SUMMARY
The gut microbiotas of zebrafish and mice share six bacterial divisions, although the specific bacteria within these divisions differ. To test how factors specific to host gut habitat shape microbial community structure, we performed reciprocal transplantations of these microbiotas into germ-free zebrafish and mouse recipients. The results reveal that communities are assembled in predictable ways. The transplanted community resembles its community of origin in terms of the lineages present, but the relative abundance of the lineages changes to resemble the normal gut microbial community composition of the recipient host. Thus, differences in community structure between zebrafish and mice arise in part from distinct selective pressures imposed within the gut habitat of each host. Nonetheless, vertebrate responses to microbial colonization of the gut are ancient: Functional genomic studies disclosed shared host responses to their compositionally distinct microbial communities and distinct microbial species that elicit conserved responses.
INTRODUCTION
Animal evolution has occurred, and is occurring, in a world dominated by microorganisms. As animals evolved to occupy different habitats (addresses) and niches (professions) in our biosphere, they have forged strategic alliances with microorganisms on their body surfaces. The genomes of microbes within these consortia encode physiologic traits that are not represented in host genomes: Microbial-microbial and host-microbial mutualism endows the resulting “super-organisms” with a fitness advantage (Ley et al., 2006b). The majority of these microbes are present in digestive tract communities where, among other things, they contribute to the harvest of dietary nutrients that would otherwise be inaccessible (Bäckhed et al., 2004; Sonnenburg et al., 2005), as well as to the education of the host’s immune system (Cebra, 1999).
The advent of massively parallel DNA sequencers provides an opportunity to define the gene content of these indigenous microbial communities with increased speed and economy. These “microbiome” sequencing projects promise to provide a more comprehensive view of the genetic landscape of animal-microbial alliances and testable hypotheses about the contributions of microbial communities to animal biology. The results should allow a number of fundamental questions to be addressed. Is there an identifiable core microbiota and microbiome associated with a given host species? How are a microbiota and its microbiome selected, and how do they evolve within and between hosts? What are the functional correlates of diversity in the membership of a microbiota and in the genetic composition of its microbiome?
Answers to these questions also require model organisms to assess how communities are assembled, to determine how different members impact community function and host biology, and to as certain the extent of redundancy or modularity within a microbiota. One approach for generating such models is to use gnotobiotics—the ability to raise animals under germ-free (GF) conditions—to colonize them at varying points in their life cycle with a single microbe or more complex collections, and to then observe the effects of host habitat on microbial community structure and function and of the community on the host. Methods for raising and propagating rodents under GF conditions have been available for 50 years (see Wostmann, 1981), although genomic and allied computational methods for comprehensively assessing microbial community composition, gene content, and host-microbial structure/function relationships have only been deployed in the last five years (e.g., Hooper and Gordon, 2001; Ley et al., 2005). Recently, we developed techniques for rearing the zebrafish(Danio rerio)under GF conditions (Rawls et al., 2004).In principle, this model organism provides a number of attractive and distinctive features for analyzing host-microbial mutualism. Zebrafish remain transparent until adulthood, creating an opportunity to visualize microbes in their native gut habitats in real time. A deep draft reference genome sequence of D. rerio is available (http://www.sanger.ac.uk/Projects/D_rerio/). In addition, forward genetic tests and chemical screens can be conducted (Patton and Zon, 2001; Peterson and Fishman, 2004) to characterize zebrafish signaling pathways regulated by microbial consortia and/or their component members.
A preliminary functional genomic study of the effects of colonizing GF zebrafish with an unfractionated microbiota harvested from adult conventionally raised (CONV-R) zebrafish revealed 59 genes whose responses were similar to those observed when GF mice were colonized with an adult mouse gut microbiota (Rawls et al., 2004). These genes encode products affecting processes ranging from nutrient metabolism to innate immunity and gut epithelial cell turnover (Rawls et al., 2004). The experiments did not distinguish whether the host responses were evolutionarily conserved and thus present in the last common ancestor of fish and mammals, or if they had been independently derived in mammals and fish. However, the fact that numerous homologous genes and shared cellular changes comprised the “common” response favors the notion of evolutionary conservation over convergence. It was also unclear whether these common host responses were elicited by the same or different bacterial signals in each host or by signals from the whole community versus from specific bacteria.
A recent comprehensive 16S rRNA sequence-based survey of the adult mouse gut disclosed that, as in humans, >99% of the bacterial phylogenetic types (phylotypes) belong to two divisions—the Firmicutes and Bacteroidetes (Ley et al., 2005). In contrast, limited surveys of different fish species indicate that their gut communities are dominated by the Proteobacteria (Cahill, 1990; Huber et al., 2004; Rawls et al., 2004; Bates et al., 2006; Romero and Navarrete, 2006). Fish and mammals live in very different environments, so it is possible that differences in their gut microbiotas arise from “legacy effects” (e.g., local environmental microbial community composition or inheritance of a microbiota from a parent). Furthermore, legacy effects might combine with “gut habitat effects” (e.g., distinct selective pressures arising from differences in anatomy, physiology, immunologic “climate,” or nutrient milieu) to shape the different community structures of fish and mammals.
In the present study, we have performed reciprocal microbiota transplantations in GF zebrafish and mice. We provide evidence that gut habitat shapes microbial community structure and that both animal species respond in remarkably similar ways to components of one another’s microbiota.
RESULTS
Comparison of the Zebrafish and Mouse Gut Microbiota: Overlapping Bacterial Divisions but Marked Differences at More Shallow Phylogenetic Resolution
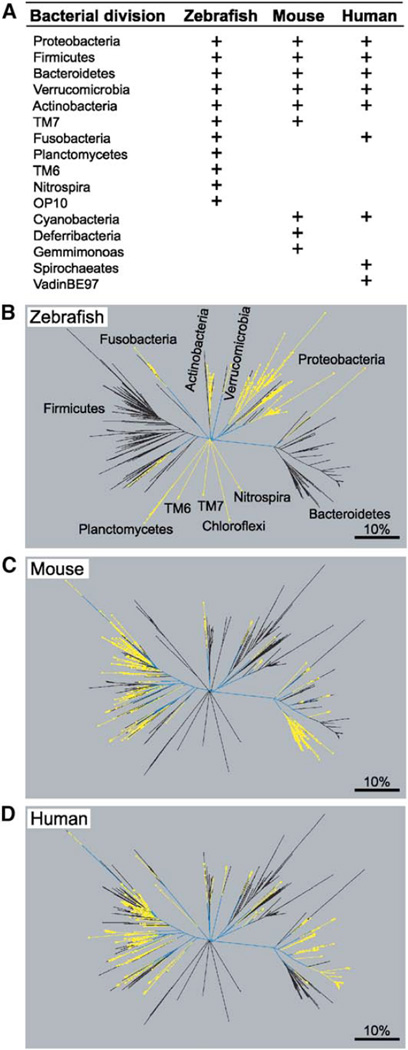
Our previous survey of the gut microbiota of adult CONV-R zebrafish was limited to 176 bacterial 16S rRNA gene sequences (Rawls et al., 2004). Therefore, we performed a more comprehensive analysis of intestinal contents pooled from 18 adult male and female C32 zebrafish (comprised of two independent pools, each containing material from 9 animals). A total of 1456 bacterial 16S rRNA sequences formed the final analyzed dataset: 616 from pool 1 and 840 from pool 2 (libraries JFR0503 and JFR0504, respectively, in Table S1 available with this article online). Phylogenetic analysis revealed 198 “species-level” phylotypes defined by 99% pairwise sequence identity. These phylotypes represented a total of 11 bacterial divisions and were dominated by the Proteobacteria (82% ± 22.9% [SD] of all clones averaged across both libraries) and the Fusobacteria (11% ± 15.2%; Figures 1 and 2). The Firmicutes, Bacteroidetes, Verrucomicrobia, Actinobacteria, TM7, Planctomycetes, TM6, Nitrospira, and OP10 divisions were minor components (3.2%–0.6%).
