Abstract
In this paper we report a systematic XAS study of a set of samples in which Cu(II) was progressively added to complexes in which Zn(II) was bound to the tetra-octarepeat portion of the prion protein. This work extends previous EPR and XAS analysis in which, in contrast, the effect of adding Zn(II) to Cu(II)–tetra-octarepeat complexes was investigated. Detailed structural analysis of the XAS spectra taken at both the Cu and Zn K-edge when the two metals are present at different relative concentrations revealed that Zn(II) and Cu(II) ions compete for binding to the tetra-octarepeat peptide by cross-regulating their relative binding modes. We show that the specific metal–peptide coordination mode depends not only, as expected, on the relative metal concentrations, but also on whether Zn(II) or Cu(II) was first bound to the peptide. In particular, it seems that the Zn(II) binding mode in the absence of Cu(II) is able to promote the formation of small peptide clusters in which triplets of tetra-octarepeats are bridged by pairs of Zn ions. When Cu(II) is added, it starts competing with Zn(II) for binding, disrupting the existing peptide cluster arrangement, despite the fact that Cu(II) is unable to completely displace Zn(II). These results may have a bearing on our understanding of peptide-aggregation processes and, with the delicate cross-regulation balancing we have revealed, seem to suggest the existence of an interesting, finely tuned interplay among metal ions affecting protein binding, capable of providing a mechanism for regulation of metal concentration in cells.
Keywords: Prion protein, XAS, Metal homeostasis
Introduction
Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative diseases in which the transmissible agent consists of a misfolded form of the prion protein (PrPC →PrPSc) Prusiner (1998). Examples of TSEs include mad cow disease, scrapie in goats and sheep, and Kuru and Creutzfeldt–Jakob diseases in humans. The normal native form of the prion protein (PrPC) is a membrane-tethered protein predominately expressed in brain tissue. Although its function is not yet fully elucidated, the ability of PrP to bind in vivo with both Cu(II) and Zn(II) ions (Brown et al. 1997) has led to the suspicion that PrP may be involved in metal homeostasis in the brain (Kenche et al. 2011). The observation that Cu and Zn homeostasis is severely altered in the brain tissue of infected organisms (Rachidi et al. 2003) compared with normal tissues has also led to the hypothesis of involvement of the two metals in genesis of the illness.
Homeostatic control is necessary to maintain essential transition metal ions at the characteristic cellular concentrations which support their physiological functions yet avoid toxic adverse effects. Prokaryotic and eukaryotic organisms have evolved the capacity to quickly adapt to a changing and challenging microenvironment in which the availability of both biologically required and non-essential transition metal ions can vary dramatically. This task is accomplished by specialised metalloregulatory proteins capable of controlling the expression of genes encoding membrane transporters and metal-trafficking proteins that collectively manage metal homeostasis and resistance. In addition to these mechanisms, as we shall argue in this paper, metal competition for protein binding is critical in the central nervous system, because metals are involved in several essential biological processes (neuro-transmission, enzymatic activity, etc.) and their imbalance is known to lead to several neuro-degenerative disorders (Faller et al. 2014; Ishara Silva and Saxena 2013; Maret 2011; Braymer and Giedroc 2014; Reyes-Caballero et al. 2011 and references therein).
With regard to the prion protein, one of the identified PrP–metal binding regions, among others, is located in the unstructured (Donne et al. 1997; Hornshaw et al. 1995; Aronoff-Spencer et al. 2000; Whittal et al. 2000; Qin et al. 2002; Wells et al. 2006) and highly conserved portion of the protein termed the octarepeat domain. This domain consists of several repeats (up to six depending on the species), each formed by the set of eight amino acids PHGGGWGQ.
Electron paramagnetic resonance (EPR) studies (Chattopadhyay et al. 2005) performed on Syrian hamster PrP have shown that at low copper concentrations a single Cu(II) ion is coordinated to the four His residues (one for each of the octarepeats) of the tetra-octarepeat domain. At high Cu concentration each octarepeat is able to bind one Cu(II) ion for a total of four Cu(II) ions per tetra-octarepeat.
The same research group (Walter et al. 2007) subsequently demonstrated that the presence of Zn(II) modulates the mode of Cu(II) coordination to the tetra-octarepeat peptide region. Changes in the Cu(II) coordination mode in the presence of Zn(II) in the prion protein have also studied by use of electron spin echo envelope modulation (Silva et al. 2014).
A parallel X-ray absorption spectroscopy (XAS) study (Stellato et al. 2011) confirmed the ability of Zn(II) to modulate Cu(II) coordination, adding the important information that a single tetra-octarepeat domain can bind Cu(II) and Zn(II) ions simultaneously. Analysis of the XAS data also showed that Zn(II) competes with Cu(II) for His-binding by directly interacting with the tetra-octarepeat region, but it is unable to completely displace Cu(II).
All these results, taken together, while suggesting that metal binding competition is important in the general context of metal homeostasis, also raise the question of whether the same occurs when Cu(II) is added to an already formed tetra-octarepeat–Zn(II) complex.
The main purpose of this investigation was to address and answer this question. By performing a systematic XAS study of a set of samples in which Cu(II) at increasing concentrations was added to tetra-octarepeat–Zn(II) complexes, we showed that the metal–peptide coordination mode depends not only, as expected, on the relative concentrations of the metals but, by comparison with the old XAS measurement of Stellato et al. (2011), that the order in which the two metal ions are added to metal–tetra-octarepeat complexes is also important. Indeed, not only is Zn(II) unable to completely remove octarepeat-bound Cu(II) ions (as was already made clear by Stellato et al. 2011) but, as we prove here, the opposite is also true, i.e. Cu(II) modifies the mode of Zn(II) coordination by partly displacing it, but fails to completely remove octarepeat-bound Zn(II) ions.
Experimental
Sample preparation
Synthetic PrP tetra-octarepeat peptides were prepared by use of the standard fluorenylmethoxycarbonyl (Fmoc) method (Burns et al. 2002; Aronoff-Spencer et al. 2000). Its complete amino acid sequence (abbreviated below to 4R8), is Ac-KKRPKPWGQ(PHGGGWGQ)4-NH2, with the initial amino acidic sequence inserted to increase solubility.
All samples were prepared by dissolving the peptide in degassed solvent containing 25 mM N-ethylmorpholine (NEM) buffer and 20 % (v/v) glycerol at pH 7.4. The latter is added to help stabilise the sample (Vagenende et al. 2009). The peptide concentration used for XAS samples was 0.2 mM. Zn and Cu were added as ZnCl2 and Cu(CH3COO)2, respectively. Cu(II) and Zn(II) concentrations in peptide samples ranged from 0 to 0.64 mM (Table 1). The concentration of Cu(II) and Zn(II) in the buffer (samples bZn and bCu in Table 1) was 2 mM.
