Significance
Stomata regulate the efficiency of photosynthesis and affect plants’ resistance to air pollutants. However, the transcriptional regulation of the genes modulating stomatal movement has not been well characterized. Using chimeric repressor gene-silencing technology (CRES-T), we identified another function of the previously studied regulators of chloroplast development GOLDEN 2-LIKE1 (GLK1) and GLK2, as positive regulators of stomatal movement and K+in channel genes. The chimeric GLK repressors induce closed stomata and tolerance to ozone exposure and reduced gene expression and activity of K+in channels in guard cells. Guard cell-specific expression of the chimeric GLK repressor may be a useful tool to confer resistance to air pollutants.
Keywords: ozone, stomatal movement, transcription factor, repressor, K+in channel
Abstract
Stomatal movements regulate gas exchange, thus directly affecting the efficiency of photosynthesis and the sensitivity of plants to air pollutants such as ozone. The GARP family transcription factors GOLDEN 2-LIKE1 (GLK1) and GLK2 have known functions in chloroplast development. Here, we show that Arabidopsis thaliana (A. thaliana) plants expressing the chimeric repressors for GLK1 and -2 (GLK1/2-SRDX) exhibited a closed-stomata phenotype and strong tolerance to ozone. By contrast, plants that overexpress GLK1/2 exhibited an open-stomata phenotype and higher sensitivity to ozone. The plants expressing GLK1-SRDX had reduced expression of the genes for inwardly rectifying K+ (K+in) channels and reduced K+in channel activity. Abscisic acid treatment did not affect the stomatal phenotype of 35S:GLK1/2-SRDX plants or the transcriptional activity for K+in channel gene, indicating that GLK1/2 act independently of abscisic acid signaling. Our results indicate that GLK1/2 positively regulate the expression of genes for K+in channels and promote stomatal opening. Because the chimeric GLK1-SRDX repressor driven by a guard cell-specific promoter induced a closed-stomata phenotype without affecting chloroplast development in mesophyll cells, modulating GLK1/2 activity may provide an effective tool to control stomatal movements and thus to confer resistance to air pollutants.
Tropospheric ozone (O3) is a major photochemical oxidant and one of the most phytotoxic air pollutants (1). High concentrations of ozone induce oxidative stress, which activates programmed cell death and significantly inhibits plant growth, causing crop losses estimated to be in the billions of dollars (2, 3). To study the mechanisms of ozone damage in plants, researchers have isolated Arabidopsis thaliana mutants that exhibit hypersensitivity to ozone exposure (4), including several mutants with higher stomatal conductance than wild type (WT), such as radical-induced cell death1 (rcd1), rcd2, and ozone sensitive1/slow anion channel-associated1 (ozs1/slac1) (5–8). In addition, the Arabidopsis ecotype Cvi-0, which has higher stomatal conductance than other ecotypes, also exhibits hypersensitivity to ozone compared with other ecotypes (9, 10). These observations indicate that stomatal movement has a strong relationship to ozone sensitivity, likely because stomata regulate the first step of ozone absorption into plant cells.
Stomatal movements are controlled by turgor pressure in guard cells in response to environmental stimuli such as CO2 concentration, light intensity, humidity, and air pollutants (11). Hyperpolarization of the plasma membrane caused by the H+-ATPases promotes stomatal opening through activation of the voltage-gated K+ inward-rectifying channels (K+in), encoded by K+ CHANNEL IN ARABIDOPSIS THALIANA1 (KAT1), KAT2, and ARABIDOPSIS K TRANSPORTER 1 (AKT1), which induce water entry into guard cells (11, 12). By contrast, depolarization of the plasma membrane activates outward-rectifying K+ channels (6–8), which induce water efflux from guard cells, resulting in stomatal closure (11, 12).
Recent studies have further elucidated the signaling pathways that regulate guard cell movement; these pathways include second messengers, plant hormones, and transcription factors (11–13). The MYB60 and MYB61 transcription factors regulate light-induced stomatal opening and dark-induced stomatal closure, respectively (14–16). MYB44, MYB15, ERF7, and NFYA5 also participate in stomatal movement in A. thaliana (17–20). The bHLH transcription factors ABA-RESPONSIVE KINASE SUBSTRATES1 (AKS1), AKS2, and AKS3 function as positive regulators of stomatal opening (21). They facilitate K+ uptake through positive regulation of KAT1, but abscisic acid (ABA) represses their transcriptional activation activity through phosphorylation (21). The aks1 aks2 mutants exhibit a weak closed-stomata phenotype and weak down-regulation of KAT1 (21). These phenotypes suggest that other, functionally redundant transcription factor(s) may regulate the expression of the genes for K+in channels and stomatal opening. Modulation of stomatal movement using transcription factors may provide a useful strategy to confer ozone tolerance, because a number of known transcription factors act as master regulators of various cellular processes. However, screens for changes in the response to ozone have yet to identify mutant lines or transcription factors that confer tolerance to ozone, possibly because of redundant functions of these key genes.
In this study, we screened a set of transgenic Arabidopsis lines expressing chimeric repressors for Arabidopsis transcription factors and found that lines expressing the chimeric repressors for GOLDEN 2-LIKE1 (GLK1) and GLK2, which regulate chloroplast development (22–26) and exhibit remarkable ozone tolerance and a closed-stomata phenotype. These transgenic plants also show down-regulation of gene expression and activity of K+in channels and of other genes involved in stomatal movement. We propose here that GLKs act as positive regulators of stomatal movement and that the guard cell-specific expression of the chimeric GLK repressor can confer tolerance to air pollutants, possibly providing a useful tool for crop protection.
Results
Plants Expressing the Chimeric Repressors for GLKs Exhibit Remarkable Tolerance to Ozone.
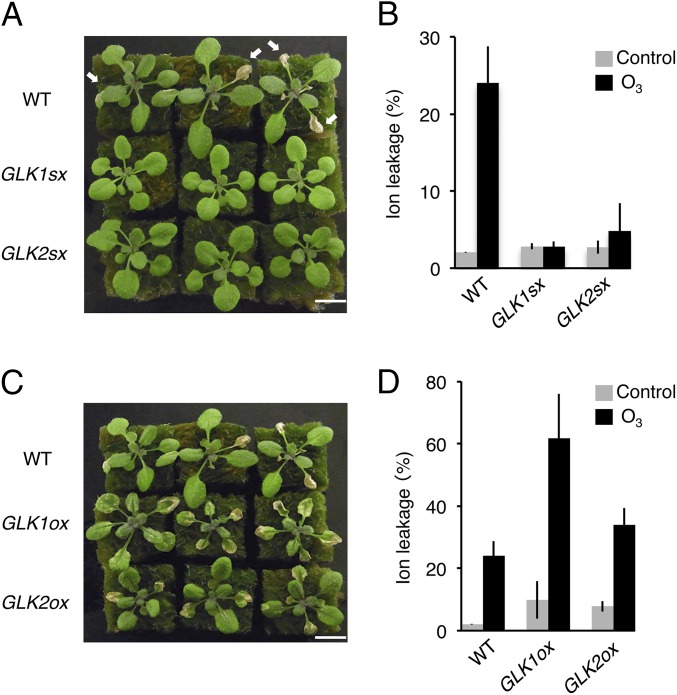
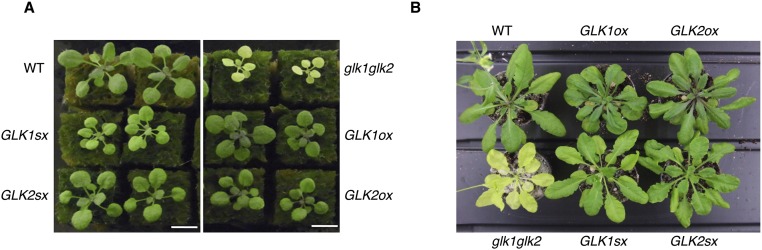
The chimeric repressor gene silencing technology (CRES-T) system converts a transcriptional activator into a strong repressor by fusion to the SRDX repression domain; this process induces a phenotype similar to loss-of-function alleles (27) and can also confer tolerance to various abiotic stresses (28). To identify a chimeric repressor that confers tolerance to ozone stress, we screened a set of transgenic Arabidopsis-expressing chimeric repressors for Arabidopsis transcription factors, termed CRES-T lines (27), exposing the plants to 0.3 ppm ozone for 7 h. We found that the CRES-T lines for the two redundant GARP family transcription factors, GOLDEN 2-LIKE1 (GLK1) and GLK2 (35S:GLK1/2-SRDX), which regulate chloroplast development (22–26), exhibited tolerance to ozone exposure, showing 12-fold less ion leakage than the WT plants (Fig. 1 A and B). By contrast, plants overexpressing GLK1 or GLK2 (35S:GLK1/2) exhibited hypersensitivity to ozone, with higher ion leakage than WT (Fig. 1 C and D).