Figure 1. Bacterial Divisions and Their Lineages Detected in the Zebrafish Digestive Tract, Mouse Cecum, and Human Colon.
(A) Summary of shared and distinct bacterial divisions in the zebrafish, mouse, and human gut microbiota (data from this study; Rawls et al., 2004; Ley et al., 2005; Eckburg et al., 2005; Bäckhed et al., 2005). Divisions found in the normal gut microbiota of each host are indicated (+). (B–D) Phylogenetic trees constructed from enumeration studies of the zebrafish digestive tract (B), mouse cecal (C), and human colonic (D) microbiotas. The zebrafish data are 1456 16S rRNA gene sequences derived from adult CONV-R C32 fish. The mouse data are 2196 sequences from adult CONV-R C57Bl/6J mice and their mothers (Ley et al., 2005). The human dataset contains 2989 bacterial 16S rRNA sequences from colonic mucosal biopsies and a fecal sample obtained from a healthy adult (Eckburg et al., 2005). Within a given panel, yellow lines indicate lineages unique to the host, blue lines indicate lineages that are shared by at least one other host, while black lines indicate lineages that are absent from the host. The scale bar indicates 10% pairwise 16S rRNA sequence divergence.
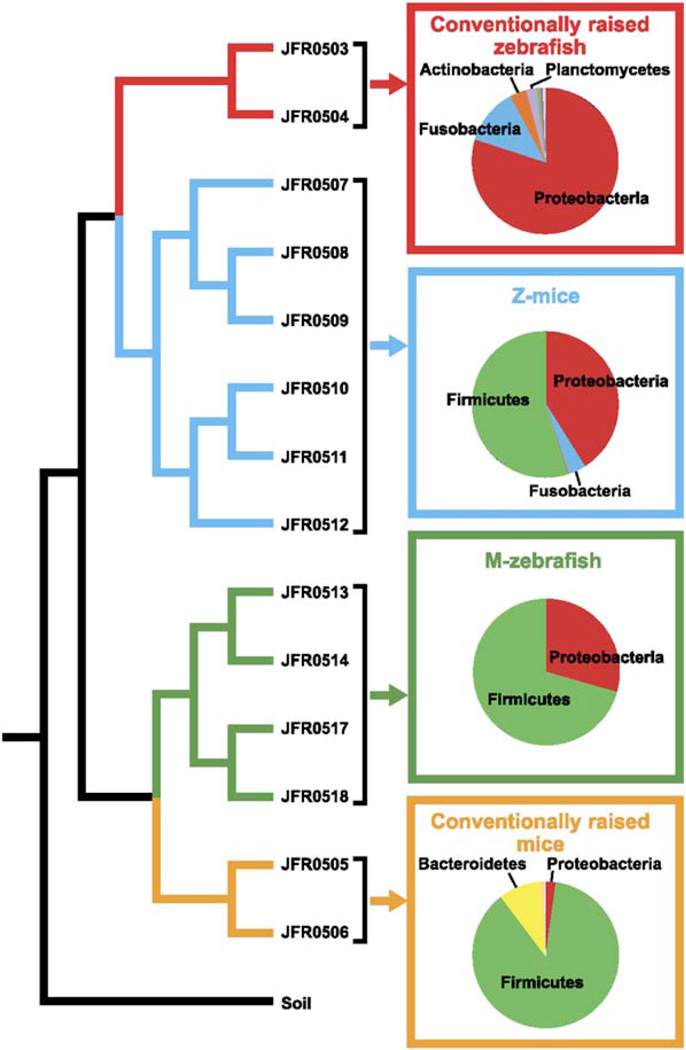
Figure 2. Comparison of Input and Output Communities following Reciprocal Transplantation of Gut Microbiotas in Gnotobiotic Zebrafish and Mice.
Tree based on pairwise differences between the following bacterial communities (weighted UniFrac metric, based on a 6379 sequence tree; Lozupone and Knight, 2005): (1) CONV-R zebrafish digestive tract microbiota (conventionally raised zebrafish, red); (2) CONV-R mouse cecal microbiota (conventionally raised mice, yellow); (3) output community from the cecal contents of ex-GF mice that had been colonized with a normal zebrafish microbiota (Z-mice, blue); (4) output community from the digestive tracts from ex-GF zebrafish that had been colonized with a normal mouse microbiota (M-zebrafish, green); and (5) a control soil community that served as an outgroup (Soil; Axelrood et al., 2002). The distance p value for this entire UniFrac tree (UniFrac P, the probability that there are more unique branches than expected by chance, using 1000 iterations) was found to be <0.001, assigning high confidence to the overall structure of the UniFrac tree. 16S rRNA library names are shown next to their respective branch (see Table S1 for additional details about these libraries). The relative abundance of different bacterial divisions within these different communities (replicate libraries pooled) is shown in pie charts with dominant divisions highlighted.
Six of the eleven bacterial divisions found in adult zebrafish are also found in mice (Ley et al., 2005); five of these are also shared by the adult human microbiota (Eckburg et al., 2005; Figure 1A). However, zebrafish community members within these shared divisions are distinct from those in mice and humans at more shallow phylogenetic resolution (Figures 1B–1D).
The Gut Selects Its Microbial Constituents
The composition of the mouse gut microbiota is affected by host genotype, as well as by legacy (it is inherited from the mother; Ley et al., 2005). To determine whether the observed differences between zebrafish and mouse microbiotas reflect host genome-encoded variations in their gut habitats versus differences in the local microbial consortium available for colonization, we colonized (1) adult GF mice with an unfractionated gut microbiota harvested from CONV-R adult zebrafish (yielding “Z-mice”) and (2) GF zebrafish larvae with a gut microbiota from CONV-R adult mice (“M-zebrafish”). By comparing the composition of the community introduced into the GF host (“input community”) with the community that established itself in the host (Z-mouse or M-zebrafish “output community”), we sought to determine whether gut microbial ecology is primarily influenced by legacy effects (the input community structure would persist in the new host) versus gut habitat effects (the representation changes when certain taxa are selected).
We introduced the pooled intestinal contents of 18 CONV-R adult zebrafish belonging to the C32 inbred strain (pools 1 and 2 above) into adult GF mice belonging to the NMRI inbred strain (n = 6, Table S1, Figure S1). The resulting Z-mice were housed in gnotobiotic isolators and sacrificed 14 days after colonization (i.e., after several cycles of replacement of the intestinal epithelium and its overlying mucus layer). Their cecal contents were harvested and provided community DNA for 16S rRNA sequence-based enumerations. The cecum was selected for this analysis because it is a well-defined anatomic structure located at the junction of the small intestine and colon, and its luminal contents can be readily and reliably recovered. It also harbors a very dense microbial population in CONV-R mice (1011–1012 organisms/ml luminal contents) that has been comprehensively surveyed (Ley et al., 2005).
