Significance
Early-diverging fungi from the phylum Microsporidia are opportunistic pathogens of humans and other animals. A genome-wide search was conducted in four species of Microsporidia and one species of Cryptomycota for genes acquired by horizontal transfer from other organisms. Up to 2% of their genes had strong evidence for horizontal transfer. We showed that many transferred nucleic acid metabolism genes lie on the boundary of the host and pathogen metabolic networks. Finally, we functionally characterized the gene encoding thymidine kinase, whose multiple parallel transfers from three different sources strongly suggest an integral adaptive role in the lifestyles of these intracellular parasites. Microsporidian thymidine kinases activate a prodrug, suggesting a treatment route for microsporidian infections.
Keywords: horizontal gene transfer, Microsporidia, Cryptomycota, thymidine kinase, metabolic networks
Abstract
Horizontal gene transfer (HGT) among bacteria, archaea, and viruses is widespread, but the extent of transfers from these lineages into eukaryotic organisms is contentious. Here we systematically identify hundreds of genes that were likely acquired horizontally from a variety of sources by the early-diverging fungal phyla Microsporidia and Cryptomycota. Interestingly, the Microsporidia have acquired via HGT several genes involved in nucleic acid synthesis and salvage, such as those encoding thymidine kinase (TK), cytidylate kinase, and purine nucleotide phosphorylase. We show that these HGT-derived nucleic acid synthesis genes tend to function at the interface between the metabolic networks of the host and pathogen. Thus, these genes likely play vital roles in diversifying the useable nucleic acid components available to the intracellular parasite, often through the direct capture of resources from the host. Using an in vivo viability assay, we also demonstrate that one of these genes, TK, encodes an enzyme that is capable of activating known prodrugs to their active form, which suggests a possible treatment route for microsporidiosis. We further argue that interfacial genes with well-understood activities, especially those horizontally transferred from bacteria or viruses, could provide medical treatments for microsporidian infections.
Horizontal gene transfer (HGT), or the nonvertical transmission of genetic information between distantly related organisms, is common in bacteria, archaea, and viruses (1–3). The importance and scale of HGT in eukaryotes, however, is a matter of debate (4–7). In particular, HGT into fungi was thought to be rare, but several examples from bacteria into fungi or between fungi have recently been described (5, 8, 9). For example, one phylum of early-diverging fungi, the Microsporidia, has had a handful of HGT events documented (5, 10–12), but these fungi are thought to have acquired relatively few genes through HGT (13).
Microsporidia are obligate intracellular parasites of animals. Their genomes are highly compact, and they have eliminated many core metabolic processes in favor of relying on their host for the synthesis of essential molecules (14). They are opportunistic pathogens that primarily infect immunocompromised individuals, such as AIDS patients and organ-transplant recipients (15). They also infect a number of economically important animals as potent zoonotic pathogens (16). Members of a related early-diverging fungal phylum, Cryptomycota, are also obligate intracellular parasites that infect algae (17), amoeboids (18), and other fungi (19), highlighting the similarity between their lifestyles and their evolutionary affinity.
Prior analysis of microsporidian genomes demonstrated that, like other microbial eukaryotes, this phylum of fungi has obtained multiple genes through HGT (20), which may have provided a portion of the raw material required for adaptation, as seen in other organisms (21). Known HGT events into Microsporidia include the following: (i) the ADP/ATP translocase gene family, originating from an HGT event that transferred the founding gene from a member of the bacterial phylum Chlamydia (10), which are known to steal energy-bearing molecules from their host; (ii) a six-gene folate synthesis pathway transferred into Encephalitozoon hellem from multiple donors, a transfer hypothesized to reduce host metabolic stress (11); and (iii) the acquisition of a glutamate-ammonia ligase from an unknown prokaryotic source by Spraguea lophii, which is thought to provide spores a mechanism for defense against the ammonia generated by the decomposing flesh in which they are embedded (12). These case studies, the first of which is shared with the Cryptomycota (19), suggest a role for HGT in the evolution of their unusual pathogenic metabolisms and indicate the need for a thorough, systematic analysis of HGT events in these early-diverging fungi.
Results
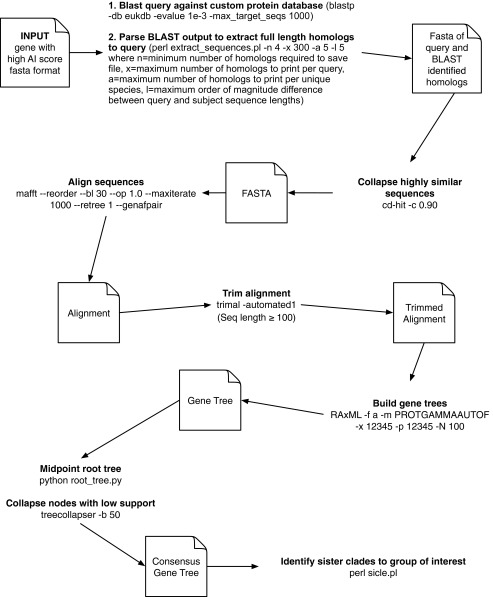
To quantify the number of HGT-derived genes in these intracellular parasites, we deployed a high-throughput analytical pipeline to analyze the mode of inheritance of 14,914 genes (Table 1) from sequenced genomes of Encephalitozoon cuniculi (22, 23), E. hellem (11), Nosema ceranae (24, 25), Nematocida parisii (26), and Rozella allomycis (19), the latter being the sole Cryptomycota genome available (Table S1). Specifically, for each gene, we first generated Alien Index (AI) scores (27) that compared the similarity of the gene between specified ingroup and outgroup taxa (e.g., fungi and bacteria, respectively; Fig. S1). We chose a more relaxed AI score cutoff than the original study because the relatively small number of genes in these organisms made manual curation of gene trees feasible. Using this AI cutoff, we were able to recover 8 of the 10 previously noted HGT events that fell within the scope of the species surveyed (5, 10, 11, 13, 22, 26, 28, 29); even using our relaxed AI threshold, two previously described HGTs were not identified, in one case narrowly (Table S2).
Table 1.
HGT events among early-diverging fungi
| Category | E. cuniculi | E. hellem | No. ceranae | N. parisii | R. allomycis |
| Total genes | 1,996 | 1,847 | 2,060 | 2,661 | 6,350 |
| Genes with AI Score | 1,296 | 1,313 | 1,004 | 1,241 | 4,406 |
| Positive AI genes | 594 | 610 | 390 | 576 | 1,240 |
| High-positive AI genes | 55 | 64 | 44 | 58 | 327 |
| Genes of interest | 50 | 56 | 39 | 39 | 268 |
| Ambiguous HGT events | 15 | 17 | 11 | 13 | 73 |
| Likely HGT events | 16 | 22 | 16 | 10 | 57 |
| Likely HGTs with nested topologies | 8 | 12 | 9 | 9 | 41 |
| Range of HGT event proportion, % | 0.40–1.55 | 0.65–2.11 | 0.44–1.31 | 0.34–0.86 | 0.65–2.04 |
Table S1.
