Abstract
In chloroplasts, the multimeric ATP synthase produces the adenosine triphosphate (ATP) that is required for photosynthetic metabolism. The synthesis of ATP is mechanically coupled to the rotation of a ring of c-subunits, which is imbedded in the thylakoid membrane. The rotation of this c-subunit ring is driven by the translocation of protons across this membrane, along an electrochemical gradient. The ratio of protons translocated to ATP synthesized varies according to the number of c-subunits (n) per oligomeric ring (cn) in the enzyme, which is organism dependent. Although this ratio is inherently related to the metabolism of the organism, the exact cause of the cn variability is not well understood. In order to investigate the factors that may contribute to this stoichiometric variation, we have developed a recombinant bacterial expression and column purification system for the c1 monomeric subunit. Using a plasmid with a codon optimized gene insert, the hydrophobic c1 subunit is first expressed as a soluble MBP-c1 fusion protein, then cleaved from the maltose binding protein (MBP) and purified on a reversed phase column. This novel approach enables the soluble expression of an eukaryotic membrane protein in BL21 derivative Escherichia coli cells. We have obtained significant quantities of highly purified c1 subunit using these methods, and we have confirmed that the purified c1 has the correct alpha-helical secondary structure. This work will enable further investigation into the undefined factors that affect the c-ring stoichiometry and structure. The c-subunit chosen for this work is that of spinach (Spinacia oleracea) chloroplast ATP synthase.
Keywords: ATP synthase, chloroplast, subunit c, subunit III, recombinant expression, maltose binding protein
1. Introduction
The photosynthetic process used by plants, algae, and cyanobacteria to convert water and carbon dioxide into biochemical energy provides the energetic foundation for nearly every living ecosystem on the planet. Photosynthesis begins with the splitting of water and proceeds with the transfer of electrons, which is coupled to the formation of a proton (H+) gradient across thylakoid membranes. These concerted processes are driven by sunlight and carried forward in a stepwise fashion by several protein complexes located in the thylakoid membranes of all photosynthetic organisms. Among these complexes is the multi-subunit ATP synthase. This enzyme uses a unique rotary mechanism to couple an electrochemical gradient-driven translocation of protons across the thylakoid membrane to the synthesis of adenosine triphosphate (ATP) from adenosine diphosphate (ADP) and inorganic phosphate (Pi). The ATP synthase is one of the key enzymes involved in the photosynthetic process, and is also similarly involved in cellular respiration. As such, it is assumed to be present in some form in all bacterial, archaeal, and eukaryotic cells. Because of the rotational nature of its structure, the ATP synthase can be regarded as the smallest molecular motor in biology [1].
1.1. Structure and functional activity
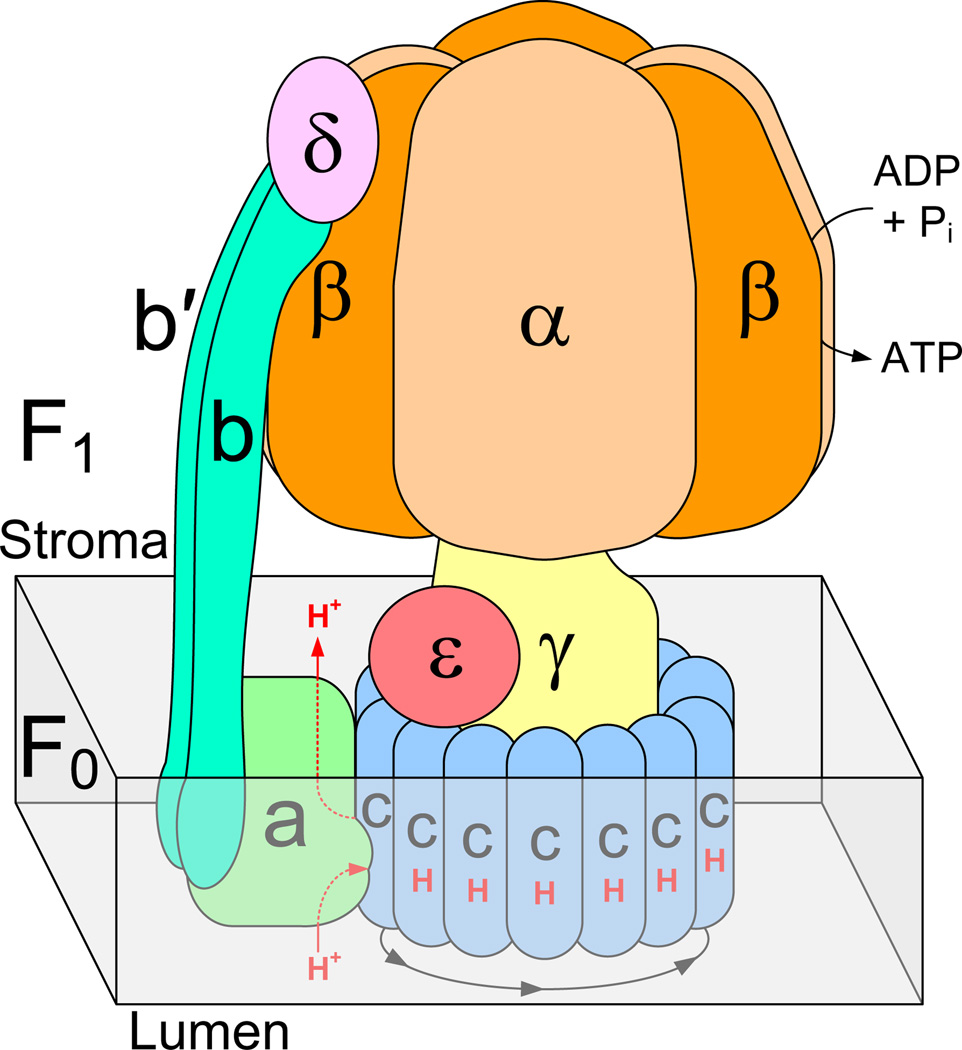
Structurally, the chloroplast ATP synthase is comprised of a membrane extrinsic ‘head’ region designated as ‘F1’, and a membrane intrinsic region designated as ‘F0’ (Figure 1). The F1 region includes the stromal subunits α3, β3, γ, δ, and ε. Subunits a, b, b’ and cn are included in the F0 region1. The two regions are connected by a rotational γ-stalk and a stationary b/b’-stalk, and thus mechanically coupled. In the chloroplast F0 region, single protons from the lumen are directed to a Glu residue on c1 (monomeric) subunits through a putative half-channel provided by the adjacent a-subunit. As (n) protons enter from the lumen and bind to (n) c-subunits, a complete 360° stepwise rotation of the cn (multimeric) ring takes place that allows the bound protons to be released individually at each step, and directed into the stroma via another putative half-channel provided by the a-subunit [2]. The rotation of the cn ring is coupled to the rotation of the γ-stalk in the F1 region, where subunit γ functions as a shaft inside the α3β3 head. The γ-rotation drives the catalysis of the ADP + Pi → ATP reaction that occurs at each of the three α-β subunit interfaces in F1 [3]. This cyclical sequence of rotation, translocation and catalysis produces 3 ATP molecules for every (n) value of protons that pass from the lumen to the stroma [3–5]. This process is reversible in the chloroplast ATP synthase and it is classified as an F-type enzyme. Archaeal (A-type) ATP synthases are also reversible, while vacuolar (V-type) ATP-ases function only as proton or ion pumps driven by ATP hydrolysis.
Figure 1.
Structural scheme of the F-type ATP synthase subunit arrangement. The gray box represents the region of cell membrane. A tetradecameric c-subunit ring is represented here, as observed in spinach chloroplast ATP synthase.
1.2. Ring stoichiometry and coupling ratio
The stoichiometry of F-type ATP synthase cn rings is known to range from c10 to c15 among the short list of organisms for which it has been determined (Table 1). Because the number of c subunits per ring (n) is organism dependent, the coupling ratio (ions transported : ATP generated) is known to range from 3.3 to 5.0 among these organisms [6, 7]. This value is entirely dependent on the variable (n), since the number of ATP generated per cn rotation is constantly held at 3 in all known ATP synthases [8]. Various hypotheses have been presented, however, the cause or purpose of the cn stoichiometric variation has not yet been defined [7, 9, 10]. Further investigations into the relationship between a monomeric c1 subunit and its respective multimeric cn ring are needed in order to understand the purpose of stoichiometric differences. The discovery of additional c-subunit ring stoichiometries in other organisms would also help explain this observation by enabling broader comparisons to be made.
Table 1.