In addition to the 1456 16S rRNA sequences representing 198 phylotypes from the input zebrafish community (libraries JFR0503 and JFR0504; see above), we obtained a total of 1836 sequences representing 179 phylotypes from the Z-mouse cecal community (libraries JFR0507–12; Figures S1 and S2). Only 12% of the phylotypes found in the Z-mouse community, representing 39% of all sequences, were detected in the input zebrafish community. The dominant division in the input zebrafish community (Proteobacteria) persisted but shrank in abundance in the Z-mouse community (82% ± 22.9% in the input versus 41.7% ± 8.9% in the output; Figure 2). The Z-mouse community only contained members of the γ- and β-Proteobacteria subdivisions, whereas the input zebrafish community had also included δ- and α-Proteobacteria. In addition, members of the Bacteroidetes detected in the input zebrafish community were not observed in the Z-mouse community. The Z-mouse community showed a striking amplification of the Firmicutes (1% ± 1.1% of the input, 54.3% ± 6.5% of the Z-mouse output; Figure 2); this amplification included members of Bacilli as well as Clostridia classes.
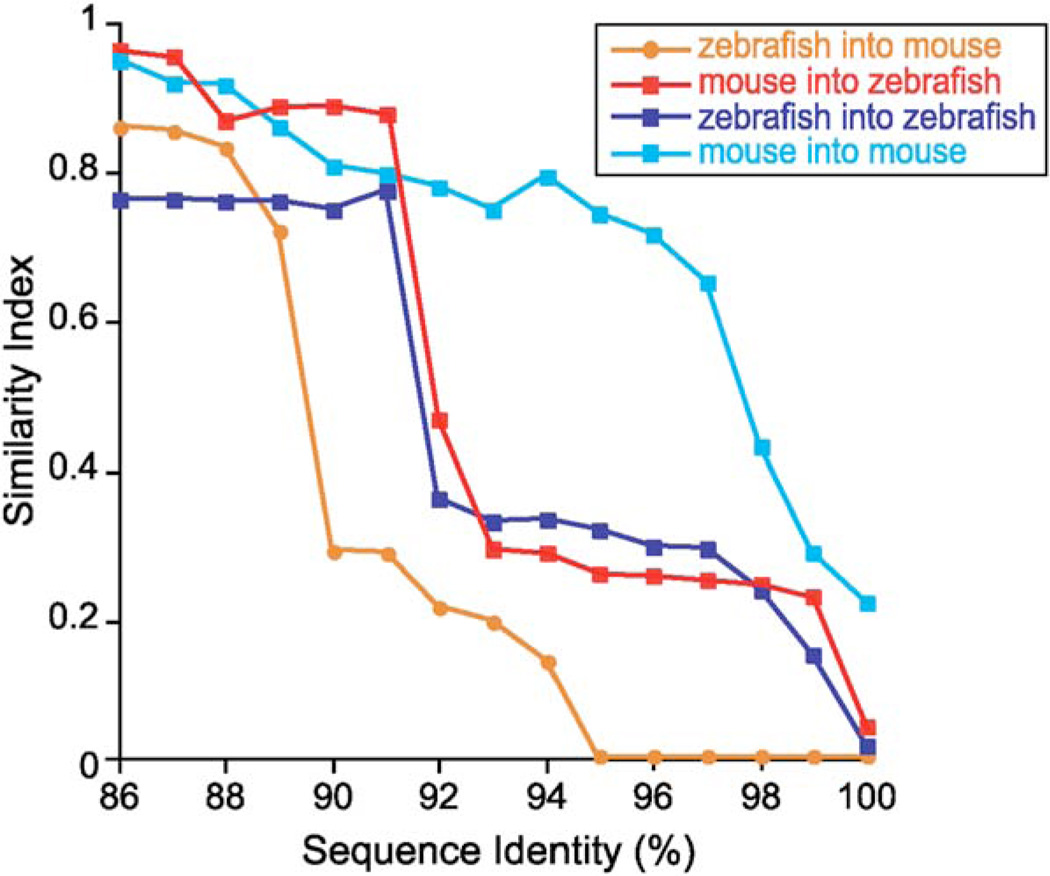
By comparing communities at multiple thresholds for pairwise percent identity among 16S rRNA gene sequences (%ID), we determined that divergence between the input zebrafish and output Z-mouse communities occurred at 89%ID and higher (Figure 3). This implies that genera represented within the zebrafish and Z-mouse gut microbiotas are different but represent the same major lineages. The analysis also demonstrated that the phylotypes that bloomed in the mouse cecum were minor constituents of the input zebrafish digestive tract community. Despite the difference in genus/species representation, the richness and diversity of the input zebrafish and Z-mouse gut communities remained similar through the shift in microbial community composition (Figure S2 and Table S1).
Figure 3. Similarity Indices for Pairwise Comparisons of Communities Defined as Assemblages of Phylotypes Computed at Levels of %ID Ranging from 86%ID to 100%ID and Compared at Each %ID Threshold using the Chao-Jaccard Abundance-Based Similarity Index.
Abbreviations: zebrafish into mouse, CONV-R zebrafish compared to Z-mouse microbiotas; mouse into zebrafish, CONV-R mouse compared to M-zebrafish microbiotas; zebrafish into zebrafish, CONV-R zebrafish compared to Z-zebrafish microbiotas (data from Rawls et al., 2004); mouse into mouse, CONV-R mouse compared to M-mouse microbiotas (data from Bäckhed et al., 2004). Similarity indices range from 0 (no overlap in composition) to 1 (identical communities).
When a similar analysis was applied to the input mouse and M-mouse communities obtained from a mouse-into-mouse microbiota transplant experiment (Bäckhed et al., 2004), we found that a high degree of similarity was maintained at levels as great as 97%ID (Figure 3). Based on these results, we concluded that (1) the difference in composition of the input zebrafish and output Z-mouse communities is not likely to be due to the microbiota transplantation procedure per se and (2) the adult mouse cecum is able to support a complex foreign microbial consortium by shaping its composition.
We performed the reciprocal experiment by colonizing recently hatched (3 days post-fertilization [dpf]) GF C32 zebrafish with the pooled cecal contents of three CONV-R adult female mice (libraries JFR0505 and JFR0506 in Table S1) and conducting surveys of the recipients’ digestive tract communities 3 or 7 days later (libraries JFR0513-18 in Table S1; Figure S1). As in the previous experiment, the dominant bacterial division in the input mouse community (Firmicutes) persisted in the output M-zebrafish community (87.3% ± 2.2% of input, 64.9% ± 41.7% of output; Figure 2). However, only members of Bacilli, the dominant Firmicute class in the zebrafish but not the normal mouse gut microbiota, were retained; other prominent members of the Firmicutes found in the input mouse library (i.e., Clostridia and Mollicutes) were no longer detected in the M-zebrafish gut. Bacteroidetes (9.8% ± 3.3% of input community) were also undetected. Proteobacteria, a minor member of the input mouse community, were amplified markedly in the M-zebrafish gut (2.2% ± 0.6% of input, 35.1% ± 41.7% of output; Figure 2).
In addition to their drastic compositional differences, we also found that the output M-zebrafish community was less rich and less diverse than the input mouse community (Table S1 and Figure S2), indicating that only a small subset of the mouse gut microbial consortium was able to establish and/or thrive in the larval M-zebrafish gut. In contrast to the reciprocal zebrafish-into-mouse experiment where the contents of the adult fish gut were gavaged directly into the stomachs of recipient GF mice, our mouse-into-zebrafish gut microbiota transplantation involved introduction of mouse cecal contents into gnotobiotic zebrafish medium (GZM) containing 3dpf fish. Therefore, environmental factors could operate to select a subset of the input mouse community prior to entry in the recipient fish gut.
