Significance
We reconstituted a simple in vitro molecular ecosystem using two self-replicating RNA molecules. The host RNA species produces an RNA-dependent RNA polymerase for their replication, and the parasitic RNA relies on the same enzyme for its replication as well. We found that only when compartmentalized into microscale droplets did the ecosystem show a typical host–parasite oscillatory dynamics. Furthermore, after a long cultivation, the ecosystem evolved to allow the coexistence of the two species. These results demonstrate that cell-size compartmentalization is essential for the ecological dynamics and evolution of host and parasite species.
Keywords: parasite, oscillation, compartment, RNA replication, evolution
Abstract
To date, various cellular functions have been reconstituted in vitro such as self-replication systems using DNA, RNA, and proteins. The next important challenges include the reconstitution of the interactive networks of self-replicating species and investigating how such interactions generate complex ecological behaviors observed in nature. Here, we synthesized a simple replication system composed of two self-replicating host and parasitic RNA species. We found that the parasitic RNA eradicates the host RNA under bulk conditions; however, when the system is compartmentalized, a continuous oscillation pattern in the population dynamics of the two RNAs emerges. The oscillation pattern changed as replication proceeded mainly owing to the evolution of the host RNA. These results demonstrate that a cell-like compartment plays an important role in host–parasite ecological dynamics and suggest that the origin of the host–parasite coevolution might date back to the very early stages of the evolution of life.
Various functions of living organisms have been reconstituted in vitro from biological molecules to understand the basic principles underlying complex biological phenomena observed in the cell or in nature (1, 2). Several types of continuous systems of self-replication of genetic information have been constituted in vitro from RNA alone (3–6), and from the combinations of RNA and proteins (7) or of DNA, RNA, and proteins (8). One of the next important challenges is the reconstitution of interacting networks of self-replicating species, including the investigation of how such interactions generate complex ecological behaviors such as those observed in nature.
Organisms in the wild self-reproduce and interact with each other to form ecosystems. Such interaction generates complex ecological behaviors such as the oscillation dynamics observed in prey–predator or host–parasite populations (9–12). The causes and results of such periodic patterning have been a focus of intensive debate for decades (13–17). Such interactions and the resultant coevolution of the interacting species are thought to be an important factor explaining the high degree of extant biodiversity and complexity (16, 18). However, to date, the in vitro reconstitution of such ecological behavior has been limited; one example is of prey–predator oscillation dynamics constructed using a combination of small hybridizing DNA fragments and several enzymes, although the DNA fragment did not evolve in this system (19).
Previously, we had constructed a simple translation-coupled RNA replication system in which an artificial genomic RNA replicated through translation of the self-encoded RNA replicase and evolved (7). This system consists of a reconstituted translation system of Escherichia coli and an RNA encoding the RNA replicase (Qβ replicase), which is composed of the catalytic β-subunit and the translational factors, EF-Tu and EF-Ts. We found that parasitic RNAs, which had lost their replicase encoding region, spontaneously appeared in this system at a high genome concentration. The parasitic RNA appears possibly through recombination of the host RNA based on the facts that the appearance of the parasitic RNA depends on the presence of the host RNA (20) and that RNAs spontaneously recombine in a nonhomologous manner (21). Once such parasitic RNAs appeared, the host RNA replication was completely inhibited because of the much higher replication rate of the parasitic RNA. Therefore, in our previous long replication experiment we had to manually adjust the genome concentration as low as possible to avoid the appearance of parasitic RNAs.
In this study, we attempted to continuously repeat the replication process of the host and parasitic RNAs under bulk conditions by serial transfer methods and to examine the effect thereon of compartmentalization of the system. Theoretically, compartmentalization is predicted to be a factor important for allowing stable replication in the presence of parasitic or selfish replicators (22–24), although this has not yet been experimentally verified. Furthermore, we examined the population dynamics of the host and parasitic RNAs.
Results
RNA Replication System.
The RNA replication system used in this study consisted of a single-stranded genomic RNA encoding the catalytic subunit of the RNA replicase Qβ (host RNA), a small, single-stranded RNA that lacked the coding region for the replicase (parasitic RNA), and a reconstituted E. coli translation system (25) (Fig. 1A). In this system, the host RNA replicates using the host-generated Qβ replicase, whereas the parasitic RNA lacking the replicase coding region also replicates by relying on the replicase translated from the host RNA. Although the sequences of parasitic RNAs vary, they are usually ∼220 nt in length and show a high degree of similarity to a previously reported small RNA template for the Qβ replicase, s222 (26). Owing to its small size, the parasitic RNA usually replicates much faster than the larger host RNA (∼2,000 nt). Therefore, once such parasitic RNAs appear, they replicate much faster than the host RNA and competitively inhibit the host replication.
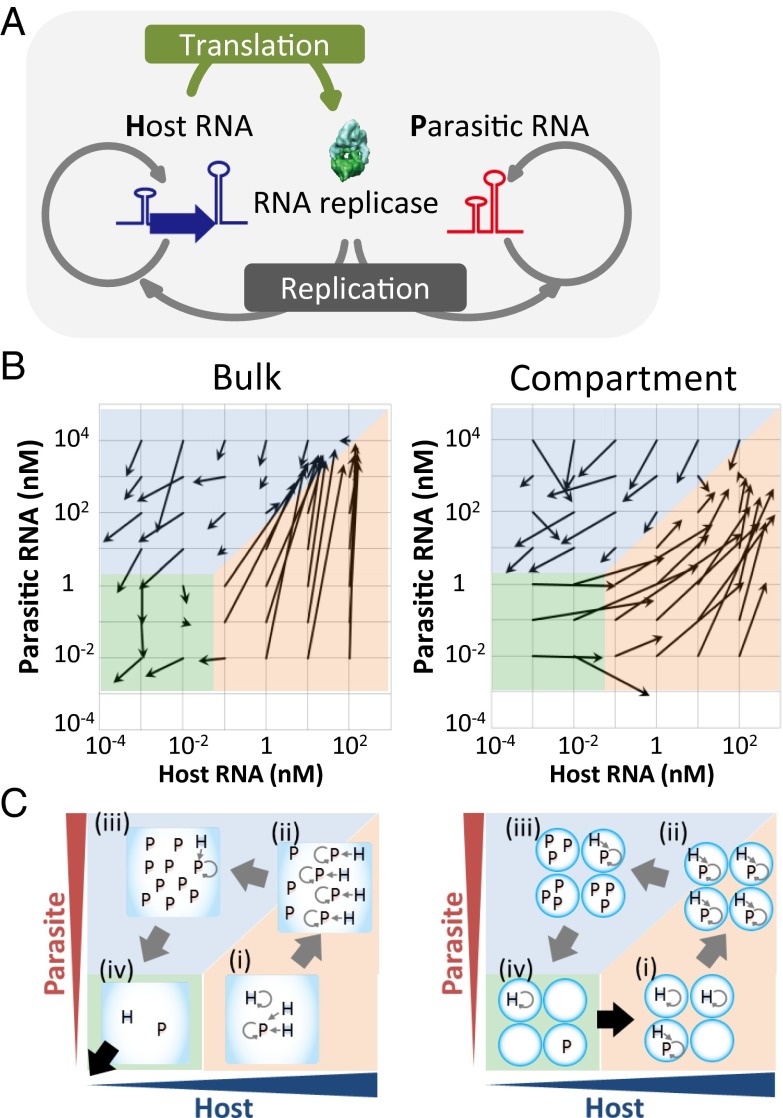
Fig. 1.
RNA replication system and single-round replication assay. (A) Replication scheme of the host and parasitic RNAs. (B) Vector plots of the host and parasitic RNA replications in a single round of the transfer experiment. The initial and averaged final concentrations are indicated by roots and heads of arrows (n = 2–4). (C) Schematic drawing of the host and parasitic RNA replications in each region.
Single-Round Replications Using Various Host and Parasitic RNA Concentrations.
Before the transfer of the host and parasitic RNAs, we first performed single-round replication assays (i.e., replication for 5 h at 37 °C and fivefold dilution) at various host and parasitic RNA concentrations to examine possible host–parasite dynamics. The reaction was performed in a bulk test tube and in water-in-oil droplets of 1.0-μm radius (Fig. S1). The initial and the final concentrations are indicated by the tails and heads of arrows, respectively, in Fig. 1B. Under bulk and compartmentalized conditions both the host and parasitic RNA concentrations increased in the orange highlighted regions and decreased (in most of the cases) in the blue highlighted regions. However, a clear difference between the bulk and compartmentalized conditions was observed in the green highlighted regions, in which the expression of both types of RNAs was constant or decreased under the bulk condition whereas the host RNA selectively increased under the compartmentalized condition. These trajectories were further confirmed by computer simulation using a simplified mathematical model and relevant parameters (Fig. S2A). The possible trajectories of the host and parasitic RNA populations in the transfer experiments were predicted by following the arrows; both the host and parasitic RNA populations were expected to become extinguished under the bulk condition, whereas they were expected to continue replicating and exhibit circulating dynamics under the compartmentalized condition.