Genomes analyzed
| Genome ID | Genome | Download date | Phylum | Reference |
| Rall | R. allomycis CSF55 v1.0 | 10-Jul-14 | Cryptomycota | James et al. (19) |
| Ecun | E. cuniculi GB-M1 v1.0 | 12-Nov-14 | Microsporidia | Katinka et al. (22); Peyretaillade et al. (23) |
| Ehel | E. hellem ATCC 50504 v1.0 | 12-Nov-14 | Microsporidia | Pombert et al. (11) |
| Npar | N. parisii ERTm1 v1.0 | 3-Feb-15 | Microsporidia | Cuomo et al. (26) |
| Ncer | No. ceranae BRL01 v1.0 | 12-Nov-14 | Microsporidia | Cornman et al. (25); Chen et al. (24) |
Downloaded from the Joint Genome Institute Genome Portal (genome.jgi.doe.gov).
Fig. S1.
Workflow for genome-wide AI score calculation. See Methods for specific details on genome-wide AI calculation.
Table S2.
Calibration of HGT calling methodology using prior works
| Gene name | Species prior works noted | Prior works where noted | This work's AI score in prior noted species | Species this work notes | Phylogenetic support for HGT? | Within scope of this study? |
| CK | E. cuniculi | Marcet-Houben & Gabaldón (5); Cuomo et al. (26) | 110.5241 | E. cuniculi, E. hellem, No. ceranae, N. parisii | Yes | Yes |
| Hemolysin III-like protein | E. cuniculi | Marcet-Houben & Gabaldón (5); | 24.92297 | E. cuniculi, E. hellem | Yes | Yes |
| Phosphoribosyltransferase | E. hellem | Pombert et al. (11) | 168.8619 | E. hellem | Yes | Yes |
| GTP cyclohydrolase | E. hellem | Pombert et al. (11) | 124.3396 | E. hellem | Yes | Yes |
| PNPs | E. hellem | Selman et al. (29) | 84.38472 | E. hellem | Yes | Yes |
| Folylpolyglutamate synthase | E. hellem | Pombert et al. (11) | 56.17833 | E. hellem | Yes | Yes |
| ADP/ATP translocase | E. cuniculi | Katinka et al. (22); Tsaousis et al. (10) | 61.4767–43.85448* | All | Yes | Yes |
| Aspartate-ammonia ligase | No. ceranae | Heinz et al. (28) | 67.46811 | No. ceranae | Yes | Yes |
| Cysteine-rich secretory proteins, antigen S, and pathogenesis-related 1 protein | E. cuniculi, N. parisii | Nakjang et al. (13) | 3.1920, 14.5334 | n/a | n/a | Yes |
| Mechanosensitive ion channel | E. cuniculi, E. hellem, No. ceranae, N. parisii | Nakjang et al. (13) | 6.83, 9.90, 0.755, -0.315 | n/a | n/a | Yes |
| Glycosyltransferase | N. parisii | Nakjang et al. (13) | 7.1309 | n/a | n/a | No |
| Folic acid synthase | E. hellem | Pombert et al. (11) | −17.50439 | n/a | n/a | No |
| Dihydrofolate synthase | E. hellem | Pombert et al. (11) | −30.22129 | n/a | n/a | No |
n/a, not applicable.
These genes have been duplicated over time, and each one has a unique AI score.
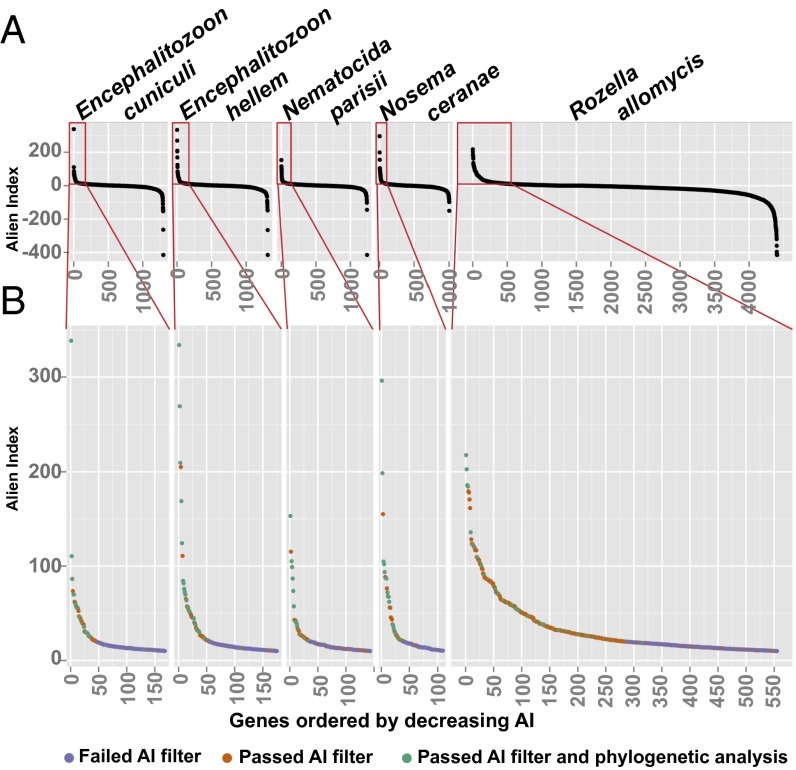
Detection of most, but not all, known HGT events suggests that our methodology is sensitive but conservative. Between 44 and 64 microsporidian genes per genome and 327 R. allomycis genes were designated as high-positive AI genes (genes with AI ≥ 20 or with 10 ≤ AI < 20 and no significant BLAST hit to other fungi; Dataset S1). The high-positive AI genes were cross-referenced with Eukaryotic Orthologous Group (KOG), Kyoto Encyclopedia of Genes and Genomes (KEGG), Gene Ontology (GO), and InterPro annotations provided by the Joint Genome Institute MycoCosm Portal (30). Between 39 and 56 microsporidian genes per genome and 268 R. allomycis genes had at least one functional annotation and were designated as genes of interest. These genes of interest were analyzed phylogenetically, and 10–22 microsporidian genes per genome and 57 R. allomycis genes had strong phylogenetic support for HGT (Table 1, Fig. 1, and Dataset S2). In addition, the majority of these likely transferred genes were nested within well-established higher-order taxa, which further supports their HGT origin. Depending on the stringency of criteria applied, the range of HGT events varied between 0.34% and 2.11% of the total genes present in a genome.
Fig. 1.
Plots of the AI for five early-diverging fungal eukaryotes. (A) AI scores were calculated for every gene in the five genomes analyzed, and they were ordered by decreasing AI score. (B) Genes with an AI score ≥ 10 were plotted with colors corresponding to whether they passed or failed the AI score or phylogenetic filters (see main text).