The similarities between input mouse and M-zebrafish communities were high, from 86%ID to 91%ID, above which the communities diverged in composition (Figure 3), i.e., different genera were representative of the same deeper phylogenetic lineages. Indeed, there was no overlap between phylotypes with threshold pairwise ≥99%ID in the datasets obtained from the input mouse and M-zebrafish communities. This was due, in part, to the limited degree of coverage (73% for the input community according to Good’s method; Good, 1953). Phylotypes that were detected only in the M-zebrafish community were identifiable in the input mouse community using PCR and phylotype-specific primers (e.g., Staphylococcus; data not shown). Compared to the reciprocal zebrafish-into-mouse transplantation experiment, the input mouse and output M-zebrafish communities diverged at a higher %ID cutoff (Figure 3), indicating that they were more similar at a higher taxonomic level than the zebrafish/Z-mouse communities. Part of the drop in similarity could be attributed to the experimental manipulation since a similar analysis of a zebrafish-into-zebrafish transplant (Rawls et al., 2004) revealed a drop in similarity at a comparable %ID (Figure 3).
The similarity indices described above are derived from phylotype abundances at different phylotype thresholds (%IDs). However, an implicit assumption underlying such an analysis is that all phylotypes are treated equally regardless of lineage, even though they may represent similar or very unrelated lineages (Lozupone and Knight, 2005). Another way to compare communities is the UniFrac analysis: In this method, the abundance of each lineage is weighted, such that the abundance of lineages is considered as well as which lineages are present (Lozupone and Knight, 2005). The UniFrac approach circumvents the problem of having to decide at what %ID level to define the phylotype units that we call “different” (the cut-off is likely to vary according to lineage).
UniFrac analysis revealed that replicate Z-mouse datasets are most similar to the input zebrafish datasets with respect to detected lineages (Figure 2). However, the abundance of the Firmicutes in Z-mice expanded to resemble the division’s abundance in CONV-R mice, indicating that the input community, although derived from a zebrafish, has been shaped to resemble a native mouse community. Similarly, the M-zebrafish communities are most similar to the mouse input communities by UniFrac, but the Proteobacteria in M-zebrafish expanded to resemble a CONV-R zebrafish community, indicating that the input mouse community has been shaped to resemble a native zebrafish microbiota (Figure 2).
Together, the results from our reciprocal microbiota transplantation experiments disclose that (1) gut habitat sculpts community composition in a consistent fashion, regardless of the input, and (2) stochastic effects are minimal (One notable exception was that γ-Proteobacteria in M-zebrafish [Escherichia, Shigella, and Proteus spp.] were more abundant in one experimental replicate [69.8% ± 20.5%] compared to the other [0.5% ± 0.6%]). The amplified taxa in both sets of transplantation experiments represented dominant divisions in the native gut microbiota of the respective host: Firmicutes in the case of teleostification (zebrafish-into-mouse), Proteobacteria in the case of murinization (mouse-into-zebrafish).
Shared Responses Elicited in Gnotobiotic Mice after Exposure to a Mouse or Zebrafish Gut Microbiota from Conventionally Raised Animals
While the studies described above indicated that the composition of the gut microbiota is sensitive to host habitat, we did not know whether the host response was sensitive to microbial community composition. Therefore, we conducted a GeneChip-based functional genomic analysis of gene expression in the distal small intestines (ileums) of mice that had been subjected to zebrafish-into-mouse (Z-mice) and mouse-into-mouse (M-mice) microbiota transplantations. All animals (n = 3–5/treatment group) were sacrificed 14 days after inoculation, RNA was prepared from the ileum of each mouse, and the cRNA target generated from each RNA sample was hybridized to an Affymetrix 430 v2 mouse GeneChip. Ingenuity Pathways Analysis software (IPA; see Supplemental Data) was then used to compare host responses to these different microbial communities. IPA software was utilized for genes that exhibited a ≥1.5-fold change (increased or decreased) in their expression compared to GF controls (false discovery rate <1%).
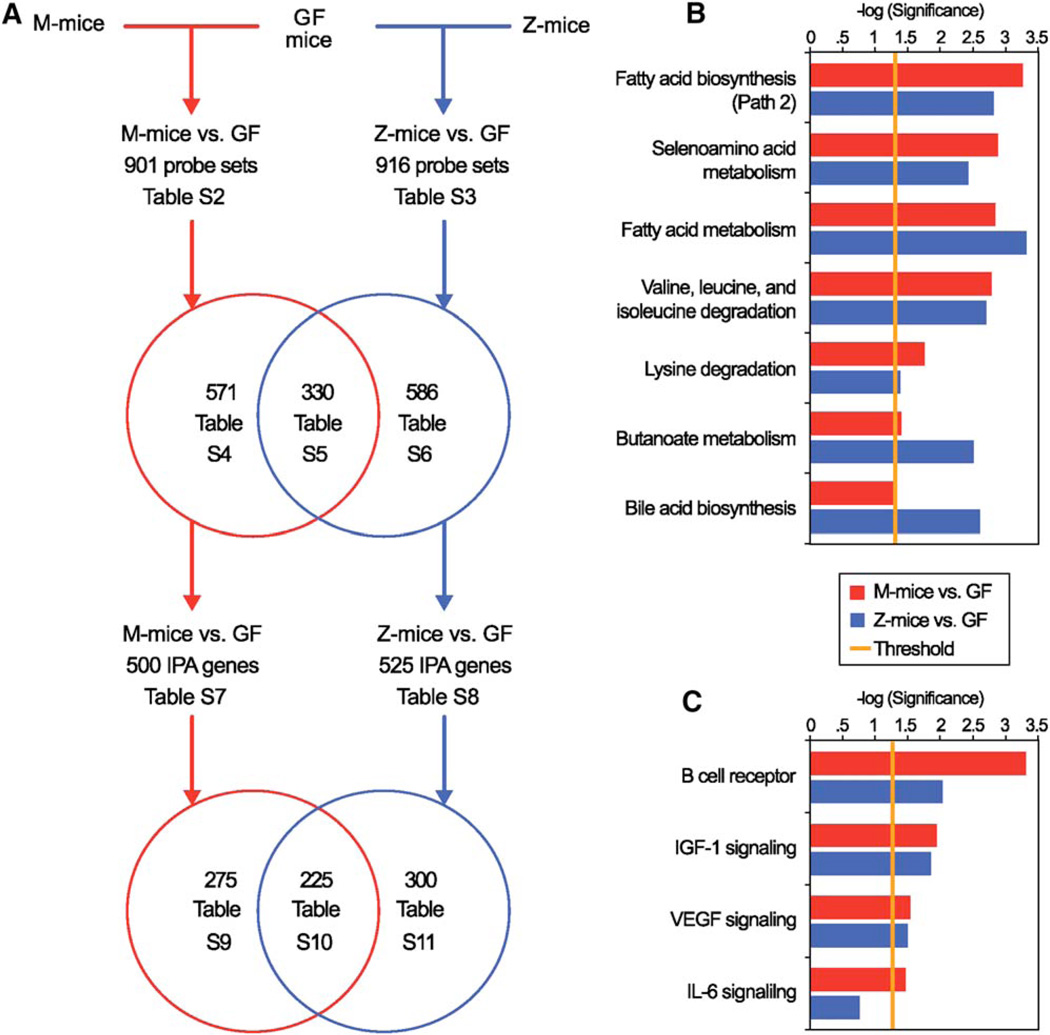
Despite the different bacterial compositions of the two input communities, their impact on the mouse was remarkably similar (Figure 4). The number of IPA-annotated mouse genes whose expression changed in response to the two microbiotas was comparable: 500 in response to the native mouse microbiota (Table S7) and 525 in response to the zebrafish microbiota (Table S8 and Figure 4A). Approximately half of the genes (225) were responsive to both microbial communities (Table S10): 217 (96.4%) were regulated in the same direction. Among the two sets of responsive genes, there was shared enrichment of IPA-annotated metabolic pathways involved in (1) biosynthesis and metabolism of fatty acids (sources of energy as well as substrates for synthesis of more complex cellular lipids in an intestinal epithelium that undergoes continuous and rapid renewal); (2) metabolism of essential amino acids (valine, isoleucine, and lysine); (3) metabolism of amino acids that contain the essential trace element selenium (selenocystine/selenomethioinine) and are incorporated into the active sites of selenoproteins such as glutathione peroxidase; (4) metabolism of butyrate (a product of polysaccharide fermentation that is a key energy source for the gut epithelium); and (5) biosynthesis of bile acids needed for absorption of lipids and other hydrophobic nutrients (Figure 4B and Table S12).