Fig. 1.
Sensitivity to ozone of GLK1/2 transgenic Arabidopsis. (A) Rosette plants of WT and 35S:GLK1/2-SRDX (GLK1sx and GLK2sx) 1 d after exposure to 0.3 ppm O3 for 7 h. Arrows indicate the damaged leaves. (B) Ion leakage of WT and GLK1/2sx plants. The gray and black bars represent plants exposed to fresh air or O3, respectively. The average of three biological replicates (three plants per replicate) is shown. Error bars represent SD. (C) Rosette plants of WT and 35S:GLK1/2 (GLK1ox and GLK2ox) 1 d after exposure to 0.3 ppm O3 for 7 h. (D) Ion leakage of WT and GLK1/2ox plants. The gray and black bars represent plants exposed to fresh air or O3, respectively. The average of three biological replicates (three plants per replicate) is shown. Error bars represent SD.
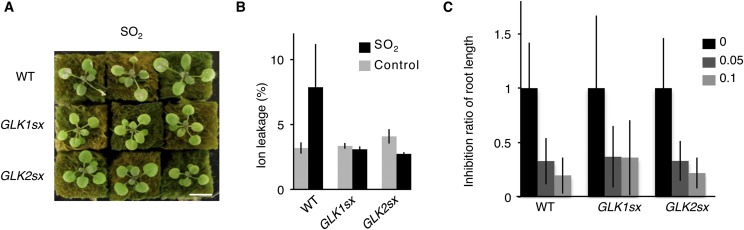
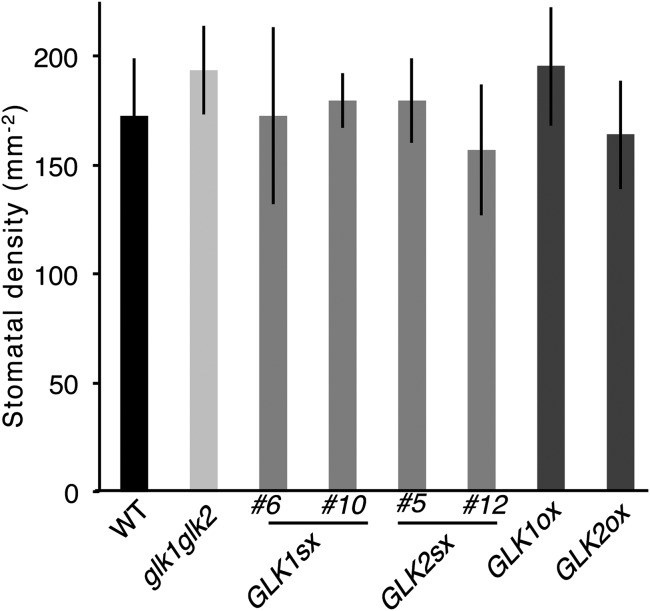
In addition to ozone, 35S:GLK1/2-SRDX seedlings also exhibited tolerance to sulfur dioxide, an oxidative stress reagent similar to ozone. However, the 35S:GLK1/2-SRDX seedlings showed no resistance to aqueous paraquat (methyl viologen) in the medium, as measured by the inhibition of root elongation (Fig. S1). These results suggest that the tolerance of 35S:GLK1/2-SRDX plants to ozone and sulfur dioxide was due to an alteration of gas absorption through stomata, not to an alteration of sensitivity to oxidative stress. The 35S:GLK1/2-SRDX plants showed a similar stomatal density as the WT plants (Fig. S2), indicating that their differing sensitivity to ozone may be attributable to an alteration of stomatal aperture.
Fig. S1.
Sensitivity to sulfur dioxide (SO2) and paraquat of WT and 35S:GLK1/2-SRDX (GLK1/2sx) plants. (A) Rosette plants of WT and GLK1sx and GLK2sx transgenic Arabidopsis 1 d after 11 h of exposure to 1.0 ppm SO2. (B) Ion leakage of WT and GLK1/2sx transgenic Arabidopsis. The gray and black bars represent plants exposed to fresh air or SO2, respectively. The average of three biological replicates (three plants per replicate) is shown. Error bars represent SD. (C) The inhibition of root elongation of WT and GLK1/2sx plants grown on MS medium containing 0, 0.05, and 0.1 µM paraquat. The average of 35–40 plants is shown. Error bars represent SD.
Fig. S2.
WT, glk mutants, and GLK1/2 transgenic plants have similar stomatal density. Stomatal density of WT, glk1 glk2 double mutants (glk1glk2), and 35S:GLK1/2-SRDX (GLK1/2sx) and 35S:GLK1/2 (GLK1/2ox) transgenic plants. The average of five replicates is shown. Error bars represent SD.
GLKs Affect Stomatal Aperture.
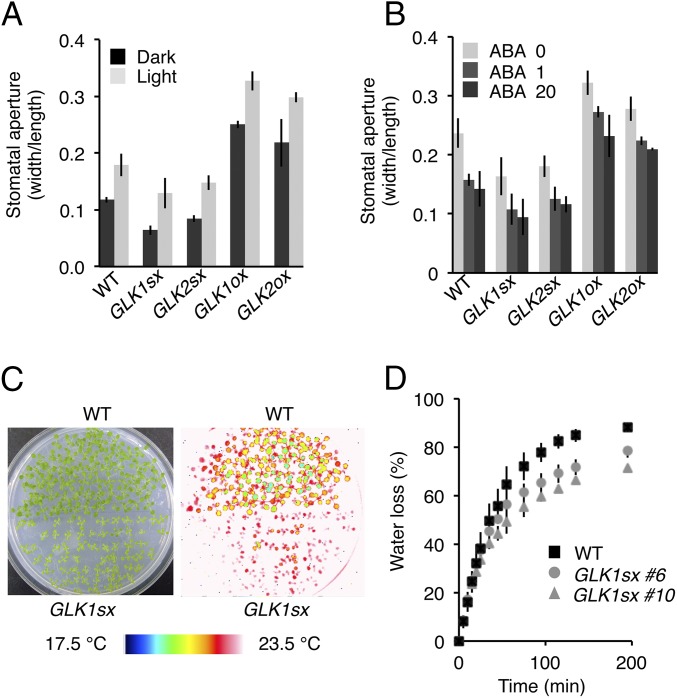
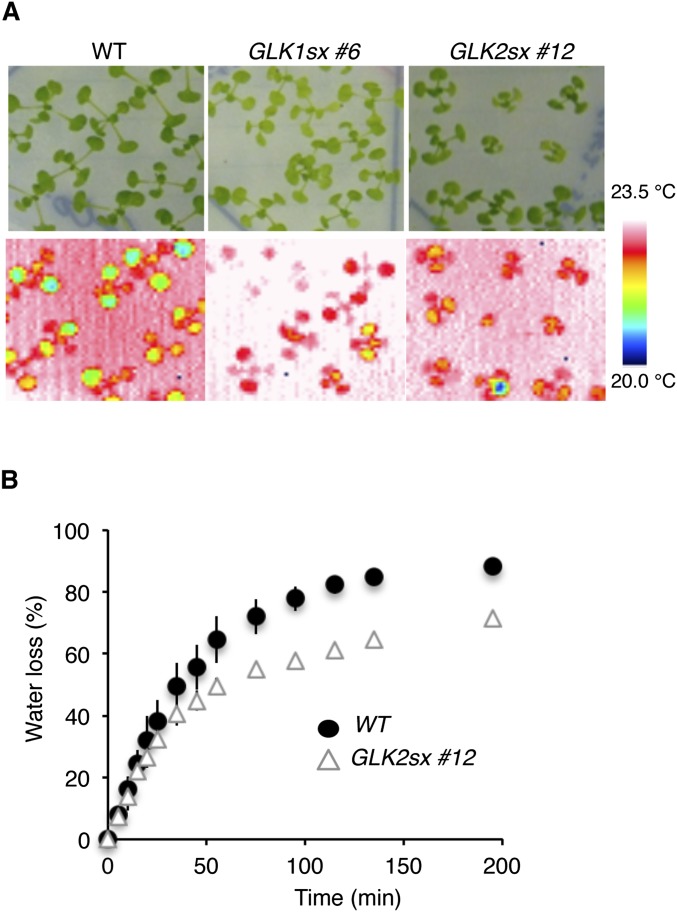
We measured the stomatal aperture (as the ratio of width/length) of each plant grown in normal light conditions or kept in dark conditions for 1 d before the experiments. We found, as expected, that the 35S:GLK1/2-SRDX plants had much smaller apertures than WT, with average apertures of 55% and 72% (for 35S:GLK1-SRDX plants) and 72% and 83% (for 35S:GLK2-SRDX plants) of that of WT under dark and light conditions, respectively (Fig. 2A). By contrast, 35S:GLK1/2 plants had much larger apertures than WT, with average apertures of 210% and 186% (for 35S:GLK1 plants) and 183% and 166% (for 35S:GLK2 plants) of that of WT under dark and light conditions, respectively (Fig. 2A). Thermography monitoring also revealed that the leaf surface temperature of 35S:GLK1/2-SRDX plants was 1.5 °C higher than that of WT and that the water loss of detached seedlings was 13% lower than WT (Fig. 2 C and D and Fig. S3), showing that the 35S:GLK1/2-SRDX plants have low transpiration rates. The stomata of 35S:GLK1/2-SRDX plants closed in response to abscisic acid (ABA) treatment, similar to WT, when the leaf epidermis was treated with 1 or 20 µM ABA for 2.5 h under light (Fig. 2B), indicating that the alteration of stomatal apertures in 35S:GLK1/2-SRDX plants does not appear to be attributable to a change in sensitivity to ABA.