Variable c subunit ring stoichiometries currently known. Stoichiometries of other cyanobacterial c-rings have been also suggested, with the hypothesis that the number may vary according to energetic requirements [9].
| Organism | Location | n (c-subunits/ring) = |
|---|---|---|
| Saccharomyces cerevisiae [40] | Mitochondria/Inner membrane | 10 |
| Escherichia coli [41] | Cell membrane | 10 |
| Ilyobacter tartaricus [42] | Cell membrane | 11 |
| Propionigenium modestum [43] | Cell membrane | 11 |
| Bacillus pseudofirmus [44] | Cell membrane | 13 |
| Synechococcus elongatus [9] | Thylakoid membrane | 13 |
| Spinacia oleracea [45] | Chloroplast/Thylakoid membrane | 14 |
| Spirulina platensis [46] | Chloroplast/Thylakoid membrane | 15 |
1.3. Recombinant approach
In the following sections, we report in detail how the ATP synthase monomeric c1 subunit from the chloroplast of spinach (Spinacia oleracea) has been produced in a recombinant Escherichia coli expression system, and subsequently purified in mg quantities. A brief description of alternate approaches which were explored at each step of the process is included to enable future modifications or new applications of the methods described. The recombinant c1 is currently being used by our lab for experiments focused on the reconstitution of the multimeric ring (cn). If reconstitution is successful, the development of this recombinant expression system will enable the use of molecular biology techniques which cannot otherwise be applied to a native cn ring. This added capability is in demand for further studies aimed at investigating the factors that influence the stoichiometric variation of the intact ring [9].
2. Materials and Methods
2.1. Codon optimized atpH gene construction
The plastid genome sequence of spinach (Spinacia oleracea) is mapped and sequenced, with the gene atpH coding for the c-subunit of ATP synthase [11, 12]. Using the known 81 amino acid sequence (UniProtKB accession no.: P69447), we have designed a synthetic atpH gene with codons optimized for E. coli expression, and terminal restriction sites added for cloning (Figure 2). Codons were chosen with the assistance of Gene Designer software by DNA2.0 [13]. The synthetic atpH gene was constructed by annealing and ligating 14 overlapping oligonucleotides (synthesized by Integrated DNA Technologies), ranging from 24 to 46 bp in length. Prior to annealing, phosphates were added to the 5’ end of all individual oligonucleotides (minus the two 5’ terminus oligonucleotides) in 10 µL reactions by mixing 100 pmol of each oligonucleotide with 1 mM ATP, 1× T4 Polynucleotide Kinase Buffer A (Fermentas, provided with the kinase), and 0.1 units T4 Polynucleotide Kinase (Fermentas). This kinase reaction was incubated for 30 minutes at 37°C, followed by a 5 minute inactivation period at 70°C. A 5 µL volume of each oligonucleotide in this solution was mixed with 5 µL of its corresponding annealing partner, heated to 80°C, and cooled to 20°C over a 60 minute period in order to produce 7 annealed duplex DNA fragments.
Figure 2.
Gene sequence of recombinant atpH gene, codon optimized for E. coli expression. The version shown here has 255 bp and is designed for insertion into the pMAL-c2x vector at the XmnI and XhoI restriction sites. The expressed c1 subunit contains 81 amino acids with a mass of 8 kDa.
The annealed duplex fragments were ligated together in sequence to form the complete atpH gene in a two step ligation process. First, 1.5 µL of three proximal duplexes from the previous annealing reaction were mixed with 1× Ligase Buffer (Invitrogen, provided with the Ligase) in a 20 µL volume. The same reaction was prepared for the other 4 proximal duplex DNA fragments, and both reaction mixtures were heated to 50°C then cooled to 20°C over a 60 minute period, followed by an additional 2 hour incubation period at room temperature with 0.5 units of T4 DNA Ligase (Invitrogen) in each reaction. The resulting ligated fragments were separated on a 3% Metaphor agarose gel (Lonza) prepared with and run in TAE Buffer (40 mM Tris-Base, 20 mM acetic acid, 1 mM EDTA; pH 8.5), and equilibrated in 1 µg/mL ethidium bromide for detection of DNA under UV light. Successfully ligated DNA was excised from the gel and purified using the Promega Wizard SV Gel and PCR Clean-Up Kit. The two purified fragments were mixed together in equimolar amounts with 1× Ligase Buffer in a 10 uL volume. The reaction was heated to 50°C and cooled to 20°C over a 60 minute period, followed by the 2 hour incubation period at room temperature with 0.5 units of T4 DNA Ligase. The resulting DNA was again separated on a 3% agarose gel, and the ligated atpH fragment was excised from the gel, and purified in the same way as described in the previous step.
2.2. Cloning of atpH into various vector constructs
The synthetic atpH gene was inserted into vectors pMAL-c2x (New England Biolabs), pET-32a(+) (Novagen), and pFLAG-MAC (Sigma-Aldrich) for the purpose of comparing alternate modes of c-subunit expression. The version of the pMAL-c2x vector we used had been previously modified with a His tag, however this modification was not used and has no relevance to the studies reported herein. Prior to inserting into each vector, the freshly ligated synthetic atpH gene was amplified by using high-fidelity PCR with the Phusion Polymerase (New England BioLabs), according to product instructions.
A synthetic atpH gene was produced with a 5’ blunt end and 3’ XhoI restriction site for insertion into the pMAL-c2x vector at the XmnI and XhoI restriction sites to produce the plasmid pMAL-c2x-malE/atpH (Supp. Figure 1). Similarly, an atpH gene was produced with 5’ NdeI and 3’ XhoI terminal restriction sites, and inserted into the pMAL-c2x vector at the corresponding sites to create pMAL-c2x-atpH. The same atpH insert was also inserted into the pET-32a(+) vector at the NdeI and XhoI restriction sites to create pET-32a(+)-atpH. And, an atpH gene was produced with 3’ HindIII and 5’ XhoI terminal restriction sites and inserted into the pFLAG-MAC vector at the corresponding sites to create vector pFLAG-atpH. Following ligation, each new vector construct was cloned by transforming DH10B E. coli cells (Invitrogen, ElectroMAX T1 Phage-Resistant) via electroporation at 1660 V. A Qiagen Miniprep kit was used to lyse DH10B cells and harvest high concentrations of plasmid DNA from viable transformant growth cultures. Nucleotide sequencing services were provided by the Core DNA Laboratory at Arizona State University, where automated methods are used with an Applied Biosystems 3730 capillary sequencer. The four resultant atpH vectors are compared in Table 2. Restriction endonucleases were obtained from New England BioLabs, and T4 DNA Ligase was obtained from Invitrogen.
Table 2.
Plasmids constructed with the atpH gene for expression of chloroplast ATP synthase c subunit (c1).
| Plasmid Construct | Promoter | Product |
|---|---|---|
| pMAL-c2x-malE/atpH | tac | MBP-c1 |
| pMAL-c2x-atpH | tac | c1 |
| pET-32a(+)-atpH | T7 | c1 |
| pFLAG-atpH | tac | FLAG-c1 |
BL21 derivative E. coli cells (T7 Express lysY/Iq, New England Biolabs) were separately transformed with each vector construct. Cells were also transformed with pMAL-c2x (no atpH) for use as a negative control for atpH expression; and co-transformed with the pMAL-c2x-malE/atpH and pOFXT7KJE3 vectors, where the latter expresses the chaperone proteins DnaK, DnaJ, and GrpE. The co-expression of these chaperone proteins has been shown to substantially increase quantities of recombinant proteins which are toxic or otherwise difficult to produce [14]. The pOFXT7KJE3 plasmid was produced and generously provided by Castanié, et al [15]. The accompanying protocol for transformation of T7 Express lysY/Iq cells was followed precisely, except with the co-transformation where 118 ng of pMAL-c2x-malE/atpH and 75 ng of pOFXT7KJE3 plasmid was used. Successful transformant clones were selected for on LB-agar plates with 50 µg/mL ampicillin. For the co-transformants, double antibiotic plates that also included 50 µg/mL spectinomycin were used.
2.3. Comparison of atpH expression with different vectors
The expression levels of ATP synthase c-subunit were compared in the E. coli transformed with pMAL-c2x-malE/atpH, pMAL-c2x-atpH, pET-32a(+)-atpH, pFLAG-atpH, pMAL-c2x negative control, and pMAL-c2x-malE/atpH + pOFXT7KJE3. 100 mL of LB-glucose expression medium (1.0% tryptone, 0.5% yeast extract, 0.4% glucose, 0.5% NaCl, 50 µg/mL ampicillin, and 50 µg/mL spectinomycin for the co-transformant) was inoculated separately with each transformant, and grown to an optical density of 0.6–0.7 at 37°C incubation and 200 RPM 1” orbital shaking. At this point, targeted expression of atpH was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to 1.0 mM concentration and incubating for an additional 30 minutes. Cell pellets were prepared by centrifugation at 6029 × g RCF for 20 minutes, and stored at −80°C. Thawed cell pellets were resuspended in 2 mL of Lysis Buffer (20 mM Tris-HCl pH 8.0, 2% v/v Protease Inhibitor Cocktail (Sigma, P8465)). Lysozyme was added to each resuspension to 1 mg/mL, and they were incubated at 4°C for 1.5 hrs. prior to sonication at 50–75 W.
A 12% polyacrylamide gel (Section 2.7) was prepared with 0.25 µL samples of total cell lysate from each transformant, along with 0.4 µg native spinach ATP synthase as a positive control for c-subunit, and 8 µL of Bio-Rad Western C standard. The gel was used for immunoblotting (Section 2.10) to confirm expression of c1. It was determined that c1 could only be expressed when fused to MBP by the vector pMAL-c2x-malE/atpH (See section 3.2), so this construct was chosen for further experimental application.
