Abstract
A novel yellow-green leaf mutant yellow-green leaf-1 (ygl-1) was isolated in self-pollinated progenies from the cross of maize inbred lines Ye478 and Yuanwu02. The mutant spontaneously showed yellow-green character throughout the lifespan. Meanwhile, the mutant reduced contents of chlorophyll and Car, arrested chloroplast development and lowered the capacity of photosynthesis compared with the wild-type Lx7226. Genetic analysis revealed that the mutant phenotype was controlled by a recessive nuclear gene. The ygl-1 locus was initially mapped to an interval of about 0.86 Mb in bin 1.01 on the short arm of chromosome 1 using 231 yellow-green leaf individuals of an F2 segregating population from ygl-1/Lx7226. Utilizing four new polymorphic SSR markers, the ygl-1 locus was narrowed down to a region of about 48 kb using 2930 and 2247 individuals of F2 and F3 mapping populations, respectively. Among the three predicted genes annotated within this 48 kb region, GRMZM2G007441, which was predicted to encode a cpSRP43 protein, had a 1-bp nucleotide deletion in the coding region of ygl-1 resulting in a frame shift mutation. Semi-quantitative RT-PCR analysis revealed that YGL-1 was constitutively expressed in all tested tissues and its expression level was not significantly affected in the ygl-1 mutant from early to mature stages, while light intensity regulated its expression both in the ygl-1 mutant and wild type seedlings. Furthermore, the mRNA levels of some genes involved in chloroplast development were affected in the six-week old ygl-1 plants. These findings suggested that YGL-1 plays an important role in chloroplast development of maize.
Introduction
The color of plant leaves can significantly vary due to differences in the content of chlorophyll in the chloroplast. This type of mutation is known as chlorophyll deficient and is also called leaf color mutation. Such mutations have been reported in many higher plants since the 1930s, including rice [1], soybean [2], maize [3], barley [4], wheat [5], tomato [6], oilseed rape [7], and Arabidopsis [8].
Mutated genes for leaf-color influence chlorophyll biosynthesis and degradation as well as chloroplast development [9–12], and are ideal plant materials to investigate molecular mechanisms that regulate such pathways. The characterization of numerous genes in recent years has yielded major advancements in understanding mechanisms of leaf color mutation. For example, elm1, elm2, vyl-Chr.1 and vyl-Chr.9 in maize [13–15]; ygl1, ygl2, ygl3, ygl7, ygl98 and YGL138(t) in rice [16–21]; and chaos, egy1, apg1, and var3 in Arabidopsis [22–25] have been cloned and characterized. The pathway of chlorophyll biosynthesis is particularly well understood in Arabidopsis, in which 27 genes for all 15 steps and 15 kinds of enzymes in this role have been cloned and characterized [26–27]. Molecular mechanisms that regulate chlorophyll degradation and chloroplast development have been defined by major genes affecting these processes in Arabidopsis [28–29].
There are various leaf color mutants known in maize, such as albino, etiolated, light green, stripe or spot, virescent (v), and zebra according to their phenotypic characteristics [30]. According to the maizegdb database (http://www.maizegdb.org/), more than 210 leaf color mutant loci have been identified, distributed over all 10 maize chromosomes. Most genes associated with leaf color mutation are just approximately located to different chromosomes or chromosomal regions, with only a subset cloned and characterized. Among the cloned genes, three encode chlorophyll biosynthesis enzymes or their related proteins. elm1 encodes a phytochromobilin synthase, and its seedling mutant plants have a pale green leaf, pronounced elongation of the mesocotyl, and disrupted de-etiolation responses to red and far-red light [31, 13]. Mature mutant plants maintain the pale green leaf phenotype and flower earlier than wild-type plants under long days [13]. Oy1 encodes the I subunit of magnesium chelatase (ZmCHLI), and its seedling mutant plants exhibit yellow-green leaves that can be enhanced by growth at low temperature (24°C) [32]. Mature, field-grown mutant plants show a pale-green phenotype consistent with chlorophyll deficiency. elm2 encodes a heme oxygenase (HO) that is required for chlorophyll biosynthesis, and its mutants spontaneously develop a yellowish phenotype during their entire life [14]. This gene was isolated by map-based cloning, and the 31 bp deletion in the coding region which reduced endogenous HO activity and disrupted the de-etiolation responses to red and far-red signal at seedling stage. Two genes involved in chloroplast development were also characterized. hcf60 encodes the chloroplast ribosomal small subunit protein 17, and its mutants show temperature and light-dependent chlorophyll deficiencies [33]. vyl encodes the ClpP5 subunit of the chloroplast Clp protease, and its mutants exhibit a yellow leaf phenotype after emergence but can recover and become indistinguishable from wild-type plants after about 2 weeks [15].
The above-mentioned leaf-color mutant loci provided useful information in unraveling the chlorophyll synthesis and chloroplast development in maize. However, its mechanisms of leaf color mutation are still far from being fully understood and new genes related to leaf color mutation are of value. Among them, chlorophyll deficient mutants related to the chloroplast signal recognition particle (cpSRP) pathway could be useful tools for studying the progress of chloroplast development in maize [34–35]. The cpSRP contains four proteins that work together: cpSRP54, cpSRP43, cpFtsY, and ALB3 [36].
In this study, we isolated a new recessive yellow-green leaf mutant ygl-1 in maize. The mutant spontaneously exhibited yellow-green leaves phenotype throughout all growth stages. By map-based cloning we isolated the ygl-1 gene. It was predicted to encode a cpSRP43 protein and acts as a chloroplast precursor. Physiological and microscopic analysis revealed that chloroplast development was affected. Transcript levels of some genes associated with chloroplast development were significantly changed in ygl-1 six-week old plants. Our findings provided direct evidence that the YGL-1 gene plays an essential role in chloroplast development of maize.
Materials and Methods
Plant materials and mapping population
The ygl-1 mutant was identified in self-pollinated progenies from the cross of two maize inbred lines Ye478 and Yuanwu02. An F2 segregating population was constructed from the cross of a ygl-1 mutant and normal green leaf variety Lx7226. A total of 231 recessive yellow-green leaf individuals from three ears were selected and used for initial mapping. Normal green plants from the same three ears were self-pollinated to generate an F3 segregating population. A total of 2247 individuals from the F3 segregating population were used for fine mapping. Later, a large F2 population was obtained from a cross of the ygl-1 mutant and normal green leaf variety Lx7226. To fine map ygl-1, 2930 individuals from the F2 population were screened. Plants with crossovers were self-pollinated to generate offspring for phenotypic verification. All the plants used for mapping ygl-1, chlorophyll content determination, transmission electron microscopy analysis and semi-quantitative RT-PCR were grown in the greenhouse of the Shandong Academy of Agricultural Sciences.