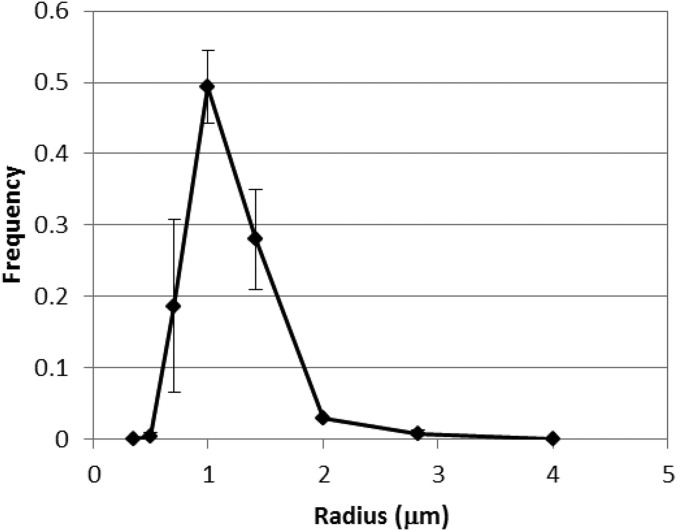
Fig. S1.
Size distribution of the water droplets used in this study. The water droplets were prepared by the same method as used for the single-round assay as shown in Fig. 1B and described in Materials and Methods. The sizes of 100 water droplets in water-in-oil emulsion were measured using an optical microscope. The error bars indicate SDs (n = 3).
Fig. S2.
Simulations of the replication of the host and parasitic RNAs. (A) We performed simulations the replication of the host and parasitic RNAs at various starting concentrations under bulk or compartmentalized conditions according to the method in SI Materials and Methods. The initial and final concentrations after concentrations after dilution are indicated by roots and heads of arrows, respectively. (B) Simulation of an oscillation in the host and parasitic RNA concentrations. We performed the simulation of replication of the host and parasitic RNA according to the method in SI Materials and Methods. We started the simulation at host = 1 nM and parasite = 10−3 nM. (C) Replot of the host and parasitic RNA concentrations on the host–parasite plane. We also started the simulation from 36 combinations of initial concentrations from 10−3 to 102 nM and found that the trajectories converged to the same.
The difference between the bulk and compartmentalized conditions in the green highlighted regions can be explained as follows: The translated replicase concentration was expected to be of the same order as the host RNA concentration according to the associated translation rate (7). Therefore, the replicase concentration under the bulk condition was less than 10−2 nM, much lower than the Km value (∼0.5–5 nM) (7); thus, it would be stochastically difficult for the replicases to associate with the RNAs (Fig. 1C, Left, iv). In contrast, under the compartmentalized condition (Fig. 1C, Right, iv), the effective concentration of the translated replicase in the compartments (1-μm radius) containing a host RNA was about 0.4 nM, which is sufficiently high for the replication of the host RNA. In addition, the host RNA in the green highlighted region has sufficient chance to be encapsulated in a parasite-free compartment and can thus replicate selectively. For example, at the middle point of the green highlighted region (0.001 nM host and 0.1 nM parasite), the expected number of host and parasitic RNAs in the compartment are 0.0025 and 0.25, respectively. At these concentrations, the possibility that a host RNA is encapsulated in a parasite-free compartment is 0.78 according to Poisson distribution.
Serial Transfer Experiments.
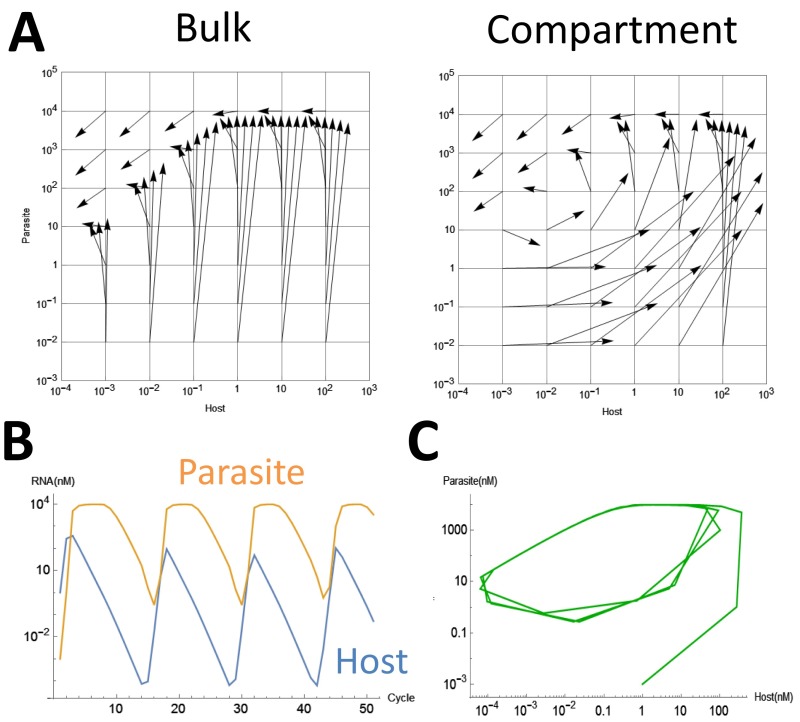
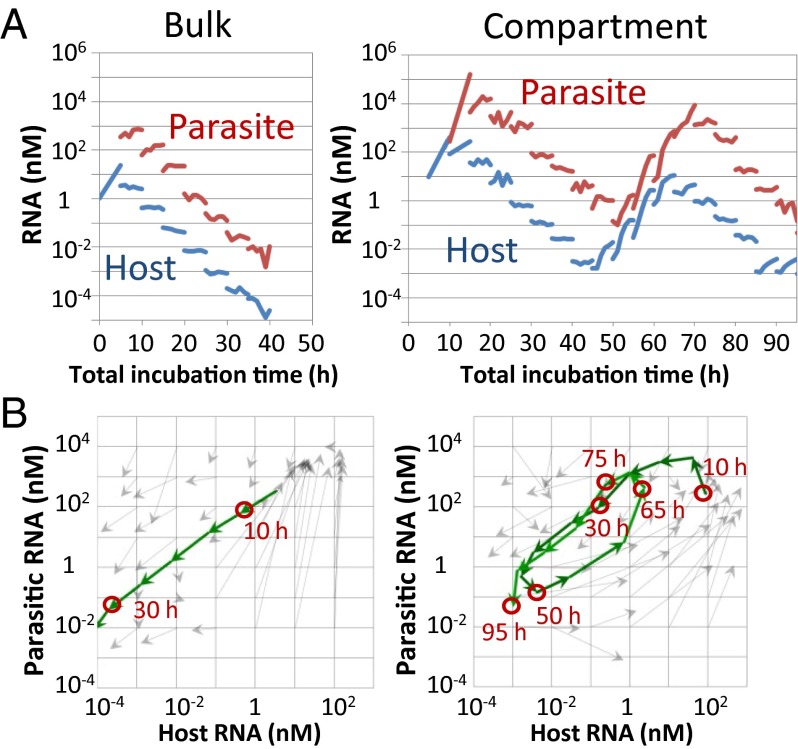
We attempted to repeat replication of the host and parasitic RNAs by serial transfer methods under bulk and compartmentalized conditions. Under both conditions, we started the reaction with 1 nM of the host RNA and performed replication for 5 h at 37 °C. An aliquot of the solution was diluted fivefold with a new solution containing the translation system. The mixture was then incubated for 5 h at 37 °C for the next round of replication. We performed the transfer experiment for 19 rounds (total incubation time of 95 h) and measured the host and parasitic RNA concentrations every hour (Fig. 2A). Under both the bulk and compartmentalized conditions, the parasitic RNA appeared after 5–10 h of incubation and then significantly amplified to a concentration ∼100 times greater than that of the host RNA. With dilution, the host and parasitic RNA concentrations declined continuously (5–40 and 15–45 h in the bulk and compartment conditions, respectively). Under the bulk condition, the concentrations of both types of RNAs continued to decrease until they were undetectable; these RNAs could not be recovered even after an additional five rounds (25 h). In contrast, under the compartmentalized condition, the concentrations of both RNA types increased again after 45 h of total incubation. The increase continued until 70 h and then the concentrations decreased again. To compare this result with the prediction made via the single-round reaction assay, we overlaid the trajectories of the host and parasitic RNA concentrations onto Fig. 1B (Fig. 2B). The results were consistent with the arrows obtained in the single-round experiments, and the oscillation dynamics were consistent with those obtained during simulation with the mathematical model (Fig. S2 B and C). Thus, these results indicated that compartments are necessary to generate the oscillation dynamics of the host and parasitic RNAs.