Fig. 2.
GLK1/2 control stomatal opening. (A) Stomatal aperture of epidermal fragments from the plants under dark conditions, in which plants were incubated 1 d before measurements (dark), and light conditions (light). (B) Stomatal aperture of epidermal fragments incubated with 0, 1, and 20 µM ABA under white light for 2.5 h. The average of three independent experiments is shown. Approximately 150 stomata in total (n = 3 independent experiments, 50 stomata per experiment) were analyzed in each line. Error bars represent SD. (C) Thermal images of WT (Upper) and GLK1sx (Lower) plants grown on MS medium, showing the higher temperature of GLK1sx plants. (D) Water loss in WT and GLK1sx plants for the evaluation of transpiration rate. The average of three biological replicates (five to seven plants per replicate) is shown. Error bars represent SD.
Fig. S3.
Transpiration rate of WT and 35S:GLK1/2-SRDX (GLK1/2sx) plants. (A) Thermography images of WT, GLK1sx (no. 6), and GLK2sx (no. 12) transgenic Arabidopsis grown on a same culture plate. (B) Time course of water loss ratio from detached WT and GLK2sx seedlings grown on MS medium. The average of three biological replicates (five to seven plants per replicate) is shown. Error bars represent SD.
GLKs Affect the Expression of Genes Related to Stomatal Movement.
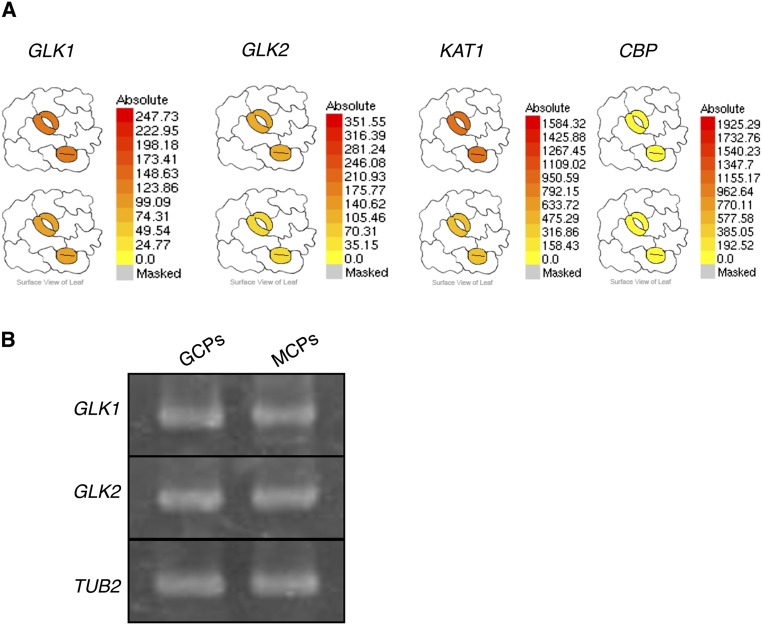
We detected the expression of GLK1 and GLK2 in guard cells and mesophyll cells (Fig. S4), and our observations were consistent with the previously reported microarray data (29). To examine the possible mechanisms of the closed-stomata phenotype of 35S:GLK1/2-SRDX plants, we performed microarray experiments to find genes regulated by GLK1. Among transcripts of numerous genes changed in abundance in 35S:GLK1-SRDX seedlings, we identified a set of genes related to stomatal movement (listed in Table S1). In 35S:GLK1-SRDX plants, we found significant down-regulation (P < 0.06) of KAT1, KAT2, and AKT1, which encode Shaker-type K+in channels (11, 12). In addition, BLUE LIGHT SIGNALING1 (BLUS1) (30), which encodes a kinase phosphorylated by phototropins and regulates blue light-induced stomatal opening through activation of H+-ATPase, and FLOWERING LOCUS T (FT) (31), which regulates stomatal opening, were also down-regulated (P < 0.07).
Fig. S4.
Expression of GLK1 and GLK2 in guard cells. (A) Relative expression of GLK1 and GLK2 in guard cells displayed by Arabidopsis eFP browser (www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). KAT1, At5g46240, leaf guard cell marker gene; CBP, At4g33050, mesophyll cell marker gene. (B) RT-PCR analyses of GLK1 and GLK2 in guard cell protoplasts (GCP) and mesophyll cell protoplasts (MCP) from WT.
Table S1.
Expression ratio of genes related to stomatal movements between 35S:GLK1-SRDX and WT seedlings in microarray analysis
| Common name | AGI code | Ratio (GLK1sx/WT) | P |
| Light-sensing | |||
| PHOT1 | AT3G45780 | 0.99 | 0.881 |
| PHOT2 | AT5G58140 | 1.20 | 0.319 |
| CRY1 | AT4G08920 | 1.19 | 0.133 |
| CRY2 | AT1G04400 | 1.21 | 0.034 |
| Circadian rhythm | |||
| ELF3 | AT2G25930 | 1.26 | 0.066 |
| FT | AT1G65480 | 0.15 | 0.046 |
| GI | AT1G22770 | 1.10 | 0.256 |
| CO | AT5G15840 | 0.77 | 0.550 |
| TSF | AT4G20370 | 1.02 | 0.875 |
| Protein kinase | |||
| BLUS1 | AT4G14480 | 0.66 | 0.063 |
| Phosphatases | |||
| ToPP1 | AT2G29400 | 0.98 | 0.789 |
| PRSL1 | AT4G40100 | 0.50 | 0.426 |
| H+ATPase | |||
| AHA1 | AT2G18960 | 1.23 | 0.013 |
| AHA2 | AT4G30190 | 0.72 | 0.101 |
| AHA5 | AT2G24520 | 1.08 | 0.33 |
| K+ and anion channels (and related genes) | |||
| KAT1 | AT5G46240 | 0.28 | 0.001 |
| KAT2 | AT4G18290 | 0.74 | 0.059 |
| AKT1 | AT2G26650 | 0.56 | 0.009 |
| AKT2 | AT4G22200 | 0.87 | 0.645 |
| KC1 | AT4G32650 | 0.92 | 0.677 |
| CIPK23 | AT1G30270 | 0.87 | 0.373 |
| CBL1 | AT4G17615 | 0.94 | 0.653 |
| CBL9 | AT5G47100 | 0.88 | 0.229 |
| SLAC1 | AT1G12480 | 0.97 | 0.769 |
| SLAH3 | AT5G24030 | 1.12 | 0.616 |
| GORK | AT5G37500 | 0.88 | 0.128 |
| ABA signalings | |||
| PYL9 | AT1G01360 | 0.94 | 0.576 |
| ABI1 | AT4G26080 | 0.92 | 0.373 |
| ABI2 | AT5G57050 | 1.16 | 0.041 |
| OST1 | AT4G33950 | 1.11 | 0.396 |
| CPK21 | AT4G04720 | 1.35 | 0.010 |
| AKS1 | AT1G51140 | 1.17 | 0.085 |
| AKS2 | AT1G05805 | 0.97 | 0.801 |
AGI, Arabidopsis Genome Initiative.