2.4. Large-scale expression and purification of MBP-c1
For preparative purposes, a 1.0 L volume of cells co-transformed with pMAL-c2x-malE/atpH + pOFXT7KJE3 was cultured for the extraction of MBP-c1. The 1.0 L culture was incubated and induced in the same manner as described for the 100 mL cultures in section 2.3. A 3.1 g bacterial cell pellet was separated from the culture by centrifugation at 6029 × g RCF for 20 minutes, and stored at −80°C. The cell pellet was later thawed, and resuspended in 50 mL of Lysis Buffer. Lysozyme was added to the resuspension at 1 mg/mL final concentration, and incubation proceeded at 4°C for 1.5 hr. to hydrolyze bacterial cell walls. Cell lysis was completed by sonicating the viscous resuspension at 150 W in intervals with cooling on ice in between, until it was no longer viscous. The cell lysate was then centrifuged for 30 minutes at 18,677 × g RCF, and the resulting supernatant was separated from the pellet. The cell lysate supernatant was diluted to a 100 mL volume in Amylose Column Buffer 1 (20 mM Tris-HCl pH 8.0, 0.2 M NaCl, 1 mM EDTA) and passed through 7.5 mL of amylose resin (New England Biolabs) on a 25 mm diameter gravity column to bind the MBP-c1 fusion protein. The flow rate was maintained at ~1 mL/min during this step. The resin was then washed of non-binding cell lysate proteins by passing 420 mL of Amylose Column Buffer 2 (20 mM Tris-HCl pH 8.0, 0.2 M NaCl) through. Finally, MBP-c1 was eluted with 50 mL Maltose Elution Buffer (20 mM Tris-HCl pH 8.0, 0.2 M NaCl, 10 mM maltose). The eluted fraction was concentrated 10× to 5 mL using a Vivspin 20 30K MWCO centrifugal concentrator with a polyethersulfone (PES) membrane (Sartorius), according to product instructions.
Samples were taken from each of the following fractions for analysis on a coomassie blue stained 12% polyacrylamide gel (Sections 2.7, 2.8): total cell lysate (3 µL), cell lysate pellet (3 µL), cell lysate supernatant (6 µL), amylose column flow through (6 µL), Amylose Column Buffer 2 wash (9 µL), and Maltose Elution Buffer wash (9 µL). Also included was 10 µL of the Bio-Rad standard.
2.5. Factor Xa protease cleavage of c1 from MBP
Optimal protease cleavage conditions were determined based on our prior experimental comparisons using variations of temperature, incubation period, and protease concentration. These factors are known to influence the activity of Factor Xa [16]. We also tested the effect of detergent as a variable factor in protease cleavage because it is needed in order to prevent aggregation of the cleaved c1 subunit product. Protease Buffers were prepared with different detergents at a range of concentrations. Sodium dodecyl sulfate (SDS, 0.01%), sodium cholate (0.01%, 0.1%, 1.0%), n-β-dodecyl-D-maltoside (βDDM, 0.001%, 0.01%, 0.1%), and β-octyl-D-glucopyranoside (βOG, 0.01%, 0.1%, 1.0%) were each added to 20 mM Tris-HCl pH 8.0, and 2 mM CaCl2. 0.3 mL of MBP-c1 (1 mg) was dialyzed against each of these Protease Buffers, and 1% w/w Factor Xa Protease (New England Biolabs) was added. The protease reactions were incubated at 4°C for 24 hours. For analytical comparison, a 12% polyacrylamide gel was prepared for immunoblotting (Section 2.10) with 1.25 µL of each protease reaction product, 0.5 µg of native spinach ATP synthase as a positive control for c1, and 8 µL of Bio-Rad Western C standard. The highest yield of c1 product occurred with n-β-dodecyl-D-maltoside in the Protease Buffer at 0.01% (1× critical micelle concentration (CMC)) and 0.10% (10× CMC), as discussed in Section 3.4. The detergent concentration was further optimized to 0.05% (5× CMC), which was used for all subsequent preparative purposes.
The 5 mL preparative sample of MBP-c1 fusion protein purified on the amylose column (Section 2.4) was dialyzed against 2 L of optimized Protease Buffer (20 mM Tris-HCl pH 8.0, 2 mM CaCl2, 0.05% n-β-dodecyl-D-maltoside) for at least 12 hours at 4°C using 13K MWCO dialysis tubing. 1% w/w Factor Xa Protease was then added to the dialyzed protein and the protease reaction was incubated at 4°C for about 24 hours with light stirring. The post-protease cleaved sample (MBP+c1) was then immediately applied to the reversed phase column in the next step.
2.6. Reversed phase HPLC purification of c1
The optimal conditions for reversed phase column (RPC) chromatography purification of c1 were chosen based on several prior small scale HPLC comparisons. A RESOURCE RPC 3 mL column (GE Healthcare) was used for these analytical trials. Purified water buffered with 20 mM Tris-HCl pH 8.0 was used for Eluent A. Eluent B was tested as the variable in separate trials with 100% methanol, 100% ethanol, or 100% 2-propanol. In each trial, the column was equilibrated by washing with 1–2 column volumes of Eluent A, then Eluent B, then Eluent A again. About 1.7 mg of protease cleaved sample (MBP+c1) was loaded onto the column following centrifugation to remove any precipitant. The column was run with 10 column volumes of Eluent A, followed by a gradient increasing to 100% Eluent B over a 100 minute period. The column continued to be washed with 100% Eluent B until protein no longer eluted from the column. A flow rate of 1 mL/min. was maintained. Immunoblot analysis (Section 2.10) was performed on fractions collected from each Eluent B trial, and those fractions which contained c1 were further analyzed on silver stained 12% polyacrylamide gels (Sections 2.7, 2.8) to assess purity. We determined that ethanol was the most favorable Eluent B solvent tested in terms of both yield and purity (Section 3.5), and so this was used in subsequent larger scale preparative column runs.
A 15 mL SOURCE HR 16/10 reversed phase column manufactured by GE Healthcare was chosen for the large scale purification of c1. This preparative column uses the same hydrophobic polystyrene/divinyl benzene bead media as the 3 mL analytical column. The Eluent A was 20 mM Tris-HCl pH 8.0 in water, and the Eluent B was 100% HPLC grade ethanol (Sigma). Both buffers were degassed thoroughly. The column was equilibrated with Eluent A, then Eluent B, then Eluent A again by washing with 1–2 column volumes of each. At the end of the 24 hour protease cleavage period, the 5 mL sample of MBP+c1 (Section 2.5) was centrifuged at 12,110 × g for 5 minutes to remove a small amount of precipitant and loaded onto the equilibrated reversed phase column. The amount of protein loaded was estimated to be about 18 mg, according to the modified Lowry assay technique [17]. Hydrophilic protein products were eluted with 10 column volumes of Eluent A. A gradient was produced increasing to 100% Eluent B over a 500 minute period, and washing continued with 100% Eluent B until a stable UV absorbance signal indicated that no more protein was eluting. A flow rate of 1 mL/min. was maintained throughout the process, and 10 mL fractions were collected.
To confirm the presence of c1 in the fractions, a 12% polyacrylamide gel was prepared for immunoblotting (Section 2.10) with 12 µL of fractions E10, E11, E12, F1 and F2, 0.375 µL of MBP+c1, 0.375 µL of MBP-c1, 0.3 µg native spinach ATP synthase as a positive control for c1, and 10 µL Bio-Rad Western C standard. To assess the purity of the c1 containing fractions, a 12% polyacrylamide gel was prepared for silver staining (Section 2.8) with 1.25 mL (27 µg c1) of combined fractions E11+E12, 2.5 µL (4.4 µg) of MBP+c1, 2.5 µL (4.4 µg) of MBP-c1, 8.25 µg native spinach ATP synthase as positive control, and 3.3 µL of Bio-Rad standard. The ethanol-containing RPC fractions were first evaporated, then resuspended in water and Sample Loading Buffer followed by heating at 95°C for 5 min. prior to loading onto the polyacrylamide gels. N-terminal amino acid sequencing services were provided by the Core Proteomics and Protein Chemistry Lab at Arizona State University, where automated Edman degradation methods are used.
2.7. SDS Polyacrylamide gel electrophoresis
Tricine SDS denaturing gel electrophoresis techniques described by Schägger were used for the qualitative analysis of various protein samples [18]. For most purposes, adequate separation of protein bands could be obtained with a 12% polyacrylamide separating gel layered with a 1 cm 4% polyacrylamide stacking gel. The Bio-Rad Mini-PROTEAN Tetra Cell electrophoresis system was used for preparing and running gels. Precision Plus Standards (Bio-Rad) were used for gels which were stained, and Precision Plus WesternC Standards (Bio-Rad) were used for gels which were immunoblotted. All samples were prepared in Sample Loading Buffer (6.67% sodium dodecyl sulfate, 33 mM Tris-HCl pH 8.0, 625 mM 2-mercaptoethanol, 6.7% glycerol, 33 µM bromophenol blue; final concentration) and heated at 95°C to denature prior to loading. Cell lysate, cell pellet, and native ATP synthase samples required 15 minutes of heating to denature, and other samples required 1–5 minutes of heating.