Fig. 2.
Transfer experiments. (A) The host and parasitic RNA concentrations during incubation in the transfer experiments. (B) Trajectory of the host and parasitic RNA concentrations on a host–parasite plane.
Changes of the Oscillation Pattern.
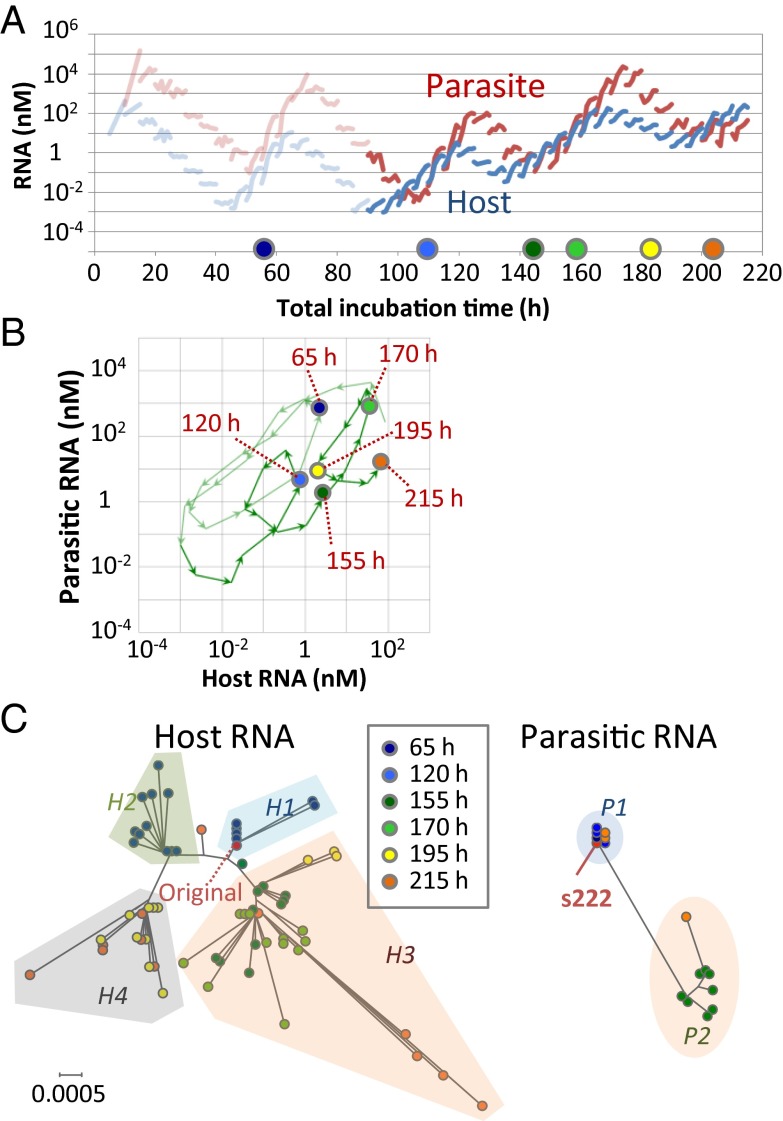
We then repeated the transfer experiments for another 24 rounds (an additional total incubation time of 120 h) under the compartmentalized condition and found that the oscillation dynamics gradually changed (Fig. 3A). For example, the host RNA concentration, which had always been lower than that of the parasitic RNA during the incubation period ranging from 10 to 90 h, was higher at 105, 150, and 200 h. In addition, the host RNA concentrations at the valleys of the oscillation dynamics changed; the concentrations were ∼10−3 nM at the first (45 h) and second (85 h) minima but increased ∼100-fold or 1,000-fold at the third (135 h) and fourth (190 h) minima, respectively. The mapping of these changes on the host–parasite plane showed that the center of rotation had moved to the right and upwards after 95 h (Fig. 3B). These results suggested that the host RNA acquired the ability to replicate in the presence of increased concentrations of parasitic RNA.
Fig. 3.
Changes of the oscillation dynamics under the compartmentalized condition. (A) The host and parasitic RNA concentrations in the continued transfer experiments. At the time indicated by the colored circles, the host and parasitic RNAs were cloned and sequenced. (B) Trajectory of the host and parasitic RNA populations on a host–parasite plane. (C) Phylogenetic trees of the host and parasitic RNA clones; the trees were drawn on the same scale. For parasitic RNA, the frequently amplified s222 RNA is also indicated.
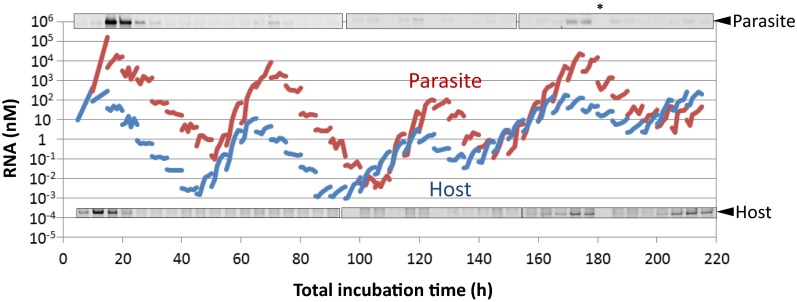
The RNA concentrations of the host and parasitic RNAs were measured by indirect methods such as quantitative PCR, which depend on exponential signal amplification, and therefore these data have a relatively large degree of uncertainty. To verify the oscillation dynamics of RNA concentrations, we performed polyacrylamide gel electrophoresis and RNA staining to directly detect the host and parasitic RNAs. In the high-concentration regions for both the host and parasitic RNAs, we observed bands of both the host and parasitic RNAs, confirming the oscillation pattern of both RNA concentrations (Figs. S3 and S4). However, we sometimes also observed a band corresponding to the parasitic RNA even though the quantified parasite concentration was low (e.g., at 105 h), indicating that the quantification method for the parasitic RNA might be underestimating the actual parasitic RNA concentrations.
Fig. S3.
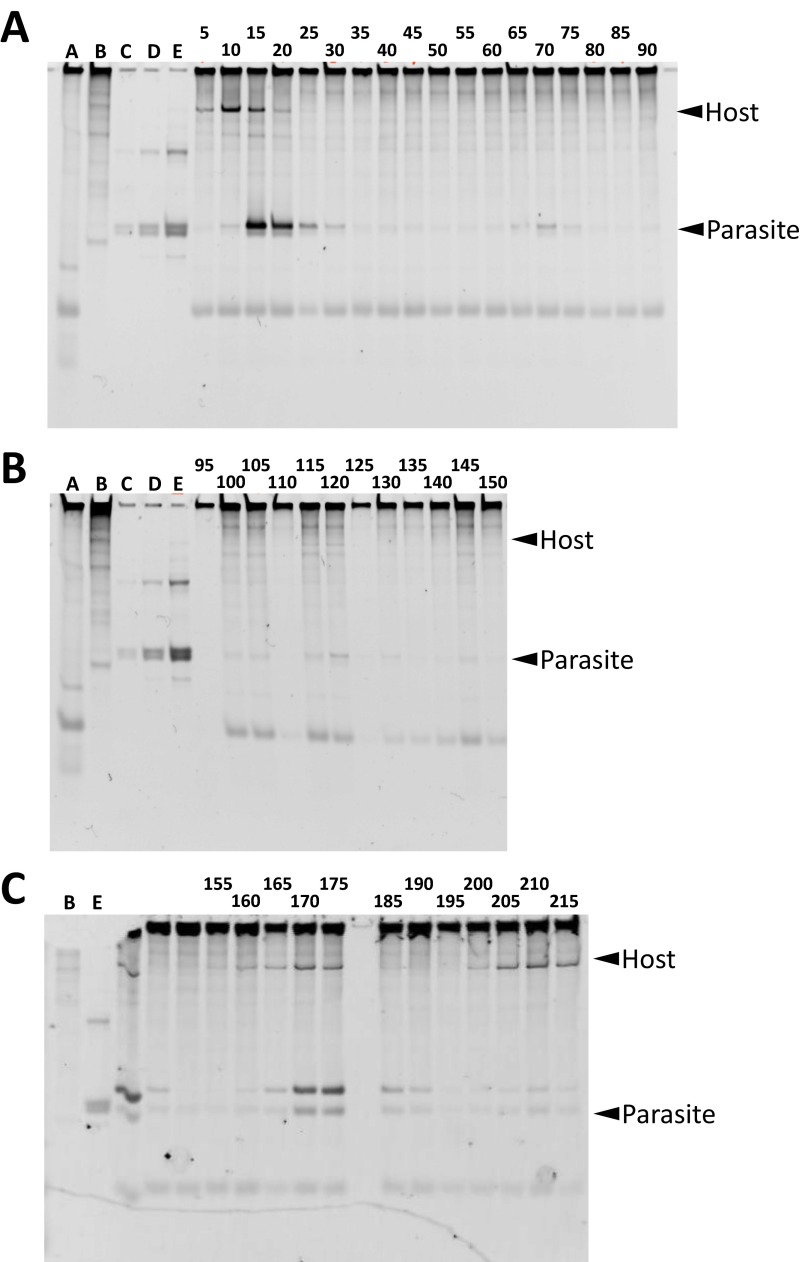
Comparison between the quantification results of the host and parasitic RNAs using RNA staining after polyacrylamide gel electrophoresis. The water droplets during the transfer experiment were collected and applied to polyacrylamide gel electrophoresis and RNAs were stained with SYBR green II. The parts of the gel corresponding to the parasite and the host were aligned with the quantitation data of Fig. 3A. *No sample was applied here because, unfortunately, the sample was lost. The original gel data are shown in Fig. S4.