Table 1.
Measured samples, concentrations (in equivalents), measured K-edges and figure colour code
| Sample | Zn | Cu | K-edge | Colour |
|---|---|---|---|---|
| S1 | 0.8 | 0 | Zn | Magenta |
| S2 | 0.8 | 0.8 | Zn, Cu | Red |
| S3 | 0.8 | 3.2 | Zn, Cu | Green |
| S4 | 2 | 0 | Zn | Orange |
| S5 | 3 | 3 | Zn, Cu | Light blue |
| bZn | 10 | 0 | Zn | Black |
| bCu | 0 | 10 | Cu | Black |
The desired amount of ZnCl2 was added to the peptide buffer solution some days in advance, whereas Cu acetate was added from a few tens of minutes to 24 h before the XAS measurements. The spectra of all samples were acquired at the Zn K-edge and, in the presence of Cu(II), also at the Cu K-edge.
X-ray data collection
XAS experiments were performed at the BM30B beamline of the European Synchrotron Radiation Facility (Grenoble, France) (Proux et al. 2005). The storage ring was operated in 7/8 + 1 bunches mode at 6 GeV with a 200 mA current. The beam energy was selected using a Si(220) double-crystal monochromator with a resolution of ~0.5 eV (Proux et al. 2006). The beam spot on the sample was approximately 300 × 200 μm2 (H × V, FWHM).
Spectra were recorded in fluorescence mode by use of a 30-element solid-state Ge detector. To avoid photo-degradation and spectra evolution during XAS measurements, all the samples were first rapidly brought from room temperature to 77 K (liquid nitrogen) and then cooled and kept at 13 K by use of a liquid helium cryostat throughout XAS measurement. In the process of data acquisition samples were systematically moved to a different position for each scan so that the radiation did not always hit the same portion of the sample. Each scan never exceeded 30 min.
Data analysis
The XAS absorption coefficient, μ(E), and the normalised signal, χ(k),1 are extracted from the raw data by following the fully standard procedure encoded in the ATHENA software (Ravel and Newville 2005).
In particular, the absorption coefficient, μ(E), is obtained by subtracting a linear background fitted through the pre-edge region data, and is normalised by requiring the difference between the extrapolated value of the pre-edge line and a post-edge quadratic fit be equal to unity at the edge energy. The definition of pre-edge and post-edge regions was kept the same in the analysis of all the spectra. The definition of χ(k) and its relationship with the absorption coefficient, μ(E) are discussed in the appendix of (Morante 2008).
XAS data are presented by separating the XANES and EXAFS regions of the spectrum.2 As is customary, the absorption coefficient is plotted when displaying the XANES region of the spectrum, whereas the normalised signal is reported when showing the EXAFS part of the spectrum.
To extract the desired structural information, the EXCURV98 package (Binsted et al. 1998) was used to fit the experimental signal, χ(k), against theory. The so-called constrained refinement strategy, which consists in treating the lateral chain of amino acid residues as a rigid body, was used. In our case, we considered the imidazole ring of His residues as undeformable. Thus, in the fitting step we refined only the distance of the bound nitrogen and the orientation of the imidazole ring plane. This enabled significant reduction of the number of free variables in the fitting procedure (Binsted et al. 1992).
Results and discussion
For easier further reading we list in Table 1 the complete set of samples (column 1) we have measured, specifying the corresponding concentrations of Zn(II) (column 2) and Cu(II) (column 3) ions in units of peptide concentration (peptide equivalents, eq). In column 4 the ion K-edge at which each spectrum was recorded is indicated. For comparison, the concentrations of Zn(II) and Cu(II) in buffer (2 mM) are also given in the units peptide equivalents. In the last column the colour code used in subsequent figures is summarised.
Qualitative data analysis
Before moving in the next section to quantitative analysis of the XAS data, it is useful to study all the spectra qualitatively, to identify the main similarities and differences and to become familiar with the models and the structural properties.
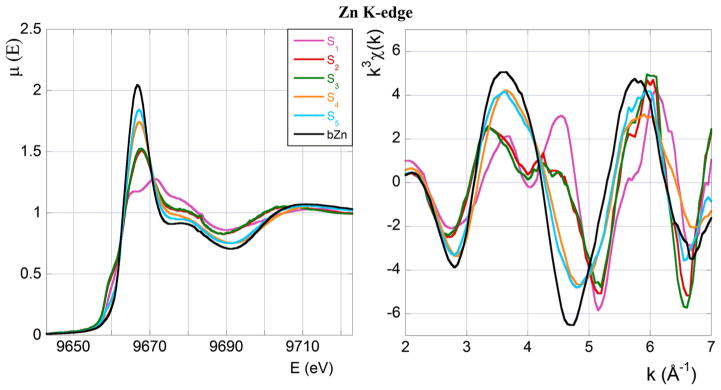
In Figs. 1 and 2 the XAS spectra at the Zn and Cu K-edge of all the measured samples (buffers included) in the XANES (left panels) and EXAFS (right panels) regions are displayed and compared. Only for the purpose of this first qualitative comparison, we concentrate on the EXAFS data belonging to the restricted region going from k ≃ 2 Å−1 to k ≃ 7 Å−1.
Fig. 1.
XAS spectra at the Zn K-edge. Left panel: XANES absorption coefficient, μ(E), as a function of the photon energy, E in eV. Right panel: normalised, weighted EXAFS signal, k3 χ(k), as a function of the wave vector, k in Å−1. Colour code and sample composition are as given in Table 1
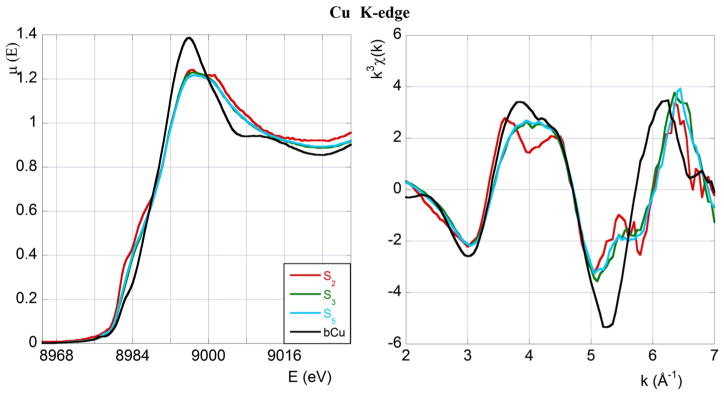
Fig. 2.