Measurement of chlorophyll and carotenoid contents and photosynthetic characteristics
Leaf samples of wild type Lx7226 and the ygl-1 mutant (of approximately 100mg fresh weight, respectively) were cut and submerged in 10ml of a 2:1 solution of 95% (V/V) acetone and ethanol for 48h at 26°C under dark conditions. The concentrations of total chlorophyll (total Chls), chlorophyll a (Chl a), chlorophyll b (Chl b), and carotenoids (Car) were measured with UH5300 UV/Vis spectrophotometer (Hitachi) at 663, 645, and 470nm, respectively. Pigment contents were calculated according to the following equations with slight modification obtained from the methods of Arnon [37].
Photosynthetic characteristics of wild-type Lx7226 and the ygl-1 mutant were measured on sunny mornings from 9:00 to 10:00 h under greenhouse condition using a Li-6400 portable photosynthesis system. The data were recorded at the middle of the ear leaves at tasseling stage.
Transmission electron microscopy analysis
Leaf samples of wild type Lx7226 and the ygl-1 mutant were harvested at different stages of development from the greenhouse. Only the top fully expanded leaves excluding the flag leaf were taken as samples. The fresh leaves were quickly cut into strips of 2mm width. The strips were fixed in a solution of 2.5% (v/v) glutaraldehyde and further fixed with 1% osmium tetroxide. The tissues were dehydrated through a series of acetone solutions and embedded in EMBed-812 prior to thin sectioning on Ultracut UCT (Leica). They were viewed using a transmission electron microscope H-7500 (Hitachi).
DNA extraction
Leaf tissue of each plant was harvested at the two-week old stage, and genomic DNA was extracted by the CTAB method [38]. At the same time, 15 green and yellow-green plants each were randomly selected from the F2 population to construct DNA pools used to screen for polymorphic markers between ygl-1 and Lx7226.
Marker development and gene prediction
SSR markers used for linkage analysis were obtained from the Maize Genetics and Genomics Database (www.maizegdb.org). To fine map the region of the target locus, additional SSR markers were obtained from previous studies [39]. Gene prediction and annotation within the located region was conducted by Softberry and according to the Maize Genetics and Genomics Database.
Gene cloning and sequence analysis
The DNA sequences of candidate gene GRMZM2G007441 were amplified from both the ygl-1 and its wild-type parents using primer pairs 21-3F (5′-CCCAAACGAACATGACCTAAAGC) and 21-6R (5′-TTCATCAAGTAATCTCTATCACCTGC). The target DNA fragments were cut from the gel and purified by the DNA Gel Extraction kit (TransGen Biotech). The fragments were connected to the pEASY-T1 cloning vector and sequenced at Invitrogen Biotech. The mutant site of the candidate gene was confirmed by the comparison and analysis of the sequencing results using DNAStar software. Multiple DNA sequence analysis was performed using DNAMAN version 5.0 software.
Multiple amino acid sequence alignment and phylogenetic analysis
The full length protein sequence of YGL-1 (GRMZM2G007441) was obtained from GenBank (http://www.ncbi.nlm.nih.gov). Its homologs in the ygl-1 mutant and 6 wild type lines were obtained according to the translation to amino acid sequences from DNA sequences using BioXM 2.6 software. BLASTP searches against the nr database were carried out with the amino acid sequence of the maize gene YGL-1 used as a query and its homologs from other species were retrieved from GenBank (http://www.ncbi.nlm.nih.gov). Multiple amino acid sequence alignment was conducted using DNAMAN version 5.0 software. Phylogenetic analysis was conducted by using MEGA version 5.1 with the rooted neighbor-joining tree using percentage identities based on a multiple sequence alignment generated with the DNAMAN 5.0 software.
Semi-quantitative RT-PCR
Total RNA was isolated using an RNA Isolation Kit (Tiangen Biotech). High-quality first-strand cDNA was generated using oligo (dT) and PrimeScript™ II (Takara). Amplification of ygl-1 and YGL-1 cDNA (GenBank accession: DAA43190.1) was performed using primer pairs (21JF: 5′-CCCAGCATCCCGAACCCAA, 21JR: 5′-AGATGGCGTTGTGGGAGGG). Specific primers of 19 genes used for semi-quantitative RT-PCR were designed using primer 5.0 or selected from previous study [31]. The maize actin gene (GRMZM2G126010) was used as a control in the experiment. The primer pairs were designed using primer 5.0 and shown in S1 Table.
Results
Characterization of the ygl-1 mutant
A yellow-green leaf mutant, designated ygl-1, was a spontaneous mutant isolated in self-pollinated maize progenies from the cross of Ye478/Yuanwu02, and exhibited yellow-green leaves throughout development (Fig 1). All F1 plants of the ygl-1 mutant crossed with wild-type inbred lines Lx7226, B73 and Chang7-2 displayed normal green leaves, and their F2 populations all showed a segregation ratio of 3:1 (green: yellow-green plants, χ2 < χ20.05 = 3.84, P>0.05; Table 1). These results indicated that the yellow-green leaf phenotype in the ygl-1 mutant was controlled by a single recessive nuclear gene.
Fig 1. Phenotypic analysis of mutant ygl-1 and wild type plants.
(a, b), Phenotypic observation of 2-week old mutant plant ygl-1 and wild type plant Lx7226. (c), Phenotypic observation of 6-week old mutant plants ygl-1 (left) and wild-type plants Lx7226 (right). (d), Phenotypic observation of mutant plants ygl-1 (left) and wild type plants Lx7226 (right) at tasseling.
Table 1. Segregation of F1 and F2 populations from three crosses.
| Combination | F1 population | F2 population | ratio | χ2 | P-value | |
|---|---|---|---|---|---|---|
| WT plants | ygl-1 plants | |||||
| Lx7226 × ygl-1 | green | 218 | 78 | 2.79 | 0.29 | 0.5–0.7 |
| B73 × ygl-1 | green | 212 | 62 | 3.41 | 0.82 | 0.3–0.5 |
| Chang7-2 × ygl-1 | green | 205 | 63 | 3.25 | 0.32 | 0.5–0.7 |
WT, wild type.
To characterize the yellow-green leaf phenotype of ygl-1, the pigment contents of total Chls, Chl a, Chl b, and Car at seedling and tasseling stages were examined (Table 2). At the seedling stage, the contents of total Chls, Chl a, Chl b, and Car were 1.65, 1.39, 0.26, and 0.34 mg g-1 in the ygl-1 mutant, significant reductions of 24.32%, 20.4%, 40.2%, and 1.96%, respectively, compared to the wild type Lx7226. At the tasseling stage, total Chls, Chl a, Chl b, and Car were further reduced to 48.43%, 35.71%, 72.79%, and 17.19% of wild type levels, respectively. At both seedling and tasseling stages, ygl-1 mutants exhibited higher Chl a/b ratios than wild type, indicating that the reduction of Chl b was greater than Chl a in the ygl-1 mutant. These results indicated that the ygl-1 mutant phenotype was mainly because of reduced levels of total Chls, Car, and an increased ratio of Chl a/b. In addition, the significantly decreased content of Chl b in ygl-1 might play an important role in its yellow-green leaf phenotype.