Fig. S4.
The native polyacylamide gel electrophoresis of the reaction mixture during the transfer experiment. The number on the top of the gels indicates the incubation time during the transfer [(A) 5–90 h, (B) 95–150 h, and (C) 155–215 h]. Lane A is the reaction mixture without any host or parasite (i.e., only the reconstituted translation system). Lane B is pure original host RNA. Lanes C, D, and E are three different amounts of the original parasitic RNA. Both the host and parasitic RNAs exhibit multiple bands caused by structural heterogeneity. The major bands indicated in Fig. S3 have been marked in this figure with arrowheads.
Sequence Analysis.
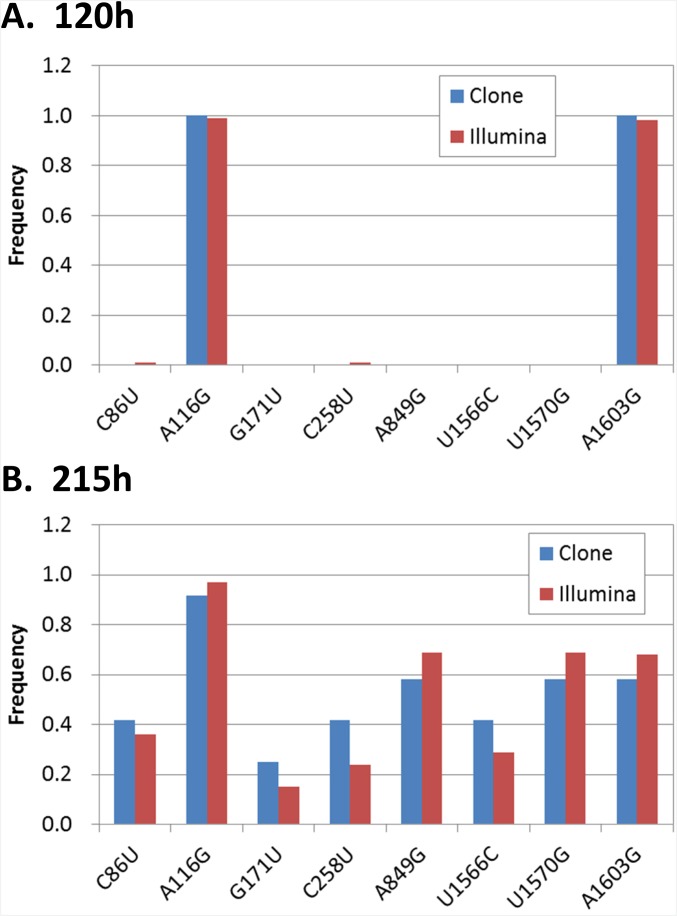
The changes in the oscillation pattern might be caused by evolution of the host and/or parasitic RNAs because in this system mutations were spontaneously introduced through replication errors, and the more highly replicable RNA could dominate the population according to the principles of Darwinian evolution (7). Therefore, to analyze the RNA sequences, we obtained 5–12 clones of the host RNA at 65, 120, 155, 170, 195, and 215 h and performed phylogenic analysis using the resulting sequences (Fig. 3C). From this, the host RNA sequences were classified into four clusters (H1–4) and the host RNA population moved back and forth among these clusters in the course of the transfer experiment. The original host RNA and all clones at 65 h belonged to the H1 cluster. The host RNA population then moved to cluster H2 at 120 h and at 155 h moved back to the H3 cluster, which is closer to the original H1 cluster. At 195 h, a major fraction of the population moved evolutionarily forward again to cluster H4, which exists beyond the H2 cluster. These movements were caused by the transient fixation of some mutations (Table S1). For example, some mutations (e.g., A116G and A1603G) that had been fixed at 120 h disappeared at 155–170 h but reappeared after 195 h. These results indicated that the evolutionary process of the host RNA was not unidirectional as had been observed in our previous study (7) performed in the absence of parasitic RNAs, but seemed to be bidirectional, which might be caused by the coexistence of the parasite that changed the environment for evolution of the host RNA. To further verify the mutation frequencies, we performed large-scale sequencing with a next-generation sequencer (Illumina Miseq) and obtained similar results in the frequencies of all of the fixed mutations at 120 h (Fig. S5A) and 215 h (Fig. S5B).
Table S1.
Mutations fixed in the host RNA
 |
Only mutations that were observed in more than half of the clones in a given time are shown, and the points at which the mutations occupied more than half the clones are highlighted.
Fig. S5.
Comparison of frequency of mutations between different sequencing methods. The frequency of each fixed mutation in the host RNA at (A) 120 h and (B) 215 h were compared between the two different methods: sequencing by Sanger method after cloning (Clone, the same data as Table S1) and a large-scale sequencing method using Illumina Miseq without cloning (Illumina). For the Illumina analysis, more than 1400 reads were obtained at all of the mutation sites, and no other fixed mutations that existed in more than 50% of the reads were found.
In contrast, we obtained relatively fewer clones (one to eight) at 65, 120, 170, and 215 h for the parasitic RNA. This lower cloning efficiency of parasitic RNA could be due to the strong RNA secondary structures. The sequences of one clone at 65 h and three clones at 120 h were the same as that of s222 (Table S2) and formed the cluster P1 in the phylogenic tree (Fig. 3C). At 155 h, eight clones commonly acquired four mutations and formed a different cluster (cluster P2), but at 215 h two of three clones had returned to the original cluster P1. These data suggest that the dominant sequences of the parasitic RNA population also continually changed, but it should be noted that the sequence results of parasitic RNA might contain larger uncertainty than that of the host RNA because of small sample number.
Table S2.
Mutations fixed in the parasitic RNA
 |
Mutations were identified comparing to the s222 RNA sequence, the most common parasitic RNA. Only mutations that were observed in more than half of the clones in a given time are shown, and the points at which the mutations occupied more than half the clones are highlighted.
Biochemical Analysis.
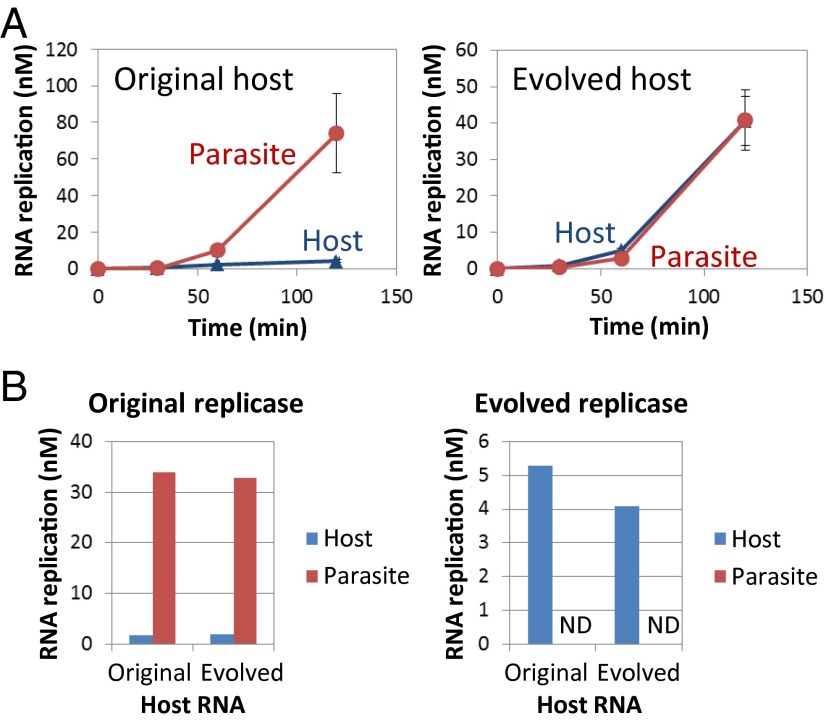
The changes in the population dynamics (Fig. 3 A and B) suggested that the host RNA acquired the ability to replicate in the presence of excess parasitic RNAs. To confirm this suggestion, we performed a competition experiment between the host and parasite RNAs. We used the original host RNA and one of the host clones obtained at 215 h (evolved host RNA) as the host RNAs and s222 as the parasitic RNA. We mixed 10 nM of the original or evolved host RNA with the parasitic RNA in 100-fold excess and incubated the mixture in the translation system. When using the original host RNA, the parasitic RNA predominantly amplified and the host RNA replication was repressed (Fig. 4A, Left). When using the evolved host RNA, both the host and parasitic RNAs replicated to almost the same amount (Fig. 4A, Right), indicating that the replication of the evolved host RNA had acquired resistance against the presence of the parasitic RNA.