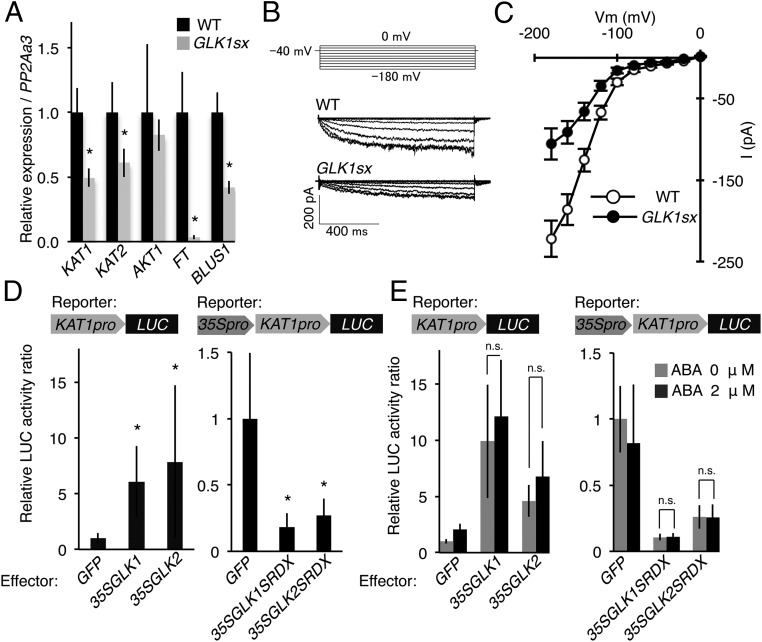
We analyzed the expression of KAT1, KAT2, AKT1, BLUS1, and FT by quantitative (q)RT-PCR using RNA isolated from stomata-rich epidermal cells and confirmed the reduction of expression of these genes (P < 0.05), except for AKT1 in 35S:GLK1-SRDX plants (Fig. 3A and Table S2). Expression of FT was remarkably suppressed in 35S:GLK1-SRDX plants (P < 0.01). These expression profiles suggest that the severe closed-stomata phenotype of 35S:GLK1/2-SRDX plants may be caused by a combination of different effects; among these effects are the reduction of blue light signaling and K+in channel activity in guard cells.
Fig. 3.
GLK1/2 regulate K+ channel genes. (A) Relative expression of KAT1, KAT2, AKT1, FT, and BLUS1 determined by qRT-PCR, using RNA extracted from guard cell-enriched epidermis. The average of four biological replicates is shown. Error bars represent SD. *P < 0.05. (B) The whole-cell inward K+ currents in response to membrane potentials by voltage protocol (Upper), stepped from a holding potential of −40 mV to pulse potentials from 0 to −180 mV in a 20-mV decrement in guard cell protoplasts of WT and GLK1sx plants. (C) Steady-state current–voltage relationship in guard cells of WT (n = 18 experiments) and GLK1sx (n = 8 experiments) plants. Error bars represent SE. (D) Relative luciferase activities after cobombardment of Arabidopsis leaves with 35S:GLK1/2 effectors and the KAT1pro:LUC reporter construct and 35S:GLK1/2-SRDX effectors and the 35S-KAT1pro:LUC reporter construct, respectively. The luciferase activity is shown as the relative ratio to the value obtained by the combination of 35S:GFP effector (control) and each reporter construct. The average of six replicates is shown. Error bars represent SD. *P < 0.05. (E) Relative luciferase activities after cobombardment of Arabidopsis protoplasts with 35S:GLK1/2 effectors and the KAT1pro:LUC reporter construct and 35S:GLK1/2-SRDX effectors and the 35S-KAT1pro:LUC reporter construct, respectively. The black and gray bars indicate samples treated with or without 2 μM ABA, respectively. All luciferase activities are shown as the relative ratio to the value obtained by the combination of 35S:GFP effector (control) and each reporter construct. The average of five replicates is shown. The relative luciferase activity of individual transient assay is shown in Table S4. Error bars represent SD. n.s., not significant (P > 0.05).
Table S2.
Individual data of qRT-PCR analysis in Fig. 3A
| Replicate | KAT1 | KAT2 | AKT1 | FT | BLUS1 |
| WT | |||||
| Biological rep.1 | 1.010 | 0.998 | 0.822 | 0.964 | 1.058 |
| Biological rep.2 | 0.864 | 0.770 | 0.691 | 0.651 | 0.884 |
| Biological rep.3 | 0.859 | 0.906 | 0.700 | 0.977 | 0.866 |
| Biological rep.4 | 1.267 | 1.326 | 1.787 | 1.409 | 1.192 |
| Average | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| SD | 0.191 | 0.237 | 0.528 | 0.311 | 0.154 |
| GLK1SRDX | |||||
| Biological rep.1 | 0.496 | 0.533 | 0.678 | 0.053 | 0.467 |
| Biological rep.2 | 0.578 | 0.632 | 0.779 | 0.024 | 0.412 |
| Biological rep.3 | 0.500 | 0.753 | 0.887 | 0.040 | 0.353 |
| Biological rep.4 | 0.405 | 0.517 | 0.953 | 0.019 | 0.452 |
| Average | 0.495 | 0.609 | 0.824 | 0.034 | 0.421 |
| SD | 0.071 | 0.109 | 0.121 | 0.016 | 0.051 |
The average of the relative expression values of each gene in WT is set as 1. rep., replicate.
Table S4.
Relative luciferase activity of each transient expression analysis shown in Fig. 3E
| Reporter and replicate | Effector | |||||
| KAT1pro:LUC | 35S:GFP | 35S:GFP | 35S:GLK1 | 35S:GLK1 | 35S:GLK2 | 35S:GLK2 |
| ABA, 2 μM | — | + | — | + | — | + |
| Rep.1 | 0.76 | 1.86 | 10.57 | 14.47 | 5.79 | 5.65 |
| Rep.2 | 1.12 | 2.57 | 2.08 | 3.42 | 4.84 | 5.96 |
| Rep.3 | 0.93 | 2.62 | 15.22 | 12.60 | 4.13 | 7.50 |
| Rep.4 | 0.87 | 1.52 | 13.08 | 16.49 | 5.85 | 11.65 |
| Rep.5 | 1.32 | 1.73 | 8.53 | 13.53 | 2.40 | 3.05 |
| Average | 1.00 | 2.06 | 9.90 | 12.10 | 4.60 | 6.76 |
| SD | 0.22 | 0.51 | 5.05 | 5.06 | 1.42 | 3.17 |
| 35S:KAT1pro:LUC | 35S:GFP | 35S:GFP | 35S:GLK1-SRDX | 35S:GLK1-SRDX | 35S:GLK2-SRDX | 35S:GLK2-SRDX |
| ABA, 2 μM | — | + | — | + | — | + |
| Rep.1 | 1.30 | 1.35 | 0.13 | 0.12 | 0.22 | 0.28 |
| Rep.2 | 1.09 | 1.24 | 0.13 | 0.15 | 0.41 | 0.41 |
| Rep.3 | 0.78 | 0.48 | 0.09 | 0.07 | 0.18 | 0.20 |
| Rep.4 | 0.70 | 0.61 | 0.07 | 0.10 | 0.23 | 0.15 |
| Rep.5 | 1.12 | 0.41 | 0.11 | 0.11 | 0.26 | 0.24 |
| Average | 1.00 | 0.82 | 0.11 | 0.11 | 0.26 | 0.26 |
| SD | 0.25 | 0.44 | 0.03 | 0.03 | 0.09 | 0.10 |
The average of the activity of 35S:GFP effector without ABA treatment is set as 1. —, without; +, with; Rep., replicate.
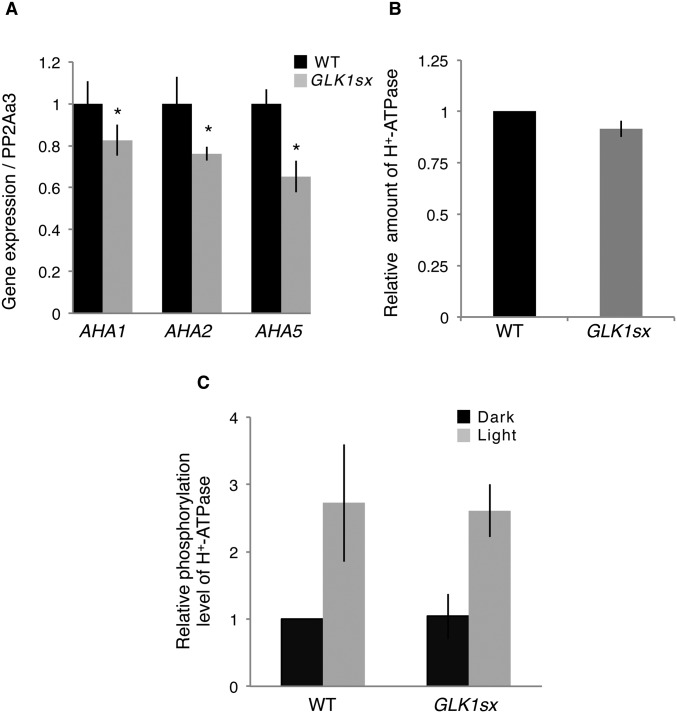
Because blus1 and ft knockout mutants showed a closed-stomata phenotype attributable to the reduction of the H+-ATPase activity (30, 31), we analyzed the expression of the H+-ATPase genes and the activity of H+-ATPases in guard cells using an immunohistochemical method (32) by monitoring the blue light-induced phosphorylation of the penultimate Thr of these H+-ATPases. Results of qRT-PCR using RNA isolated from stomata-rich epidermal cells showed that the expression of three H+-ATPase genes, Arabidopsis H+-ATPase 1 (AHA1), AHA2, and AHA5, which are mainly expressed in guard cells (33), was slightly but significantly reduced in 35S:GLK1-SRDX plants (P < 0.05; Fig. S5A). By contrast, the level of blue light-induced phosphorylation and the protein levels of guard cell H+-ATPases did not significantly differ between 35S:GLK1-SRDX and WT plants (Fig. S5 B and C), implying that the activities of H+-ATPases may not be directly involved in the closed-stomata phenotype of 35S:GLK1/2-SRDX plants.