Table 2. Pigment contents in leaves of wild-type Lx7226 and ygl-1 mutant, in mg g-1 fresh weight.
| Lines | Total Chls | Chl a | Chl b | Chl a/b ratio | Car | Growth Stage |
|---|---|---|---|---|---|---|
| WT | 2.18±0.02 | 1.75±0.00 | 0.43±0.00 | 4.06±0.01 | 0.35±0.00 | seedling |
| ygl-1 | 1.65±0.01 | 1.39±0.00 | 0.26±0.00 | 5.40±0.01 | 0.34±0.00 | seedling |
| Compared with WT | -24.32%** | -20.4%** | -40.2%** | 33.11%** | -1.96%* | |
| WT | 2.45±0.04 | 1.61±0.05 | 0.84±0.01 | 1.92±0.07 | 0.29±0.02 | Tasseling |
| ygl-1 | 1.26±0.05 | 1.03±0.04 | 0.23±0.01 | 4.53±0.04 | 0.24±0.01 | Tasseling |
| Compared with WT | -48.43%** | -35.71%** | -72.79%** | 136.25%** | -17.19%* |
WT, wild type of Lx7226.
**, Significantly different at P≤0.01
*, Significantly different at P≤0.05.
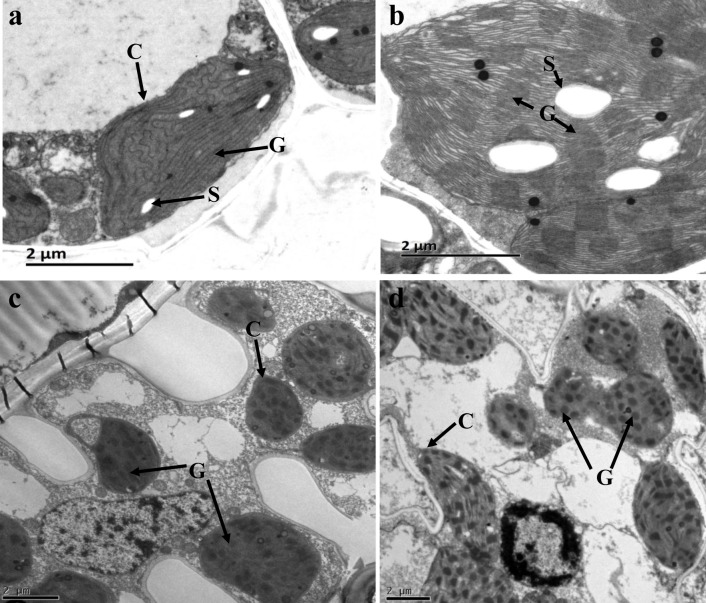
To determine whether the yellow-green leaf phenotype of the ygl-1 mutant affected chloroplast development, wild-type line Lx7226 and the ygl-1 mutant leaf sections at the seedling and tasseling stages were compared using transmission electron microscopy. At the seedling stage, the ygl-1 mutant had less non-appressed granal stacks (Fig 2A), while the wild-type inbred line Lx7226 had more and larger granal stacks (Fig 2B). Rare and less dense granal stacks appeared in the ygl-1 mutant at tasseling (Fig 2C). The wild-type inbred line Lx7226 had more, well-ordered and dense granal stacks in the chloroplast (Fig 2D). However, there was no apparent difference in the structure of the chloroplast in the ygl-1 mutant and wild-type. These results indicated that chloroplast development was suppressed in the ygl-1 mutant.
Fig 2. Transmission electron microscopy (TEM) analysis of chloroplast ultrastructure.
(a, b), mutant plant ygl-1 and wild type plant Lx7226 at the seedling stage, respectively. (c, d), mutant plant ygl-1 and wild type plant Lx7226 at the tasseling stage, respectively. Bar equals 2 um. C, chloroplast; S, starch granule; G, granum.
We compared the photosynthetic characteristics between wild-type line Lx7226 and the ygl-1 mutant at tasseling stage. As shown in Table 3, the net photosynthesis rate (Pn), the stomatal conductance (Cs), the intercellular CO2 concentration (Ci) and the transpiration rate (Tr) were significantly deceased in the ygl-1 mutant compared with wild-type Lx7226. The results indicated that the capacity of photosynthesis was impaired in the ygl-1 mutant, which might be consistent with its chlorophyll deficiency and impaired development of chloroplasts.
Table 3. Photosynthetic characteristics in the wild-type Lx7226 and ygl-1 mutant at tasseling stage.
| Materials | Pn (umol m-2s-1) | Cs | Ci (uL/L) | Tr (mmol m-2s-1) |
|---|---|---|---|---|
| Lx7226 | 15.58±1.18 | 0.13±0.03 | 258.75±17.40 | 1.54±0.29 |
| ygl-1 | 5.13±0.57** | 0.05±0.01* | 155.5±1.29** | 0.76±0.11* |
Pn, net photosynthesis rate; Cs, stomatal conductance; Ci, intercellular CO2 concentration; Tr, transpiration rate.
**, Significantly different at P≤0.01
*, Significantly different at P≤0.05.