XAS spectra at the Cu K-edge. As in Fig. 1, left and right panels display XANES and EXAFS data, respectively. Colour code and sample composition are as given in Table 1
The quantitative EXAFS data analysis illustrated in the section “EXAFS region data analysis” will be performed for a wider photon energy range, namely from k ≃ 3 Å−1 to k ≃ 11 Å−1 for the spectra taken at the Zn K-edge and from k ≃ 3 Å−1 to k ≃ 12 Å−1 for the spectra taken at the Cu K-edge. The reason for this difference in the choice of the k range will be explained in the subsection “Zn K-edge”).
Zn K-edge
We now present a few general observations that will guide successive quantitative data analysis.
We start by noticing that the spectra of all the Si (i = 1, . . . , 5) samples visibly differ from the Zn-buffer (bZn) spectrum, clearly indicating that at least some fraction of Zn(II) ions is bound to the peptide. Second, the sample that is mostly different from the buffer is S1, which was prepared at low (0.8 eq) Zn(II) concentration and in the absence of Cu(II). Addition of a sub-stoichiometric amount of Cu(II), as in sample S2, is sufficient to drastically modify the spectrum shape, making the S2 spectrum significantly different from that of S1.
Furthermore, samples S2 and S3, that differ in Cu(II) concentration only (0.8 eq in S2, and 3.2 eq in S3; Table 1), give essentially undistinguishable XANES and EXAFS spectra, indicating that a further increase of the Cu(II) concentration above 0.8 eq has no appreciable consequence on the mode of Zn(II) coordination.
Sample S4, similar to S1, does not contain Cu(II), but at variance with the latter it was prepared at a much higher Zn(II) concentration (2 eq). Their spectra differ because of the contribution to the XAS spectrum of sample S4 from the Zn present in excess in the solution. In the subsection “XANES region data analysis” we will provide a quantitative argument in support of this interpretation.
A similar situation occurs if one compares the spectrum of sample S5 (in which both Zn(II) and Cu(II) are present at high concentration, 3 eq) with that of sample S3. In this case also the difference between the two spectra should be ascribed to the presence of excess of Zn in the solution (as discussed in the subsection “XANES region data analysis”).
Finally we observe that the most striking feature of the χ (k) behaviour of samples S1, S2 and S3 in the EXAFS region (right panel of Fig. 1), is the appearance of a double peak in the wave number range between k ≃ 3.5 Å−1 and k ≃ 4.5 Å−1. The double peak is known to be indicative of bound His (Strange et al. 1987). The progressive disappearance of this spectral feature, which should be thus interpreted as a decrease in the number of Zn(II)-bound His residues, is seen to be related to an increase of the Cu(II) concentration. In the section “Cu K-edge” we will return to this question, and provide a structural explanation of this behaviour.
Cu K-edge
In Fig. 2 XANES and EXAFS spectra acquired at the Cu K-edge are displayed and compared. Again, for the purpose of this first qualitative comparison only, we show EXAFS data in the restricted region from k ≃ 2 Å−1 to k ≃ 7 Å−1.
As for the Zn K-edge, all the spectra significantly differ from that of the Cu-buffer (bCu), meaning that some fraction of Cu(II) is always bound to the peptide irrespective of Zn(II) concentration. As already shown by Chattopadhyay et al. (2005), the Cu(II) coordination mode depends on the [Cu]:[4R8] ratio.
In the EXAFS region (right panel of Fig. 2) we notice again in sample S2 the presence of a double peak in the wave number range between k ≃ 3.5 Å−1 and k ≃ 4.5 Å−1. The double peak disappears at higher Cu(II) concentrations (samples S3 and S5) indicating that the number of Cu(II) bound His residues decreases with increasing Cu(II) concentration, in agreement with the EPR results of Chattopadhyay et al. (2005).
Finally, the XAS spectra of samples S3 and S5 are very similar in both the XANES and the EXAFS regions (the green S3 spectrum is hardly visible below the light blue S5 spectrum), despite the fact that the two samples were prepared starting from substantially different Zn(II) concentrations (0.8 and 3 eq, respectively). This observation shows that the Cu(II) coordination mode does not depend on Zn(II) concentration. Notice that, as seen in Fig. 1, the corresponding Zn K-edge spectra are, instead, definitely not superimposable.
Quantitative data analysis
We now present quantitative analysis of the measured XAS spectra of the systems listed in Table 1 and provide detailed structural information about the atomic environment around the metals at the different concentrations we have studied.
XANES region data analysis
One of the conclusions emerging from the qualitative analysis presented in the previous section is that when the [Zn]:[4R8] concentration is large (over-stoichiometric), as for samples S4 and S5, a substantial fraction of Zn is not bound to the peptide and it is found free in solution. This means that the measured XAS spectrum at the Zn K-edge receives contributions from two kinds of Zn(II) geometry, that of Zn bound to the peptide and that of Zn in solution.
This simple interpretation of the spectral features of the S4 and S5 samples can be checked in a model-independent way if one treats the above considerations mathematically by saying that the XAS measured signal is a linear combination, with appropriate coefficients, of the signal from the Zn bound to the peptide and that from Zn in solution. The strength of this approach is that it is not necessary to make recourse to any specific model for the Zn geometry in the different atomic environments in which the metal is supposed to exist, if one assumes that the XAS signal from the Zn in solution can be identified with the Zn-buffer signal and the signal from the Zn bound to the peptide with the S1 spectrum in the absence of Cu and with the S3 spectrum when the Cu concentration is large (3 eq), respectively.
We now discuss in turn the details of the analysis of the S4 and S5 XANES spectra along the lines outlined above.
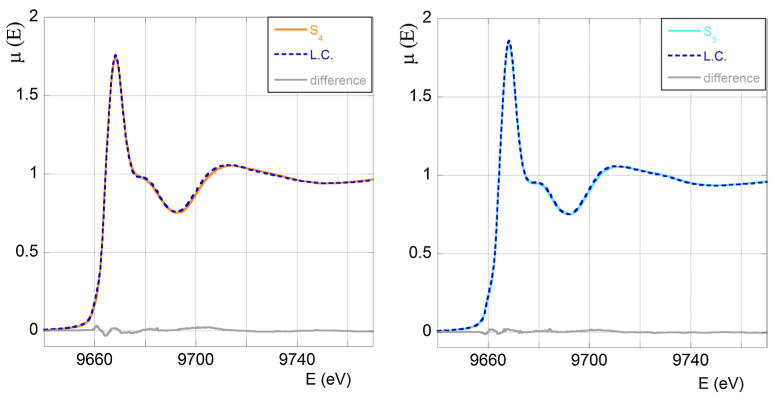
• Sample S4
If we call 0 ≤ p ≤ 1 the fraction of Zn(II) ions in the S1-like coordination mode, then (1 − p) will represent the fraction of Zn(II) in the bZn-like coordination. According to our simple assumptions we can determine the best value of p by minimising the “residual function”
| (1) |
where the sum is extended over the energies Ei at which XANES data were taken and μA is the absorption coefficient of sample A (A = S4, S1 and bZn).