2.8. Polyacrylamide gel staining
Protein bands were detected either with Coomassie Blue Staining or Silver Staining methods. Coomassie Blue Staining was done by equilibrating the gel in heated Coomassie Blue Stain (0.1% CBB 250, 30% methanol, 10% acetic acid) for 5 minutes, followed by equilibration in heated Destain I (50% methanol, 10% acetic acid) for 5 minutes, and final equilibration in Destain II (5% methanol, 10% acetic acid) until bands were adequately resolved. Silver Staining was done by equilibrating the gel in 12.5% glutaraldehyde solution for 60 minutes, followed by a 5 minute rinse in H2O and equilibration in a 1.0% AgNO3 solution for 60 minutes, also followed with a 5 minute rinse in H2O. The gel was then transferred to Developer Solution (0.25% formaldehyde, 6.25% Na2CO3) and equilibrated until bands could be distinctly resolved, at which point the gel was immediately transferred to Fixing Solution (10.0% w/v glycerol, 10% v/v acetic acid) to preserve development. Silver staining has proven to be the most practical means for visualizing the hydrophobic 8 kDa c1 subunit, while coomassie blue staining is sufficient for observation of the 50 kDa MBP-c1 fusion protein.
2.9. Antibody production
A polyclonal antibody was produced for the native spinach chloroplast ATP synthase c14 by Agrisera in collaboration with our lab. The native c14 ring was extracted and purified from spinach leaves in our lab as published previously [19], and provided to Agrisera for use as an antigen. Resulting antibody samples produced from serum of 6 rabbits was provided to us periodically by Agrisera for comparative screening. The resulting antibody sample with the strongest and most specific signal after several weeks of screening was chosen for commercial production (Agrisera, AS05 071), and used to produce the immunoblot results reported herein. This antibody has proven to be very sensitive and specific for the c1 subunit and c14 ring of spinach chloroplast ATP synthase.
2.10. Western immunoblot detection
Protein samples separated on a 12% polyacrylamide SDS tricine gel as described in section 2.7 were blotted onto a PVDF membrane at 100 mA for 85 minutes in a Tricine Transfer Buffer (25 mM Tris-Base, 192 mM tricine, 20% v/v methanol). The Bio-Rad Mini Trans-Blot Module was used for the electrophoretic transfer. Blotted PVDF membranes (Bio-Rad) were equilibrated in blocking solution (50 mM Tris-Base, 150 mM NaCl, 0.05% Tween 20, 5% w/v Non-fat Dry Milk) for 30 minutes. Then the polyclonal primary antibody against c1 subunit (Agrisera) was added in a 1:1000 dilution and equilibration continued for an additional 60 minutes. Two 5 minute washes in Tween-Tris Buffered Saline (TTBS) Buffer (50 mM Tris-Base, 150 mM NaCl, 0.05% Tween 20) followed. Then the PVDF membrane was transferred to 20 mL TTBS Buffer treated with a 1:5000 dilution of goat anti-rabbit IgG-HRP conjugate (Santa Cruz Biotechnology) and 1:5000 StrepTactin-HRP conjugate (Bio-Rad) and equilibrated for 75 minutes. Two 20 minute washes of the PVDF membrane in TTBS Solution followed. Immun-Star HRP substrate (Bio-Rad) was used to activate secondary antibody luminescence. Chemiluminescent images of the bound conjugates were taken with a Kodak Gel Logic 440 CCD camera using a 2 minute dark exposure period.
2.11. Circular dichroism spectroscopy
A Jasco J-710 Spectropolarimeter was used for measuring the circular dichroism (CD) spectrum of the purified recombinant c1 subunit. The purified c1 sample was prepared for CD measurement as described, with the exception that a 10 mM phosphate buffer at pH 8.1 was used for Eluent A during the reversed phase column purification (Section 2.6). Under these conditions, c1 eluted on the gradient at about 83% ethanol (Eluent B) and 17% Eluent A. A buffer of this composition was used for the blank reference measurement. The eluted c1 was concentrated to approximately 0.1 mg/mL using a 5000 MWCO Vivaspin 2 concentrator with a Hydrosart membrane (Sartorius). The protein concentration was determined according to modified Lowry methods [17]. CD spectra were measured from 195–260 nm at room temperature (25°C) in a 0.1 cm quartz cuvette. Parameters were set at 0.2 nm data pitch, continuous scan mode, 50 nm/min scan speed, 4 second response, and 1 nm bandwidth. Output data was generated from an accumulation of 3 scans. Values of mean residue ellipticity ([θ]) and molar CD (Δε) were calculated as described by Greenfield [20]. The algorithm CDSSTR [21] was used with the CDPro [22] software program to estimate the secondary structure content of the sample by comparing the measured data with the data set SMP50, which contains 13 membrane proteins and 37 soluble proteins [23].
3. Results
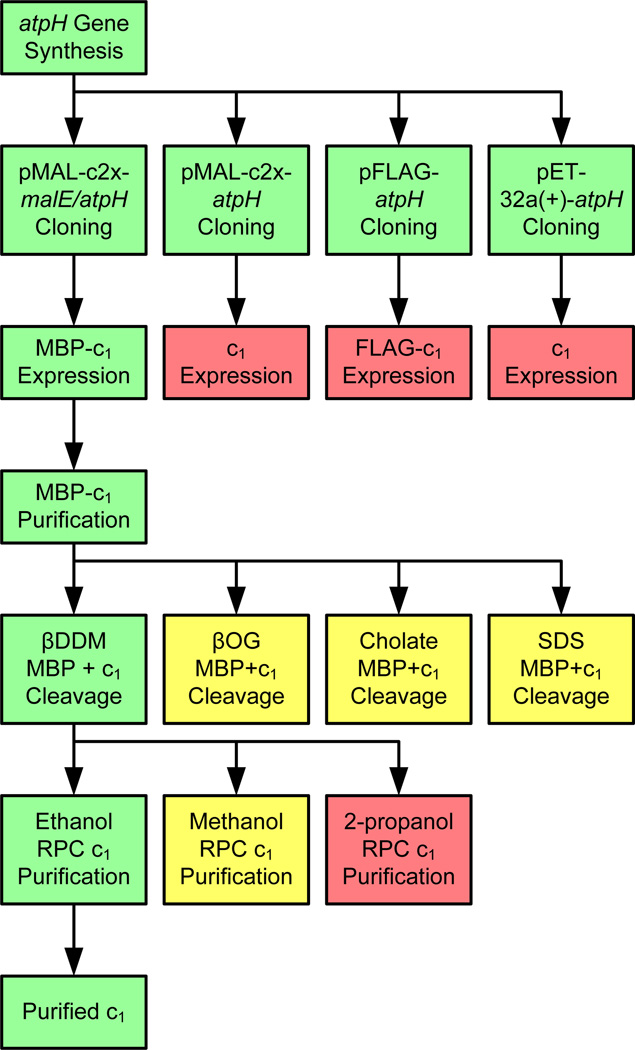
The sequence of methods used to produce the following results is summarized in the schematic diagram in Figure 3. Alternate approaches to some steps which did not produce optimal results are included as well for technical comparison.
Figure 3.
Developmental scheme of the reported approach for expression and purification of recombinant subunit c1. Steps highlighted in green were optimal and used to advance the process. Those highlighted in yellow were sub-optimal, and those in red were non-functional.
3.1. Four atpH expression vectors successfully cloned
The codon optimized synthetic atpH gene was successfully assembled and inserted into different vectors in order to compare different modes of expression. The four resultant atpH vectors are shown in Table 2, with their corresponding promoters and intended expression products. These vectors are designed to express the c-subunit alone under control of a T7 or tac promoter; and to express the c-subunit with an amino-terminus FLAG or MBP tag under control of a tac promoter. Nucleotide sequencing confirmed that all bases on atpH were accounted for, and that atpH was inserted at the intended restriction sites and within the correct reading frame on each vector.