Initial mapping of ygl-1
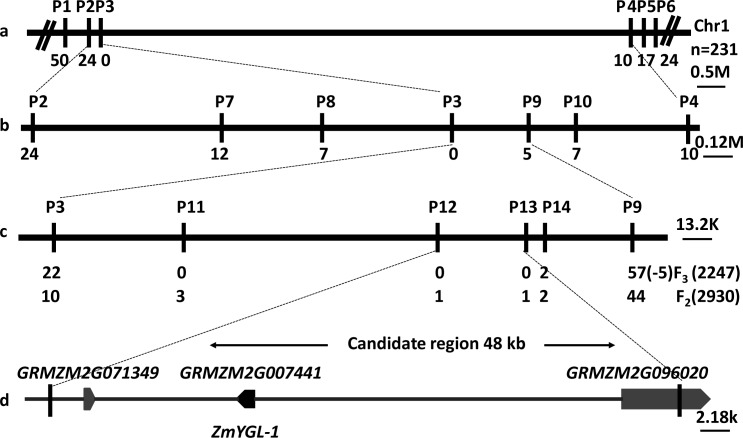
Preliminary mapping of ygl-1 was conducted using 231 recessive individuals showing yellow-green leaf phenotype from an F2 segregating population obtained from the cross of ygl-1/Lx7226. A total of 224 pairs of SSR markers distributed over the 10 maize chromosomes, available from the public database maizegdb, were screened by polyacrylamide gel electrophoresis. We found only P3 in bin 1.01 on the short arm of this chromosome to exhibit polymorphism between the two parents and the DNA pools from the randomly selected 15 green and 15 yellow-green plants, respectively (see methods) (Table 4). A total of 23 SSR primers located in bin 1.01 available from maizegdb were detected by polyacrylamide gel electrophoresis between the two parents and the two DNA pools. Five of the 23 primers had polymorphisms both between the two parents and the two DNA pools. All six polymorphic markers were used to screen the 231 recessive individuals of F2. Finally, two of them, P1 and P2, were positioned on one side of ygl-1 toward the telomeric end of chromosome 1, with 50 and 24 recombinants, respectively; the other three markers, P4, P5 and P6 were located on the other side of ygl-1 toward the centromeric region, with 10, 17 and 24 recombinants, respectively (Table 4, Fig 3A). No recombinant was screened out by marker P3, suggesting that this marker was closely linked to ygl-1. These results indicated that the locus of ygl-1 was between markers P2 and P4, which delineated an interval of 2.6 Mb according to the reported B73 whole genome sequence (Fig 3A). Four new SSR markers showing polymorphisms between two parents and two DNA pools were used to screen the 34 recombinant individuals detected by markers P2 and P4. A total of 12, 7, 5 and 7 plants with crossovers were found with P7, P8, P9 and P10, respectively. Thus, ygl-1 was mapped to an interval of about 0.86 Mb between P8 and P9 (Table 4, Fig 3B).
Table 4. Markers used for genetic mapping of ygl-1.
| BAC accession number | Marker | Primer sequence (5′-3′) | Product size (bp) |
|---|---|---|---|
| AC194148.5 | P1 | F:TCTTCATCTCTCTATCAAACTGACA | 230 |
| R:TGGCACATCCACAAGAACAT | |||
| AC193998.4 | P2 | F:GAAGTGGGGAACATGGTTAATGTC | 157 |
| R:TCACGGTTCAGACAGATACAGCTC | |||
| AC190890.6 | P7* | F:TTGTCCCTGCTTGCATGACA | 155 |
| R:TGGCTCGATCAACTTCCCTG | |||
| AC193473.4 | P8* | F:ACGAACAGGAGAACATGCGT | 212 |
| R:CATGGCAGCCCACATTTGTT | |||
| AC191330.4 | P3 | F:CGCCTGTGATTGCACTACAC | 161 |
| R:CACGCTGTTTCAGACAGGAA | |||
| AC191330.4 | P11* | F:CTTCCCAAAAGCCACCCAGA | 198 |
| R:GTGGATGCTTGCATGACGAC | |||
| AC201968.5 | P12* | F:CCTCAACTTCCCCATCTCCG | 199 |
| R:TGCGCCTAACCTTCGAAGTT | |||
| AC195193.4 | P13* | F:CTCATTTTGTTCCAGACCCGC | 144 |
| R:ACTGGTACCTTTCAGGGCAA | |||
| AC195193.4 | P14* | F:GTGCTGTGCATGCGTATCTG | 204 |
| R:GTACCACCGACCATCCCATC | |||
| AC195193.4 | P9* | F:TTTCAGTTCGGCGTCGATCA | 227 |
| R:GGGCCCGTGTACATGTTACA | |||
| AC195884.4 | P10* | F:GAGATCACCAGCCGTTCCTC | 191 |
| R:GACGATAGGCGGTTCTCGTG | |||
| AC190859.4 | P4 | F:TATATTAGAGGCACCTCCCTCCGT | 377 |
| R:AGCTGCTTCAGCGACTTTGG | |||
| AC190706.4 | P5 | F:GTGAGAATCCTTCAGCGGAG | 182 |
| R:CTGTGGCAGATGTGGTATGG | |||
| AC205622.1 | P6 | F:CGTTTGATATGATGTGGAGATTCG | 135 |
| R:AAGCTTGTGAATGTTCTGGATGTC |
*, These eight SSR markers came from previous study [39].
Fig 3. Fine mapping of the ygl-1 locus.
(a), The ygl-1 locus was delimited to an interval flanked by markers P2 and P4 in bin 1.01 on the short arm of chromosome 1 using 231 yellow-green individuals of an F2 segregating population. (b), Utilizing 4 new polymorphic markers, ygl-1 was mapped to a region between two markers P8 and P9. (c), The ygl-1 locus was finally delimited to an interval of about 48 kb flanked by P12 and P13 using 2,247 individuals of F3 and 2,930 individuals of F2 populations, respectively. (d), There are three annotated genes in the candidate region according to the maizegdb database (www.maizegdb.org). The numbers below the horizontal lines represent recombinational events. The scale bars represent physical distance.
Fine mapping of ygl-1
To fine map ygl-1, four new SSR markers polymorphic between the two parents and two DNA pools were identified (Table 4). Meanwhile, 2247 and 2930 plants of F3 and F2 segregating populations, respectively, were screened to obtain more recombinant individuals. A total of 32 plants were confirmed with crossovers by P3 located on the side of P8, including 22 and 10 from F3 and F2 populations, respectively. A total of 96 recombinant plants were detected by P9, including 52 and 44 from F3 and F2 populations, respectively. All 133 (32 + 96 + 5) recombinant individuals screened by P3 and P9 were genotyped by new polymorphic SSR markers P11, P12, P13 and P14. Finally, only one recombinant was identified both by marker P12 and P13, on the opposite side of ygl-1. Thus, ygl-1 was narrowed down to a genomic region of about 48 kb in the B73 genome sequence, flanked by markers P12 and P13 (Fig 3C).