Figure 4. Identifying a Common Response of the Germ-free Mouse Distal Small Intestine to Colonization with Mouse and Zebrafish Gut Microbial Communities.
(A) Summary of results of GeneChip analysis of the ileal transcriptome in GF mice versus mice colonized for 14 days with a mouse cecal microbiota (M-mice versus GF; red lines) or a normal zebrafish digestive tract microbiota (Z-mice versus GF; blue lines). Note that only a subset of all Affymetrix GeneChip probe sets are annotated by Ingenuity Pathway Analysis (IPA). Supplemental tables containing GeneChip probe set and IPA gene information are indicated. IPA reveals metabolic pathways (panel B; Table S12) and molecular functions (panel C; Table S13) that are significantly enriched (p < 0.05) in the host response to each community. The seven most significant metabolic pathways and the four most significant signaling pathways from the M-mice versus GF mice comparison (red bars) are shown along with corresponding data from the Z-mice versus GF mice comparison (blue bars). (Not shown: the 275 IPA-annotated mouse genes regulated by the mouse microbiota but unchanged by the zebrafish digestive tract microbiota were significantly enriched for components of ERK/MAPK, SAPK/JNK, antigen presentation, and the pentose phosphate pathways [Table S9]. In contrast, the 300 IPA-annotated mouse genes regulated by the zebrafish microbiota but unchanged by the mouse microbiota were enriched for components of glutamate and arginine/proline metabolism, ketone body synthesis/degradation, plus β-adrenergic signaling pathways [Table S11]).
Both communities altered expression of a similar set of genes involved in insulin-like growth factor-1 (Igf-1), vascular endothelial growth factor (Vegf), B cell receptor, and interleukin-6 (Il-6) signaling pathways (Figure 4C and Table S13). These results are intriguing: Previous mouse-into-mouse and zebrafish-into-zebrafish transplantations revealed that the microbiota-directed increase in proliferative activity of gut epithelial lineage progenitors is a shared host response (Rawls et al., 2004). The underlying mechanisms are not known. However, we recently found that components of Igf-1, Vegf, B cell receptor, and Il-6 signaling pathways were significantly enriched in mouse small intestinal epithelial progenitors (Giannakis et al., 2006). Thus, it is tempting to speculate that these pathways may be involved in mediating the microbiota’s effect on mouse intestinal epithelial renewal.
Taken together, these results reveal a commonality in the transcriptional responses of the mouse to two microbial communities with shared divisions represented by different lineages at a finer phylogenetic resolution (Figure 1). This common response to a microbiota may reflect as yet unappreciated shared functional properties expressed by the two compositionally distinct communities and/or a core response, evolved by the mouse gut to distinct microbial communities.
Comparison of Zebrafish Host Responses to a Zebrafish versus a Mouse Gut Microbiota
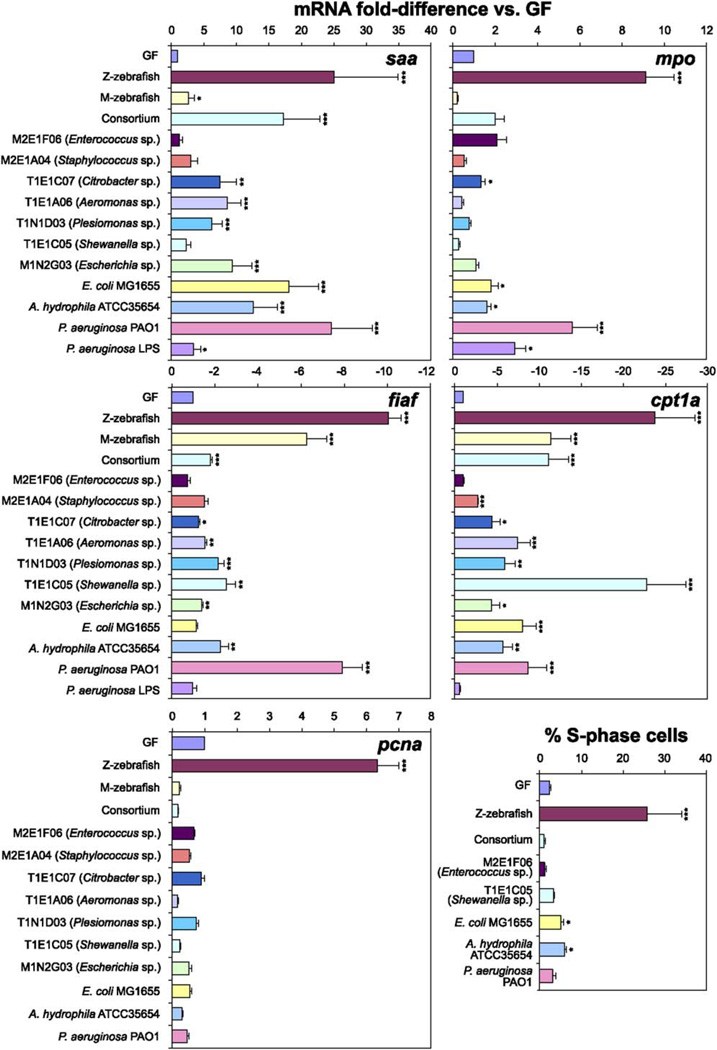
Analysis of zebrafish 3 days after colonization with either a zebrafish or a mouse microbiota at 3dpf also demonstrated shared features of the host response to both microbial communities. To quantify these responses, we selected biomarkers identified from our comparisons of 6dpf GF, CONV-R, and Z-zebrafish (Rawls et al., 2004). Quantitative real-time RT-PCR (qRT-PCR) of biomarkers of lipid metabolism, including fasting-induced adipose factor (fiaf; circulating inhibitor of lipoprotein lipase, Bäckhed et al., 2004), carnitine palmitoyltransferase 1a (cpt1a), and the trifunctional enzyme hydroxyacylCoA dehydrogenase/3-ketoacylCoA thiolase/enoyl CoA hydratase a (hadha), revealed that the mouse microbiota was able to largely recapitulate the effect of the zebrafish microbiota (Figures 5 and S3). In contrast, the zebrafish microbiota, but not the mouse microbiota, prominently increased host expression of (1) innate immune response biomarkers (serum amyloid a [saa], myeloperoxidase [mpo; Lieschke et al., 2001; Figure 5], and complement component factor b [bf; Figure S3]) and (2) proliferating cell nuclear antigen (pcna; biomarker of epithelial cell renewal; Figure 5).