Fig. S5.
Gene expression and level of protein amount and phosphorylation of guard cell H+-ATPase. (A) Relative expression of AHA1, AHA2, and AHA5 by real-time RT-PCR, using RNA extracted from guard cell-enriched epidermis. The average of four replicates is shown. Error bars represent SD. *P < 0.05. (B) Relative amount of H+-ATPase by quantification of fluorescence images of stomata in the epidermis using anti–H+-ATPase. The average of three independent experiments is shown (n = 30). Error bars represent SD. (C) Relative phosphorylation level of H+-ATPase by quantification of fluorescence images of stomata in the epidermis using anti-pThr. The average of three independent experiments is shown (n = 30). Error bars represent SD.
GLKs Regulate K+in-Related Genes.
The expression of major K+in channel genes was reduced in 35S:GLK1-SRDX plants (Fig. 3A); therefore, we analyzed the activity of K+in channels using patch-clamp experiments with protoplasts isolated from guard cells. The average of the steady-state whole-cell current at −180 mV was −221.9 ± 22.1 pA in WT but was −105.8 ± 18.9 pA (P = 0.0071) in 35S:GLK1-SRDX plants (Fig. 3 B and C), indicating that the activity of K+in channels of 35S:GLK1-SRDX plants was 52.3% lower than the activity in WT. Our results indicate that GLK1-SRDX down-regulates the gene expression and activity of K+in channels.
The A. thaliana KAT1 locus contains three GLK-binding sites in the 3-kb 5′ region upstream of the translation initiation site. Those are two CACGTG, which is also recognized as the G-box (25), and CCAATC. Transient expression analyses in Arabidopsis leaves revealed that the luciferase reporter gene (LUC) driven by the 5′ upstream region of KAT1 (KAT1pro:LUC) was up-regulated when coexpressed with 35S:GLK1/2 (Fig. 3D; P < 0.05). Also, coexpression with 35S:GLK1/2-SRDX down-regulated the activity of the 35S-KAT1pro:LUC reporter gene, in which the enhancer region of the CaMV 35S promoter was fused to the upstream region of KAT1pro:LUC (Fig. 3D; P < 0.05). These results suggest that KAT1 may be a direct target of GLK1/2.
To analyze whether ABA affects GLK1 activity, we treated protoplasts with 2 µM ABA after cotransformation with the effector plasmid and reporter gene and found that ABA did not affect the activation by 35S:GLK1/2 or the repression by 35S:GLK1/2-SRDX of the KAT1pro:LUC or 35S-KAT1pro:LUC reporter genes (Fig. 3E; P > 0.05). These results suggest that GLKs regulate KAT1 expression independently from ABA signaling.
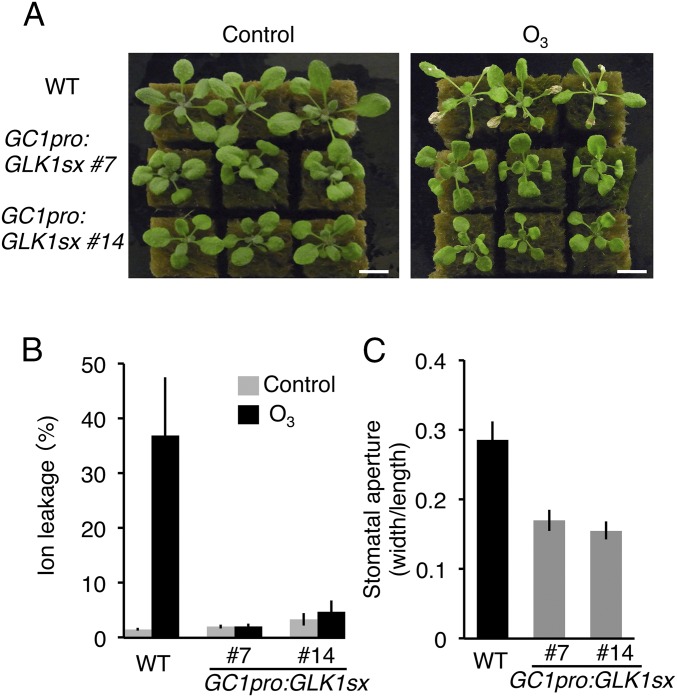
Guard Cell-Specific Expression of GLK1-SRDX Induces Ozone Tolerance and a Closed-Stomata Phenotype.
The glk1 glk2 double mutants exhibit a pale green phenotype due to inhibition of chloroplast development (23). The 35S:GLK1/2-SRDX plants were paler green than WT but much darker and larger than the glk1 glk2 mutants (Fig. S6). To limit the activity of GLK1-SRDX specifically to guard cells, we expressed GLK1-SRDX under the control of the GC1 (At1g22690) promoter (GC1pro:GLK1-SRDX), which shows strong activity in guard cells (34). GC1pro:GLK1-SRDX plants exhibited a closed-stomata phenotype and tolerance to ozone, similar to the 35S:GLK1/2-SRDX plants (Fig. 4). However, unlike the 35S:GLK1-SRDX plants or glk1 glk2 mutants, the GC1pro:GLK1-SRDX plants exhibited a green color similar to WT, showing that GC1pro:GLK1-SRDX did not affect chloroplast development in mesophyll cells (Fig. 4A). In addition, these results indicate that the closed-stomata phenotype of 35S:GLK1-SRDX plants was not attributable to a defect in the mesophyll chloroplasts.
Fig. S6.
Morphological phenotype of transgenic Arabidopsis expressing GLKs. Two-week-old (A) and 5-wk-old (B) WT, glk1 glk2, 35S:GLK1/2-SRDX (GLK1/2sx) and 35S:GLK1/2 (GLK1/2ox) transgenic Arabidopsis grown on rock wool (A) or soil (B).
Fig. 4.
Phenotype of GC1pro:GLK1-SRDX plants. (A) WT and two independent lines of GC1pro:GLK1-SRDX (GC1pro:GLK1sx) 1 d after exposure to fresh air (Left) or 0.3 ppm O3 (Right) for 7 h. (B) Ion leakage of WT and GC1pro:GLK1sx plants exposed to fresh air and 0.3 ppm O3 for 7 h. The gray and black bars represent plants exposed to fresh air or O3, respectively. The average of three biological replicates (three plants per replicate) is shown. Error bars represent SD. (C) Stomatal aperture of WT and GC1pro:GLK1sx plants. The average of three independent experiments is shown. Approximately 150 stomata in total (n = 3 independent experiments, 50 stomata per experiment) were analyzed in each line. Error bars represent SD.
Discussion
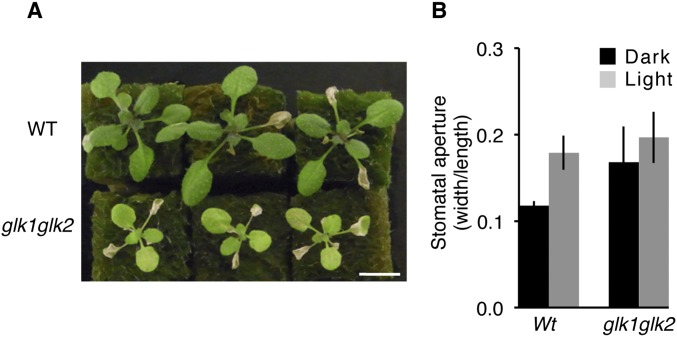
In this study, we showed that transgenic Arabidopsis plants that expressed 35S:GLK1/2-SRDX exhibited a closed-stomata phenotype and tolerance to ozone exposure, whereas 35S:GLK1/2 plants exhibited an open-stomata phenotype and hypersensitivity to ozone. We demonstrated that the activity of K+in channels was down-regulated in 35S:GLK1-SRDX plants, and KAT1 appears to be a direct target of GLKs (Fig. 3), suggesting that GLKs act as positive regulators of K+in channel genes and stomatal movement. Similar to the GLKs, the AKSs transcription factors positively regulate stomatal opening and activate KAT1 expression (21). The aks1 aks2 double mutants have reduced K+in channel activity and lower rates of light- or fusicoccin (Fc)-dependent stomata opening; AKSs also function in an ABA-dependent manner and are regulated by phosphorylation (21). In contrast to AKSs, ABA does not appear to affect the transcriptional activity of GLK1 (Fig. 3). Although GLKs and AKSs may act redundantly in the regulation of genes for K+in channels, each of the transcription factors appears to be regulated by a different signaling pathway. This putative functional redundancy might explain why aks1 aks2 mutants exhibit a weak closed-stomata phenotype (21) and why glk1 glk2 double mutants do not exhibit a clear closed-stomata phenotype or ozone tolerance (Fig. S7). On the other hand, 35S:GLK1/2-SRDX plants exhibit a severe closed-stomata phenotype. This observation is probably because the chimeric repressor dominantly suppresses the target genes even in the presence of endogenous or functionally redundant transcription factors (27).