Many of the putatively HGT-derived genes identified in this screen are annotated as aminoacyl-tRNA synthetases, DNA repair enzymes, and nucleic acid anabolic enzymes (Dataset S2). The latter group of genes is particularly interesting because it includes previously characterized HGTs. For example, purine nucleotide phosphorylases (PNPs) and phosphoribosyltransferases (PRTs) are hypothesized to play roles in guanine metabolism in E. hellem (11), whereas ADP:ATP translocases transferred from Chlamydia are found in all Microsporidia (10). In addition, cytidylate kinase (CK) was previously noted as a possible HGT (5, 26), but its role in Microsporidian biology and its connection to other HGTs is uncharacterized. These, together with the newly identified nucleic acid salvage gene thymidine kinase (TK), may enable the harvest of host nucleosides and nucleotides.
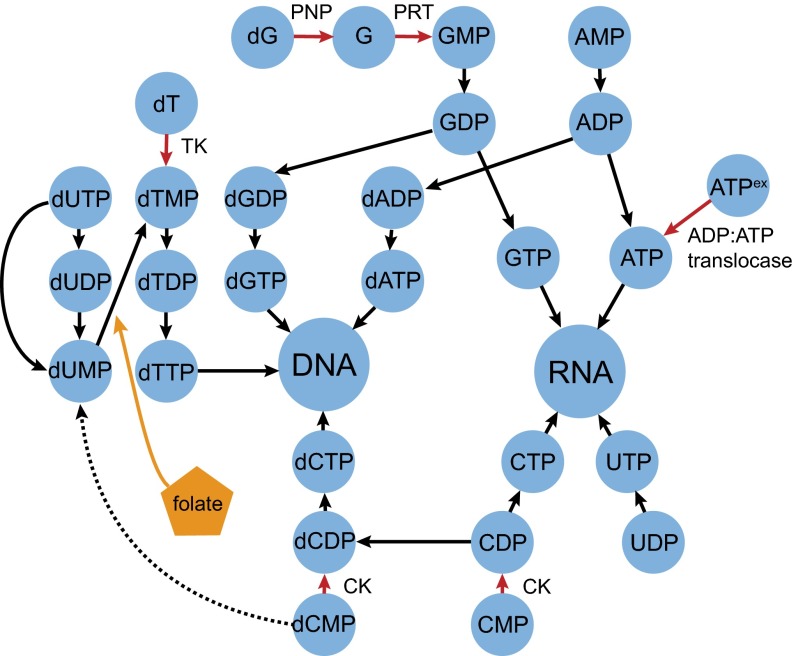
Closer examination of the nucleic acid subpathway in the Microsporidia E. hellem revealed it to be completely disconnected from the core metabolic network, an observation conserved in the other microsporidians examined (Fig. 2). Thus, many, if not all, Microsporidia are unable to synthesize nucleic acid components de novo and must rely on scavenging free nucleic acid components from the host. Remarkably, each of the HGT-derived genes in the E. hellem metabolic subpathway lies on or near the perimeter. The only HGT-derived gene (PNP) that is removed by one step coexists with an HGT-derived gene (PRT) that directly connects it to the perimeter of the subpathway (11, 29). Thus, each HGT provides a new interface that connects the pathogen's nucleic acid subpathway to the host metabolic network. To test the hypothesis that HGT-derived metabolic nodes tend to lie at the host–pathogen interface, we permuted the E. hellem nucleic acid metabolic subpathway, independently evaluating enzymatic steps and genes for the number of steps remaining to the perimeter. We found that the observed subnetwork was among the most extreme permutations (P = 0.0027 and P = 0.019, respectively; Dataset S3), leading us to focus on the genes at the perimeter of this network.
Fig. 2.
Early-diverging fungal HGT events and nucleic acid metabolism. A schematic of the E. hellem nucleic acid anabolic subpathway indicates that HGT-derived genes are found near the host–parasite interface, which provides the parasite with additional sources of nucleosides and nucleotides. HGT-derived enzymatic functions are denoted by red arrows (10, 11, 29), whereas the dCMP deaminase present in some other Microsporidia (but absent in E. hellem) is denoted by a dashed black arrow. HGTs are also contained within the E. hellem folate biosynthesis pathway (orange), which donates a methyl group to convert dUMP to dTMP. CK, cytidylate kinase; PNP, purine nucleotide phosphorylase; PRT, phosphoribosyltransferase; TK, thymidine kinase.
The transfer of TK into microsporidians would enable the direct phosphorylation of thymidine from the host into dTMP for further synthesis by the pathogen, relieving shortages that might be encountered at various stages of the host’s cell cycle. In particular, TK reduces the need for folate by microsporidians, the ultimate source of which is the diet of the animal host. Folate donates a methyl group to convert dUMP to dTMP, and this particular stress has been invoked previously to explain the horizontal transfer of several genes encoding folate synthesis enzymes into E. hellem (11). Interestingly, not all microsporidians possess the bacterial TK, such as Vavraia culicis and Trachipleistophora hominis. Instead, these organisms have retained an ancestral gene encoding a deoxycytidine monophosphate deaminase, which produces dUMP and, ultimately, dTMP from dCMP (Fig. 2). Thus, microsporidians have evolved three distinct, but sometimes overlapping, strategies for the production of dTMP, suggesting that production of this nucleotide is central to their intracellular parasitic lifestyle.
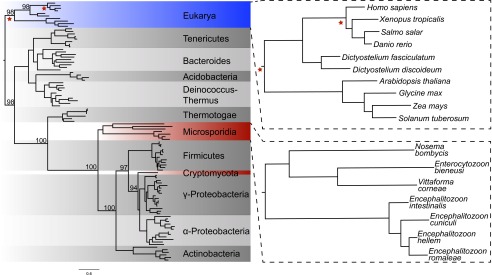
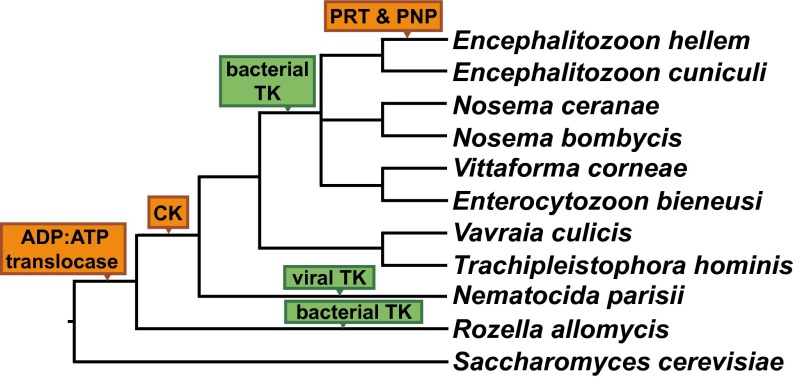
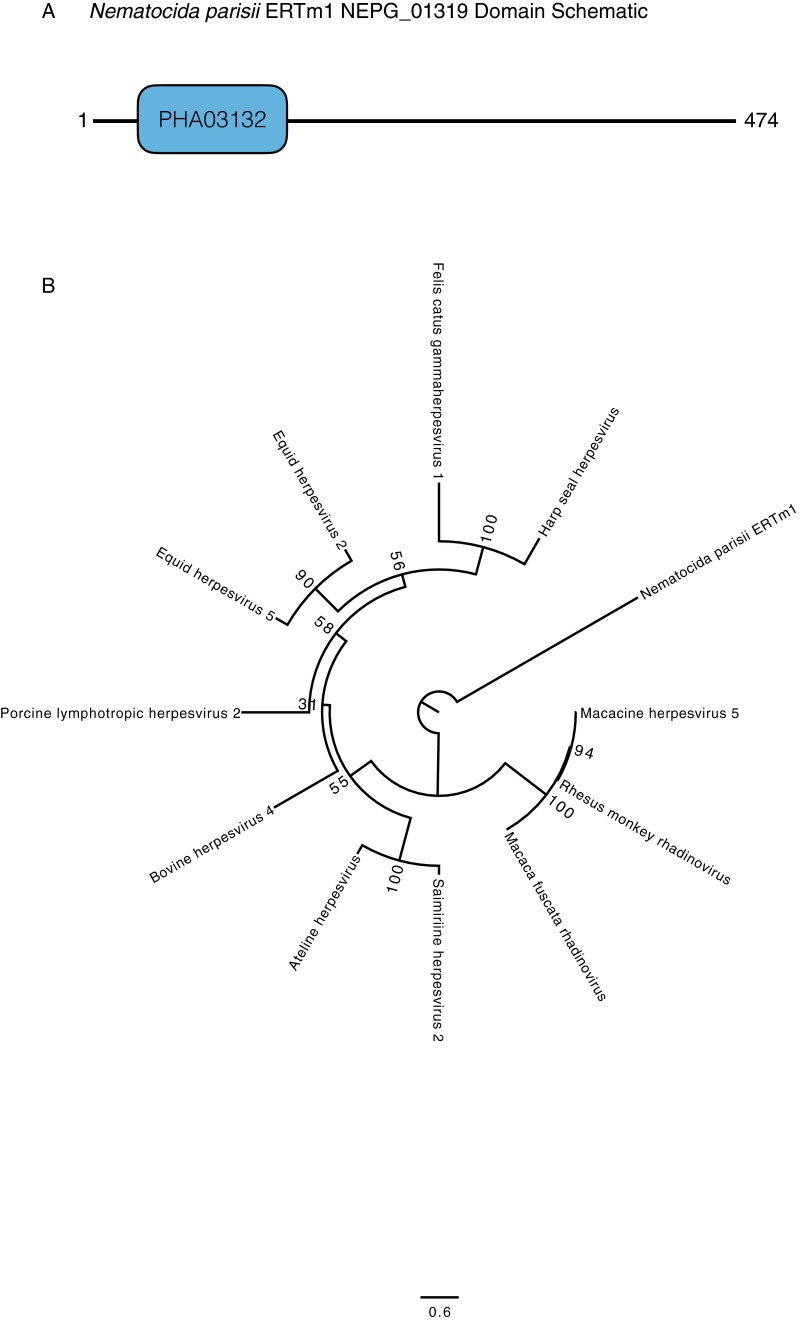