Figure 5. qRT-PCR Assays of the Responses of Germ-free Zebrafish to Colonization with Individual Culturable Members of the Zebrafish and Mouse Gut Microbiotas.
Expression levels of serum amyloid a (saa), myeloperoxidase (mpo), fasting-induced adipose factor (fiaf), carnitine palmitoyltransferase 1a (cpt1a), and proliferating cell nuclear antigen (pcna) were assessed using RNA extracted from the pooled digestive tracts of 6dpf zebrafish inoculated since 3dpf with a CONV-R zebrafish microbiota (Z-zebrafish), a CONV-R mouse microbiota (M-zebrafish), a consortium of seven primary isolates (Consortium), a primary Enterococcus isolate (M2E1F06), a primary Staphylococcus isolate (M2E1A04), a primary Citrobacter isolate (T1E1C07), a primary Aeromonas isolate (T1E1A06), a primary Plesiomonas isolate (T1N1D03), a primary Shewanella isolate (T1E1C05), a primary Escherichia isolate (M1N2G03), an Escherichia coli type strain (E. coli MG1655), an Aeromonas hydrophila type strain (A. hydrophila ATCC35654), a Pseudomonas aeruginosa type strain (P. aeruginosa PAO1), or 0.1 µg/ml P. aeruginosa LPS (P. aeruginosa LPS). Data from biological duplicate pools (≥10 animals per pool) were normalized to 18S rRNA levels and results expressed as mean fold-difference compared to GF controls ± SEM. S phase cells were quantified in the intestinal epithelium of 6dpf zebrafish colonized since 3dpf with a CONV-R zebrafish microbiota (Z-zebrafish), a consortium of seven primary isolates (Consortium), or individual species. The percentage of all intestinal epithelial cells in S phase was scored using antibodies directed against BrdU, following incubation in BrdU for 24 hr prior to sacrifice. Data are expressed as the mean of two independent experiments ± SEM (n = 9–15 five micron-thick transverse sections scored per animal, ≥7 animals analyzed per experiment). ***, p < 0.0001; **, p < 0.001; *, p < 0.05.
Selecting Readily Culturable Microbial Species that Are Useful Models for Translating Information about Host-Bacterial Mutualism from Zebrafish to Mice
In order to use gnotobiotic zebrafish as a surrogate for studying the mechanisms underlying host-microbial mutualism in the mammalian gut, we sought culturable bacterial species that were capable of (1) efficiently colonizing the digestive tracts of GF zebrafish and mice and (2) eliciting evolutionarily conserved host responses in both hosts. Therefore, we performed culture-based bacterial surveys of the Z-mouse and M-zebrafish output communities in parallel with our culture-independent 16S rRNA surveys. 16S rRNA sequence-based analysis of 160 different bacterial isolates from the communities of six Z-mice yielded 47 different phylotypes (defined at 97%ID) representing four divisions (Proteobacteria, Fusobacteria, Actinobacteria, and Firmicutes). Similarly, an analysis of 303 isolates recovered from the communities of 18 M-zebrafish yielded 41 phylotypes representing the Proteobacteria and Firmicutes (Tables S1 and S14).
We selected seven primary isolates from the transplantation experiments representing the Firmicutes (Enteroccoccus and Staphylococcus spp.) and the Proteobacteria (Shewanella, Aeromonas, Citrobacter, Plesiomonas, Escherichia spp.). Three laboratory strains of γ-Proteobacteria (Aeromonas hydrophila ATCC35654, Pseudomonas aeruginosa PAO1, and E. coli MG1655) were used as controls (Table S15). These primary isolates and lab strains were selected based on the relative abundance of their phylotypes in our culture-based surveys of input and output communities (Table S14).
3dpf GF zebrafish were exposed to 104 CFU of each primary isolate or strain per milliliter of gnotobiotic zebrafish medium (GZM); all reached similar densities in the digestive tract by 6dpf (104–105 CFU/gut). These densities are similar to those documented in age-matched CONV-R or Z-zebrafish (Rawls et al., 2004).
An epidermal degeneration phenotype that develops in fed (but not fasted) GF zebrafish beginning at 9dpf (Rawls et al., 2004) was ameliorated by colonization with nine of the ten bacterial strains at 3dpf. The Enterococcus isolate M2E1F06 was the only tested strain that did not have any detectable effect (Figure S4). We found that epidermal degeneration could also be prevented by placing a mesh bag, containing an autoclaved mixture of activated carbon and cation exchange resin, into the GZM (Figure S4). This latter finding suggests that rescue by most of the tested bacterial strains involves bioremediation of toxic compounds that accumulate when GF zebrafish are exposed to food. Our subsequent analysis of the impact of the Firmicutes (i.e., Enterococcus and Staphylococcus isolates) on gut gene expression was performed using zebrafish raised in the presence of activated carbon and resin.
qRT-PCR analysis of biomarkers of lipid metabolism, including fiaf, cpt1a, and hadha, revealed that five of the seven primary isolate strains and all of the type strains tested were able to at least partially recapitulate the response obtained after exposure to an unfractionated zebrafish microbiota. Colonization with T1E1C05 (Shewanella sp.) and P. aeruginosa PAO1 had the largest effects (Figures 5 and S3). Two biomarkers of innate immune responses, saa and bf, were also responsive to the majority of these strains, but the granulocyte-specific marker mpo was relatively specific for P. aeruginosa PAO1 (Figures 5 and S3). None of the tested individual bacterial strains, including PAO1, were able to recapitulate the degree of stimulation of cell division in the intestinal epithelium of 6dpf zebrafish seen in the presence of an unfractionated zebrafish microbiota harvested from CONV-R donors, whether judged by qRT-PCR assays of pcna expression or by immunohistochemical analysis of the incorporation of BrdU administered 24 hr prior to sacrifice (Figure 5).
We also assessed the host response to colonization with a consortium consisting of an equal mixture of all seven primary isolates (n = 2 groups of 20 GF zebrafish colonized at 3dpf and sacrificed at 6dpf). qRT-PCR indicated that this model microbiota was able to partially recapitulate the nutrient metabolic and innate immune (but not epithelial proliferative) responses to the normal zebrafish microbiota. Importantly, the response to the consortium was a nonadditive representation of the responses to each component strain, and not equivalent to what was observed with a complete microbiota from CONV-R zebrafish (Figures 5 and S3).
qRT-PCR assays established that treatment of 6dpf zebrafish larvae with lipopolysaccharide (LPS) purified from P. aeruginosa was able to partially recapitulate innate immune responses seen with live P. aeruginosa (Figure 5). In contrast, LPS treatment did not affect expression of biomarkers of nutrient metabolism (Figure 5). This notion of distinct bacterial signaling mechanisms for innate immune and metabolic responses is supported by the observation that some of the tested isolates (e.g, T1E1C05, a Shewanella sp.) are able to induce robust nutrient metabolic responses without eliciting innate immune responses (Figures 5 and S3). Moreover, we found that all three classes of host response (innate immunity, nutrient metabolism, and cell proliferation) are strongly attenuated in the absence of an exogenous nutrient supply (See Figure S5).