Fig. S7.
Sensitivity to ozone of glk1 glk2 plants. (A) Rosette plants of WT and glk1 glk2 double mutants (glk1glk2) 1 d after exposure to 0.3 ppm O3 for 7 h. (B) Stomatal aperture of WT and glk1 glk2 plants grown on solid MS medium. Stomatal aperture of epidermal fragments from the plants under dark conditions, in which plants were incubated 1 d before measurements (dark), and light conditions (light). Approximately 150 stomata in total (n = 3 independent experiments, 50 stomata per experiment) were analyzed in each line. Error bars represent SD.
The kat1 single mutant does not affect or only slightly inhibits Fc-dependent stomatal opening (21, 35). By contrast, the dominant negative form of KAT1 or KAT2 impaired the activity of K+in channels in guard cells and clearly inhibited light-dependent stomata opening (36, 37), indicating that the activity of K+in channels in guard cells is essential and might be much higher than the threshold necessary for stomata opening (35–38). Although the severe closed-stomata phenotype of 35S:GLK1/2-SRDX plants might result from combinatorial effects of multiple genes, we suggest that their closed-stomata phenotype in light conditions was attributable, at least in part, to a decrease in the activity of K+in channels and a reduction of the expression of K+in channel genes in guard cells as shown in aks1 aks2 mutants (21). We found that the expression of numerous genes was affected in 35S:GLK1-SRDX seedlings. However, we did not find obvious differences in the expression of ABA-responsive genes in our microarray data, probably because ABA sensitivity is not altered in 35S:GLK1-SRDX plants (Figs. 2B and 3E). Further analysis of the genes affected by GLK1-SRDX may provide new insights into the mechanisms of stomatal movements regulated by GLK1/2.
The mechanisms of the transcriptional regulation of stomatal aperture are not fully understood. We propose that GLK1/2 positively regulate stomata opening, likely by activating the expression of K+in channel genes. Light induces the expression of GLK1 and GLK2 (23) and ABA decreases the expression of GLK2 in guard cells (39), similar to the behavior of KAT1 and consistent with the movements of stomata. Because an increase in K+in channel activity does not produce additional stomatal opening (21, 36, 37, 40), the open-stomata phenotype of 35S:GLK1/2 plants indicates that GLK1/2 may regulate the expression of genes necessary for stomatal opening other than those for K+in channels (Fig. 3A), which may induce the more-severe closed-stomata phenotype of 35S:GLK1/2-SRDX plants. Unknown factor(s) that act downstream of GLKs may regulate stomatal movement, because the activity of H+-ATPases does not seem to be directly involved in the closed-stomata phenotype of 35S:GLK1/2-SRDX plants (Fig. S5). Guard cell chloroplasts are essential for blue light-dependent stomatal opening (41), implying a relationship between the activity of GLKs and the regulation of stomatal movements. Further study of the functions of GLK1/2 in stomatal movement will be necessary to provide new insights into the relationship between chloroplast development and transpiration, which affect photosynthetic efficiency.
Stomata regulate transpiration rate and absorption of gasses. Modification of stomatal movements could improve both the efficiency of photosynthesis and the tolerance to air pollutants. Transcription factors can be useful tools for the manipulation of plant traits because transcription factors regulate multiple genes and some act as master regulators of phenotype. Regulation of the expression of GLK1-SRDX specifically in guard cells could prove useful to create crops that are tolerant to air pollutants.
Materials and Methods
Plant Materials.
A. thaliana ecotype Col-0 were grown at 23–25 °C, with a photoperiod of 16 h/8 h light/dark and 14 h/10 h light/dark, on solid Murashige and Skoog (MS) medium (containing 0.8% agar, 0.5% sucrose, and 0.5 g/L Mes, pH 5.7 by KOH) and on rock wool, respectively. The glk1 glk2 double mutant and 35S:GLK1/2 Arabidopsis lines were provided by Langdale (23, 24).
Construction of Plasmids.
The protein-coding regions and the 5′ upstream promoter regions of genes were amplified from a cDNA library or from genomic DNA of A. thaliana with the appropriate primer sets (Table S3). The GLK1-SRDX and GLK2-SRDX transgenes were constructed as described previously (42). To prepare the GC1pro:GLK1-SRDX construct, 1,745 bp of the GC1 promoter region and the GLK1 protein coding region were cloned into the pDONRG_P4P1R and pDONR207 vectors (42), respectively, using the Gateway BP reaction (Life Technologies), and then were assembled in the R4pGWB5_SRDX multisite gateway vector (43) by the Gateway LR reaction. To prepare the 35S:GLK1, 35S:GLK2, 35S:GLK1-SRDX, and 35S:GLK2-SRDX effector plasmids for transient effector-reporter experiments, protein-coding regions of GLK1 and GLK2 were cloned into the SmaI site of p35SG or p35SSRDXG (42). To prepare the KAT1pro:LUC and 35S:KAT1pro:LUC reporter plasmids, 2,900 bp of 5′ upstream region of the translation initiation site of KAT1 was cloned into pDONRG_P4P1R (43) using the Gateway BP reaction (Life Technologies) and transferred by the Gateway LR reaction into R4L1pDEST190LUC and 35S_R4L1pDEST190LUC, which has a R4L1 recombination cassette upstream of the TATA box of the p190LUC vector (44) without or with the CaMV 35S enhancer region further upstream, respectively.
Table S3.
Primers used in this study
| Primer name | Target gene name | Sequences (5′– 3′) |
| For semi-qRT-PCR | ||
| AT2G20570-RT-s | GLK1 | CCTCTTCAGCTTCTTCCAAGAAC |
| AT2G20570-RT-as | GLK1 | GCGGTGCTCTAAATCTCGTAG |
| AT5G44190-RT-s | GLK2 | ACAAAACGGTGCGTCGAG |
| AT5G44190-RT-as | GLK2 | ATGCCATACCATATCCCTGTG |
| PP2AA3-F | PP2Aa3 | GCCATTGTAGAACTTGCTGAAGACAGG |
| PP2AA3-R | PP2Aa3 | TCCACCAAGCATGGCCGTATCATG |
| For RT-PCR | ||
| 5G46240Rf1 | KAT1 | GACGCTGAGTATTTCCCACCAA |
| 5G46240Rr1 | KAT1 | GAAGTCCACTGCTCCTGACA |
| 4G18290fRT2 | KAT2 | GAAGCTCCTACTGACCTCTACAT |
| 4G18290rRT2 | KAT2 | TCTCTCCAAATGCATCGCCAACAA |
| 2G26650Rf1 | AKT1 | TGCGCTCCATACCGCTGTAT |
| 2G26650Rr1 | AKT1 | ATCCGCACCTTGCTCCAGAA |
| 1G65480Rf1 | FT | CTGCTACAACTGGAACAACCTTTG |
| 1G65480Rr1 | FT | TTTGCCTGCCAAGCTGTCGAA |
| 4G14480fRT | BLUS1 | TTTTCTGATTACGAGATCAATACGA |
| 4G14480rRT | BLUS1 | GCTTTTGAGAACTTCTTGTTACCC |
| 2G18960Rf1 | AHA1 | GCTTGGGCCAGCTTGTTTGA |
| 2G18960Rr1 | AHA1 | TTTGGCTGCAGACCGTGCAA |
| 4G30190Rf1 | AHA2 | GCAAGCTAAGAGAAGAGCTGAGA |
| 4G30190Rr1 | AHA2 | ACAGTGTAGTGACTGGGAGTTTCA |
| AT2G24520qRTf | AHA5 | GGCTGTTGCAAGACAGGAA |
| AT2G24520qRTr | AHA5 | CGGAGGATCAAAAAGAGGTAAA |
| 1G13320Rf2 | PP2Aa3 | GACCAAGTGAACCAGGTTATTGG |
| 1G13320Rr2 | PP2Aa3 | TACTCTCCAGTGCCTGTCTTCA |
| For plasmid construction | ||
| AT2G20570N | GLK1 | gATGTTAGCTCTGTCTCCGGCGACAAGAGA |
| AT2G20570.1SC | GLK1 | TCAGGCACAAGACGCGGTCGGAGGAACCTC |
| AT5G44190N | GLK2 | gATGTTAACTGTTTCTCCGGCTCCAGTACT |
| AT5G44190.1SC | GLK2 | TCAAGGAAGAGGAGGAACATTAGAAACTCC |
| At1g22690pF1725 | GC1 | GGGGACAACTTTGTATAGAAAAGTTGCTACAAGAAGAGTAAAGATTCAGTAAC |
| At1g22690pR | GC1 | GGGGACTGCTTTTTTGTACAAACTTGGATTTCTTGAGTAGTGATTTTGAAGTAG |
| AT5G46240pF2700 | KAT1 | GGGGACAACTTTGTATAGAAAAGTTGCCAGAAAAGAAAAAAAATTGTAGTGGA |
| AT5G46240pR | KAT1 | GGGGACTGCTTTTTTGTACAAACTTGGCTTTTTTTCTTTTTGGTTGAGAATTTG |
qRT-PCR, quantitative RT-PCR.