3.2. c1 expresses when fused to maltose binding protein
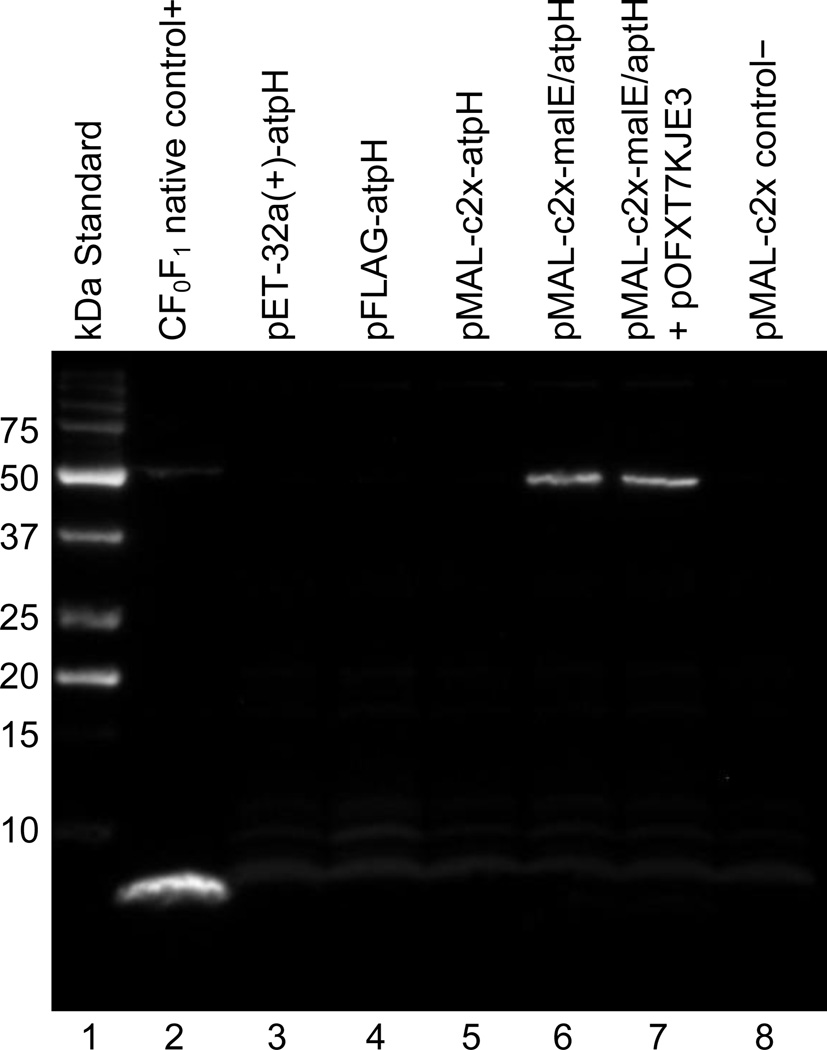
A comparison of the expression products from each of the four atpH vectors showed that c1 expression was only observed when the maltose binding protein (MBP) was fused to the amino-terminus. This expression is clearly evident in lanes 6 and 7 of the immunoblot shown in Figure 4; while no expression of c1 is observed as a stand-alone protein under control of the T7 or tac promoter (lanes 3 and 5, respectively), and no expression of c1 is observed with the FLAG tag on the amino-terminus (lane 4). The MBP and FLAG tag expression vectors both used a tac promoter with an amino-terminus tag; however, the 42 kDa MBP tag is about 5 times larger in mass than the 8 kDa c1, whereas the 1.1 kDa FLAG tag is much smaller than c1. The size advantage of MBP is apparently necessary for stabilizing c1 as it is translated and conferring solubility to it thereafter. The MBP has been shown in other studies to increase the solubility and proper folding of various polypeptides to which it has been fused [24]. For this reason, we chose to test it in these experiments as a candidate to enable the expression of the otherwise insoluble and unstable recombinant c1. Although these results were obtained using the T7 Express lysY/Iq E. coli strain, we have observed similar results with standard BL21(DE3) E. coli strains as well.
Figure 4.
Chemiluminescent immunoblot showing detection of the expressed 50 kDa MBP-c1 in lanes 6 and 7. No detection of the 8 kDa c1 is observed without a tag in lanes 3 or 5, and no detection of the 9.1 kDa FLAG-c1 is observed in lane 4.
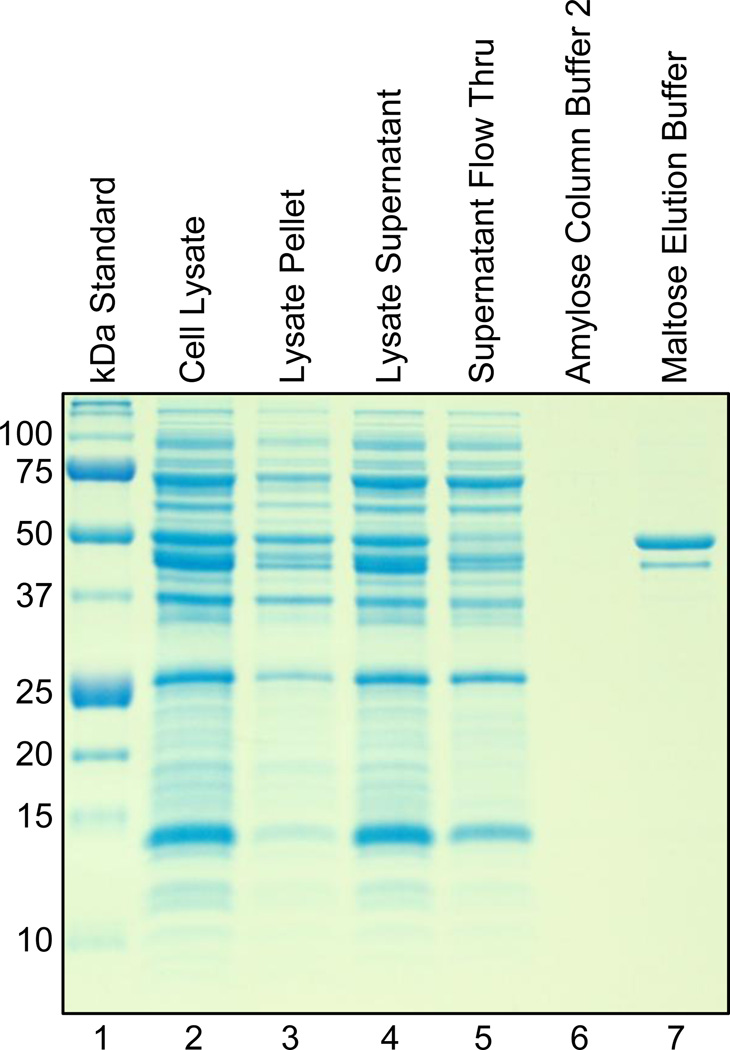
3.3. MBP-c1 purifies on an amylose affinity column
The expressed MBP-c1 fusion protein has been purified directly from the soluble fraction of cell lysate using affinity column chromatography with amylose resin. In the gel shown in Figure 5, the 50 kDa MBP-c1 product is distinctly present in the maltose fraction, and the only other visible contaminant is the 41 kDa DnaJ chaperone. These results demonstrate that in addition to lending stability and solubility to c1, the MBP also confers the ability to bind amylose resin for the purpose of purification on a column. This method has been used successfully with a variety of other proteins [25–27], and for this reason we chose to test MBP as a carrier for c1 expression and purification.
Figure 5.
Coomassie Blue stained 12% polyacrylamide gel showing the results of the amylose column purification scheme. The 50 kDa MBP-c1 is purified from the cell lysate supernatant in the Maltose Elution Buffer fraction (lane 7).
We have obtained up to 10 mg of this fairly pure MBP-c1 amylose column purification product from 1 L of cell culture. However, our results indicate that this yield can be increased up to nearly two fold when MBP-c1 is co-expressed with the chaperone proteins (DnaK, DnaJ, GrpE). And so for large scale expression and purification of MBP-c1, we have chosen to use cells co-transformed with the chaperone vector pOFXT7KJE3 and pMAL-c2x-malE/atpH. Although the co-expression may cause an increase in the DnaJ contaminant after the amylose column purification step, DnaJ can easily be removed during a later reversed phase HPLC purification step (Section 2.6).