Fig. 4.
Competition experiments between the host and parasitic RNAs. (A) Translation-coupled replication competition experiments. The error bars indicate the SD (n = 3). (B) Competition experiments with purified replicases. When using the evolved replicase, the parasitic RNA was not detected (ND) with either the original or evolved host RNAs.
We identified two possible causes of the parasite resistance: improved activity of the evolved host RNA as a replication template or increased specificity of the replicase encoded by the evolved host RNA. To examine these possibilities, we purified the replicase encoded in the evolved host RNA (evolved replicase) and performed competition experiments using the purified replicase instead of the internally translated replicase. The evolved replicase had acquired four nonsynonymous mutations (K208E, M300T, L448R, and Q459R) compared with the original replicase used in this study. We mixed the original or evolved replicase (10 nM) with the original or evolved host RNA (100 nM) and the parasitic RNA (100 nM) in a translation system in which tyrosine and cysteine had been omitted to inhibit internal replicase translation and incubated the mixture at 37 °C for 5 h. When using the original replicase, the parasitic RNA predominantly replicated when using either the original or evolved host RNAs (Fig. 4B, Left). In contrast, when using the evolved replicase, the host RNAs replicated to a certain amount, but the parasitic RNA was below the detection limit regardless of whether the original or evolved host RNA had been used (Fig. 4B, Right). These results demonstrated that the parasite resistance of the evolved host RNA can be mostly attributed to a functional change of the replicase encoded by the evolved host RNA.
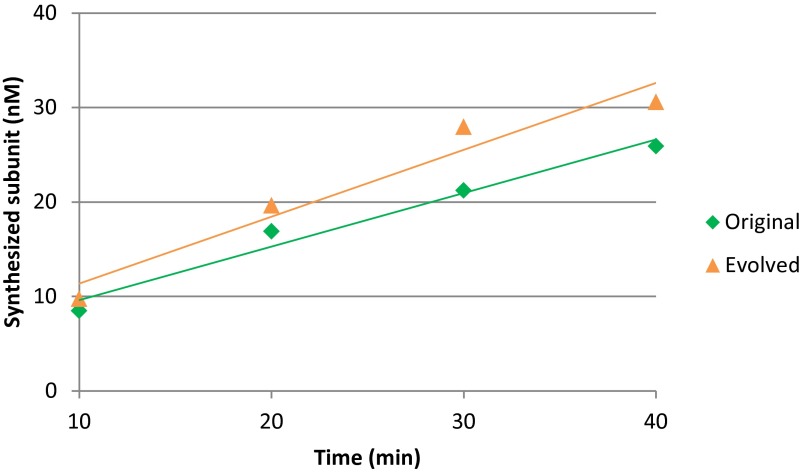
To quantitatively evaluate the evolution of the replicase, we measured the kinetic parameters of replication and translation. We first measured the translation rate of the replicase generated from the original and evolved host RNAs (Fig. S6). The translation rate of the evolved host RNA was 0.26 proteins per min, which was not significantly changed from that of the original host RNA at 0.35 proteins per min. We next measured the kinetic parameters of replication using cognitive pairs of host and replicase (i.e., the original host and original replicase or the evolved host and evolved replicase). The kcat/Km values of the evolved pair was 1/74-fold lower than that of the original pair (Table 1), which was attributed to 1/2.3-fold lower kcat and 3.2-fold higher Km in the evolved pair. We then measured the kinetic parameters of parasitic RNA replication using the original parasite (s222) and the original or evolved replicases. The kcat/Km values of the evolved replicase was 1/2,100-fold lower than that of the original replicase, which was attributed to the 1/91-fold lower kcat and 23-fold higher Km values with the evolved replicase. In summary, the activity of host replication decreased to a certain extent, but that of parasitic RNA replication decreased even more, resulting in parasite resistance.
Fig. S6.
Translation of the evolved host RNA. To estimate the kinetic parameter of translation of the replicase, we performed translation with the original or evolved host RNAs in the translation system containing [35S]methionine. In this experiment, we omitted UTP to prevent replication of the host RNA. After incubation for the indicated periods, aliquots were subjected to SDS polyacrylamide gel electrophoresis, and the synthesized β-subunit of the replicase was measured after autoradiography as described previously (20). The estimated translation rates were shown in Table 1.
Table 1.
Kinetic parameters
| RNA | Original pair | Evolved pair | Ratio (evolved/original) |
| Host RNA | |||
| kcat, per min | 0.22 (±0.016) | 0.094 (±0.017) | 1/2.3 |
| Km, nM | 0.76 (±0.32) | 2.4 (±1.2) | 3.2 |
| kcat/Km, nM/min | 0.29 | 0.0039 | 1/74 |
| Original replicase | Evolved replicase | ||
| Parasitic RNA | |||
| kcat, per min | 3.1 (±0.15) | 0.034 (±0.0021) | 1/91 |
| Km, nM | 2.1 (±0.56) | 48 (±8.1) | 23 |
| kcat/Km, nM/min | 1.5 | 0.00071 | 1/2,100 |
Fig. S7.
Estimation of kinetic parameters of replication. We performed the replication reaction with several combinations of the purified replicases and the host and parasitic RNAs to estimate the kinetic parameters of replication, kcat and Km, shown in Table 1. We mixed the original or evolved purified replicase with the original or evolved hosts or the parasitic RNA in the translation system containing [32P]UTP to trace the amplification of both the RNAs. In this experiment, we omitted tyrosine and cysteine to prevent the translation of the replicase. After incubation for the indicated periods, aliquots were subjected to polyacrylamide gel electrophoresis, and the synthesized RNA was measured by autoradiography as described in SI Materials and Methods. The initial RNA synthesis rates were plotted and fitted by Michaelis–Menten curve (gray lines) to estimate kcat and Km values.
Discussion
In this study we demonstrated that the parasitic RNAs that spontaneously appear in the translation-coupled replication system collapse host RNA replication under the bulk condition, whereas when the system is compartmentalized the two types of RNA coreplicate continuously and exhibit oscillating population dynamics. These results indicate that a cell-like compartment is important for continuous host–parasite coreplication and for ecological oscillation in the case that the hosts and parasites competitively use the same replicase and the parasites replicate faster than the hosts. The appearance of parasitic or selfish replicators have been considered as large hurdles for the development of primitive life forms into more complex forms (22, 23, 27, 28), and theoretically compartmentalization might represent a possible solution (22–24). To the best of our knowledge, this study is the first to experimentally demonstrate that a replication system can stably function in the presence of parasitic species under a compartmentalized condition. In addition, the low concentrations of both the host and parasitic RNAs in the “green regime” in Fig. 1 are similar to the origin-of-life scenario in ancient earth. Compartmentalization that allows reamplification of the host and parasite from such low concentrations might have played an important role in the accumulation of genetic molecules in the prebiotic world. Furthermore, we observed that the continuous replication of this simple host–parasite replication system allows the evolution of both RNA species, suggesting that the origin of the host–parasite coevolution might date back to the very early stages of the evolution of life.
The evolution of the host RNA observed in this study is different from that observed in our previous study (7), which was performed under different selection pressure. In the previous evolution, we attempted to circumvent the appearance of the parasitic RNA by adjusting the initial host RNA concentrations as low as possible in every round. In that method, a host RNA that replicated most rapidly in the absence of parasitic RNA should be selected. As expected, the final evolved host RNA acquired the ability to rapidly replicate by increasing the kcat values (krep_plus and krep_minus) two- to threefold through improving its function as a replication template. In this study, we performed replication of the host RNA in the presence of the parasitic RNA. Under this condition, the host RNA that could selectively replicate even among excess parasitic RNA should be selected. Accordingly, the final evolved host RNA acquired a replicase that selectively replicated the host RNA at the expense of a modest reduction of the kcat/Km value.
Accumulating evidence has suggested that the existence of parasites is not only an obstacle for host replicators but also plays an important role in host evolution by maintaining the genetic diversity of the host species (16, 18). In this study, we found that the evolution of the host RNA in the presence of parasitic RNA was bidirectional (Fig. 3C), which contrasted with the unidirectional evolution that occurred in the absence of parasites in our previous study (7). The existence of the parasitic RNA might increase the diversity of the host RNA by increasing the number of possible evolutionary pathways. In addition, the existence of parasites is proposed as one of the driving forces of host evolution in the Red Queen hypothesis (29), in which the evolution of host species becomes accelerated by rapidly evolving parasitic species through an evolutionary arms race. In this study, we found that the host RNA acquired a certain level of parasite resistance, and thereby the parasitic RNA replicated at a lower concentration than the host RNA in the last few rounds (200–215 h, Fig. 3A). It remains to be determined whether the parasitic RNA might evolve to be replicable again by the evolved replicase after further rounds of replication. Overall, this host–parasite replication system might be a useful experimental model to investigate host and parasite coevolution processes in detail.