O3 and SO2 Treatments and Measurement of Ion Leakage.
Two-week-old seedlings grown on solid MS medium (containing 0.8% agar, 0.5% sucrose, and 0.5 g/L Mes, pH 5.7 by KOH) in 150-mm Petri dishes or rock wool were exposed to 0.3 ppm ozone for 7 h or 1.0 ppm sulfur dioxide for 11 h in a growth chamber at 25 °C and 70% relative humidity under continuous light of 350 µmol m−2 s−1 (photosynthetic photon flux density). Ion leakage was measured with detached second leaves from three individual plants exposed to O3 or SO2 or fresh air as a control, as described previously (6, 45).
Inhibition of Root Elongation by Methyl Viologen.
Transgenic seeds were sown on solid MS medium [containing 1.2% (wt/vol) agar, 0.5% sucrose, and 0.5 g/L Mes, pH 5.7 by KOH] containing various concentrations of methyl viologen hydrate (paraquat; Nakalai tesque). The root length of 14-d-old seedlings was measured.
Analysis of Stomatal Responses.
The transpiration rate of GLK transgenic plants grown on solid MS medium (containing 0.8% agar, 0.5% sucrose, and 0.5 g/L Mes, pH 5.7 by KOH) in 150-mm Petri dishes was evaluated by thermoimaging using a thermal video system (TVS-8500; Nippon Avionics). Analysis of water loss rate was performed using 2-wk-old seedlings detached from MS medium by measuring the fresh weight at the indicated periods of time in a plant growth room at 23 °C under 40% relative humidity. For analyses of stomatal aperture, second rosette leaves of 2-wk-old seedlings grown on solid MS medium (containing 0.8% agar, 0.5% sucrose, and 0.5 g/L Mes, pH 5.7 by KOH) in 150-mm dish were used. The leaf epidermis samples, isolated by blender, were incubated in buffer (5 mM Mes/bis-Tris propane, 50 mM KCl, and 0.1 mM CaCl2, pH 6.5) (46–48) with/without ABA for 2.5 h at 23 °C under white light conditions. All measurements were conducted between 3 and 5 h after turning on the light in the growth chamber.
Isolation of RNA and Analysis of RNA Expression.
Total RNA was isolated from 2-wk-old seedlings grown on MS medium [containing 0.8% agar, 3% sucrose, 1 mL/L Gamborg's vitamin solution (Sigma) and 0.5 g/L Mes, pH 5.7 by KOH], using an RNeasy Plant Mini kit (Qiagen). Epidermis-enriched cells were isolated from blender-treated leaf samples. For qRT-PCR analyses, 0.5 µg of total RNA was subjected to first-strand cDNA synthesis using the PrimeScript RT Master Mix (Takara Bio). qRT-PCR was performed by the SYBR green method using the GoTaq qPCR Master Mix (Promega) and ABI 7500 real-time PCR system (Applied Biosystems), with the appropriate primers (Table S3). Relative amounts of transcripts were calculated by an absolute quantification method using the PP2AA3 gene as an internal control.
Immunohistochemical Staining of the Guard Cell H+-ATPase.
Detection of the phosphorylated H+-ATPase and total H+-ATPase in guard cells was performed according to the immunohistochemical method (32), with some modifications. Epidermal fragments from rosette leaves were fixed with 4% (wt/vol) paraformaldehyde for 2 h at room temperature and then digested with 2% (wt/vol) Driselase (Sigma) and 0.5% (wt/vol) Macerozyme R-10 (Yakult) for 45 min at 37 °C. The tissues were permeabilized with 3% (wt/vol) Triton X-100 for 30 min at room temperature. After the blocking with 3% BSA (Gibco), the samples were treated with anti-phosphorylated penultimate Thr of the H+-ATPase (anti-pThr) or anti-catalytic domain of the H+-ATPase at a dilution of 1:500 as a primary antibody. The stained signals were visualized using Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) as a secondary antibody. The signal intensities were quantified and expressed according to the previous method (32).
Patch-Clamp Analysis.
Guard cell protoplasts were prepared from 4- to 6-wk-old Arabidopsis plants, and the patch-clamp experiments were carried out as described previously (21). The pipette solution contained 30 mM KCl, 70 mM K-Glu, 2 mM MgCl2, 6.7 mM EGTA, 3.35 mM CaCl2, 5 mM ATP, and 10 mM Hepes⋅Tris (pH 7.1). The bath solution contained 30 mM KCl, 40 mM CaCl2, 2 mM MgCl2, and 10 mM Mes⋅Tris (pH 5.5). Osmolality was adjusted to 500 mmol/kg (pipette solution) and 485 mmol/kg (bath solution) with d-sorbitol. The voltage protocol was stepped from a holding potential of −40 mV to pulse potentials from 0 to −180 mV in 20-mV decrements. Leak currents were not subtracted.
Transient Effector–Reporter Analysis.
For transient expression analysis using rosette leaves, 0.8 µg of reporter plasmid, 0.6 µg of effector plasmid, and 0.4 µg of reference plasmids (Renilla LUC gene) were transiently introduced into rosette leaves of 3- to 4-wk-old plants grown on soil with a photoperiod of 12 h/12 h light/dark by particle bombardment. For transient expression analysis using protoplasts, 2.0 µg of reporter plasmid, 3.0 µg of effector plasmid, and 0.4 µg of reference plasmids (Renilla LUC gene) were transiently introduced into protoplasts isolated from 4-wk-old plants by the PEG method (49). Relative luciferase activity was quantified and normalized as described previously (50).
Microarray Analysis.
The microarray experiments were performed using the Agilent Arabidopsis 3 (44,000) microarray (Agilent Technologies) according to the manufacturer’s instructions. Total RNA was isolated from 2-wk-old seedlings grown on MS medium (containing 0.8% agar, 0.5% sucrose, and 0.5 g/L Mes, pH 5.7 by KOH) using an RNeasy Plant Mini kit (Qiagen) and used for microarray experiments. Four biological replicates were tested with a one-color method. Spot signal values were calculated with Feature Extraction version 9.1 software (Agilent). The quality control (QC) value was defined as 1 when a spot passed the “FeatNonUnifOL” filter and as 2 when the spot further passed the “FeatPopnOL” filter. The detection value was defined as 1 when a spot passed the “IsPosAndSignif” filter and as 2 when the spot further passed the “IsWellAboveBG” filter. All signal values were divided by the median value among spots with a QC of 2 to enable comparison with other microarray data. Spot-to-gene conversion was accomplished based on a table provided by The Arabidopsis Information Resource (TAIR) (ftp://ftp.arabidopsis.org/home/tair/Microarrays/Agilent/agilent_array_elements-2010-12-20.txt). The average values were used for the genes corresponding to two or more probes. All data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo) under accession no. GSE42545.
Acknowledgments
We thank Dr. Langdale (Department of Plant Sciences, University of Oxford) for providing seeds of GOLDEN 2-LIKE overexpression lines and glk1 glk2 double-mutant Arabidopsis and N. Ujiie, M. Watanabe, Y. Kimura, M. Oonuki, Y. Kiguchi, S. Takahashi, Y. Sugimoto, A. Kuwazawa, K. Kigoshi, F. Tobe, T. Ishizuka, and Y. Takiguchi for skilled technical assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE42545).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1513093113/-/DCSupplemental.