Of the three strategies for making dTMP, the HGT of TK stands out because the gene is thought to have been lost in the fungal lineage shortly after it diverged from animals (31). The phylogeny of TK sequences supports two independent horizontal transfers of bacterial TK genes into several Microsporidia taxa and into R. allomycis (Fig. 3). Likelihood ratio tests rejected alternate tree topologies where the Microsporidia TKs were sister to either Animalia or Eukarya TKs; likewise, alternate topologies in which the R. allomycis TK was sister to either Animalia or Eukarya TKs were rejected. Finally, an alternate topology in which R. allomycis and the Microsporidia TKs were grouped together was also rejected, further supporting the independent transfer of TK into these phyla (Table 2). In addition to these two HGT events, we found an independent HGT of a putative, viral-like TK into N. parisii (Fig. 4 and Fig. S2), documenting a third parallel transfer of TK into these fungal intracellular parasites.
Fig. 3.
TK has been transferred from bacteria into Microsporidia and Cryptomycota by independent HGT events. Top BLASTP hits for E. cuniculi, No. ceranae, and R. allomycis TK were aligned along with TK sequences from representative eukaryotes and other bacterial phyla, and the resulting alignment was used to infer a maximum-likelihood phylogeny. Red stars indicate the location on the tree where likelihood-ratio tests were performed to evaluate the relationship of the sequences in question with Animalia or other Eukarya representatives. Bootstrap values for key nodes are noted. The N. parisii TK results from a third independent HGT, but is absent from the phylogenetic tree because of its distinctive viral domain structure and amino acid sequence (Fig. S2). Note that the No. ceranae TK has also been excluded due its long branch length (Dataset S2), but Nosema bombycis is shown.
Table 2.
Likelihood ratio tests support two independent transfers into Microsporidia and Cryptomycota
| Description | Likelihood | Difference | P value |
| Best tree from Fig. 3 | −23,232.36221 | n/a | n/a |
| Rozella and Microsporidia grouped | −23,276.41079 | −44.048579 | 5.57E-07 |
| Microsporidia sister to Animalia | −23,354.81342 | −122.451214 | 2.36E-23 |
| Microsporidia sister to Eukarya | −23,314.16858 | −81.80637 | 5.89E-15 |
| Rozella sister to Animalia | −23,438.13523 | −205.773023 | 1.15E-46 |
| Rozella sister to Eukarya | −23,349.71471 | −117.352501 | 2.40E-27 |
n/a, not applicable.
Fig. 4.
Schematic of nucleic acid metabolism HGT events in the Microsporidia and Cryptomycota. A cladogram of representative members of early-diverging fungi highlighting HGT-derived genes that function in the nucleic acid subpathway. Green boxes denote three independent horizontal transfers of TK, while orange boxes denote other horizontal transfers of nucleic acid metabolism genes (10, 11, 27).
Fig. S2.
N. parisii has a putative viral TK. (A) Domain analysis of the N. parisii gene suggests the presence of a viral-like TK gene similar to those found in herpesviruses from various animals. (B) Phylogenetic analysis suggests a loose relationship between this hypothetical N. parisii gene and herpesvirus TK.
The absolute dependence of microsporidians on their hosts for nucleic acid components suggests that this subpathway could be a viable target for antimicrosporidian drugs. TK is an especially promising target for disruption because of the apparent importance placed on dTMP synthesis during evolution and the wide range of TK-activated prodrugs currently used as antiviral therapies. First, we confirmed that TK is expressed by analyzing a previously published E. cuniculi transcriptome dataset (32); TK was in the top quartile of expressed genes at all three postinfection time points (Table S3 and Dataset S4). We then evaluated the predicted TK proteins of E. cuniculi, N. parisii, and R. allomycis in an assay in which the nucleoside kinase activity of TK would activate the prodrug 5-fluoro-2-deoxyuridine (FUdR), converting it into the toxic compound fluorodeoxyuridine monophosphate and killing the tester strain of Saccharomyces cerevisiae (31). The growth of strains expressing Homo sapiens, E. cuniculi, and N. parisii TK were completely inhibited when FUdR was present, whereas strains expressing either predicted version of the R. allomycis TK behaved identically to the blank vector control strain, perhaps due to genome annotation or heterologous expression issues. Indeed, we also noted that the N. parisii TK strain did not grow as robustly as the other four TK-expressing strains or the blank vector control strain in any media tested, suggesting that N. parisii TK is detrimental to S. cerevisiae growth (Table 3). Thus, we conclude that at least two of the three TK proteins that underwent HGT—one from viruses and one from bacteria—are active and capable of activating known prodrugs.