Materials and Methods
Materials.
For this study, we used the reconstituted translation system of E. coli [PURE System (25)], the composition of which is customized for RNA replication (7). This system contains all of the components required for translation and RNA replication including ribosomes, amino acids, tRNAs, and NTPs. The original and evolved replicases encoded in the original and evolved host RNAs, respectively, were purified according to the previous study (30).
RNA Preparation.
As the original host RNA we used a clone from round 128 in our previous study (30). As the evolved host RNA we used a clone taken at 215 h of incubation in this study. The whole sequences of these host RNAs are shown in SI Materials and Methods. As the parasitic RNA for the competition experiments, we used s222 RNA, a commonly appearing parasitic RNA (20). The host and parasitic RNAs were prepared by in vitro transcription using T7 RNA polymerase and the template plasmids pUC-N96(+) for the original host RNA, pUC-Bansho-R43-8 for the evolved host RNA, and pUC-s222A for the parasitic RNA after digestion with SmaI and were purified with an RNeasy column (Qiagen).
Transfer Experiments.
We initiated the transfer experiments using the original host RNA (1 nM) and no parasitic RNA in the translation system. For the single-round assay shown in Fig. 1B, we used the indicated concentrations of the original host and parasitic RNAs. Under the bulk condition, the reaction mixture (10 μL) was incubated at 37 °C for 5 h and one-fifth (2 μL) of the solution was transferred to 8 μL of the reconstituted translation system. The mixture was then incubated at 37 °C for 5 h for the next round of replication. Under the compartmentalized condition, the reaction mixture (10 μL) was vigorously mixed with the buffer-saturated oil phase (1 mL) prepared according to the previous study (7) using a homogenizer (POLYTRON PT-1300D; KINEMATICA) with a plastic disposable shaft at 16,000 rpm for 1 min on ice. The size distribution of the water droplets is shown in Fig. S1. The emulsion was incubated at 37 °C for 5 h and one-fifth (200 μL) of the emulsion was transferred to a new emulsion (800 μL) prepared with the translation system (8 μL) as a water phase by the same method as described above. The added emulsion was then mixed with the homogenizer at 16,000 rpm for 1 min on ice to completely mix the contents of the water droplets and incubated at 37 °C for 5 h for the next round of replication. At each hour during the incubation, aliquots were collected and the host and parasitic RNA concentrations were measured after 10,000-fold dilution for the bulk condition or 100-fold dilution for the compartmentalized condition with 1 mM EDTA (pH 8.0).
Measurement of Host and Parasitic RNA Concentrations.
To determine the host RNA concentration, the RNA samples diluted with 1 mM EDTA were heated at 95 °C for 5 min and subjected to quantitative PCR after reverse transcription using the PrimeScript One Step RT-PCR Kit (TaKaRa) and primers 5′ GCTGCCTAAACAGCTGCAAC 3′ and 5′ CGCTCTTGGTCCCTTGTATG 3′. The host RNA concentration was determined using the original host RNA as a standard.
To determine the parasitic RNA concentration, we performed exponential amplification of the parasitic RNA and compared the amplification curve with those of a series of dilution gradients of the known initial parasitic s222 RNAs. This method is principally the same as quantitative PCR except that RNA replicase was used instead of DNA polymerase. The RNA samples (2 μL) diluted with 1 mM EDTA were mixed into 18 μL of a reaction mix containing 1 µM wild-type Qβ RNA replicase (30), 125 mM Tris⋅HCl (pH 7.5), 10 mM MgCl, 1.25 mM each NTP, 0.01% BSA, 0.4× ROX (Roche), 1× SYBR Green II (Takara), and 0.5% Triton X-100 (Sigma). The mixture was incubated at 30 °C and the fluorescence of the amplified RNA was measured every 15 s for 30 min. Note that although this method also amplifies the host RNA, the much smaller parasitic RNA is preferentially detected because this experiment was performed with an excess amount of replicase and thus the RNAs replicate exponentially at rates correlated with the RNA size (26).
Sequence Analysis.
The host RNA was reverse-transcribed using PrimeScript reverse transcriptase (Takara) and a reverse-transcription (RT) primer (5′ CCGGAAGGGGGGGACGAGG 3′) and then PCR-amplified using the RT primer and a PCR primer (5′ GGGTCACCTCGCGCAGC 3′). The PCR products were subjected to agarose gel electrophoresis and the bands corresponding to the host or parasitic RNA were extracted. The extracted DNA fragment was ligated into a plasmid as described (7). The parasitic RNA was first purified by 8% polyacrylamide gel electrophoresis followed by excision and extraction using QIAEX II (Qiagen). The purified parasitic RNA was ligated to a plasmid in the same method as the host RNA. For the large-scale sequencing, the host PCR products before gel extraction was subjected to large-scale sequencing with Illumina Miseq. The analysis procedure was the same as reported previously (31). The minimum read number was 1,400 for all mutational sites.
Competition Experiments.
For the translation-coupled replication competition experiments (Fig. 4A), the original or evolved host RNAs (10 nM) were mixed with the parasitic s222 RNA (1,000 nM) in a translation system containing [32P]UTP under the bulk condition. After incubation at 37 °C for the indicated time, the mixtures were subjected to 8% polyacrylamide gel electrophoresis and the synthesized host and parasitic RNA concentrations were measured by autoradiography as described previously (7). In the competition experiments with purified replicases (Fig. 4B), the original or evolved host RNAs (100 nM) were mixed with one the s222 parasitic RNA (100 nM) in a translation system containing the original or evolved purified replicase (10 nM) and [32P]UTP but lacking cysteine and tyrosine under the bulk condition. After incubation at 37 °C for 5 h, the host or parasitic RNA concentrations were measured by autoradiography as described above.
Polyacrylamide Gel Electrophoresis.
The water phases were collected from the emulsion samples during the transfer experiments and RNAs were purified with a spin column (PureLink; Life technologies). The purified RNA samples were subjected to 8% polyacrylamide gel electrophoresis with 0.1% SDS in TBE buffer (pH 8.4) containing Tris(hydroxymethyl)aminomethane (100 mM), boric acid (90 mM), and EDTA (1 mM), followed by staining with SYBR Green II (Takara).
Simulation.
The method used for simulating the host and parasite replication is described in SI Materials and Methods.
SI Materials and Methods
Simulation Method.
The numbers in parentheses are values used for the simulation. These values are obtained from our previous study (7) or this study (Table 1).
KH: Dissociation constant of the host RNA and the replicase (0.76 nM)
KP: Dissociation constant of the parasitic RNA and the replicase (3.1 nM)
kH: Rate constant of the host replication from the host–replicase complex
kP: Rate constant of the parasitic RNA replication from the parasite–replicase complex
kR: Rate constant of replicase translation (0.0035/min per 1 μM ribosome)
kHmax: Rate constant of host replication from host–replicase complex at initial NTP concentration (0.21/min)
kPmax: Rate constant of parasite replication from parasite–replicase at initial NTP concentration (3.1/min)
LH: Length of host RNA (nt)
LP: Length of parasite RNA (nt)
NTP0: NTP concentration before incubation
C: Concentration conversion coefficient [nanomolar/(molecule/emulsion)]
: Mean concentration of host RNA (molecule/compartment)
: Mean concentration of parasite RNA (molecule/compartment)
[H]total: Total concentration of the host RNA
[P]total: Total concentration of the parasitic RNA
[R]total: Total concentration of the replicase
[H]: Concentration of the free host RNA
[P]: Concentration of the free parasite RNA
[HR]: Concentration of the host–replicase complex
[PR]: Concentration of the parasite–replicase complex.
The simulation is based on a mathematical model of the replication of the host and parasitic RNAs, which consist of seven types of reactions as described below.
-
i)
Translation:
Host RNA → host RNA + replicase
-
ii)
Association of replicase on the host RNA:
Host RNA + replicase → host–replicase complex
-
iii)
Association of replicase on the parasitic RNA:
Parasitic RNA + replicase → parasite–replicase complex
-
iv)
Dissociation of replicase on the host RNA:
Host RNA + replicase → host–replicase complex
-
v)
Dissociation of replicase on the parasitic RNA:
Parasitic RNA + replicase → parasite–replicase complex
-
vi)
Replication of the host RNA:
Host-replicase complex → 2 host RNA + replicase
-
vii)
Replication of the parasitic RNA:
Parasite–replicase complex → 2 parasitic RNA + replicase.