The function Q is found to have a minimum at p = 0.33 ± 0.01 meaning that in the S4 sample 1/3 of Zn(II) is bound to the peptide and 2/3 is in solution. The agreement between the measured S4 XANES spectrum and the optimum combination of the S1 and bZn signals shown in Fig. 3 (left panel) is extremely good. For convenience we also report the difference between the experimental data and the theoretical fit. A similar analysis for the EXAFS data gives a comparably good fit, yielding an optimum value of p consistent with that determined from the XANES fit (data not shown), confirming the validity of our assumptions and the robustness of the result.
Fig. 3.
In the left panel we show the S4 experimental XANES spectrum (orange full line) compared with the optimum linear combination (LC, blue broken line) of the S1 and bZn spectra. In the right panel we show the S5 experimental XANES spectrum (light blue full line) compared with the optimum linear combination (LC, blue broken line) of the S3 and bZn spectra obtained. In grey we draw the difference between the experimental signal and the theoretical fit
We can thus conclude that the total amount of Zn(II) in S4 in term of equivalents (Table 1) is distributed as follows:
| (2) |
| (3) |
where with [Zn]S1 and [Zn]bZn we denote the concentration of bound to the peptide Zn(II) in the configuration it takes in S1 and free in solution, respectively. The interesting interpretation of the fact that the proportionality factor between the concentration of bound Zn(II) and that of 4R8 equals 2:3 is that small aggregates are formed, where, on average, triplets of tetra-octarepeats are cross-linked by two Zn(II) bridges. This quite remarkable result is reminiscent of a similar structural arrangement found in systems in which β-amyloid peptides are complexed with Zn(II) ions (Giannozzi et al. 2012).
• Sample S5
A completely similar analysis of the XANES S5 data in terms of the measured XANES signals of the S3 sample and of the Zn-buffer gives p = 0.35 ± 0.06. The high quality of the fit can be judged by looking at the right panel of Fig. 3, where the difference between the experimental signal and the theoretical fit (grey curve) is also reported. Again a similar analysis of the EXAFS region of the spectrum gives a rather good fit and an optimum value of p consistent with the previous one (data not shown).
We conclude that the total amount of Zn(II) in S5 in terms of equivalents (Table 1) is distributed as follows:
| (4) |
| (5) |
with similar meaning as before for [Zn]S3 and [Zn]bZn. These numbers tell us that each 4R8 peptide binds one Zn(II) ion. Actually, as we shall see in the section “Cu K-edge”, the other three His residues of the peptide each bind a Cu(II) ion.
EXAFS region data analysis
We now proceed to analysis of the EXAFS region of the spectra to obtain quantitative structural information about the geometry of the different kinds of metal site occurring in the samples we have considered. We will separately discuss Zn(II) and Cu(II) K-edge data and compare our results with the complementary study by Stellato et al. (2011) in which successive addition of Zn(II) to a Cu(II)–4R8 bound complex was studied.
Zn K-edge
Among the spectra collected at the Zn K-edge (Table 1) only the following three must be analysed and fitted:
S1–0.8 eq of Zn, fully bound to 4R8, and no Cu
S3–0.8 eq of Zn, fully bound to 4R8, with the addition of 3.2 eq of Cu
bZn–Zn in buffer
The reasons for this choice are as follows. As seen in Fig. 1, samples S2 and S3 give essentially identical spectral features both in the XANES and the EXAFS regions. Furthermore, as we remarked when we discussed the XANES data, the (rather similar) EXAFS spectra of samples S4 and S5 can also be well represented by linear superposition of the EXAFS signal of the S1 sample with that of Zn in buffer, and that of the S3 sample again with Zn in buffer, respectively. Thus, all the structural information of interest can be obtained by analysing the three samples listed above.
The energy range of the Zn K-edge EXAFS spectra (in contrast with the Cu K-edge spectra discussed below) was limited to approximately 11 Å−1 because of the unfavourable signal-to-noise-ratio of data above this point.3
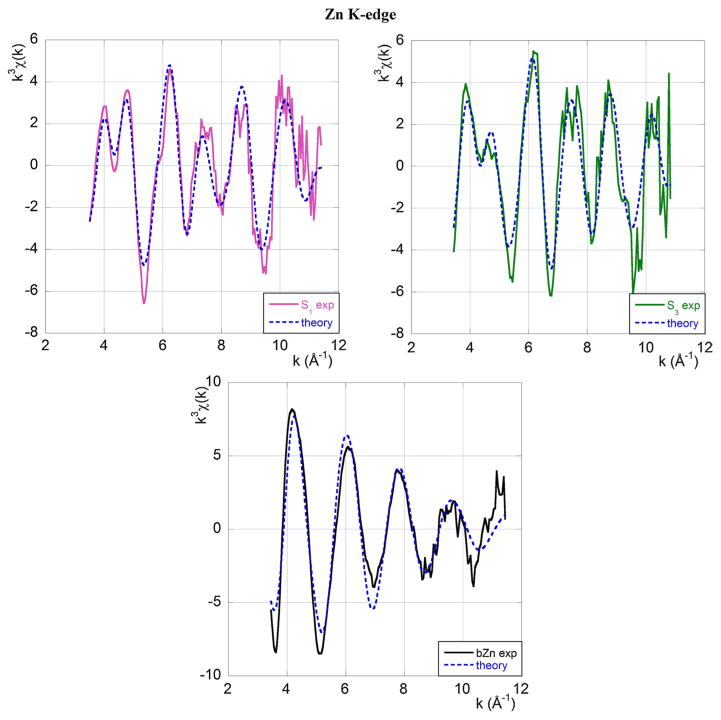
The Zn coordination modes are determined from the best-fits shown in Fig. 4 that also include the relevant multiple scattering contributions. The detailed structural information emerging from the fitting procedure is summarised in Table 2.
Fig. 4.
Experimental EXAFS data at the Zn K-edge (full line) and the best-fit theoretical model (broken blue line). The top left panel refers to sample S1, the top right panel to sample S3 and the central panel to Zn in buffer. Colour code and sample composition are as given in Table 1
Table 2.