3.4. Factor Xa cleaves c1 in the presence of detergents
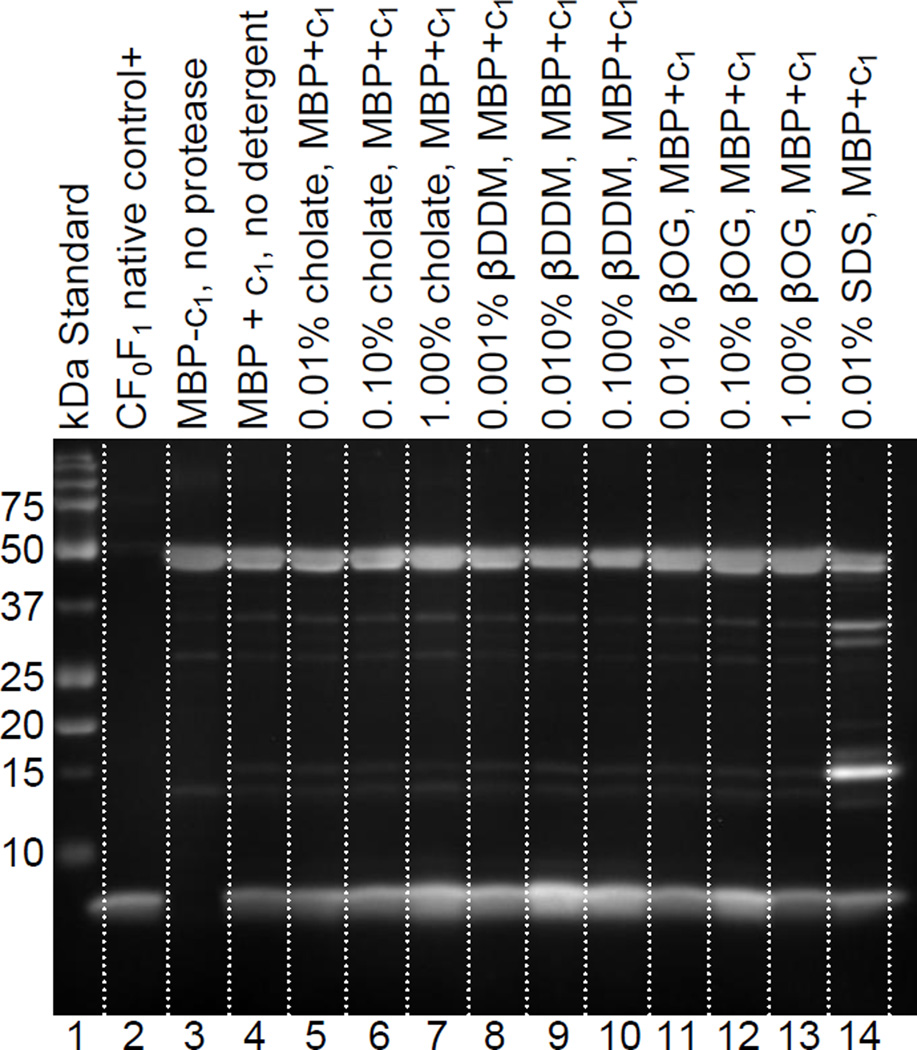
A systematic comparison of Factor Xa cleavage conditions revealed that the protease can effectively cleave c1 from MBP in the presence of various detergents, which were included to prevent precipitation of the cleaved c1 product. The immunoblot shown in Figure 6 shows that the type and concentration of detergent used can influence the activity of the Factor Xa protease. In fact, these results show that in some cases the detergent can actually improve the ability of the protease to cleave c1. This likely occurs because the detergent causes a slight denaturation of the fusion protein, which increases accessibility of the intended cleavage site. The sample with 0.01% β-dodecyl-D-maltoside appeared to produce the highest yield of c1 product among the samples shown in Figure 6. There was also no significant precipitation observed in the reaction products with β-dodecyl-D-maltoside. Many of the other samples shown in this immunoblot yielded comparable cleavage products; with the exception of the SDS sample. SDS produced a high level of secondary MBP-c1 cleavage product, which is problematic for RPC purification of c1 in the following step. As noted in section 2.5, the results with β-dodecyl-D-maltoside were further optimized to a concentration of 0.05% for preparative purposes. Protease cleavage results at this concentration can be seen in lane 4 of the gel and immunoblot shown in Figures 7b and 7c.
Figure 6.
Comparison of Factor Xa protease cleavage in the presence of different detergents. Optimal cleavage products are observed with n-β-dodecyl-D-maltoside (lanes 9,10). Lines were added to the image between lanes for distinction.
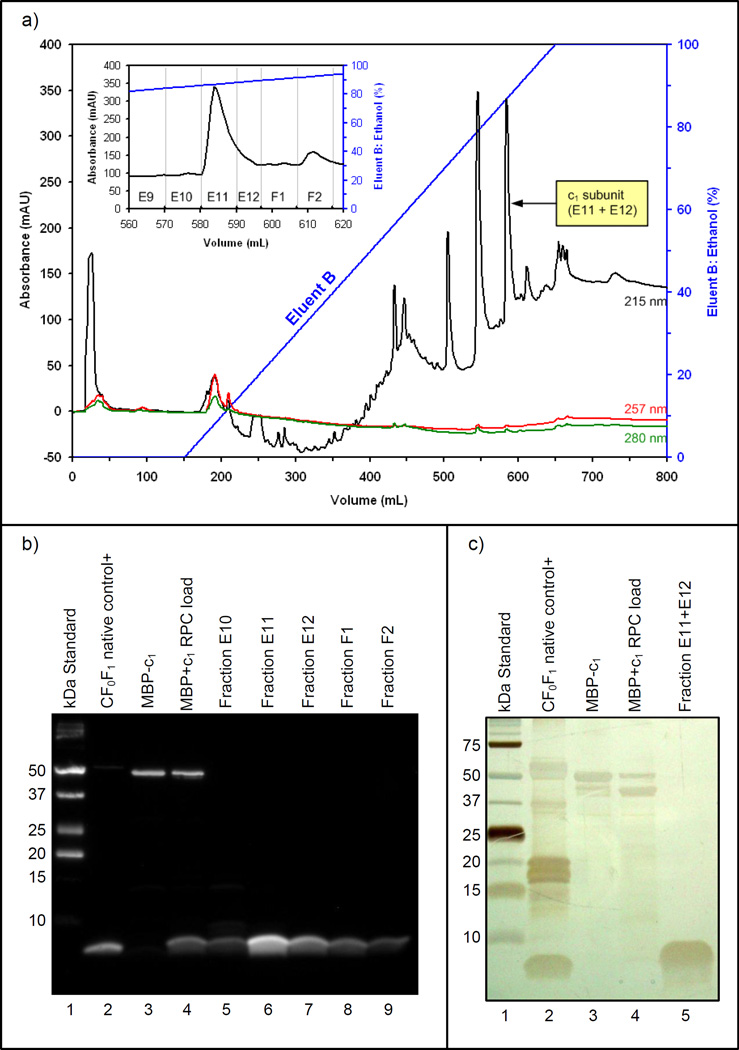
Figure 7.
(a) Chromatogram of the reversed phase HPLC column separation produced with 215, 257, and 280 nm UV absorbance data. A detail of the c1 eluting fractions is included. (b) The immunoblot confirms the elution of c1 at approximately 87% ethanol in fractions E11 and E12. (c) The 12% polyacrylamide gel stained with AgNO3 shows the purity of c1 in combined fractions E11+E12.
3.5. c1 purifies on a reversed phase column with ethanol
Purification of the c1 subunit from the MBP and other post-cleavage products has been successfully accomplished using reversed phase column chromatography. The results of the preparative purification with ethanol are shown in Figure 7. The chromatogram in Figure 7a shows that the absorbance peak which corresponds to the elution of the c1 subunit in fractions E11 and E12 occurs when the Eluent B gradient approached 87% ethanol. The presence of c1 in these fractions was confirmed by the immunoblot results shown in Figure 7b, and the purity of the confirmed c1 product is clearly evident in lane 5 of the silver stained gel shown in Figure 7c. It was estimated by the modified Lowry assay [17] that 0.4 mg of highly purified c1 subunit was recovered from the 18 mg of MBP+c1 post-cleavage product initially loaded onto the reversed phase column. This amounts to an approximate 14% theoretical recovery of c1, where losses are attributed to the protease cleavage and the reversed phase column purification steps.
RPC chromatography methods were selected for this final purification step because they are designed to separate smaller molecules according to their hydrophobicity by eluting with organic solvents. Formerly referred to as the ATP synthase proteolipid [28], the c-subunit structure is notoriously characterized by its strong hydrophobic character which allows it to be stable and soluble in organic solvents [29, 30]. Whereas this property is a source of difficulties with regards to recombinant expression and detection, we show here that it can be conveniently exploited for the sake of purification.
3.6. Purified recombinant c1 is highly alpha-helical
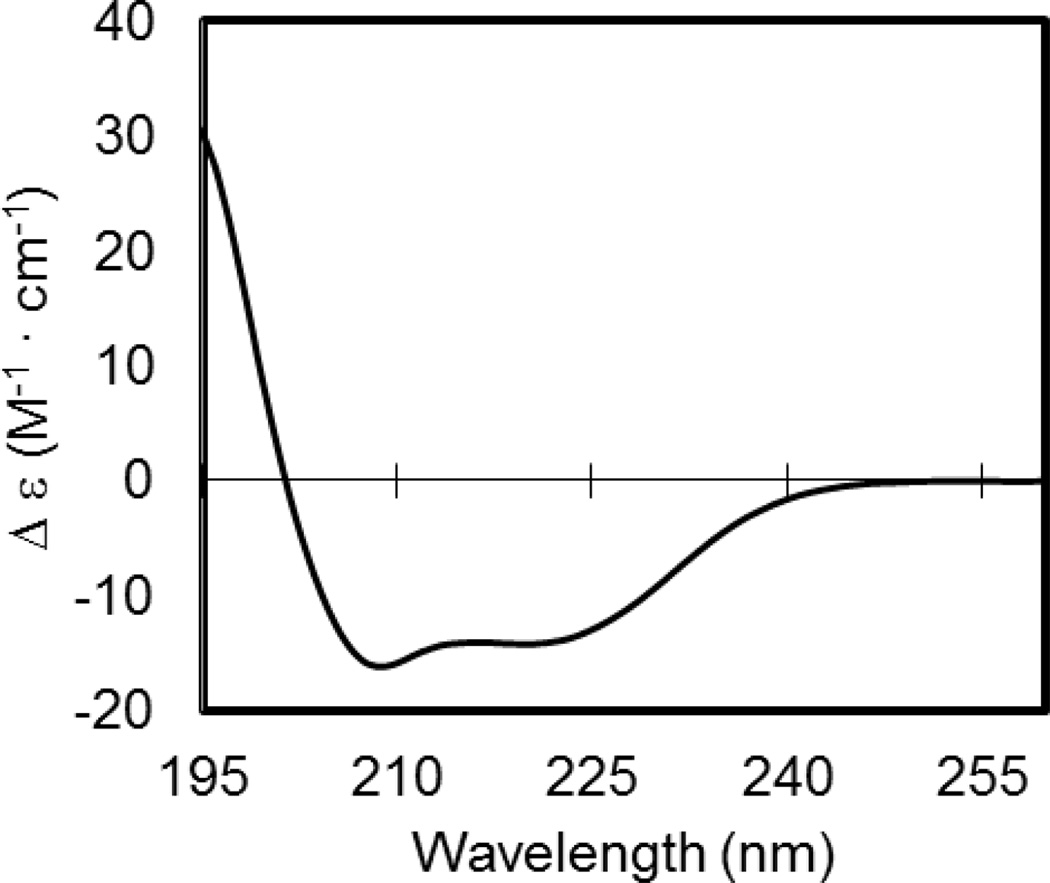
The CD spectrum of the purified recombinant c1 subunit is shown in Figure 8. The resulting curve shows the characteristic peaks associated with alpha-helical proteins, with two minima at 208.6 nm and 220.2 nm. Software analysis with CDSSTR produced a deconvoluted estimation of 85.5% alpha-helices, 6.0% beta sheets, 3.9% turns, and 4.5% of residues in random coil conformations. The NRMSD value of the assignments is 0.052. This proportion of alpha-helices is consistent with the alpha-helical content as derived from NMR structures of monomeric c1 subunits from Bacillus and E. coli (PDB ID: 1WU0, 1C0V, 1A91) [31]. The alpha-helical content is also similar to what is predicted by secondary structure prediction software programs. The program APPSPS2 [32] predicts 82.7% alpha-helices. The agreement of our CD results with the structure prediction programs and the NMR structures thereby provides strong evidence that the purified recombinant c1 subunit is correctly folded.