Candidate gene annotation and sequence analysis
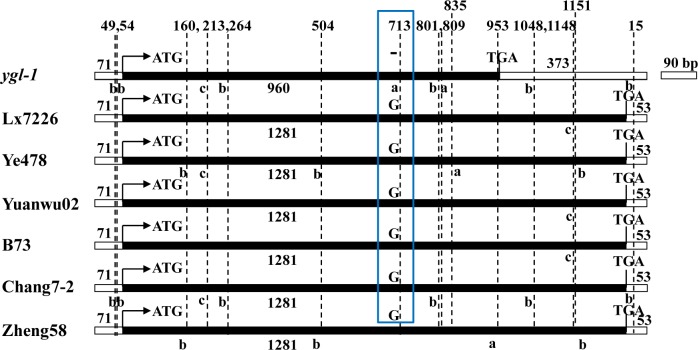
Gene prediction in the 48 kb region of the maize genome inferred to contain ygl-1 revealed three genes (Fig 3D, S2 Table). One of the three genes, GRMZM2G007441 comprises only one exon and was predicted to encode a signal recognition particle 43 kDa (SRP43) protein and act as a chloroplast precursor. This protein has been reported to confer a pale green leaf phenotype in Arabidopsis [22]. Motivated by these results, we sequenced the GRMZM2G007441 gene from amplified genomic DNA of mutant line ygl-1 and six wild-type inbred lines, Lx7226, Ye478, Yuanwu02, B73, Chang7-2 and Zheng58, using primer pairs 21-3F and 21-6R, respectively. A single-nucleotide was deleted at the position 713 bp (-/G) from the ATG start codon in ygl-1, which resulted in a frame shift mutation (Fig 4). The resulting 960 bp truncated translational region encodes a predicted protein of 319 amino acids in ygl-1 compared to 1281 bp ORFs (open reading frame) encoding predicted proteins of 426 amino acids in all 6 wild-type lines (Fig 4 and S1 Fig). The conserved region of ANK_2 and ANK changed and CHROMO almost disappeared in ygl-1 compared to the 6 wild-type lines (S1 Fig). To our knowledge, the other two candidate genes GRMZM2G071349 and GRMZM2G096020 were not reported to participate in Chl metabolism or chloroplast development (S2 Table).
Fig 4. Schematic diagram of YGL-1 genomic structure.
Black boxes with numbers indicate ORFs; blank boxes with numbers indicate sequences upstream/downstream of the ORFs; light blue box indicates 1-bp (-/G) deletion at 713 bp position from the ATG start codon in ygl-1. a-c, a single SNP at specific position upstream/downstream or in the ORFs that can divide the seven lines into two different groups, which distinguish one material, two materials or three materials from other lines, respectively. Numbers above the dashed lines indicate the positions of SNPs that upstream, downstream or in the ORFs.
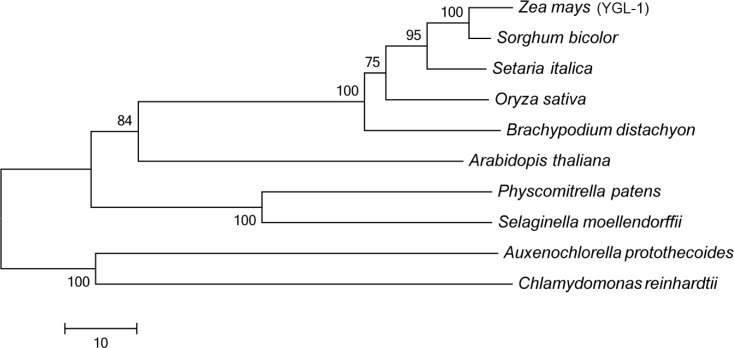
BLAST search of the maize genome database revealed that YGL-1 (GRMZM2G007441) is a single-copy gene. We aligned the amino acid sequences of YGL-1 and its related proteins from Sorghum bicolor, Setaria italica, Oryza sativa (LOC_Os03g03990), Brachypodium distachyon, Arabidopsis thaliana (AT2G47450), Physcomitrella patens, Selaginella moellendorffii, Auxenochlorella protothecoides and Chlamydomonas reinhardtii. YGL-1 exhibits most sequence similarity to the related protein in Sorghum bicolor, with 87% identity. YGL-1 also exhibits 81%, 76%, 71%, 54%, 47%, 46%, 43% and 36% sequence similarity to proteins in Setaria italica, Oryza sativa, Brachypodium distachyon, Arabidopsis thaliana, Physcomitrella patens, Selaginella moellendorffii, Auxenochlorella protothecoides and Chlamydomonas reinhardtii, respectively (S2 Fig). We also investigated the possible phylogenetic relationships between YGL-1 and these proteins mentioned above. The results indicated that the proteins from higher plants including monocotyledonous plants (Z. mays, S. italica, O. sativa and B. distachyon), dicotyledonous plant (A. thaliana), bryophyte (P. patens) and pteridophyte (S. moellendorffii) are divided clearly into one subgroup, while the proteins from lower plants (A. protothecoides and C. reinhardtii) form another subgroup (Fig 5). These data indicate that the YGL-1 (GRMZM2G007441) is conserved in plants.
Fig 5. Phylogenetic anaysis of the YGL-1 protein and its related proteins.
Accession numbers of the protein sequence were as follows: Zea mays [YGL-1, DAA43190.1]; Sorghum bicolor [XP_002465920.1]; Setaria italica [XP_004985832.1]; Oryza sativa [NP_001048866.1]; Brachypodium distachyon [XP_003562136.1]; Arabidopsis thaliana [NP_566101]; Physcomitrella patens [XP_001771412.1]; Selaginella moellendorffii [XP_002989974.1]; Auxenochlorella protothecoides [XP_011399198.1]; Chlamydomonas reinhardtii [AGC59877.1].
Expression analysis of YGL-1
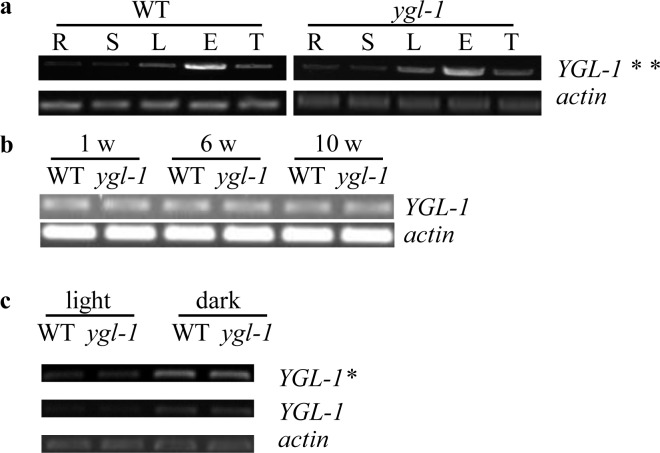
To compare YGL-1 expression profiles, total RNA was extracted from roots, stems, leaves, ears and tassels of the ygl-1 mutant and wild-type plants, respectively. Semi-quantitative RT-PCR analysis was performed. YGL-1 mRNA was expressed in all tested tissues with relatively higher levels in ears than in the other four organs (Fig 6A). Expression levels of the YGL-1 transcript were similar in the ygl-1 mutant and wild type from early to mature stages (Fig 6B). These results indicated that the premature termination mutation in the coding region of ygl-1 did not result in variation of its own mRNA expression. Furthermore, the effect of light and dark growth conditions on the expression of YGL-1 was conducted. Transcript levels were higher when ygl-1 or wild-type plants were grown under dark than light conditions, indicating that light intensity regulates the YGL-1 or ygl-1 mRNA expression (Fig 6C).