Zn coordination geometry in S1, S3 samples and bZn buffer
| Zn K-edge
| |||||
|---|---|---|---|---|---|
| Atom | Multiplicity | r ± δr (Å) | σ 2 ± δσ 2 (Å2) | Ef (eV) | R (%) |
| S1 sample | |||||
| N(His) | 2 | 2.22 ± 0.02 | 0.007 ± 0.003 | ||
| O | 2 | 1.95 ± 0.01 | 0.003 ± 0.001 | −6 ± 2 | 38 |
| Cl | 2 | 2.75 ± 0.01 | 0.004 ± 0.001 | ||
| S3 sample | |||||
| N(His) | 1 | 2.24 ± 0.04 | 0.001 ± 0.001 | ||
| O | 1 | 1.93 ± 0.02 | 0.001 ± 0.001 | −4 ± 2 | 39 |
| O | 2 | 2.11 ± 0.04 | 0.004 ± 0.003 | ||
| Cl | 3 | 2.75 ± 0.02 | 0.006 ± 0.001 | ||
| bZn buffer | |||||
| O(H2O) | 4 | 1.99 ± 0.02 | 0.006 ± 0.001 | 5 ± 1 | 27 |
| Cl | 2 | 2.27 ± 0.01 | 0.005 ± 0.002 | ||
In the first column of Table 2 we report the identities of Zn-bound atoms. For nitrogen atoms we explicitly indicate when they belong to a His residue. Because there is a single His residue in each octarepeat, the number of His residues also denotes the number of octarepeats to which the metal is bound. We are very confident about this counting of bound His residues, because the XAS technique is known to be especially sensitive to this number (Morante 2008; Strange et al. 1987; Penner-Hahn 2005).
As shown in Table 2, the number of bound His residues decreases from two (in sample S1) to one (in sample S3) as a result of adding 3.2 eq Cu(II). The ensuing structural modification is an indication of the ability of Cu(II) to compete for 4R8 binding.
In the second column we report the multiplicity of each type of scatterer. The third and fourth columns provide the absorber–scatterer distances and the Debye–Waller factors with their statistical errors. In the last two columns the Fermi energy shift, Ef, and the value of the fit quality factor, R, are displayed. The R factors we obtain should all be considered adequate for complex biological systems such as those we are studying here (Binsted et al. 1998) (they are all <40 %).
We recall that in the subsection “XANES region data analysis” we suggested a simple model to explain the features of the S4 XAS spectrum. This model works under the assumption that the S4 spectrum is a linear combination, in the appropriate ratio, of the signal from sample S1 and the signal from Zn in solution. The resulting 1:2 proportionality factor leads to the [Zn(II)]:[4R8 ] = 2:3 concentration ratio of Eq. 2. From the EXAFS analysis just presented we found, on the other hand, that Zn(II) in the S1 sample is coordinated to two His residues. As we said before, the two results taken together suggest the existence of an interesting Zn(II)–4R8 coordination mode of the type shown in the left-upper panel of Fig. 6, in which two Zn(II) ions bridge three tetra-octarepeats.
Fig. 6.
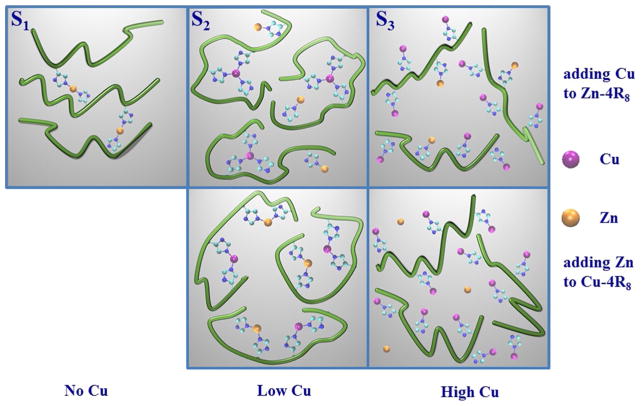
Schematic illustration of the suggested effect of adding Cu (Zn) to the Zn–(Cu)–4R8-complex. Cu(II) and Zn(II) ions are drawn as magenta and yellow dots, respectively. Only the bonds between metals and His residues imidazole rings are indicated (short blue segments). In the first (second) row we display the metal–peptide arrangements identified in this work (in Stellato et al. 2011). The three columns show the effect of ending with a system with zero, low, and high Cu concentration. In this work Cu was added to a sample already containing Zn, whereas Stellato et al. (2011) prepared samples at different Cu concentrations and then added Zn at a later stage. The number of magenta (Cu) and yellow (Zn) dots per tetra-octarepeat shown in the different panels is indicative only and does not represent the real proportion among tetra-octarepeats, Cu and Zn
Addition of Cu(II) to this kind of aggregate modifies the Zn(II) coordination mode, as is seen by studying the spectral features of sample S3. Specifically, when 3.2 eq Cu(II) is titrated into a sample in which Zn(II) at a low concentration (0.8 eq) is fully bound to 4R8 peptides, the number of Zn(II)-bound His residues changes from two to one. We conclude that Cu(II) is able to disrupt the pre-existing bridging arrangement (second and third upper panel of Fig. 6) with apparently no significant release of Zn(II) into solution.
These results reveal an interesting cross-regulation pattern between metals in which the small [Zn(II)]2–(4R8)3 aggregates are ready to accept Cu(II) binding, offering a useful mechanism for “neutralising” (“sterilising”) possibly dangerous free Cu ions within the cell, without liberating in solution potentially dangerous Zn(II) ions.
Cu K-edge
Among the spectra collected at the Cu K-edge (Table 1) the following three have been analysed and fitted:
S2–0.8 eq of Zn, fully bound to 4R8, with the addition of 0.8 eq of Cu
S3–0.8 eq of Zn, fully bound to 4R8, with the addition of 3.2 eq of Cu
bCu–Cu in buffer
We no longer considered the S5 sample because, as already noticed, at the Cu K-edge its EXAFS spectrum is essentially indistinguishable from that of sample S3. We stress that samples S2 and S3 differ only in the amount of Cu(II) added to a sample in which 0.8 eq Zn(II) was bound to 4R8 peptides.