DISCUSSION
There is considerable interest in how communities assemble at the microbial scale, and how the environment (e.g., local chemistry) and legacy effects (e.g., microbes available to colonize) interact to predict the composition of a community (Hughes-Martiny et al., 2006). Some host-associated microorganisms exhibit patterns of genetic differentiation that are related to the geographic distribution of their hosts (Bala et al., 2003; Falush et al., 2003). This raises the question of how much of the variation is due to habitat differences that correlate with geographic separation, versus the legacy of past communities. Our study directly tests the effect of habitat in assembling a community: We constrained the legacy effect by presenting empty GF hosts with a known microbial community so that observed changes in diversity could be correlated with factors specific to host gut habitat (e.g., either direct effects of the niche space or indirect effects on intercommunity dynamics).
UniFrac showed the output community of the Z-mouse to be made up of zebrafish-specific lineages, but the proportional representation of the divisions was more similar to what is typical of a mouse gut community. Conversely, the M-zebrafish digestive tract community was “teleostified” by a change in the proportions of divisions from the mouse input. Moreover, all ten of the individual cultured strains introduced into the GF host guts took up residence. These results show that the host will “work” with what it gets: We constrained the input by presenting the empty host with a constrained microbiota, and the resulting community took on a relative divisional abundance characteristic of the recipient host’s naturally occurring community.
What determines the host’s relative abundance of divisions? Its reproducibility regardless of the provenance of the input community underscores the presence of very powerful organizing principles in community composition that have yet to be fully explored. A simple interpretation of these findings is that members of the Firmicutes and Proteobacteria possess division-wide properties that allow them to succeed in the mouse and zebrafish gut, respectively; thus, even distantly related members within a division will respond similarly to habitat effects. If so, the implication is that there is considerable functional and/or physiological redundancy within lineages that are selected for in specific host gut habitats. One obvious difference between the Gram-positive Firmicutes and the Gram-negative Proteobacteria is their cell wall structure, which could be a target for selection. Another trait that may differentiate gut Firmicutes from Proteobacteria is their oxygen tolerence: The larval and adult zebrafish gut is predicted to have higher levels of oxygen than the mouse cecum and might exclude Firmicutes, whose members are more likely to be strictly anaerobic than the Proteobacteria. However, generalizations about division-level traits are conjecture and almost certainly prone to exceptions, particularly since they are based on a severely limited knowledge of the genomic features and phenotypes of gut bacteria. This is highlighted by our observation that the Firmicutes amplified in the ceca of Z-mice were only from the classes Bacilli and Clostridia, while the Proteobacteria amplified in M-zebrafish digestive tracts were only from the γ-Proteobacteria class.
The bacteria that establish themselves in a new host do not necessarily need to be identical by 16S rRNA %ID to be functionally similar ecotypes and to have similar genome content. Closely related phylotypes that form polytomies (i.e., star phylogenies) are common in the environment and in the animal gut (Acinas et al., 2004; Eckburg et al., 2005; Ley et al., 2006a, 2006b): Whole-genome comparisons of gut-dwelling Bacteroidetes species show that their proteomes have similar functional profiles, although they can differ in 16S rRNA %ID by as much as 12% (Xu et al., 2003; J. Xu, M.A.M, R.E.L., and J.I.G., unpublished data).
Curtis and Sloan (2004) state that when a new community is formed, it must be initiated by drawing from the available microbes at random. Two random samples from a log-normal distribution can have quite different compositions. Therefore, physically identical habitats (in this study, genetically identical hosts) will have different communities if they are formed at random from large seeding communities and will only be similar if the seeding community is small enough that the same bacteria arrive by chance (Curtis and Sloan, 2004). However, the input communities (mouse and zebrafish) each contained hundreds of species, making it unlikely that the same bacteria would establish by chance in each recipient GF animal.
In addition to host habitat factors, dynamics within microbial communities will interact with the host habitat to shape the final community. The relative abundance of divisions can be viewed as a simple emergent property of the community that belies underlying, highly complex organizational principles. Community-level interactions such as competition, cooperation, predation, and food web dynamics will all interact to shape a community (Ley et al., 2006b). The host provides the habitat and a basic niche space that the microbial community expands by its physical presence and metabolic activities. It is remarkable that such complex interactions can result in the predictable community structure that we observed at the division level. The shared host response to reciprocally transplanted zebrafish and mouse gut microbiotas suggests that this predictability of community composition also extends to the functions encoded in their microbiomes.
EXPERIMENTAL PROCEDURES
Animal Husbandry
All experiments using zebrafish and mice were performed using protocols approved by the Washington University Animal Studies Committee.
Conventionally Raised Animals
CONV-R zebrafish belonging to the C32 inbred strain were maintained under a 14 hr light cycle and given a diet described in an earlier publication (Rawls et al., 2004). CONV-R Swiss-Webster mice were purchased from Taconic Labs and fed an irradiated PicoLab chow diet (Purina) ad libitum. Mice were reared in a specific pathogen-free state, in a barrier facility, under a 12 hr light cycle.
Germ-free Animals
Zebrafish were derived as GF and reared using established protocols and diets (Rawls et al., 2004). GF zebrafish were maintained at 28.5°C in plastic gnotobiotic isolators at an average density of 0.3 individuals/ml gnotobiotic zebrafish medium (GZM; Rawls et al., 2004). GF mice belonging to the NMRI inbred strain were housed in plastic gnotobiotic isolators and fed an autoclaved chow diet (B&K Universal) ad libitum (Hooper et al., 2002). GF zebrafish and mice were kept under a 12 hr light cycle and monitored routinely for sterility (Rawls et al., 2004).
Colonization
GF zebrafish were conventionalized at 3dpf with a digestive tract microbiota harvested from CONV-R C32 donors, using established protocols (Rawls et al., 2004). To colonize zebrafish with individual bacterial species, or with defined consortia (see below), cultures were added directly to GZM containing 3dpf GF zebrafish (final density 104 CFU/ml). Colonization with members of the Firmicutes was coupled with addition of a cotton mesh bag containing 15 ml of ammonia-removing resin and activated carbon (AmmoCarb, Aquarium Pharmaceuticals) per 100 ml GZM at 3dpf.
To colonize zebrafish with a mouse gut microbiota, cecal contents were pooled from three adult CONV-R Swiss-Webster female mice under aerobic conditions, diluted 1:1200 in PBS, and added directly (1:100 dilution) to GZM containing 3dpf GF zebrafish (final density: 102 CFU/ml [aerobic culture]; 103 CFU/ml [anaerobic culture], as defined by incubation on BHI-blood agar for 2 days at 28°C).
GF NMRI mice were colonized at 7–11 weeks of age with a microbiota harvested from the cecal contents of adult CONV-R female Swiss-Webster mice (Bäckhed et al., 2004). To colonize mice with a zebrafish microbiota, the pooled digestive tract contents of 18 CONV-R adult C32 zebrafish were diluted 1:4 in sterile PBS under aerobic conditions and a 100 µl aliquot was introduced, with a single gavage (5 × 103 CFU/mouse, as defined by anaerobic and aerobic culture on BHI-blood agar and tryptic soy agar for 2 days at 37°C).
Other Treatments of Zebrafish
GF 3dpf animals were immersed in filter-sterilized GZM containing 0.1 mg/ml LPS purified from Pseudomonas aeruginosa ATCC27316 (Sigma, L8643). Sterility during this treatment was monitored routinely by culturing the aquaculture medium under a variety of conditions (Rawls et al., 2004).
To quantify cellular proliferation in the intestinal epithelium, 5dpf zebrafish were immersed in a solution of 5-bromo-2′-deoxyuridine (BrdU; 160 µg/ml of GZM) and 5-fluoro-2′-deoxyuridine (16 µg/ml GZM) for 24 hr prior to sacrifice. S phase cells were detected and scored as described (Rawls et al., 2004).