Fig 6. Semi-quantitative RT-PCR analysis of YGL-1.
(a), Expression profiles of YGL-1 in root (R), stem (S), leaf (L), ear (E) and tassel (T) of WT and ygl-1 mutant. (b), YGL-1 expression in WT and ygl-1 mutant leaves of 1-, 6- and 10-week-old plants. (c), YGL-1 expression in WT and ygl-1 mutant leaves of 1-week-old plants grown under light or dark conditions. actin was amplified 24 cycles and YGL-1 was amplified 28 cycles because the gene expression was too low to be visible when amplified 24 cycles. *, YGL-1 was amplified 30 cycles; **, YGL-1 was amplified 32 cycles. WT, wild type plants of Lx7226.
The effect of the ygl-1 mutation on the expression of other genes
To investigate whether the ygl-1 mutation affected the transcript levels of genes associated with Chl biosynthesis, photosynthesis and chloroplast development, semi-quantitative RT-PCR primers for 19 genes related to these pathways were designed (S1 Table). Semi-quantitative RT-PCR analysis was conducted in six-week old leaves of ygl-1 and wild type plants. The expression of genes involved in Chl biosynthesis, such as elongated mesocotyl1 (elm1) and elongated mesocotyl2 (elm2), showed no difference between the ygl-1 mutant and wild type [13–14] (Fig 7). rbcS is a nuclear gene encoding the small subunit of Rubisco [40]. psbA and rbcL1 are chloroplast genes encoding reaction center polypeptides and the large subunit of Rubisco [41–42], respectively. Their mRNA levels were also not significantly different between the ygl-1 mutant and wild type (Fig 7). 7 nuclear genes (lhca1, lhcb1, lhcb2, lhcb3, lhcb6, lhcb7 and lhcb9), encoding the light-harvesting chlorophyll proteins (LHC-PI or LHC-PII) which are synthesized in the cytoplasm and after import into chloroplast, then they are targeted and inserted into thylakoid membrane, their transcript levels were also compared. 6 genes (lhca1, lhcb1, lhcb2, lhcb6, lhcb7 and lhcb9) were at a similar transcript level in the ygl-1 mutant and wild type (Fig 7). lhcb3, encoding the light harvesting chlorophyll a/b binding protein of PSII, was altered in the ygl-1 mutant, with no expression or too low to be visible (Fig 7). Interestingly, 6 other genes participating in chloroplast development or function were also significantly different in the ygl-1 mutant and the wild type. vyl-Chr.1 and vyl-Chr.9, encoding the ClpP5 subunit of Clp protease which was important for chloroplast development or function [15, 43], were slightly decreased in the ygl-1 mutant (Fig 7). csr1 (chloroplast SRP receptor1) encoding cpFtsY important for efficient thylakoid targeting of LHCPs [35], showed severely reduced mRNA level in the ygl-1 mutant (Fig 7). X1, which was predicted to encode Zea mays signal recognition particle 54 kDa protein and act as a chloroplast receptor (GenBank), showed significantly increased expression in the ygl-1 mutant (Fig 7). hcf60, a nuclear gene, encodes the chloroplast ribosomal small subunit protein 17 [33]. hcf106, another nuclear gene, is required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize [44]. Their transcript levels were also significantly increased in the ygl-1 mutant (Fig 7). hsp70, encoding a stromal chaperone protein which facilitates protein folding and plays a role in protein translocation across membranes, did not appear to be significantly affected in ygl-1 [45] (Fig 7). Overall the ygl-1 mutation might mainly affect the mRNA levels of some genes involved in chloroplast development but not genes associated with Chl biosynthesis or photosynthesis. These results suggested that the expression of these genes associated with chloroplast development might be regulated at the translational or post-translational levels, since the mutation of YGL-1 did not alter its own mRNA expression in ygl-1 (Fig 6A and 6B).
Fig 7. Expression analysis of nineteen genes associated with Chl biosynthesis, photosynthesis or chloroplast development by semi-quantitative RT-PCR analysis.
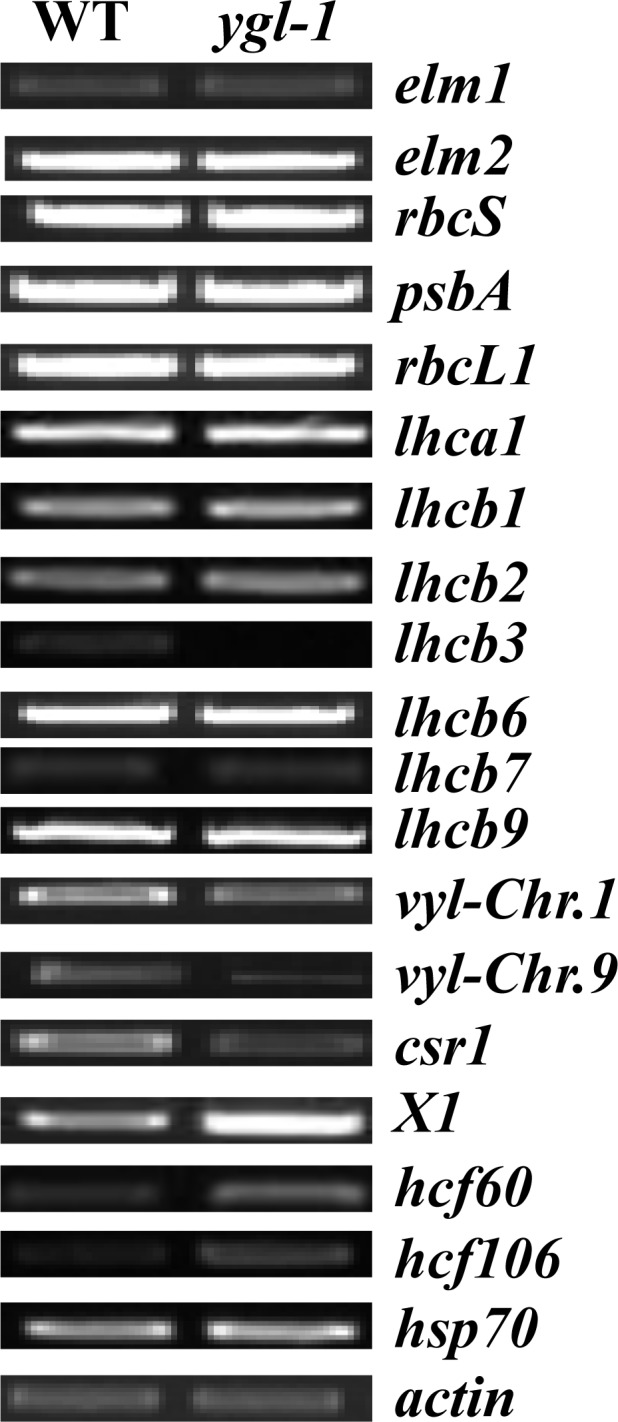
Total RNA was isolated from WT and ygl-1 leaves of six-week old plants. All these genes were amplified 24 cycles. actin was amplified as a control. WT, wild type plants of Lx7226.