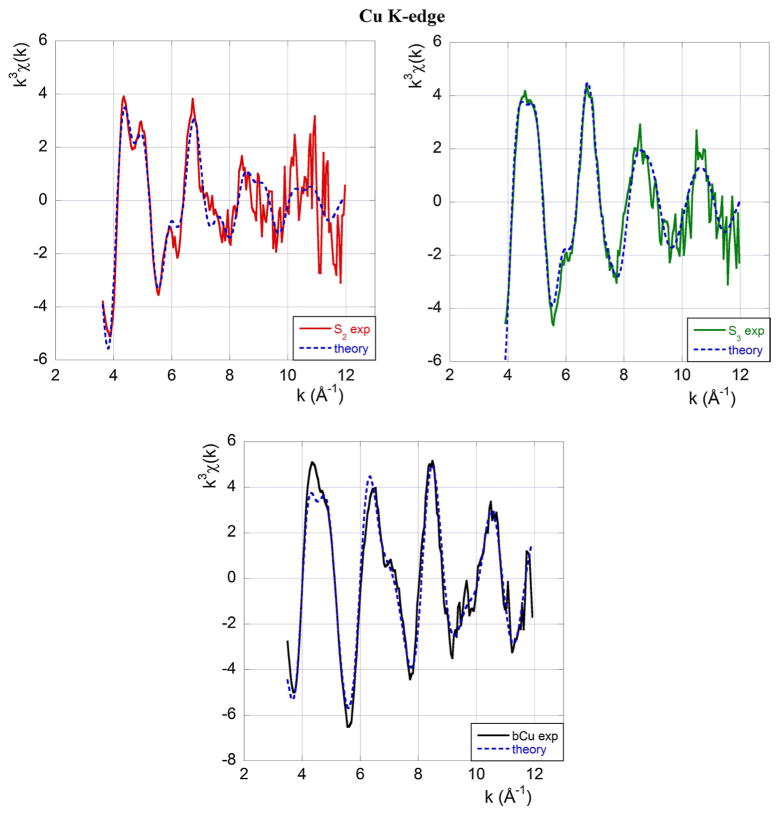
A standard EXAFS analysis of the kind performed at the Zn K-edge, including multiple scattering contributions, yields the best-fit curves shown in Fig. 5. The initial geometrical model used to fit the S3 spectrum was taken from the PDB data of Burns et al. (2002).4
Fig. 5.
Experimental EXAFS data at the Cu K-edge (full line) and best-fit theoretical model (broken blue line). The top left panel refers to sample S2, the top right panel to sample S3 and the central panel to Cu in buffer. Colour code and sample composition are as given in Table 1
The resulting structural information concerning the Cu(II) binding site in the S2, S3 samples and the Cu-buffer, which we report for completeness, are summarised in Table 3.
Table 3.
Cu coordination geometry in S2, S3 samples and bCu buffer
| Cu K-edge
| |||||
|---|---|---|---|---|---|
| Atom | Multiplicity | r ± δr (Å) | σ 2 ± δσ 2 (Å2) | Ef (eV) | R (%) |
| S2 sample | |||||
| N(His) | 3 | 1.97 ± 0.02 | 0.003 ± 0.002 | ||
| O | 1 | 1.80 ± 0.03 | 0.003 ± 0.002 | −5 ± 1 | 38 |
| O | 1 | 2.14 ± 0.03 | 0.003 ± 0.002 | ||
| S3 sample | |||||
| N(His) | 1 | 1.96 ± 0.03 | 0.006 ± 0.001 | ||
| N(Gly) | 2 | 1.94 ± 0.02 | 0.006 ± 0.001 | 4 ± 1 | 25 |
| O(Gly) | 1 | 1.92 ± 0.02 | 0.006 ± 0.001 | ||
| bCu buffer | |||||
| O(H2O) | 4 | 1.95 ± 0.02 | 0.005 ± 0.001 | 3 ± 1 | 23 |
At low Cu(II) concentration (0.8 eq—S2 sample) each tetra-octarepeat binds one Cu(II) ion, as indicated by the multiplicity of His residues in Table 3.
At high Cu(II) concentration (3.2 eq—S3 sample) each tetra-octarepeat binds three Cu(II) ions. The amino acids in parentheses in Table 3 are the first four of the crystallised pentapeptide sequence HGGGW.
Taken together these results indicate that Cu(II) is not able to fully displace Zn(II), not even at high concentrations (as in sample S3). Increasing [Cu(II)] from zero to 0.8 eq (low concentration) in samples in which an equally low (0.8 eq) concentration of Zn(II) is bound to 4R8 peptides gives rise to coordination modes similar to those found by Stellato et al. (2011), despite the fact that the metals are added in reverse order. In both cases the initially bound metal, be it Zn(II) here or Cu(II) in Stellato et al. (2011), remains fully bound to the peptide.
In contrast, at high Cu(II) concentration the way in which the two metals are coordinated to the 4R8 peptide is substantially different from that observed in our previous study (Stellato et al. 2011). In fact, we see in this work that, starting from a situation in which Zn is bound to the peptide, by progressively adding Cu(II) (i.e., by passing from the S2 to the S3 sample) one arrives at a situation in which Zn(II) remains bound by one His bond and, on the basis of the absence of a contribution to the EXAFS signal from Cu(II) in solution, one concludes that each of the remaining three tetra-octarepeat His residues binds a Cu(II) ion.
We recall that in the situation investigated by Stellato et al. (2011) it was shown that when Zn(II) is added to the Cu–tetra-octarepeat complex the former is able to bind to the peptide at low, but not at high, Cu concentration.
In Fig. 6 we suggestively illustrate the different metal–peptide binding modes we have identified at different Zn(II) and Cu(II) concentrations.
Putting together old and new results, the picture that emerges is that the process of adding a second metal to an already formed metal–peptide complex is not reversible (of course within reasonable time and activation energy supply), in the sense that the final coordination state of the system depends on which metal was first bound to the peptide. In particular, we have seen that the Zn(II)–4R8 complexes formed at not too high Zn concentration (e.g., 0.8 peptide eq) are ready to bind three Cu(II) ions, thus functioning as a sort of Cu(II) buffer for the cell but with no release of Zn(II) into solution.
Conclusions
The main results of this work can be synthetically summarised in four statements.
Zn(II) and Cu(II) compete for tetra-octarepeat peptide binding as proved by the fact that they are able to reciprocally affect their coordination mode.
In general, the way in which one metal acts on the coordination mode of the other depends on the order in which they are added to the peptide solution and their relative concentration.
Zn(II) at low concentration (less than [Zn]:[4R8] = 1:1) promotes the formation of small aggregates in which the Zn(II) to tetra-octarepeat concentration ratio is 2:3. Similarly to the behaviour already observed for β-amyloids (Giannozzi et al. 2012), this result hints at the formation of small aggregates in which two Zn(II) ions bridge three peptides.
When Cu(II) is added to this system, the small aggregates are disrupted with Cu(II) partially replacing Zn(II) as an His ligand.
These conclusions should be compared with those of Stellato et al. (2011), who studied the situation in which Zn(II) was added to an already formed Cu(II)–4R8 complex. In that situation it was found that Zn(II) can bind the peptide only in the presence of a low Cu(II) concentration. At high Cu concentration all metal binding sites are occupied by Cu(II) ions, thus they are not available to Zn(II).
The fact that Cu(II) can replace Zn(II) in the Zn–peptide complex, but Zn(II) is unable to displace Cu(II) in the preformed Cu–peptide complex, is consistent with considerations based on the Irving–Williams series (1953) from which it seems that Cu(II) interacts more strongly than Zn(II) with the peptide.