Phylogenetic and Diversity Analyses
Bulk DNA was obtained from the digestive tracts of zebrafish and the ceca of mice by solvent extraction and mechanical disruption (Ley et al., 2005; Rawls et al., 2004). The DNA was used in replicate PCRs using Bacteria-specific 16S rRNA gene primers. Amplicons from replicate PCRs were pooled and cloned prior to sequencing (See Supplemental Data).
16S rRNA gene sequences were edited and assembled into consensus sequences using PHRED and PHRAP aided by XplorSeq (Daniel Frank, University of Colorado, Boulder, personal communication); bases with a PHRAP quality score of <20 were trimmed. Contiguous sequences with at least 1000 >Q20 bp were checked for chimeras and then aligned to the 16S rRNA prokMSA database using the NAST server (http://greengenes.lbl.gov/cgi-bin/nph-NAST_align.cgi). The resulting multiple sequence alignments were incorporated into a curated Arb alignment (Ludwig et al., 2004) available at http://gordonlab.wustl.edu/supplemental/Rawls/Gut_Micro_Transplant.arb.
Assignment of the majority of sequences to their respective divisions was based on their position after parsimony insertion to the Arb dendrogram (omitting hypervariable portions of the 16S rRNA gene using lanemaskPH provided with the database). Chloroplast sequences were identified in CONV-R zebrafish libraries and removed (i.e., 8 sequences from library JFR0503 and 59 sequences from library JFR0504). Sequences that did not fall within described divisions were characterized as follows. Phylogenetic trees including the novel sequences and reference taxa were constructed by evolutionary distance (using PAUP* 4.0 [Swofford, 2003], a neighbor-joining algorithm with either Kimura two-parameter correction or maximum-likelihood correction with an empirically determined γ distribution model of site-into-site rate variation and empirically determined base frequencies). Bootstrap resampling was used to test the robustness of inferred topologies.
Distance matrices generated in Arb (with hypervariable regions masked, and with Olsen correction [Ley et al., 2006a]) were used to cluster sequences into operational taxonomic units (OTU’s) by pair-wise identity (%ID) with a furthest-neighbor algorithm and a precision of 0.01 implemented in DOTUR (Schloss and Handelsman, 2005). We use “phylotype” to refer to bins of sequences with ≥99% pairwise identity. Collector’s curves, Chao1 diversity estimates, and Simpson’s diversity index were calculated using DOTUR and Chao-Jaccard Abundance-based diversity indices using EstimateS 7.5 (Colwell, 2005). The percentage of coverage was calculated by Good’s method with the equation (1 − [n/N]) × 100, where n is the number of phylotypes in a sample represented by one clone (singletons) and N is the total number of sequences in that sample (Good, 1953).
To cluster the communities from each treatment, we used the UniFrac computational tool (Lozupone and Knight, 2005). To do so, the masked Arb alignment containing 5527 sequences from this study plus 852 sequences obtained from soil (Axelrood et al., 2002) was used to construct a neighbor-joining tree. The neighbor-joining tree was annotated according to the treatment from which each sequence was derived, and the fraction of tree branch length unique to any one treatment in pairwise comparisons (the UniFrac metric) was calculated. The p value for the tree, reflecting the probability that the there are more unqiue branch lengths than expected by chance alone, was calculated by generating 1000 random trees (Lozupone and Knight, 2005).
Functional Genomics
Analyses of gene expression in the mice and zebrafish using Affymetrix GeneChips, quantitative real-time RT-PCR, and Ingenuity Pathways Analysis were performed using methods described in previous publications (Giannakis et al., 2006; Hooper and Gordon, 2001; Rawls et al., 2004). For additional details, see Supplemental Data.
Supplementary Material
Acknowledgments
We are grateful to David O’Donnell and Maria Karlsson for assistance with husbandry of gnotobiotic animals, Sabrina Wagoner and Jill Manchester for valuable technical help, plus Fredrik Bäckhed, Peter Crawford, Marios Giannakis, and Justin Sonnenburg for many helpful discussions. This work was supported by the Ellison Medical Foundation, the W.M. Keck Foundation, and the NIH (DK30292, DK62675, DK073695).
Footnotes
Supplemental Data
Supplemental Data include Experimental Procedures, 5 Figures, and 16 tables and can be found with this article online at http://www.cell.com/cgi/content/full/127/2/423/DC1/.
Accession Numbers
16S rRNA sequences have been deposited in GenBank under accession numbers DQ813844–DQ819377. GeneChip datasets have been deposited in Gene Expression Omnibus under accession number GSE5198.
REFERENCES
- Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. Fine-scale phylogenetic architecture of a complex bacterial community. Nature. 2004;430:551–554. doi: 10.1038/nature02649. [DOI] [PubMed] [Google Scholar]
- Axelrood PE, Chow ML, Radomski CC, McDermott JM, Davies J. Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can. J. Microbiol. 2002;48:655–674. doi: 10.1139/w02-059. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bala A, Murphy P, Giller KE. Distribution and diversity of rhizobia nodulating agroforestry legumes in soils from three continents in the tropics. Mol. Ecol. 2003;12:917–929. doi: 10.1046/j.1365-294x.2003.01754.x. [DOI] [PubMed] [Google Scholar]
- Bates JM, Mittge E, Kuhlman J, Baden KN, Cheesman SE, Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Cahill MM. Bacterial flora of fishes: a review. Microb. Ecol. 1990;19:21–41. doi: 10.1007/BF02015051. [DOI] [PubMed] [Google Scholar]
- Cebra JJ. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 1999;69:1046S–1051S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- Colwell RK. EstimateS: Statistical estimation of species richness and shared species from samples ( http://purl.oclc.org/estimates. 2005
- Curtis TP, Sloan WT. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 2004;7:221–226. doi: 10.1016/j.mib.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Giannakis M, Stappenbeck TS, Mills JC, Leip DG, Lovett M, Clifton SW, Ippolito JE, Glasscock JI, Arumugam M, Brent MR, Gordon JI. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J. Biol. Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Mills JC, Roth KA, Stappenbeck TS, Wong MH, Gordon JI. Combining gnotobiotic mouse models with functional genomics to define the impact of the microflora on host physiology. Mol. Cell. Microbiol. 2002;31:559–589. [Google Scholar]
- Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, Gram L. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhynchus mykiss, Walbaum) J. Appl. Microbiol. 2004;96:117–132. doi: 10.1046/j.1365-2672.2003.02109.x. [DOI] [PubMed] [Google Scholar]
- Hughes-Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Harris JK, Wilcox J, Spear JR, Miller SR, Bebout BM, Maresca JA, Bryant DA, Sogin M, Pace NR. Unexpected diversity and complexity from the Guerrero Negro hyper-saline microbial mat. Appl. Environ. Microbiol. 2006a;72:3685–3695. doi: 10.1128/AEM.72.5.3685-3695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006b;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98:3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2001;2:956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- Peterson RT, Fishman MC. Discovery and use of small molecules for probing biological processes in zebrafish. Methods Cell Biol. 2004;76:569–591. doi: 10.1016/s0091-679x(04)76026-4. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero J, Navarrete P. 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch) Microb. Ecol. 2006;51:422–430. doi: 10.1007/s00248-006-9037-9. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer Associates; 2003. ( http://paup.csit.fsu.edu/) [Google Scholar]
- Wostmann BS. The germfree animal in nutritional studies. Annu. Rev. Nutr. 1981;1:257–279. doi: 10.1146/annurev.nu.01.070181.001353. [DOI] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.