Table S3.
Expression analysis of E. cuniculi horizontally transferred and control genes
| Gene name | JGI gene no. | 24 h | 48 h | 72 h | |||
| A | B | A | B | A | B | ||
| Actin | 43 | 13,724.9 | 14,143.4 | 10,083.2 | 10,461.3 | 9,906.21 | 10,043 |
| TK (HGT) | 72 | 985.193 | 965.304 | 758.042 | 764.095 | 799.44 | 754.058 |
| RNA Pol II, second subunit | 183 | 354.053 | 337.362 | 289.602 | 289.471 | 263.585 | 271.615 |
| ADP:ATP translocase (HGT) | 210 | 374.823 | 388.014 | 306.662 | 304.801 | 294.997 | 297.993 |
| 60S ribosomal protein L35 | 555 | 2,949.59 | 2,933.83 | 2,362.19 | 2,214.58 | 2,292.86 | 1,939.53 |
| CK (HGT) | 840 | 577.16 | 528.322 | 547.497 | 564.53 | 539.359 | 481.178 |
Table 3.
Endpoint analysis of optical density of TK-expressing strains relative to control cultures
| Media condition | Blank vector | H. sapiens TK | E. cuniculi TK | N. parisii TK* | R. allomycis TK A | R. allomycis TK B |
| SC-MSG | 11.0 ± 0.21 | 10.6 ± 0.01 | 10.3 ± 0.35 | 2.8 ± 0.07 | 10.8 ± 0.21 | 10.6 ± 0.35 |
| SC-MSG +FUdR | 9.4 ± 0.16 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | 9.2 ± 0.29 | 9.0 ± 0.41 |
N. parisii TK expression inhibits growth; data gathered after 168 h. Other strains were evaluated after 60 h.
Discussion
We have shown that members of the early-diverging fungal phyla Microsporidia and Cryptomycota have acquired dozens of genes by HGT, the vast majority of which are, to our knowledge, reported here for the first time. These findings are inconsistent with the hypothesis that intracellular parasites possess few HGT-derived genes (13). Despite their prevalence, the mechanism by which these genes are transferred into early-diverging fungi is still unclear, and previously proposed eukaryotic HGT mechanisms, such as through viral intermediates (7, 33), seem unlikely for intracellular parasites. One possible mechanism is that phagocytosis of spores by host cells into intracellular vesicles (34) may bring nucleic acids from lysed environmental bacteria into contact with receptive spores. Because microsporidians reside in the host cytoplasm, another possibility is that host mRNA molecules are reverse-transcribed and incorporated into microsporidian genomes. Whatever the molecular mechanism of the HGTs, their impact on the microsporidian genome and metabolism has been widespread.
Nucleic acid anabolic genes, in particular, have been transferred multiple times into the Cryptomycota and Microsporidia. These HGTs provide the intracellular parasites with novel sources of nucleosides and nucleotides, while simultaneously reducing metabolic stress on their hosts. This evolutionary strategy has been described in the intracellular Apicomplexan parasite Cryptosporidium parvum (35), which, like many microsporidians, has acquired a bacterial TK to diversify its nucleotide sources. Herpes viruses also rely on TK to increase dTMP levels, and TK strongly affects its pathogenicity (36).
Previous work has shown that N. parisii replication can be slowed by FUdR treatment, although whether this effect was a function of host or parasite metabolism was unclear (37). Here we show that some Microsporidia TK genes, including N. parisii TK, indeed encode active nucleoside kinases, providing a likely drug target with a known mechanism of action (38, 39). Thus, antiviral prodrugs that are activated by TK, such as FUdR or acyclovir, may offer a path to a viable treatment option for microsporidiosis. These drugs are already used to treat diseases, such as herpes simplex virus (40), HIV (41), and certain types of cancers (42). In the latter cases, cancer cells are preferentially targeted because a viral exogenous TK has been inserted into the cell that has lower specificity than the endogenous TK (38), or native human TK expression has been increased due to uncontrolled cellular division (43). Although we did not detect any activity for R. allomycis TK, a negative result in a heterologous gene expression assay is not definitive. R. allomycis TK may require a chaperone that S. cerevisiae does not have; it may require a posttranslational modification that S. cerevisiae is unable to perform; it may function in a narrow range of cellular conditions; or the intron–exon boundaries may have been misannotated.
The treatment of fungal pathogens of animals has traditionally proven challenging due to the shared physiological traits of these sister kingdoms. Because of its inability to synthesize nucleic acid components, nucleotide analogs that are activated by the existing microsporidian nucleic acid subnetwork could provide new treatments for microsporidiosis in humans and livestock. For example, bacterial CK prefers CMP or dCMP as a substrate, whereas the UMP/CMP kinase found in animals and fungi prefers CMP or UMP as a substrate (44, 45). Because Microsporidia possess the bacterial CK, they may be much more susceptible to dCMP analogs than their host cells, a treatment strategy that has been used against HIV (46). Many other pathogenic fungi that infect animals, such as Aspergillus fumigatus, have also acquired an appreciable number of genes through HGT (47), including entire secondary metabolism gene clusters (48). Using HGT events to identify novel candidate drug targets may thus prove a promising general strategy across diverse clades of pathogenic fungi and other recalcitrant pathogens whose physiology is similar to humans.
Methods
AI Analysis of Early-Diverging Fungal Genomes.
All predicted proteins from five early-diverging fungal genomes (Table S1) were queried by using BLAST (E-value = 0.001, max target seqs = 1,000) against a custom database consisting of the National Center for Biotechnology Information’s (NCBI’s) nonredundant protein database (last updated November 24, 2014), as well as additional protein sequences from 411 fungal and plant genome assemblies (Dataset S5). These additional genomes served to increase the representation of close relatives to microsporidians in the database, which is critical when searching for HGTs based on relative BLAST scores. They also allowed the taxonomic extent of HGTs into the Microsporidia to be assessed without risking false positives that could be caused by genomes of variable quality (e.g., bacterial contamination). An AI approach was used to screen these genomes for genes with significantly better BLAST hits to distantly related organisms (e.g., metazoans or bacteria) than to closely related ones (e.g., other fungi) and thereby identify HGT candidates. Two taxonomic lineages were first specified: the recipient lineage into which possible HGT events may have occurred (i.e., Microsporidia or Cryptomycota, depending on the query) and a larger group of related taxa (i.e., Fungi). When parsing the BLAST output, all hits to the recipient lineage were skipped. The AI score is given by the formula:
where bbhG is the E-value of the best BLAST hit to a species within the group lineage (i.e., the best nonmicrosporidian or, in the case of R. allomycis, noncryptomycotan fungal match) and bbhO is the E-value of the best blast hit to a species outside of the group lineage (i.e., the best nonfungal match). In cases where there were no significant BLAST hits, the corresponding bbhG or bbhO was set to 1. AI can range from 461 to −461, and AI is greater than zero if the gene has a better BLAST hit to a species outside of the group lineage. Contamination in the genome assembly could also result in a positive AI score; therefore, any assembly contigs containing only genes with positive AI scores were flagged as potential contamination and eliminated from the analysis.