All our results are summarised pictorially in Fig. 6. The existence of a metal competition mechanism is clear from the observation that, although in three of the five panels the 4R8 peptide is able to bind both metals, in the four panels in which both metals are present, the Cu(II) and Zn(II) coordination modes seem to be quite different, meaning that their coordination mode crucially depends on the order in which metals are added to the peptide solution and their relative concentrations.
It is, in particular, worth noticing that the small Zn(II)–4R8 aggregates sketched in the upper-left panel of Fig. 6 are progressively disrupted as the Cu(II) concentration is increased, because of the increasing number of His residues that can bind Cu. The result is that the Zn(II)–4R8 aggregates, by modifying their structure, are able to sequestrate Cu(II), possibly providing a workable and finely tuned mechanism for metal homeostasis in the cell. As we said, similarly structured aggregates have been proved to exist in Zn–β-amyloid complexes also, with, again, two Zn ions cross-linking three β-amyloid peptides (Giannozzi et al. 2012).
One can speculate that the sophisticated scenario of metal binding processes uncovered by EPR (Chattopadhyay et al. 2005; Silva et al. 2014) and XAS (this paper and Stellato et al. 2011) studies can be at the basis of a general mechanism for metal cross-regulation in living organisms selectively evolved to avoid cell damage from such highly reactive chemicals as metal ions. Indeed, a synthetic way of summarizing our results is to say that Zn–peptide complexes can rearrange the metal coordination sphere to accommodate Cu(II) but free Zn(II) cannot replace Cu(II) in preformed Cu–peptide complexes. This is in agreement with the biological observation that free Zn(II) is not as damaging as free Cu(II) so there is a need to sequester Cu(II) more than Zn(II).
Acknowledgments
We acknowledge SUMA—INFN (Italy) for financial support.
Footnotes
We recall that the relationship between wave number, k, and E is , where Eth is the edge energy and me is the electron mass.
The X-ray absorption near edge structure (XANES) region is conventionally defined as the region of the spectrum corresponding to a photon energy from approximately 30–50 eV before the edge to approximately 50–100 eV after the edge, the extended X-ray absorption fine structure (EXAFS) region as the region from the end of the XANES region up to approximately 1,000 eV above the edge.
The reason for this signal deterioration is contamination in the counting of the Zn Kα fluorescence photons from the nearby Cu Kβ fluorescence line. Indeed, the relevant Zn Kα fluorescence lines are centred at 8,615 and 8,638 eV (α1 and α2), whereas the Cu Kβ line is at 8,905 eV. Because solid-state detector energy resolution is of the order of ~250–300 eV (full width half maximum value), photons from the Cu Kβ line can to be counted in the Zn background fluorescence spectrum, making it rather noisy above 10 Å−1.
We did not include in the model a distant O(H2O) present in the PDB because, as was shown by the abinitio simulations of Guerrieri et al. (2009), this water molecule was in a quite unstable configuration. We checked, anyway, that this oxygen has very little effect on the quality of the fit.
Contributor Information
Francesco Stellato, Department of Physics and INFN, University of Rome Tor Vergata, Via della Ricerca Scientifica, 00133 Rome, Italy.
Velia Minicozzi, Department of Physics and INFN, University of Rome Tor Vergata, Via della Ricerca Scientifica, 00133 Rome, Italy.
Glenn L. Millhauser, Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA 95064, USA
Marco Pascucci, Department of Physics and INFN, University of Rome Tor Vergata, Via della Ricerca Scientifica, 00133 Rome, Italy.
Olivier Proux, Observatoire des Sciences de l’Univers de Grenoble, CNRS and Université Joseph Fourier, BP 53, 38041 Grenoble Cedex 9, France.
Giancarlo C. Rossi, Department of Physics and INFN, University of Rome Tor Vergata, Via della Ricerca Scientifica, 00133 Rome, Italy. Centro Studi e Ricerche “Enrico Fermi”, Piazza del Viminale 1, 00184 Rome, Italy
Ann Spevacek, Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA 95064, USA.
Silvia Morante, Email: morante@roma2.infn.it, Department of Physics and INFN, University of Rome Tor Vergata, Via della Ricerca Scientifica, 00133 Rome, Italy.
References
- Aronoff-Spencer E, Burns CS, Avdievich NI, Gerfen GJ, Peisach J, Antholine WE, Millhauser GL. Identification of the Cu2+ binding sites in the n-terminal domain of the prion protein by epr and cd spectroscopy. Biochemistry. 2000;39(45):13760–13771. doi: 10.1021/bi001472t. http://www.ncbi.nlm.nih.gov/pubmed/9414160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binsted N, Strange RW, Hasnain SS. Constrained and restrained renement in exafs data analysis with curved wave theory. Biochemistry. 1992;31(48):12117–12125. doi: 10.1021/bi00163a021. http://pubs.acs.org/doi/abs/10.1021/bi00163a021. [DOI] [PubMed] [Google Scholar]
- Binsted N, Gurman SJ, Campbell JW. Daresbury Laboratory EXCURV98 Program. CLRC Daresbury Laboratory; Warrington: 1998. [Google Scholar]
- Braymer JJ, Giedroc DP. Recent developments in copper and zinc homeostasis in bacterial pathogens. Curr Op Chem Biol. 2014;19:59–66. doi: 10.1016/j.cbpa.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Qin K, Herms JW, Madlung A, Manson J, Strome R, Fraser PE, Kruck T, von Bohlen A, Schulz-Schaeffer W, Giese A, Westaway D, Kretzschmar H. The cellular prion protein binds copper in vivo. Nature. 1997;390(6681):684–687. doi: 10.1038/37783. http://www.nature.com/nature/journal/v390/n6661/full/390684a0.html. [DOI] [PubMed] [Google Scholar]
- Burns CS, Aronoff-Spencer E, Dunham CM, Lario P, Avdievich NI, Antholine WE, Millhauser GL. Molecular features of the copper binding sites in the octarepeat domain of the prion protein. Biochemistry. 2002;41(12):3991–4001. doi: 10.1021/bi011922x. http://pubs.acs.org/doi/abs/10.1021/bi011922x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay M, Walter ED, Newell DJ, Jackson PJ, Aronoff-Spencer E, Peisach J, Gerfen GJ, Bennet B, Antholine WE, Millhauser GL. The octarepeat domain of the prion protein binds cu(ii) with three distinct coordination modes at ph 7.4. J Am Chem Soc. 2005;127(36):12647–12656. doi: 10.1021/ja053254z. http://pubs.acs.org/doi/abs/10.1021/ja053254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Structure of the recombinant full-length hamster prion protein prp: the n terminus is highly exible. Proc Natl Acad Sci USA. 1997;94(25):13452–13457. doi: 10.1073/pnas.94.25.13452. http://www.pnas.org/content/94/25/13452.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller P, Hureau C, La Penna G. Metal ions and intrinsically disordered proteins and peptides: from cu/zn amyloid- to general principles. Acc Chem Res. 2014;47(8):2252–2259. doi: 10.1021/ar400293h. [DOI] [PubMed] [Google Scholar]