Discussion
In recent years, researchers have paid increasing attention to leaf-color mutations, with important insights into the mechanisms of these mutations gained by study of several organisms. In maize, the molecular mechanisms of leaf-color mutation and the related responsible loci are still far from being fully understood. Mutant analysis is an effective approach to explore the function of genes in chloroplast development.
SRP43 confers the yellow-green mutation in ygl-1
Taking a positional cloning approach, the ygl-1 gene was mapped to an interval of 48 kb in bin 1.01 on the short arm of chromosome 1. To our knowledge, only vyl-Chr.1 related to leaf-color mutation has been previously mapped and cloned on the same chromosome [15]. Most homozygous vyl-Chr.1 plants died within 3 weeks of emergence and the surviving plants were pale-yellow and very small. In contrast, homozygous ygl-1 plants were normal and can grow to maturity. In addition, the vyl-Chr.1 gene is located in bin 1.03, not in the 48 kb region in bin 1.01 of ygl-1, so we assert that ygl-1 is a novel gene conferring leaf-color mutation in maize.
The chloroplast signal recognition particle (cpSRP), composed of one cpSRP43 dimer and one cpSRP54 monomer, is required for binding the light-harvesting chlorophyll protein (LHCP) to form a cpSRP/LHCP transit complex which targets LHCP to the thylakoid membrane [46–47]. Mutants in SRP54 have been isolated in Arabidopsis and rice. The mutant ffc, which was generated by ethyl methane sulfonate (EMS) in Arabidopsis, had yellow first true leaves that subsequently turned pale green [48]. A mutant in SRP54 of rice, designated as YGL138(t), derived from Nipponbare treated by EMS, showed yellow-green leaf phenotype throughout development [21]. Could a mutant in SRP43 also lead to chlorophyll deficient mutation? The chaos mutant of SRP43 in Arabidopsis, resulting from insertion of an enhancer trap Ds element, showed pale leaves throughout the vegetative cycle [49, 22]. The double mutant ffc/chaos had pale yellow leaves from early stage to maturity [50]. Mutations in CrcpSRP43 would lead to a low Chls content, high Chl a/b ratio and leaf-color phenotype in C. reinhardtii [51]. The two cpSRP43 mutants in Arabidopsis and C. reinhardtii showed a similar and specific decline of LHCPs in the developed chloroplast, indicating that the cpSRP43 protein plays a conserved function in green microalgae and higher plants, which is also supported by our phylogenic analysis. To our knowledge, no mutant in SRP43 has yet been reported in model monocotyledonous plants including rice and maize. We postulated that mutation in cpSRP43 of maize will result in low levels of Chls and Car, higher Chl a/b ratio and a leaf-color phenotype. In our study, a leaf-color mutant ygl-1 was isolated and it developed yellow-green leaves at all stages. The ygl-1 gene was fine mapped to a 48 kb region in the maize genome. Three candidate genes were identified in the 48 kb chromosomal region of ygl-1. One of these, GRMZM2G007441, was predicted to encode a cpSRP43 protein and act as a chloroplast precursor. We suspect that some mutational site in the GRMZM2G007441 genomic sequence which leads to mutation in the encoded product exists in ygl-1. Sequencing results revealed that 1-bp was deleted in the coding region of this gene in ygl-1, and the single nucleotide deletion led to a frame shift mutation. ygl-1 mutants showed significantly reduced contents of Chls, Car, and higher Chl a/b ratios than wild-type plants at both seedling and tasseling stages. We presumably assume that the defect in ygl-1 lead to the chlorophyll deficiency in the ygl-1 mutant, which makes it to exhibit yellow-green leaf phenotype throughout the growth period. In addition, higher Chl a/b ratios in ygl-1 might make the significantly decreased content of Chl b to play an important role in its yellow-green leaf phenotype. These results provided further evidence that GRMZM2G007441 was the gene responsible for ygl-1 in maize.
SRP43 is an important component of the SRP protein and plays a central role during chloroplast development
The cpSRP is a collection of four proteins that work together: cpSRP54, cpSRP43, cpFtsY and ALB3 [36]. These SRP proteins are believed to be involved in the proper folding of the LHCPS and their targeting to the thylakoid membrane [52]. In the chloroplast, cpSRP uses two mechanisms to target both chloroplast- and nuclear- encoded LHCPS to the thylakoid membrane [53]. The first pathway was called co-translational targeting. This system, which targets chloroplast-encoded proteins to the thylakoid membrane, requires cpSRP54 and the conserved SRP RNA of the bacterial and cytosolic ribonucleoprotein SRPs. The second pathway called post-translational targeting, targets nuclear-encoded proteins to the thylakoid membrane using cpSRP54 together with cpSRP43, but lacks the conserved SRP RNA of the bacterial and cytosolic ribonucleoprotein SRPs.
In the latter targeting pathway, the cpSRP43 works as a novel chaperone specific for light-harvesting chlorophyll a, b-binding proteins and its role cannot be taken over by other stromal chaperones [54–55]. This protein alone is sufficient to form a soluble complex with LHCPs and to prevent aggregation [54]. This supports the idea that cpSRP43 alone is sufficient to targeting of LHCPs to the thylakoid membrane [56]. The cpSRP43 protein is also able to dissolve aggregated LHCPs [54]. This ability of cpSRP43 might be advantageous if LHCP import exceeds the capacity of cpSRP43 to form the transit complex.