Systematic Identification of Likely HGT Events.
Genes with AI ≥ 20 or genes with 20 > AI ≥ 10 and with no significant BLAST hit to other Fungi, were classified as high-positive AI genes. These genes were cross-referenced with KOG, KEGG, GO, and InterPro annotations provided by the Joint Genome Institute (30), and all genes with at least one functional annotation were classified as genes of interest and analyzed by using a phylogenetic pipeline (Fig. S1). For each HGT candidate, a Perl script based publicly available software (49) extracted up to 400 homologs from the custom database (referenced above) based on BLAST similarity, allowing up to five orders of magnitude difference between query and subject lengths, and extracting up to five sequences per species. To reduce the number of sequences per gene tree, highly similar sequences were collapsed with CD-HIT using default parameters (50). Sequences were aligned with MAFFT by using the E-INS-i strategy (51). The resulting alignment was trimmed with TRIMAL by using the automated1 strategy (52). All genes with trimmed alignments <100 amino acids were discarded. Phylogenetic trees were constructed by using RAxML (53) with the PROTGAMMAAUTOF amino acid model of substitution and 100 bootstrap replicates. Trees were midpoint-rooted, and branches with <50% bootstrap support were collapsed by using TreeCollapseCL4 (emmahodcroft.com/TreeCollapseCL.htm). The resulting phylogenies were manually inspected to assess each gene’s mode of transmission. First, the presence or absence of fungal BLASTP hits was determined. Second, the nearest neighbor node to the query was identified as prokaryotic, eukaryotic, opisthokont, and/or fungal (multiple nearest neighbors are possible because poorly supported nodes were collapsed). Next, if the nearest node was opisthokont, the likelihood of parallel loss to explain the topology was assessed. Finally, we noted whether the query was nested within a well-established alien clade. A gene’s mode of transmission was labeled as a likely HGT event if and only if its phylogeny was inconsistent with the organismal phylogeny (19) and any parallel-loss scenario (e.g., a protein sequence from E. cuniculi being nested within a clade of Proteobacteria instead of sister to the Fungi). If a phylogeny could be explained by simple parallel loss or any mechanism other than horizontal transfer, its mode of transmission was labeled as unlikely HGT. If the phylogeny provided no clear support for either vertical or horizontal transmission, its mode was labeled as ambiguous. Finally, some phylogenies were labeled as “no call” because they lacked an interpretable topology (e.g., “star phylogeny”). All phylogenetic trees and calls are available in Dataset S2.
Permutation Analyses of the E. hellem Nucleic Acid Subpathway.
The enzymatic steps at the periphery of the complete E. hellem nucleic acid subpathway interface directly with the metabolism of the host (Fig. 2). These steps had a distance to the host of zero, whereas other steps were assigned a distance corresponding to the smallest number of steps from their nearest peripheral metabolites. The observed average distance to the host for enzymatic steps was 0.167 for steps experiencing HGTs, whereas the average for the complete subpathway was 1.394. Because CK (and some non-HGT enzymes) can perform multiple steps, we also considered genes in a separate analysis by averaging the values of each step they encode, yielding an observed average distance of 0.2 for genes experiencing HGTs vs. an average of 1.163 for the complete subpathway. To assess significance, 1 million random permutations were simulated, and the proportion of permuted networks that were at least as extreme as the observed network was determined. Complete documentation is provided in Dataset S3.
Phylogenetic Analysis of TK.
The E. cuniculi and R. allomycis TK sequences were used as BLASTP queries via the NCBI BLAST website (www.ncbi.nlm.nih.gov/BLAST), and the top hits were downloaded along with the TK sequences of representatives from eukaryotic and prokaryotic clades. These protein sequences were aligned in MUSCLE (54) by using default settings, and maximum-likelihood analysis was performed by using the PROTGAMMALG model of evolution in multithreaded RAxML (53). To evaluate alternative possibilities for the source of these sequences, five different alternate trees were drawn, also by using the PROTGAMMALG model in RAxML, and the likelihood ratio was calculated for each tree compared with the best tree (55). P values were calculated from this ratio in the Python Scipy library χ2 probability module (56).
Plasmid and Strain Construction.
The protein sequence for soluble TK of H. sapiens was downloaded from NCBI (BAG70082.1), and the TK protein sequences for E. cuniculi (ECU01_0740im.01) (22), N. parisii (Nempa11372) (26), and R. allomycis (Rozal12947) (19) were downloaded from JGI; note that the R. allomycis constructs were designed twice, once without and once with predicted intron sequences present, resulting in R. allomycis TK versions A and B, respectively. These protein sequences were codon-optimized for expression in S. cerevisiae by using the Integrated DNA Technologies (IDT) webtool (www.idtdna.com/CodonOpt). Although critical to ensuring proper heterologous protein expression (57), this approach necessarily eliminates any RNA regulation that may occur in E. cuniculi, N. parisii, and R. allomycis. gBlocks for the H. sapiens and E. cuniculi sequences, as well as plasmids containing synthesized sequence for the N. parisii and both versions of R. allomycis, were ordered from IDT. Primers (Table S4) were designed to amplify each gBlock or synthesized gene, while adding ∼40 bp of sequence on both ends to enable homology repair (58) of the gBlock sequence into the plasmid pRS426-HygMX under the control of the TDH3 promoter and the CYC1 terminator. This cloning was performed in the wild diploid strain M22 of S. cerevisiae (59) by transforming it using linearized pRS426-HygMX and PCR product via the lithium acetate/PEG method (60). Plasmid repair and yeast transformation was selected for on yeast extract/peptone/dextrose (YPD) +Hyg (10 g/L yeast extract, 20 g/L peptone, 20 g/L glucose, 18 g/L agar, and 200 mg/L hygromycin B) after 3 h of recovery in liquid YPD. Resulting yeast colonies had their total DNA extracted, which was used to transform Escherichia coli by electroporation, and the recovered plasmids had their insertions Sanger-sequenced. Once the gene-insertion sequences were confirmed, plasmids were again transformed into M22 by selection on YPD +Hyg plates as described above.
Table S4.