References
- 1.Pinto DM, Blande JD, Souza SR, Nerg AM, Holopainen JK. Plant volatile organic compounds (VOCs) in ozone (O3) polluted atmospheres: The ecological effects. J Chem Ecol. 2010;36(1):22–34. doi: 10.1007/s10886-009-9732-3. [DOI] [PubMed] [Google Scholar]
- 2.Ludwikow A, Sadowski J. Gene networks in plant ozone stress response and tolerance. J Integr Plant Biol. 2008;50(10):1256–1267. doi: 10.1111/j.1744-7909.2008.00738.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson S, Mills G, Illidge R, Davies WJ. How is ozone pollution reducing our food supply? J Exp Bot. 2012;63(2):527–536. doi: 10.1093/jxb/err317. [DOI] [PubMed] [Google Scholar]
- 4.Kangasjarvi J, et al. Signalling and cell death in ozone-exposed plants. Plant Cell Environ. 2005;28(8):1021–1036. [Google Scholar]
- 5.Ahlfors R, et al. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell. 2004;16(7):1925–1937. doi: 10.1105/tpc.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saji S, et al. Disruption of a gene encoding C4-dicarboxylate transporter-like protein increases ozone sensitivity through deregulation of the stomatal response in Arabidopsis thaliana. Plant Cell Physiol. 2008;49(1):2–10. doi: 10.1093/pcp/pcm174. [DOI] [PubMed] [Google Scholar]
- 7.Vahisalu T, et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature. 2008;452(7186):487–491. doi: 10.1038/nature06608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Negi J, et al. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature. 2008;452(7186):483–486. doi: 10.1038/nature06720. [DOI] [PubMed] [Google Scholar]
- 9.Brosché M, et al. Natural variation in ozone sensitivity among Arabidopsis thaliana accessions and its relation to stomatal conductance. Plant Cell Environ. 2010;33(6):914–925. doi: 10.1111/j.1365-3040.2010.02116.x. [DOI] [PubMed] [Google Scholar]
- 10.Monda K, et al. Environmental regulation of stomatal response in the Arabidopsis Cvi-0 ecotype. Planta. 2011;234(3):555–563. doi: 10.1007/s00425-011-1424-x. [DOI] [PubMed] [Google Scholar]
- 11.Sirichandra C, Wasilewska A, Vlad F, Valon C, Leung J. The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J Exp Bot. 2009;60(5):1439–1463. doi: 10.1093/jxb/ern340. [DOI] [PubMed] [Google Scholar]
- 12.Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hubbard KE, Siegel RS, Valerio G, Brandt B, Schroeder JI. Abscisic acid and CO2 signalling via calcium sensitivity priming in guard cells, new CDPK mutant phenotypes and a method for improved resolution of stomatal stimulus-response analyses. Ann Bot (Lond) 2012;109(1):5–17. doi: 10.1093/aob/mcr252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cominelli E, et al. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15(13):1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 15.Liang YK, et al. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15(13):1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 16.Cominelli E, Galbiati M, Tonelli C. Transcription factors controlling stomatal movements and drought tolerance. Transcription. 2010;1(1):41–45. doi: 10.4161/trns.1.1.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung C, et al. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008;146(2):623–635. doi: 10.1104/pp.107.110981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Z, et al. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J Genet Genomics. 2009;36(1):17–29. doi: 10.1016/S1673-8527(09)60003-5. [DOI] [PubMed] [Google Scholar]
- 19.Song CP, et al. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell. 2005;17(8):2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li WX, et al. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell. 2008;20(8):2238–2251. doi: 10.1105/tpc.108.059444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi Y, et al. bHLH transcription factors that facilitate K⁺ uptake during stomatal opening are repressed by abscisic acid through phosphorylation. Sci Signal. 2013;6(280):ra48. doi: 10.1126/scisignal.2003760. [DOI] [PubMed] [Google Scholar]
- 22.Hall LN, Rossini L, Cribb L, Langdale JA. GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell. 1998;10(6):925–936. doi: 10.1105/tpc.10.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitter DW, Martin DJ, Copley MJ, Scotland RW, Langdale JA. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002;31(6):713–727. doi: 10.1046/j.1365-313x.2002.01390.x. [DOI] [PubMed] [Google Scholar]
- 24.Waters MT, Moylan EC, Langdale JA. GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 2008;56(3):432–444. doi: 10.1111/j.1365-313X.2008.03616.x. [DOI] [PubMed] [Google Scholar]
- 25.Waters MT, et al. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell. 2009;21(4):1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura H, et al. Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol. 2009;50(11):1933–1949. doi: 10.1093/pcp/pcp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003;34(5):733–739. doi: 10.1046/j.1365-313x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- 28.Mito T, Seki M, Shinozaki K, Ohme-Takagi M, Matsui K. Generation of chimeric repressors that confer salt tolerance in Arabidopsis and rice. Plant Biotechnol J. 2011;9(7):736–746. doi: 10.1111/j.1467-7652.2010.00578.x. [DOI] [PubMed] [Google Scholar]
- 29.Bates GW, et al. A comparative study of the Arabidopsis thaliana guard-cell transcriptome and its modulation by sucrose. PLoS One. 2012;7(11):e49641. doi: 10.1371/journal.pone.0049641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takemiya A, et al. Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat Commun. 2013;4:2094. doi: 10.1038/ncomms3094. [DOI] [PubMed] [Google Scholar]
- 31.Kinoshita T, et al. FLOWERING LOCUS T regulates stomatal opening. Curr Biol. 2011;21(14):1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi M, Inoue S, Takahashi K, Kinoshita T. Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 2011;52(7):1238–1248. doi: 10.1093/pcp/pcr072. [DOI] [PubMed] [Google Scholar]
- 33.Ueno K, Kinoshita T, Inoue S, Emi T, Shimazaki K. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 2005;46(6):955–963. doi: 10.1093/pcp/pci104. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Costa A, Leonhardt N, Siegel RS, Schroeder JI. Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods. 2008;4:6. doi: 10.1186/1746-4811-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szyroki A, et al. KAT1 is not essential for stomatal opening. Proc Natl Acad Sci USA. 2001;98(5):2917–2921. doi: 10.1073/pnas.051616698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwak JM, et al. Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in arabidopsis. Plant Physiol. 2001;127(2):473–485. [PMC free article] [PubMed] [Google Scholar]
- 37.Lebaudy A, et al. Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc Natl Acad Sci USA. 2008;105(13):5271–5276. doi: 10.1073/pnas.0709732105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nieves-Cordones M, Caballero F, Martínez V, Rubio F. Disruption of the Arabidopsis thaliana inward-rectifier K+ channel AKT1 improves plant responses to water stress. Plant Cell Physiol. 2012;53(2):423–432. doi: 10.1093/pcp/pcr194. [DOI] [PubMed] [Google Scholar]
- 39.Wang RS, et al. Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics. 2011;12:216. doi: 10.1186/1471-2164-12-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ichida AM, Pei ZM, Baizabal-Aguirre VM, Turner KJ, Schroeder JI. Expression of a Cs(+)-resistant guard cell K+ channel confers Cs(+)-resistant, light-induced stomatal opening in transgenic arabidopsis. Plant Cell. 1997;9(10):1843–1857. doi: 10.1105/tpc.9.10.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suetsugu N, et al. Guard cell chloroplasts are essential for blue light-dependent stomatal opening in Arabidopsis. PloS One. 2014;9(9):e108374. doi: 10.1371/journal.pone.0108374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitsuda N, et al. Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol J. 2006;4(3):325–332. doi: 10.1111/j.1467-7652.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 43.Oshima Y, et al. Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T) Plant Biotechnol. 2011;28(2):201–210. [Google Scholar]
- 44.Mitsuda N, et al. Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol. 2010;51(12):2145–2151. doi: 10.1093/pcp/pcq161. [DOI] [PubMed] [Google Scholar]
- 45.Tamaoki M, et al. Differential ozone sensitivity among Arabidopsis accessions and its relevance to ethylene synthesis. Planta. 2003;216(4):552–560. doi: 10.1007/s00425-002-0894-2. [DOI] [PubMed] [Google Scholar]
- 46.de Carbonnel M, et al. The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 2010;152(3):1391–1405. doi: 10.1104/pp.109.150441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong SG, et al. The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses. Plant J. 2007;51(5):862–873. doi: 10.1111/j.1365-313X.2007.03187.x. [DOI] [PubMed] [Google Scholar]
- 48.Kinoshita T, et al. Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature. 2001;414(6864):656–660. doi: 10.1038/414656a. [DOI] [PubMed] [Google Scholar]
- 49.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 50.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12(3):393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]