We assumed that the association and dissociation of the replicase with both the host and parasitic RNA is equilibrated to obtain
| [S1] |
and
| [S2] |
According to these reactions, the deviation of each component is described as follows:
| [S3] |
| [S4] |
| [S5] |
| [S6] |
We assumed that the concentrations of the free host and free parasitic RNA are much higher than those of the host–replicase and parasite–replicase complexes, respectively, and obtained
| [S7] |
and
| [S8] |
Using Eqs. S1, S2, and S6–S8, the free replicase concentration, [R], is written as
| [S9] |
Using Eqs. S1, S2, S7, and S8, Eqs. S3 and S4 are written as
| [S10] |
| [S11] |
Substituting Eqs. S5, S10, and S11 with Eqs. S7 and S9 we obtained
| [A] |
| [B] |
| [C] |
Here kH and kP are defined as follows:
where LH = 2,074 nt, LP = 221 nt, and NTP0 = 8.75× 106 nM.
We found the numerical solutions of Eqs. A, B, and C by the NDSolve function of Mathematica (Wolfram), and the concentrations of the host and parasitic RNAs after a 5-h reaction are defined as the functions and , respectively, where h and p are initial numbers of the host and parasitic RNAs in one compartment (molecule per compartment), respectively.
We then simulated the dynamics of concentrations of the host and parasitic RNAs by assuming “compartment” and “dilution” effect as the following process:
-
i)
and values are calculated under all of the combination of h and P values.
-
ii)
Frequencies of each compartment that contains different number of the host and parasitic RNAs are calculated according to Poisson distribution and , where λH and λP are mean enclosed number of the host and parasitic RNAs, respectively, in a compartment.
-
iii)
The total concentration of the host and parasitic RNAs after 5 h of reaction in compartments are calculated according to the following equation.
-
iv)
The first terms indicate the host and parasitic RNAs amplified in the compartments that contained less than 11 molecules of both the host and parasitic RNAs. The second terms indicate the host and parasitic RNAs that were amplified from the compartments that contained more than 10 molecules of both the host and parasitic RNAs, where we assumed the concentrations were the same as the average concentrations, λH and λP. -
v)
These values are divided by the dilution rate, 5, and the values are defined as λH and λP in the next step.
-
vi)
We repeated procedures ii–v for 50 times, and plotted λH and λP in Fig. S2.
It is notable that in this simulation we used the following simplifications:
-
i)
The replicase is not transferred to the next round of replication. The transferable molecules are the host and parasitic RNAs.
-
ii)
There is neither degradation nor inactivation of the components.
-
iii)
The translated β-subunit immediately becomes active replicase.
-
iv)
The differences between plus and minus strands of the host or parasite are neglected.
Sequence of the Original Host RNA (N96-1).
GGGAACCCCCCTTCGGGGGGTCACCTCGCGCAGCGGGCTACGCGAGGGAGCCACGCTGCGAAGCAGCGTGGCGGTTCTCGTGCGTCACCGAAACGCACGAAGGTCGCGCCTCTTCACGAGGCGTCACCTGGGAGAGCGCGAAAGCGCTAGCCCGTGCTCTAGCTCTAGAAGGTCTCGAGATCTCCTCTAGAGATAATTTTGTTTAACTCTAAGAAGGAGATATACACATGCCTAAGACAGCATCTTCGCGTAACTCTCTCAGCGCACAATTGCGCCGAGCCGCGAACACAAGAATTGAGGCTGAAGGTAACCTCGCACTTTCCATTGCCAACGATTTACTGTTGGCCTATGGTCAGTCGCCATTTAACTCTGAGGCTGAGTGTATTTCATTCAGCCCGAGATTCGACGGGACCCCGGATGACTTTAGGATAAATTATCTTAAAGCCGAGATCATGTCGAAGTATGACGACTTCAGCCTAGGTATTGATACCGAAGCTGTTGCCTGGGAGAAGTTCCTGGCAGCAGAGGCTGAATGTGCTTTAACGAACGCTCGTCTCTATAGGCCTGACTACAGTGAGGATTTCAATTTCTCACTGGGCGAGTCATGTATACACATGGCTCGTAGAAAAATAGCCAAGCTAATAGGAGATGTTCCGTCCGTTGAGGATATGTTGCGTCACTGCCGATTTTCTGGCGGTGCTACAACAACGAATAACCGTTCGTACAGTCATCCGTCCTTCAAGTTTGCACTTCCGCAAGCGTGTACGCCTCGGGCTTTGAAGTATGTTTTAGCTCTCAGAGCTTCTACACATTTCGATATCAGAATTTCTGATATTAGCCCTTTTAATAAAGCAGTTACCGTACCTAAGAACAGTAAGACAGATCGTTGTATTGCTATCGAACCTGGTTGGAATATGTTTTTCCAACTGGGTATCGGTGGCATTCTACGCGATCGGTTGCGTTGCTGGGGTATCGATCTGAATGATCAGACGATAAATCAGCGCCGCGCTCACGAAGGCTCCGTTACTAATAACTTAGCAACGGTTGATCTCTCAGCGGCAAGCGATTCTATGTCTCTTGCCCTCTGTGAGCTCTTATTGCCCCGAGGCTGGTTTGAGGTTCTTATGGACCTCAGATCACCTAAGGGGCGATTGCCTGACGGTAGTGTTGTTACCTACGAGAAGATTTCTTCTATGGGTAACGGTTACACATTCGAGCTCGAGTCGCTTATTTTTGCTTCTCTCGCTCGTTCCGTTTGTGAGATACTGGACTTAGACTCGTCTGAGGTCACTGTTTACGGAGACGATATTATTTTACCGTCCCGTGCAGTCCCTGCCCTCCGGGAAGTTTTTAAGTATGTTGGTTTTACGACCAATACTAAAAAGACTTTTTCCGAGGGGCCGTTCAGAGAGTCGTGCGGCAAGCACTACTATTCTGGCGTAGATGTTACTCCCTTTTACATACGTCACCGTATAGTGAGTCCTGCCGATTTAATACTGGTTTTGAATAACCTATATCGGTGGGCCACTATTGACGGCGTATGGGATCCTAGGGCCCATTCTGTGTACCTCAAGTATCGTAAGTTGCTGCCTAAACAGCTGCAACATAATACTATACCTGACGGTTACGGTGATGGTGCCCTCGTCGGATCGGTCCTAATCAATCCTTTCGCGAAAAACCGCGGGTGGATCCGGTACGTACCGGTGATTACGGACCATACAAGGGACCAAGAGCGCGCTGAGTTGGGGTCGTATCTCTACGACCTCTTCTCGCGTTGTCTCTCGGAAAGTAACGATGGGTTGCCTCTTAGGGGTCCATCGGGTTGCGATTTTGCTGATCTATTTGCCATCGATCAGCTTATCTGTAGGAGTGATCCTACGAAGATAAGCAGGCCTACCGGTAAATTCGATATACAGTACATCGCGTGCTGTTGTTCGGGTTGTTGTTAGGCTTGCGGCCGCACTCGAGAGATCTAGAGCATCACGGTCGAACTCCCGTACGAGGTGCCCGCACCTCGTCCCCCCCTTCCGGGGGGGTCCCC.
Sequence of the Evolved Host RNA (R43-8).