Primers used in this study
| Number | Name | Sequence | Use |
| oHWA126 | pRS426 F WGA | CCAGGGTTTTCCCAGTCACGACG | Amplify and sequence insertion site of pRS426-HygMX |
| oHWA127 | pRS426 F WGA | CTCACTCATTAGGCACCCCAGGC | Amplify and sequence insertion site of pRS426-HygMX |
| oHWA745 | TDH3 F WGA | GTAATACGACTCACTATAGGGCGAATTGGGTACGTTTATCATTATCAATACTGCCATTTC | Amplifies TDH3 promoter from M22 |
| oHWA746 | TDH3 R WGA | TTTGTTTGTTTATGTGTGTTTATTCG | Amplifies TDH3 promoter from M22 |
| oHWA747 | CYC1 F WGA | GATCATGTAATTAGTTATGTCACGCTTACA | Amplifies CYC1 terminator from M22 |
| oHWA748 | CYC1 R WGA | CAAGCGCGCAATTAACCCTCACTAAAGGGAACAAAAGCTGCGCAAATTAAAGCCTTCGAG | Amplifies CYC1 terminator from M22 |
| oHWA749 | Ecun-TK F WGA | GAACTTAGTTTCGAATAAACACACATAAACAAACAAAATGACTAGAGGTACTTTAAATTT | Amplifies E. cuniculi gBlock sequence for cloning into functional test vector |
| oHWA750 | Ecun-TK R WGA | AGGGCGTGAATGTAAGCGTGACATAACTAATTACATGATCTTAACGATCAACTGGGATAA | Amplifies E. cuniculi gBlock sequence for cloning into functional test vector |
| oHWA751 | Hsap-TK F WGA | AAGAACTTAGTTTCGAATAAACACACATAAACAAACAAAATGTCTTGTATCAACTTACCA | Amplifies H. sapiens gBlock sequence for cloning into functional test vector |
| oHWA752 | Hsap-TK R WGA | AGGGCGTGAATGTAAGCGTGACATAACTAATTACATGATCTTAGTTAGCTGGAGAACATT | Amplifies H. sapiens gBlock sequence for cloning into functional test vector |
| oHWA753 | Npar-TK F WGA | CAAGAACTTAGTTTCGAATAAACACACATAAACAAACAAAATGGAGAAGGTCATCTCCTC | Amplifies N. parisii gBlock sequence for cloning into functional test vector |
| oHWA754 | Npar-TK R WGA | AGGGCGTGAATGTAAGCGTGACATAACTAATTACATGATCTTAGATGTTGGACTTGGAGG | Amplifies N. parisii gBlock sequence for cloning into functional test vector |
| oHWA755 | Rall-TK-B F WGA | CAAGAACTTAGTTTCGAATAAACACACATAAACAAACAAAATGAATGCCGGTAAGTCTAC | Amplifies R. allomycis version B gBlock sequence for cloning into functional test vector |
| oHWA756 | Rall-TK-B R WGA | GCGTGAATGTAAGCGTGACATAACTAATTACATGATCTTATCTATCATTACCACCAATTT | Amplifies R. allomycis version B gBlock sequence for cloning into functional test vector |
| oHWA757 | Rall-TK-A F WGA | CAAGAACTTAGTTTCGAATAAACACACATAAACAAACAAAATGAATGCCGGTAAGTCTAC | Amplifies R. allomycis version A gBlock sequence for cloning into functional test vector |
| oHWA758 | Rall-TK-A R WGA | GGCGTGAATGTAAGCGTGACATAACTAATTACATGATCTTATCTATCATTACCACCAATT | Amplifies R. allomycis version A gBlock sequence for cloning into functional test vector |
Functional Assay of TK.
Single colonies of the six plasmid-bearing strains of S. cerevisiae (Table S5) were grown overnight in SC-MSG +Hyg (1.72 g/L yeast nitrogen base, 2 g/L Complete Drop-Out Mix, 1 g/L monosodium glutamate, 20 g/L glucose, and 200 mg/L hygromycin B) liquid medium. A total of 30,000 cells of each strain were used to inoculate 3 mL of SC-MSG +Hyg +100 mg/L FUdR in quadruplicate; control medium tubes containing SC-MSG +Hyg without FUdR were also inoculated with 30,000 cells for each strain in duplicate. All cultures, except for the N. parisii TK-expressing strain, were grown for 60 h, and their optical density was measured with a 600-nm photometer (Implen GmbH). The N. parisii cultures were measured after 168 h.
Table S5.
Strains used in this study
| Name | Genotype | Use |
| M22 | MATα/MATa | Gap repair strain |
| yHWA464 | MATα/MATa [pRS426-HygMX::PTDH3:Hsap-TK:TCYC1] | H. sapiens TK tester strain |
| yHWA465 | MATα/MATa [pRS426-HygMX::PTDH3:Ecun-TK:TCYC1] | E. cuniculi TK tester strain |
| yHWA466 | MATα/MATa [pRS426-HygMX::PTDH3:Rall-TK-A:TCYC1] | R. allomycis version A TK tester strain |
| yHWA467 | MATα/MATa [pRS426-HygMX::PTDH3:Rall-TK-B:TCYC1] | R. allomycis version B TK tester strain |
| yHWA468 | MATα/MATa [pRS426-HygMX::PTDH3:Npar-TK:TCYC1] | N. parisii TK tester strain |
| yHWA470 | MATα/MATa [pRS426-HygMX] | Blank vector control strain |
Supplementary Material
Acknowledgments
We thank Issac Knoflicek, Michael G. Mangiore, and Dana Opulente for computational support and advice and Amanda B. Hulfachor for artwork. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University. This material is based upon work supported by National Science Foundation Grants IOS-1401682 (to J.H.W.), DEB-1442113 (to A.R.), and DEB-1253634 and DEB-1442148 (to C.T.H.); in part by the DOE Great Lakes Bioenergy Research Center through DOE Office of Science Grant BER DE-FC02-07ER64494; and USDA (United States Department of Agriculture) National Institute of Food and Agriculture Hatch Project 1003258. C.T.H. is an Alfred Toepfer Faculty Fellow and Pew Scholar in the Biomedical Sciences, supported by the Alexander von Humboldt Foundation and Pew Charitable Trusts, respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517242113/-/DCSupplemental.
