19 genome was divided into 500-nt windows, and the repair coverage for each window was calculated for each sample by using bedtools. The 62,743 windows with the highest score on each strand and at each time point were taken as the top 1%. Genomic distribution of reads and overlap with annotated genes was calculated by using the UCSC refGene.txt gene annotation, as described in Hu et al. (11). Bedtools intersect was used to calculate all overlaps, including those to peak intervals and mutations. For comparison and to calculate P values, we conducted the same analysis on 50 random datasets of 62,743 intervals of 500 nt from the hg19 reference genome. P values for comparison of distributions were calculated as [number of times distribution of experimental and control data overlapped]/[total number of control data tests].
Chromatin State Analysis.
Repair over the genomic intervals for each of the 15 predicted chromatin states defined by the ChromHMM algorithm was calculated by using bedtools coverage. Values were normalized per million mapped reads and per kilobase of interval length.
Cancer Mutation Analysis.
Cancer-linked somatic mutation datasets, which were filtered for variations found in normal cells and tissues, were obtained from Alexandrov et al. (21). For melanoma, these include exome-only datasets. The number of mutations overlapping the top 1% of repair windows at each time point was calculated with bedtools intersect. The results were normalized per kilobase of top windows that overlapped exons. As a control, the same analysis was performed with a random set of 500-nt windows from the hg19 reference genome. To calculate average repair over mutation sites, a 400-bp interval centered on the mutation site was created by using bedtools slop, and coverage was calculated with bedtools coverage, normalizing to 1 million mapped reads.
Plotting Average XR-Seq Profiles.
Gene lists, grouped into expression quartiles based on RNA-seq in NHDF cells, were taken from our previous analysis (11). For average XR-seq profiles relative to the annotated TSS, we limited the gene list to genes that do not have overlapping or neighboring genes for at least 6,000 bp upstream or downstream on either strand and were at least 10,000 bp in length. Read counts were calculated from the aligned .bam files by using bedtools coverage and normalized to 10 million mapped reads.
Supplementary Material
Acknowledgments
We thank Dr. Sebastian Pott for fruitful discussion and suggestions. This work was supported by National Institutes of Health Grants GM32833 and GM31082 (to A.S.) and HG006787 (to J.D.L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE76391).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603388113/-/DCSupplemental.
References
- 1.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79(79):183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 3.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272(38):23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 4.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17(20):2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugasawa K, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2(2):223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 6.Wakasugi M, Sancar A. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci USA. 1998;95(12):6669–6674. doi: 10.1073/pnas.95.12.6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9(12):958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 8.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 9.Huang JC, Svoboda DL, Reardon JT, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89(8):3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mu D, et al. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270(6):2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 11.Hu J, Adar S, Selby CP, Lieb JD, Sancar A. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015;29(9):948–960. doi: 10.1101/gad.261271.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaddameedhi S, et al. Similar nucleotide excision repair capacity in melanocytes and melanoma cells. Cancer Res. 2010;70(12):4922–4930. doi: 10.1158/0008-5472.CAN-10-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu J, et al. Nucleotide excision repair in human cells: Fate of the excised oligonucleotide carrying DNA damage in vivo. J Biol Chem. 2013;288(29):20918–20926. doi: 10.1074/jbc.M113.482257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett E, et al. The role of DNA polymerase eta in translesion synthesis past platinum-DNA adducts in human fibroblasts. Cancer Res. 2004;64(18):6469–6475. doi: 10.1158/0008-5472.CAN-04-1328. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell DL. The relative cytotoxicity of (6-4) photoproducts and cyclobutane dimers in mammalian cells. Photochem Photobiol. 1988;48(1):51–57. doi: 10.1111/j.1751-1097.1988.tb02785.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 17.Consortium EP. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gospodinov A, Herceg Z. Shaping chromatin for repair. Mutat Res. 2013;752(1):45–60. doi: 10.1016/j.mrrev.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim TK, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell. 2015;162(5):948–959. doi: 10.1016/j.cell.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative ICGC Breast Cancer Consortium ICGC MMML-Seq Consortium ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell DL, Nguyen TD, Cleaver JE. Nonrandom induction of pyrimidine-pyrimidone (6-4) photoproducts in ultraviolet-irradiated human chromatin. J Biol Chem. 1990;265(10):5353–5356. [PubMed] [Google Scholar]
- 23.Gale JM, Smerdon MJ. UV induced (6-4) photoproducts are distributed differently than cyclobutane dimers in nucleosomes. Photochem Photobiol. 1990;51(4):411–417. doi: 10.1111/j.1751-1097.1990.tb01732.x. [DOI] [PubMed] [Google Scholar]
- 24.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol. 2002;22(19):6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Hara R, Singh G, Sancar A, Lippard SJ. Nucleosomes inhibit nucleotide excision repair of site-specific platinum-DNA adducts. Biochemistry. 2003;42(22):6747. doi: 10.1021/bi034264k. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22(9):1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Powell JR, et al. 3D-DIP-Chip: A microarray-based method to measure genomic DNA damage. Sci Rep. 2015;5:7975. doi: 10.1038/srep07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zavala AG, Morris RT, Wyrick JJ, Smerdon MJ. High-resolution characterization of CPD hotspot formation in human fibroblasts. Nucleic Acids Res. 2014;42(2):893–905. doi: 10.1093/nar/gkt912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes AL, Rando OJ. Mechanisms underlying nucleosome positioning in vivo. Annu Rev Biophys. 2014;43:41–63. doi: 10.1146/annurev-biophys-051013-023114. [DOI] [PubMed] [Google Scholar]
- 30.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol Cell Biol. 2000;20(24):9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polo SE, Almouzni G. Chromatin dynamics after DNA damage: The legacy of the access-repair-restore model. DNA Repair (Amst) 2015;36:114–121. doi: 10.1016/j.dnarep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schick S, et al. Dynamics of chromatin accessibility and epigenetic state in response to UV damage. J Cell Sci. 2015;128(23):4380–4394. doi: 10.1242/jcs.173633. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci USA. 2005;102(24):8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleaver JE. Defective repair replication of DNA in xeroderma pigmentosum. Nature. 1968;218(5142):652–656. doi: 10.1038/218652a0. [DOI] [PubMed] [Google Scholar]
- 37.DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol. 2012;132(3 Pt 2):785–796. doi: 10.1038/jid.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polak P, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32(1):71–75. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haradhvala NJ, et al. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell. 2016;164(3):538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeifer GP, Besaratinia A. UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem Photobiol Sci. 2012;11(1):90–97. doi: 10.1039/c1pp05144j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heffernan TP, et al. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22(24):8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.