Figure 8.
Molar Circular Dichroism (Δε) values plotted vs. the spectra 195–260 nm for the purified recombinant c1 subunit. The resulting curve is indicative of a protein with high alpha-helical secondary structure.
4. Discussion
4.1. Method development and limitations
By nature, eukaryotic membrane proteins are not easily expressed in recombinant bacterial systems [33, 34]. As expected, this has also been the case with the c-subunit. Difficulties with c1 were not only limited to expression, but also detection. In addition to its hydrophobic nature, the small size (8 kDa) of c1 makes it difficult to reliably discern on SDS polyacrylamide gels unless silver staining is used in combination with a higher concentration of semi-pure protein. Also, because c1 has no tryptophan residues and only 1 tyrosine residue, it can only be detected at UV wavelengths less than the standard 280 nm. Prior to our work on this project, no commercial antibody was available for detecting the ATP-synthase c1 subunit or c14 ring from spinach chloroplast (or similar organism) for use in immunoblotting methods. The methods we have described here have been designed to overcome these limitations.
4.2. Previously published recombinant approaches
Attempts to express the c1 subunit or cn ring have been made in the past by other research groups using different approaches. Expression of a recombinant spinach chloroplast ATP-synthase c1 subunit (or possible cn ring) was reported by Burkovski, et al. in 1990 [35]. In this approach, an uncE knockout strain of E. coli was used and atpH expression was directed to the membrane in an unsuccessful attempt to produce a hybrid ATP synthase complex. More recently, the c11 ring of the Ilyobacter tartaricus bacterium and the nearly identical c11 ring of the Propionigenium modestum bacterium were expressed recombinantly in standard BL21(DE3) E. coli cells. In this experiment, the expression was also directed to the membrane [36]. In contrast to these approaches, our methods use a codon optimized gene with a fusion tag, which permits the soluble expression of an eukaryotic c1 subunit in a prokaryotic expression system. This will theoretically enable straightforward production and purification of a c1 subunit from other organisms in the absence of cell membranes, post-translational modifications, and eukaryotic co-facotrs or chaperones – given that the c-subunit amino acid sequence is known.
4.3. Intended applications
The availability of a recombinant monomeric c-subunit can benefit the investigation of many questions not yet answered regarding ATP synthase ring stoichiometry. Thus, it is our intended objective to reconstitute a cn ring from the recombinant c1 subunit. Methods for the in vitro reconstitution of the oligomeric ring from monomeric c-subunits using organic solvents and liposomes have been established for the native E. coli c10 ring [37], and successfully applied to the c11 ring of Ilyobacter tartaricus [30]. Because our final RPC monomeric c1 isolation step is performed in an organic solvent (ethanol), and because the purified protein is shown to have the requisite alpha-helical secondary structure, the incorporation of this in vitro reconstitution technique will likely be facilitated.
Successful reconstitution of a recombinant ring will enable molecular biology based investigations of the influence of various factors on c-ring stoichiometry in vitro. As a notable example, we have observed that chlorophyll and carotenoids co-purify with the native spinach chloroplast c14 ring and are present in c14 ring crystals, and we hypothesize their presence may have some degree of structural or functional influence on the c-ring of chloroplast ATP synthase [19]. We intend to examine the effect of these pigments, and other variable factors such as membrane environment and amino acid sequence on c-ring stoichiometry and structure.
If reconstitution yields a recombinant c-ring with the same c14 stoichiometry as the native ring, we will also use the resulting c-ring complex for x-ray crystallographic structure studies. In our lab, diffraction data has been collected from crystals of the native spinach chloroplast c14 ring at a resolution of 2.8 Å [19], however experimental phase data is lacking thus making the structure determination dependent on known cn ring structures. Recombinant expression methods also enable the use of in vivo heavy atom labeling techniques which have been established for the purpose of determining the phases using the multi-wavelength anomalous dispersion (MAD) technique [38]. We are interested in determining the experimental phases by using this heavy atom labeling technique in combination with the native diffraction data that we have already collected. If successful, the end result will improve upon the recently published 3.8 Å structure of spinach chloroplast c14 [39]. This 3.8 Å structure was determined by using a symmetry-based computational model for phasing rather than direct experimental phase data, and thus can be further improved in this regard as well.
If this bacterial expression system is capable of producing a spinach chloroplast c14 ring that can be used for stoichiometric and x-ray crystallographic studies, it may also be applied to investigating the unknown structures of other cn rings as well. Also, the application can likely be further extended to other eukaryotic membrane proteins as well. In particular, we also intend to apply the recombinant MBP-c1 methods to the ATP synthase subunit a, which is only sparsely understood in terms of its structural and functional relationship with the adjacent cn ring [7].
5. Conclusions
The combination of methods reported here has produced significant quantities of purified recombinant spinach chloroplast ATP synthase c1 subunit. We have shown that approximately 0.4 mg of highly purified monomeric c subunit can routinely be obtained from 1 to 2 L of cell culture – a volume that can easily be scaled up as needed. Most critical to achieving this end result was the use of the maltose binding protein for expression and initial purification, the use of 0.05% n-β-dodecyl-D-maltoside in the protease cleavage reaction, and the use of a reversed phase column in combination with ethanol as an eluent for final purification. Also important was the careful use of the analytical techniques described. The utility of these methods can likely be extended to other c-subunits, both eukaryotic and prokaryotic. Because the development of our reported methods has been primarily process driven, it is possible that some steps can be further improved to optimize product yield. However, each step as reported has yielded reproducible results, and the quantity, quality, and purity of the final protein obtained is sufficient to provide a foundation for the intended experimental applications.
Supplementary Material
Vector map of pMAL-c2x-malE/atpH, including genetic detail of the interface between malE and atpH. Factor Xa protease will cleave after the Arg at this interface, indicated by the arrow.
Acknowledgments
This research has been supported by funding from the National Institutes of Health grant R01 6M081490-01. We would also like to acknowledge Joanna Porankiewicz-Asplund of Agrisera for facilitating our collaboration which produced the essential antibody for the spinach chloroplast c-subunit.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
In chloroplasts these subunits have traditionally been denoted as IV(a), I(b), II(b’), and IIIn(cn). However, this paper will use the a, b, b’, cn notation in order to facilitate comparison with other ATP synthases.