References
- 1.Pál C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37(12):1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 2.Treangen TJ, Rocha EPC. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 2011;7(1):e1001284. doi: 10.1371/journal.pgen.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreira D, López-García P. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol. 2009;7(4):306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 4.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 5.Marcet-Houben M, Gabaldón T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26(1):5–8. doi: 10.1016/j.tig.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Huang J. Horizontal gene transfer in eukaryotes: The weak-link model. BioEssays. 2013;35(10):868–875. doi: 10.1002/bies.201300007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schönknecht G, Weber APM, Lercher MJ. Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. BioEssays. 2014;36(1):9–20. doi: 10.1002/bies.201300095. [DOI] [PubMed] [Google Scholar]
- 8.Wisecaver JH, Slot JC, Rokas A. The evolution of fungal metabolic pathways. PLoS Genet. 2014;10(12):e1004816. doi: 10.1371/journal.pgen.1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wisecaver JH, Rokas A. Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front Microbiol. 2015;6:161. doi: 10.3389/fmicb.2015.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsaousis AD, et al. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453(7194):553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 11.Pombert J-F, et al. Gain and loss of multiple functionally related, horizontally transferred genes in the reduced genomes of two microsporidian parasites. Proc Natl Acad Sci USA. 2012;109(31):12638–12643. doi: 10.1073/pnas.1205020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell SE, et al. The genome of Spraguea lophii and the basis of host-microsporidian interactions. PLoS Genet. 2013;9(8):e1003676. doi: 10.1371/journal.pgen.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakjang S, et al. Reduction and expansion in microsporidian genome evolution: New insights from comparative genomics. Genome Biol Evol. 2013;5(12):2285–2303. doi: 10.1093/gbe/evt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeling PJ, Fast NM. Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annu Rev Microbiol. 2002;56:93–116. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 15.Didier ES. Microsporidiosis: An emerging and opportunistic infection in humans and animals. Acta Trop. 2005;94(1):61–76. doi: 10.1016/j.actatropica.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Santín M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. 2011;90(3):363–371. doi: 10.1016/j.rvsc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Ishida S, Nozaki D, Grossart H-P, Kagami M. Novel basal, fungal lineages from freshwater phytoplankton and lake samples. Environ Microbiol Rep. 2015;7(3):435–441. doi: 10.1111/1758-2229.12268. [DOI] [PubMed] [Google Scholar]
- 18.Corsaro D, et al. Rediscovery of Nucleophaga amoebae, a novel member of the Rozellomycota. Parasitol Res. 2014;113(12):4491–4498. doi: 10.1007/s00436-014-4138-8. [DOI] [PubMed] [Google Scholar]
- 19.James TY, et al. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol. 2013;23(16):1548–1553. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- 20.Alsmark C, et al. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 2013;14(2):R19. doi: 10.1186/gb-2013-14-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall C, Brachat S, Dietrich FS. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell. 2005;4(6):1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414(6862):450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 23.Peyretaillade E, et al. Identification of transcriptional signals in Encephalitozoon cuniculi widespread among Microsporidia phylum: Support for accurate structural genome annotation. BMC Genomics. 2009;10:607–619. doi: 10.1186/1471-2164-10-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YP, et al. Morphological, molecular, and phylogenetic characterization of Nosema ceranae, a microsporidian parasite isolated from the European honey bee, Apis mellifera. J Eukaryot Microbiol. 2009;56(2):142–147. doi: 10.1111/j.1550-7408.2008.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cornman RS, et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5(6):e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuomo CA, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. 2012;22(12):2478–2488. doi: 10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320(5880):1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 28.Heinz E, et al. The genome of the obligate intracellular parasite Trachipleistophora hominis: New insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 2012;8(10):e1002979. doi: 10.1371/journal.ppat.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selman M, et al. Acquisition of an animal gene by microsporidian intracellular parasites. Curr Biol. 2011;21(15):R576–R577. doi: 10.1016/j.cub.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grigoriev IV, et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42(Database issue):D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander WG, Doering DT, Hittinger CT. High-efficiency genome editing and allele replacement in prototrophic and wild strains of Saccharomyces. Genetics. 2014;198(3):859–866. doi: 10.1534/genetics.114.170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grisdale CJ, Bowers LC, Didier ES, Fast NM. Transcriptome analysis of the parasite Encephalitozoon cuniculi: An in-depth examination of pre-mRNA splicing in a reduced eukaryote. BMC Genomics. 2013;14:207. doi: 10.1186/1471-2164-14-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, et al. Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J Virol. 2010;84(22):11876–11887. doi: 10.1128/JVI.00955-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibley LD. Invasion and intracellular survival by protozoan parasites. Immunol Rev. 2011;240(1):72–91. doi: 10.1111/j.1600-065X.2010.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Striepen B, et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci USA. 2004;101(9):3154–3159. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Efstathiou S, Kemp S, Darby G, Minson AC. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J Gen Virol. 1989;70(Pt 4):869–879. doi: 10.1099/0022-1317-70-4-869. [DOI] [PubMed] [Google Scholar]
- 37.Bakowski MA, et al. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10(6):e1004200. doi: 10.1371/journal.ppat.1004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kokoris MS, Black ME. Characterization of herpes simplex virus type 1 thymidine kinase mutants engineered for improved ganciclovir or acyclovir activity. Protein Sci. 2002;11(9):2267–2272. doi: 10.1110/ps.2460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 40.Simmons A. Clinical manifestations and treatment considerations of herpes simplex virus infection. J Infect Dis. 2002;186(Suppl 1):S71–S77. doi: 10.1086/342967. [DOI] [PubMed] [Google Scholar]
- 41.Furman PA, Barry DW. Spectrum of antiviral activity and mechanism of action of zidovudine. An overview. Am J Med. 1988;85(2A):176–181. [PubMed] [Google Scholar]
- 42.Power DG, Kemeny NE. The role of floxuridine in metastatic liver disease. Mol Cancer Ther. 2009;8(5):1015–1025. doi: 10.1158/1535-7163.MCT-08-0709. [DOI] [PubMed] [Google Scholar]
- 43.Munch-Petersen B, Cloos L, Jensen HK, Tyrsted G. Human thymidine kinase 1. Regulation in normal and malignant cells. Adv Enzyme Regul. 1995;35:69–89. doi: 10.1016/0065-2571(94)00014-t. [DOI] [PubMed] [Google Scholar]
- 44.Hsu C-H, Liou JY, Dutschman GE, Cheng YC. Phosphorylation of cytidine, deoxycytidine, and their analog monophosphates by human UMP/CMP kinase is differentially regulated by ATP and magnesium. Mol Pharmacol. 2005;67(3):806–814. doi: 10.1124/mol.104.006098. [DOI] [PubMed] [Google Scholar]
- 45.Briozzo P, et al. Structures of Escherichia coli CMP kinase alone and in complex with CDP: A new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity. Structure. 1998;6(12):1517–1527. doi: 10.1016/s0969-2126(98)00150-6. [DOI] [PubMed] [Google Scholar]
- 46.Ray AS, et al. Probing the mechanistic consequences of 5-fluorine substitution on cytidine nucleotide analogue incorporation by HIV-1 reverse transcriptase. Antivir Chem Chemother. 2003;14(3):115–125. doi: 10.1177/095632020301400301. [DOI] [PubMed] [Google Scholar]
- 47.Mallet LV, Becq J, Deschavanne P. Whole genome evaluation of horizontal transfers in the pathogenic fungus Aspergillus fumigatus. BMC Genomics. 2010;11:171. doi: 10.1186/1471-2164-11-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell MA, Rokas A, Slot JC. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol Evol. 2012;4(3):289–293. doi: 10.1093/gbe/evs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeBlasio D, Wiscaver J. 2013. SICLE: A high-throughput tool for extracting evolutionary relationships from phylogenetic trees. arXiv:1303.5785.
- 50.Li W, Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 51.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 54.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 56.Oliphant T. Python for scientific computing. Comput Sci Eng. 2007;13:10–20. [Google Scholar]
- 57.Presnyak V, et al. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160(6):1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weir M, Keeney JB. PCR mutagenesis and gap repair in yeast. Methods Mol Biol. 2014;1205:29–35. doi: 10.1007/978-1-4939-1363-3_3. [DOI] [PubMed] [Google Scholar]
- 59.Mortimer RK. Evolution and variation of the yeast (Saccharomyces) genome. Genome Res. 2000;10(4):403–409. doi: 10.1101/gr.10.4.403. [DOI] [PubMed] [Google Scholar]
- 60.Gietz RD. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol Biol. 2014;1205:1–12. doi: 10.1007/978-1-4939-1363-3_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.