- Giannozzi P, Jansen K, La Penna G, Minicozzi V, Morante S, Rossi GC, Stellato F. Zn induced structural aggregation patterns of b-amyloid peptides by rst-principle simulations and xas measurements. Metallomics. 2012;4:156–165. doi: 10.1039/c2mt00148a. [DOI] [PubMed] [Google Scholar]
- Guerrieri F, Minicozzi V, Morante S, Rossi G, Furlan S, La Penna G. Modeling the interplay of glycine protonation and multiple histidine binding of copper in the prion protein octarepeat subdomains. J Biol Inorg Chem. 2009;14(3):361. doi: 10.1007/s00775-008-0454-8. [DOI] [PubMed] [Google Scholar]
- Hornshaw MP, McDermott JR, Candy JM, Lakey JH. Copper binding to the n-terminal tandem repeat region of mammalian and avian prion protein: structural studies using synthetic peptides. Biochem Biophys Res Commun. 1995;214(3):993–999. doi: 10.1006/bbrc.1995.2384. [DOI] [PubMed] [Google Scholar]
- Irving H, Williams RJP. The stability of transition-metal complexes. J Chem Soc. 1953 doi: 10.1039/JR9530003192. [DOI] [Google Scholar]
- Ishara Silva K, Saxena S. Zn(ii) ions substantially perturb cu(ii) ion coordination in amyloid- at physiological ph. J Phys Chem B. 2013;117(32):9386–9394. doi: 10.1021/jp406067n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenche VB, Barnham KJ. Alzheimer’s disease & metals: therapeutic opportunities. Br J Farmacol. 2011;163(2):211–219. doi: 10.1111/j.1476-5381.2011.01221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret W. Metals on the move: zinc ions in cellular regulation and in the coordination dynamics of zinc proteins. Biometals. 2011;24:411–418. doi: 10.1007/s10534-010-9406-1. [DOI] [PubMed] [Google Scholar]
- Morante S. The role of metals in beta-amyloid peptide aggregation: X-ray spectroscopy and numerical simulation. Curr Alz Res. 2008;5:508–524. doi: 10.2174/156720508786898505. [DOI] [PubMed] [Google Scholar]
- Penner-Hahn JE. Characterization of “spectroscopically quiet” metals in biology. Coord Chem Rev. 2005;249(1):161–177. [Google Scholar]
- Proux O, Biquard X, Lahera E, Menthonnex J, Prat A, Ulrich O, Soldo Y, Trévisson P, Kapoujvan G, Perroux G, Taunier P, Grand D, Jeantet P, Deleglise M, Roux JP, Hazemann JL. Fame: a new beamline for x-ray absorption investigations of very-diluted systems of environmental, material and biological interests. Physica Scripta. 2005;115(T115):970–973. http://stacks.iop.org/1402-4896/2005/i=T115/a=292. [Google Scholar]
- Proux O, Nassif V, Prat A, Ulrich O, Lahera E, Biquard X, Men-thonnex JJ, Hazemann JL. Feedback system of a liquid-nitrogen-cooled double-crystal monochromator: design and performances. J Synchrotron Radiat. 2006;13(Pt 1):59–68. doi: 10.1107/S0909049505037441. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95(1):13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin K, Yang Y, Mastrangelo P, Westaway D. Mapping cu(ii) binding sites in prion proteins by diethylpyrocarbonate modication and matrix-assisted laser desorption ionization-time of ight (malditof) mass spectrometric footprinting. J Biol Chem. 2002;277(3):1981–1990. doi: 10.1074/jbc.M108744200. http://www.jbc.org/content/277/3/1981.abstract. [DOI] [PubMed] [Google Scholar]
- Rachidi W, Mange A, Senator A, Guiraud P, Riondel J, Benboubetra M, Lehmann S. Prion infection impairs copper binding of cultured cells. J Biol Chem. 2003;278(17):14595–14598. doi: 10.1074/jbc.C300092200. http://www.jbc.org/content/278/17/14595.abstract. [DOI] [PubMed] [Google Scholar]
- Ravel B, Newville M. Athena, artemis, hephaestus: data analysis for x-ray absorption spectroscopy using ifet. J Synchrotron Radiat. 2005;12(4):537–541. doi: 10.1107/S0909049505012719. http://dx.doi.org/10.1107/S0909049505012719. [DOI] [PubMed] [Google Scholar]
- Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: Metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva K, Michael B, Geib S, Saxena S. Eseem analysis of multi-histidine cu(ii)-coordination in model complexes, peptides, and amyloid- J Phys Chem B. 2014;118(30):8935. doi: 10.1021/jp500767n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellato F, Spevacek A, Proux O, Minicozzi V, Millhauser GL, Morante S. Zinc modulates copper coordination mode in prion protein octa-repeat subdomains. Eur Biophys J. 2011;40:1259–1270. doi: 10.1007/s00249-011-0713-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RW, Blackburn N, Knowles P, Hasnain S. X-ray absorption spectroscopy of metal-histidine coordination in metalloproteins. exact simulation of the exafs of tetrakis(imidazole)copper (ii) nitrate and other copper-imidazole complexes by the use of a multiple-scattering treatment. J Am Chem Soc. 1987;109:7157–7162. [Google Scholar]
- Vagenende V, Yap MG, Trout BL. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48(46):11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- Walter ED, Stevens DJ, Visconte MP, Millhauser GL. The prion protein is a combined zinc and copper binding protein:Zn2+ alters the distribution of Cu2+ coordination modes. J Am Chem Soc. 2007;129(50):15440–15441. doi: 10.1021/ja077146j. http://pubs.acs.org/doi/abs/10.1021/ja077146j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells MA, Jelinska C, Hosszu LLP, Craven CJ, Clarke ARR, Collinge J, Waltho JP, Jackson GS. Multiple forms of copper (ii) coordination occur throughout the disordered n-terminal region of the prion protein at ph 7.4. Biochem J. 2006;400(3):501–510. doi: 10.1042/BJ20060721. http://www.biochemj.org/bj/400/bj4000501.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittal RM, Ball HL, Cohen FE, Burlingame AL, Prusiner SB, Baldwin MA. Copper binding to octarepeat peptides of the prion protein monitored by mass spectrometry. Protein Sci. 2000;9(2):332–343. doi: 10.1110/ps.9.2.332. http://dx.doi.org/10.1110/ps.9.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]