GGGAACCCCCCTTCGGGGGGTCACCTCGCGCAGCGGGCTACGCGAGGGAGCCACGCTGCGAAGCAGCGTGGCGGTTCTCGTGCGTCACCGAAACGCACGAAGGTCGCGCCTCTTCGCGAGGCGTCACCTGGGAGAGCGCGAAAGCGCTAGCCCGTGCTCTAGCTCTAGAAGGTCTCGAGATCTCCTCTAGAGATAATTTTGTTTAACTCTAAGAAGGAGATATACACATGCCTAAGACAGCATCTTCGCGTAACTCTCTCAGCGCACAATTGCGCCGAGCCGCGAACACAAGAATTGAGGCTGAAGGTAACCTCGCACTTTCCATTGCCAACGATTTACTGTTGGCCTATGGTCAGTCGCCATTTAACTCTGAGGCTGAGTGTATTTCATTCAGCCCGAGATTCGACGGGACCCCGGATGACTTTAGGATAAATTATCTTAAAGCCGAGATCATGTCGAAGTATGACGACTTCAGCCTAGGTATTGATACCGAAGCTGTTGCCTGGGAGAAGTTCCTGGCAGCAGAGGCTGAATGTGCTTTAACGAACGCTCGTCTCTATAGGCCTGACTACAGTGAGGATTTCAATTTCTCACTGGGCGAGTCATGTATACACATGGCTCGTAGAAAAATAGCCAAGCTAATAGGAGATGTTCCGTCCGTTGAGGATATGTTGCGTCACTGCCGATTTTCTGGCGGTGCTACAACAACGAATAACCGTTCGTACAGTCATCCGTCCTTCAAGTTTGCACTTCCGCAAGCGTGTACGCCTCGGGCTTTGAAGTATGTTTTAGCTCTCAGAGCTTCTACACATTTCGATATCAGAATTTCTGATATTAGCCCTTTTAATGAAGCAGTTACCGTACCTAAGAACAGTAAGACAGATCGTTGTATTGCTATCGAACCTGGTTGGAATATGTTTTTCCAACTGGGTATCGGTGGCATTCTACGCGATCGGTTGCGTTGCTGGGGTATCGATCTGAATGATCAGACGATAAATCAGCGCCGCGCTCACGAAGGCTCCGTTACTAATAACTTAGCAACGGTTGATCTCTCAGCGGCAAGCGATTCTATGTCTCTTGCCCTCTGTGAGCTCTTATTGCCCCGAGGCTGGTTTGAGGTTCTTACGGACCTCAGATCACCTAAGGGGCGATTGCCTGACGGTAGTGTTGTTACCTACGAGAAGATTTCTTCTATGGGTAACGGTTACACATTCGAGCTCGAGTCGCTTATTTTTGCTTCTCTCGCTCGTTCCGTTTGTGAGATACTGGACTTAGACTCGTCTGAGGTCACTGTTTACGGAGACGATATTATTTTACCGTCCCGTGCAGTCCCTGCCCTCCGGGAAGTTTTTAAGTATGTTGGTTTTACGACCAATACTAAAAAGACTTTTTCCGAGGGGCCGTTCAGAGAGTCGTGCGGCAAGCACTACTATTCTGGCGTAGATGTTACTCCCTTTTACATACGTCACCGTATAGTGAGTCCTGCCGATTTAATACTGGTTTTGAATAACCTATATCGGTGGGCCACTATTGACGGCGTATGGGATCCTAGGGCCCATTCTGTGTACCGCAAGTATCGTAAGTTGCTGCCTAAACAGCTGCGACATAATACTATACCTGACGGTTACGGTGATGGTGCCCTCGTCGGATCGGTCCTAATCAATCCTTTCGCGAAAAACCGCGGGTGGATCCGGTACGTACCGGTGATTACGGACCATACAAGGGACCAAGAGCGCGCTGAGTTGGGGTCGTATCTCTACGACCTCTTCTCGCGTTGTCTCTCGGAAAGTAACGATGGGTTGCCTCTTAGGGGTCCATCGGGTTGCGATTTTGCTGATCTATTTGCCATCGATCAGCTTATCTGTAGGAGTGATCCTACGAAGATAAGCAGGCCTACCGGTAAATTCGATATACAGTACATCGCGTGCTGTTGTTCGGGTTGTTGTTAGGCTTGCGGCCGCACTCGAGAGATCTAGAGCATCACGGTCGAACTCCCGTACGAGGTGCCCGCACCTCGTCCCCCCCTTCCGGGGGGGTCCCC.
Acknowledgments
We thank N. Kamimura (Miki), H. Komai, R. Otsuki, T. Sakamoto, Y. Fujii, and E. Furushima for technical assistance. This work was partly supported by Japan Society for the Promotion of Science KAKENHI Grants 15H04407, 15H01534, and 15KT0080 and the Takano Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524404113/-/DCSupplemental.
References
- 1.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409(6818):387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 2.Forster AC, Church GM. Towards synthesis of a minimal cell. Mol Syst Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323(5918):1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng LK, Unrau PJ. Closing the circle: Replicating RNA with RNA. Cold Spring Harb Perspect Biol. 2010;2(10):a002204. doi: 10.1101/cshperspect.a002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaidya N, et al. Spontaneous network formation among cooperative RNA replicators. Nature. 2012;491(7422):72–77. doi: 10.1038/nature11549. [DOI] [PubMed] [Google Scholar]
- 6.Attwater J, Wochner A, Holliger P. In-ice evolution of RNA polymerase ribozyme activity. Nat Chem. 2013;5(12):1011–1018. doi: 10.1038/nchem.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichihashi N, et al. Darwinian evolution in a translation-coupled RNA replication system within a cell-like compartment. Nat Commun. 2013;4:2494. doi: 10.1038/ncomms3494. [DOI] [PubMed] [Google Scholar]
- 8.Wright MC, Joyce GF. Continuous in vitro evolution of catalytic function. Science. 1997;276(5312):614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 9.Elton C, Nicholson M. The ten-year cycle in numbers of the lynx in Canada. J Anim Ecol. 1942;11:215–244. [Google Scholar]
- 10.Huffaker CB. Experimental studies on predation: Dispersion factors and predator-prey oscillations. Hilgardia. 1958;27(14):343–383. [Google Scholar]
- 11.Utida S. Cyclic fluctuations of population-density intrinsic to the host-parasite system. Ecology. 1957;38(3):442–449. [Google Scholar]
- 12.Hudson PJ, Dobson AP, Newborn D. Prevention of population cycles by parasite removal. Science. 1998;282(5397):2256–2258. doi: 10.1126/science.282.5397.2256. [DOI] [PubMed] [Google Scholar]
- 13.Lotka AJ. Analytical note on certain rhythmic relations in organic systems. Proc Natl Acad Sci USA. 1920;6(7):410–415. doi: 10.1073/pnas.6.7.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volterra V. Fluctuations in the abundance of a species considered mathematically. Nature. 1926;118:558–560. [Google Scholar]
- 15.Blasius B, Huppert A, Stone L. Complex dynamics and phase synchronization in spatially extended ecological systems. Nature. 1999;399(6734):354–359. doi: 10.1038/20676. [DOI] [PubMed] [Google Scholar]
- 16.Vandermeer J. Oscillating populations and biodiversity maintenance. Bioscience. 2006;56(12):967–975. [Google Scholar]
- 17.Rabajante JF, et al. Red Queen dynamics in multi-host and multi-parasite interaction system. Sci Rep. 2015;5:10004. doi: 10.1038/srep10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mouritsen KN, Poulin R. Parasitism, community structure and biodiversity in intertidal ecosystems. Parasitology. 2002;124(Suppl):S101–S117. doi: 10.1017/s0031182002001476. [DOI] [PubMed] [Google Scholar]
- 19.Fujii T, Rondelez Y. Predator-prey molecular ecosystems. ACS Nano. 2013;7(1):27–34. doi: 10.1021/nn3043572. [DOI] [PubMed] [Google Scholar]
- 20.Bansho Y, et al. Importance of parasite RNA species repression for prolonged translation-coupled RNA self-replication. Chem Biol. 2012;19(4):478–487. doi: 10.1016/j.chembiol.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 21.Chetverin AB, Chetverina HV, Demidenko AA, Ugarov VI. Nonhomologous RNA recombination in a cell-free system: Evidence for a transesterification mechanism guided by secondary structure. Cell. 1997;88(4):503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bresch C, Niesert U, Harnasch D. Hypercycles, parasites and packages. J Theor Biol. 1980;85(3):399–405. doi: 10.1016/0022-5193(80)90314-8. [DOI] [PubMed] [Google Scholar]
- 23.Niesert U, Harnasch D, Bresch C. Origin of life between Scylla and Charybdis. J Mol Evol. 1981;17(6):348–353. doi: 10.1007/BF01734356. [DOI] [PubMed] [Google Scholar]
- 24.Boerlijst MC, Hogeweg P. Spiral wave structure in pre-biotic evolution - Hypercycles stable against parasites. Physica D. 1991;48(1):17–28. [Google Scholar]
- 25.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19(8):751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 26.Hosoda K, et al. Kinetic analysis of the entire RNA amplification process by Qbeta replicase. J Biol Chem. 2007;282(21):15516–15527. doi: 10.1074/jbc.M700307200. [DOI] [PubMed] [Google Scholar]
- 27.Eigen M, Schuster P. Hypercycle - Principle of natural self-organization. B. Abstract hypercycle. Naturwissenschaften. 1978;65(1):7–41. doi: 10.1007/BF00450633. [DOI] [PubMed] [Google Scholar]
- 28.Smith JM. Hypercycles and the origin of life. Nature. 1979;280(5722):445–446. doi: 10.1038/280445a0. [DOI] [PubMed] [Google Scholar]
- 29.Valen LV. A new evolutonary law. Evol Theory. 1973;1:1–30. [Google Scholar]
- 30.Ichihashi N, Matsuura T, Hosoda K, Yomo T. Identification of two forms of Qβ replicase with different thermal stabilities but identical RNA replication activity. J Biol Chem. 2010;285(48):37210–37217. doi: 10.1074/jbc.M110.117846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichihashi N, Aita T, Motooka D, Nakamura S, Yomo T. Periodic pattern of genetic and fitness diversity during evolution of an artificial cell-like system. Mol Biol Evol. 2015;32(12):3205–3214. doi: 10.1093/molbev/msv189. [DOI] [PubMed] [Google Scholar]