References
- 1.Yoshida M, Muneyuki E, Hisabori T. ATP synthase - A marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2001;2:669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 2.Fillingame RH, Angevine CM, Dmitriev OY. Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett. 2003;555:29–34. doi: 10.1016/s0014-5793(03)01101-3. [DOI] [PubMed] [Google Scholar]
- 3.Nakamoto RK, Scanlon JAB, Al-Shawi MK. The rotary mechanism of the ATP synthase. Archives Biochem. Biophys. 2008;476:43–50. doi: 10.1016/j.abb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter ML, Samra HS, He F, Giessel AJ, Kuczera KK. Coupling proton movement to ATP synthesis in the chloroplast ATP synthase. J. Bioenerg. Biomembr. 2005;37:467–473. doi: 10.1007/s10863-005-9493-9. [DOI] [PubMed] [Google Scholar]
- 5.Turina P, Samoray D, Graber P. H+/ATP ratio of proton transport-coupled ATP synthesis and hydrolysis catalysed by CF0F1 liposomes. EMBO J. 2003;22:418–426. doi: 10.1093/emboj/cdg073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller DJ, Dencher NA, Meier T, Dimroth P, Suda K, Stahlberg H, Engel A, Seelert H, Matthey U. ATP synthase: constrained stoichiometry of the transmembrane rotor. FEBS Lett. 2001;504:219–222. doi: 10.1016/s0014-5793(01)02708-9. [DOI] [PubMed] [Google Scholar]
- 7.von Ballmoos C, Wiedenmann A, Dimroth P. Essentials for ATP synthesis by F1F0 ATP synthases. Annu Rev. Biochem. 2009;78:649–672. doi: 10.1146/annurev.biochem.78.081307.104803. [DOI] [PubMed] [Google Scholar]
- 8.Cross RL, Muller V. The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio. FEBS Lett. 2004;576:1–4. doi: 10.1016/j.febslet.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 9.Pogoryelov D, Reichen C, Klyszejko AL, BrunisholZ R, Muller DJ, Dimroth P, Meier T. The oligomeric state of c rings from cyanobacterial F-ATP synthases varies from 13 to 15. J. Bacteriol. 2007;189:5895–5902. doi: 10.1128/JB.00581-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tittingdorf J, Rexroth S, Schafer E, Schlichting R, Giersch C, Dencher NA, Seelert H. The stoichiometry of the chloroplast ATP synthase oligomer III in Chlamydomonas reinhardtii is not affected by the metabolic state. Biochim. Biophys. Acta Biomembr. 2004;1659:92–99. doi: 10.1016/j.bbabio.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Hudson GS, Mason JG, Holton TA, Koller B, Cox GB, Whitfeld PR, Bottomley W. A gene cluster in the spinach and pea chloroplast genomes encoding one CF1 and 3 CF0 subunits of the H+ ATP synthase complex and the ribosomal protein S2. J. Mol. Biol. 1987;196:283–298. doi: 10.1016/0022-2836(87)90690-5. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG, Mache R. The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- 13.Welch M, Govindarajan S, Ness JE, Villalobos A, Gurney A, Minshull J, Gustafsson C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007;7 doi: 10.1186/1472-6750-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castanie HP, Berges H, Oreglia J, Prere MF, Fayet O. A set of pBR322-compatible plasmids allowing the testing of chaperone-assisted folding of proteins overexpressed in Escherichia coli. Anal Biochem. 1997;254:150–152. doi: 10.1006/abio.1997.2423. [DOI] [PubMed] [Google Scholar]
- 16.Jenny RJ, Mann KG, Lundblad RL. A critical review of the methods for cleavage of fusion proteins with thrombin and Factor Xa. Protein Expr. Purif. 2003;31:1–11. doi: 10.1016/s1046-5928(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 17.Markwell MAK, Haas SM, Bieber LL, Tolbert NE. Modification of Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 18.Schagger H. Tricine-SDS-PAGE. Nat. Protocol. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 19.Varco-Merth B, Fromme R, Wang MT, Fromme P. Crystallization of the c(14)-rotor of the chloroplast ATP synthase reveals that it contains pigments. Biochim. Biophys. Acta Bioenerg. 2008;1777:605–612. doi: 10.1016/j.bbabio.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson WC. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 22.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 23.Sreerama N, Woody RW. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 2004;13:100–112. doi: 10.1110/ps.03258404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapust RB, Waugh DS. Escherichia coli maltose-binding protein is uncommonly effective at promoting the solubility of polypeptides to which it is fused. Protein Sci. 1999;8:1668–1674. doi: 10.1110/ps.8.8.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hering TM, Kollar J, Huynh TD, Varelas JB. Purification and characterization of decorin core protein expressed in Escherichia coli as a maltose-binding protein fusion. Anal. Biochem. 1996;240:98–108. doi: 10.1006/abio.1996.0335. [DOI] [PubMed] [Google Scholar]
- 26.Motejadded H, Altenbuchner J. Construction of a dual-tag system for gene expression, protein affinity purification and fusion protein processing. Biotechnol. Lett. 2009;31:543–549. doi: 10.1007/s10529-008-9909-9. [DOI] [PubMed] [Google Scholar]
- 27.Sonezaki S, Kondo A, Oba T, Ishii Y, Kato Y, Nakayama H. Overproduction and purification of Lon protease from Escherichia coli using a maltose-binding protein fusion system. Applied Microbiol. and Biotechnol. 1994;42:313–318. doi: 10.1007/BF00902735. [DOI] [PubMed] [Google Scholar]
- 28.Deckershebestreit G, Altendorf K. The F0 complex of the proton-translocating F-type ATPase of Escherichia coli. J. Exp. Biol. 1992;172:451–459. doi: 10.1242/jeb.172.1.451. [DOI] [PubMed] [Google Scholar]
- 29.Fillingame RH. Purification of carbodiimide-reactive protein component of ATP energy-transducing system of Escherichia coli. J. Biol. Chem. 1976;251:6630–6637. [PubMed] [Google Scholar]
- 30.Wehrle F, Appoldt Y, Kaim G, Dimroth P. Reconstitution of F0 of the sodium ion translocating ATP synthase of Propionigenium modestum from its heterologously expressed and purified subunits. Eur. J. Biochem. 2002;269:2567–2573. doi: 10.1046/j.1432-1033.2002.02923.x. [DOI] [PubMed] [Google Scholar]
- 31.Girvin ME, Rastogi VK, Abildgaard F, Markley JL, Fillingame RH. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- 32.Raghava GPS. A combination method for protein secondary structure prediction based on neural network and example based learning. CASP5. 2002;A-132 [Google Scholar]
- 33.Ferreira GC, Pedersen PL. Overexpression of higher eukaryotic membrane-proteins in bacteria - Novel Insights obtained with the liver mitochondrial proton phosphate symporter. J. Biol. Chem. 1992;267:5460–5466. [PubMed] [Google Scholar]
- 34.Kiefer H. In vitro folding of alpha-helical membrane proteins. Biochim. Biophys. Acta Biomembr. 2003;1610:57–62. doi: 10.1016/s0005-2736(02)00717-4. [DOI] [PubMed] [Google Scholar]
- 35.Burkovski A, Deckershebestreit G, Altendorf K. Expression of subunit-III of the ATP synthase from spinach chloroplasts in Escherichia coli. FEBS Lett. 1990;271:227–230. doi: 10.1016/0014-5793(90)80412-c. [DOI] [PubMed] [Google Scholar]
- 36.Meier T, Yu JS, Raschle T, Henzen F, Dimroth P, Muller DJ. Structural evidence for a constant c(11) ring stoichiometry in the sodium F-ATP synthase. FEBS J. 2005;272:5474–5483. doi: 10.1111/j.1742-4658.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 37.Dmitriev OY, Altendorf K, Fillingame RH. Reconstitution of the F0 complex of Escherichia coli ATP synthase from isolated subunits - Varying the number of essential carboxylates by co-incorporation of wild-type and mutant subunit-c after purification in organic-solvent. Eur. J. Biochem. 1995;233:478–483. doi: 10.1111/j.1432-1033.1995.478_2.x. [DOI] [PubMed] [Google Scholar]
- 38.Strub MP, Hoh F, Sanchez JF, Strub JM, Bock A, Aumelas A, Dumas C. Selenomethionine and selenocysteine double labeling strategy for crystallographic phasing. Structure. 2003;11:1359–1367. doi: 10.1016/j.str.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Vollmar M, Schlieper D, Winn M, Buchner C, Groth G. Structure of the c(14) rotor ring of the proton translocating chloroplast ATP synthase. J Biol. Chem. 2009;284:18228–18235. doi: 10.1074/jbc.M109.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stock D, Leslie AGW, Walker JE. Molecular architecture of the rotary motor in ATP synthase. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 41.Jiang WP, Hermolin J, Fillingame RH. The preferred stoichiometry of c subunits in the rotary motor sector of Escherichia coli ATP synthase is 10. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4966–4971. doi: 10.1073/pnas.081424898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science. 2005;308:659–662. doi: 10.1126/science.1111199. [DOI] [PubMed] [Google Scholar]
- 43.Meier T, Matthey U, von Ballmoos C, Vonck J, von Nidda TK, Kuhlbrandt W, Dimroth P. Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. J. Mol. Biol. 2003;325:389–397. doi: 10.1016/s0022-2836(02)01204-4. [DOI] [PubMed] [Google Scholar]
- 44.Preiss L, Yildiz O, Hicks DB, Krulwich TA, Meier T. A New Type of Proton Coordination in an F1Fo-ATP Synthase Rotor Ring. PLoS. Biol. 8:10. doi: 10.1371/journal.pbio.1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seelert H, Dencher NA, Muller DJ. Fourteen protomers compose the oligomer III of the proton-rotor in spinach chloroplast ATP synthase. J Mol. Biol. 2003;333:337–344. doi: 10.1016/j.jmb.2003.08.046. [DOI] [PubMed] [Google Scholar]
- 46.Pogoryelov D, Yu JS, Meier T, Vonck J, Dimroth P, Muller DJ. The c(15) ring of the Spirulina platensis F-ATP synthase: F1/F0 symmetry mismatch is not obligatory. EMBO Rep. 2005;6:1040–1044. doi: 10.1038/sj.embor.7400517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Vector map of pMAL-c2x-malE/atpH, including genetic detail of the interface between malE and atpH. Factor Xa protease will cleave after the Arg at this interface, indicated by the arrow.