The cpSRP43 consists of ANK repeat domains and CHROMO domains [54]. The cpSRP43 ANK repeat domains provide the binding site for the conserved DPLG motif in LHCPs and are sufficient for the chaperone function, whereas the chromo domains are dispensable. In our study, the cpSRP43 protein was 319 amino acids long in the ygl-1 mutant, in which the ANK repeat domains changed a lot and the CHROMO domain almost disappeared due to the 1-bp deletion in the coding region which led to a premature termination codon. We hypothesize that the deficiency chlorophyll, the impaired development of chloroplasts, the decreased capacity of photosynthesis and the yellow-green leaf phenotype of ygl-1 might be due to the changed ANK repeat domains of cpSRP43, which might be incapable of binding the conserved DPLG motif in LHCPs to function as chaperone correctly. In our semi-quantitative RT-PCR results, lhcb3, encoding light harvesting chlorophyll a/b binding protein of PSII, was specifically suppressed in ygl-1 plants compared with the wild-type plants, whereas all other components of PSI (lhca1) and PSII (lhcb1, lhcb2, lhcb6, lhcb7 and lhcb9) remained unchanged. These findings indicate that at least lhcb3 is dependent on the cpSRP43 for targetting, other 6 proteins are either less dependent or independent of cpSRP43. This observation supports the idea that mutations in cpSRP have specific effects on a subset of LHCPs [48]. hcf106, encoding an integral membrane protein which is required in △pH pathway for targeting proteins to the thyakoid membranes [44, 57–59], showed significantly increased expression in the ygl-1 mutant. The increase in hcf106 might occur in response to the defect of functional cpSRP43. hsp70, a nuclear gene encoding a stromal chaperone protein [45], remained a similar transcript level in ygl-1 and wild type. This result is in consistent with previous reports in Arabidopsis ffc and chaos mutants [48]. In addition, the stable level of some gene transcripts (participating in Chl biosynthesis such as elm1 and elm2, or photosynthesis such as rbcS, psbA and rbcL1) and the differently affected level of other gene transcripts (involving in chloroplast development such as vyl-Chr.1, vyl-Chr.9, csr1, X1 and hcf60), probably caused by a feedback mechanism, suggest that ygl-1 mainly altered the transcript levels of genes involved in chloroplast development. The ygl-1 mutant had less non-appressed granal stacks and rare and less dense granal stacks at the seedling and tasseling stages, respectively. The suppressed development of chloroplasts and yellow-green leaf phenotype in the ygl-1 mutant is likely due to the apparent up/down-regulations of genes (hcf106, vyl-Chr.1, vyl-Chr.9, csr1, X1 and hcf60) that participated in chloroplast development, which response to the defect of ygl-1 function that are responsible for the normal chloroplast development. However, transcript levels of genes that involved in Chl biosynthesis and photosynthesis were stable, which makes why the contents of Chls and Car were reduced and the capacity of photosynthesis was decreased is still not very clear.
In the higher plant post-translational pathway, the cpSRP43 binds to the imported LHCPs, then cpSRP54 binds to the cpSRP43-LHC protein complex. This complex subsequently binds to a membrane-bound cpFtsY, and finally this LHC-cpSRP43-cpSRP54-cpFtsY complex is guided to the ALB3 protein [34]. In the co-translational pathway, cpSRP54 binds to a chloroplast-encoded protein that is newly synthesized by a ribosome, then cpFtsY guides this complex to the ALB3 protein that works in tandem with cpFesY. Our semi-quantitative RT-PCR results revealed that csr1, which encodes cpFtsY [35], showed severely suppressed transcript level in the ygl-1 mutant. X1, which was predicted to encode cpSRP54 and act as a chloroplast receptor (GenBank) showed significantly increased expression in the ygl-1 mutant. These results suggest that there might be alternative explanations for the respective interactions of these genes with cpSRP43. In addition, the ygl-1 mutant does not show a developmental effect on chlorophyll content, chloroplast ultrastructure, or yellow-green leaf phenotype. The transcript level of YGL-1 remained at a similar level from 1-week to 10-week stages both in ygl-1 and wild type, indicating that the requirement for SRP43 is not dependent on plant age. Taken together, we suggest that the cpSRP43 protein plays a central role in the post-translational targeting pathway during chloroplast development.
Conclusion
In this study, a novel yellow-green leaf mutant ygl-1 was isolated from maize. The mutant spontaneously showed a distinguishable yellow-green leaf phenotype throughout the growing period and was controlled by a single recessive nuclear gene. The YGL-1 locus was delimited within a 48 kb region in the maize genome harboring three candidate genes. A single base pair deletion discovered in cpSRP43 might lead to reduced contents of Chls and Car, an increased ratio of Chl a/b, defective chloroplast development and declined capacity of photosynthesis, which conferred the yellow-green character in ygl-1. The mutation in ygl-1 might regulate the expression levels of some genes associated with chloroplast development at translational or post-translational levels, while its own mRNA level was not affected. Our results are a valuable foundation for further study and functional characterization of ygl-1, which will facilitate dissection of the mechanism of cpSRP43 protein involvement in chloroplast development of maize.
Supporting Information
DNAMAN 5.0 software was used for sequence alignment. The light blue and red lines represent ANK_2 and ANK region, respectively, and the dark blue line represents CHROMO region.
(TIF)
The light blue and red lines represent ANK_2 and ANK region in YGL-1, respectively, and the dark blue line represents CHROMO region in YGL-1. Accession numbers of the protein sequence were as follows: Zea mays [YGL-1, DAA43190.1]; Sorghum bicolor [XP_002465920.1]; Setaria italica [XP_004985832.1]; Oryza sativa [NP_001048866.1]; Brachypodium distachyon [XP_003562136.1]; Arabidopsis thaliana [NP_566101]; Physcomitrella patens [XP_001771412.1]; Selaginella moellendorffii [XP_002989974.1]; Auxenochlorella protothecoides [XP_011399198.1]; Chlamydomonas reinhardtii [AGC59877.1]. DNAMAN software was used for the sequence alignment.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank R G, Q X, and H Z (Shandong Academy of Agricultural Sciences, China) for their cordial help in DNA extraction.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Post-doctoral Innovative Projects of Shandong Province, China (201403017). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hu Z, Peng LP, Cai YH. (1981) A yellow-green nucleus mutant of rice. Acta Genet Sin (Chinese Version) 8: 256–261. [Google Scholar]
- 2.Ghirardi ML, Melis A. (1988) Chlorophyll b deficiency in soybean mutants. I. Effects on photosystem stoichiometry and chlorophyll antenna size. Biochim Biophys Acta 932: 130–137. [Google Scholar]
- 3.Greene BA, Allred DR, Morishige DT, Staehelin LA. (1988) Hierarchical response of light harvesting chlorophyll proteins in a light-sensitive chlorophyll b-deficient mutant of maize. Plant Physiol 87(2): 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krol M, Spangfort MD, Huner NP, Oquist G, Gustafsson P, Jansson S. (1995) Chlorophyll a/b binding proteins, pigment conversions and early light induced proteins in a chlorophyll b-less barley mutant. Plant Physiol 107: 873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falbel TG, Meehl JB, Staehelin LA. (1996) Severity of mutant phenotype in a series of chlorophyll-deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol 112(2): 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falbel TG, Staehelin LA. (1996) Partial block in the early steps of the chlorophyll synthesis pathway: A common feature of chlorophyll b-deficient mutants. Physiol Plant 97(2): 311–320. [Google Scholar]
- 7.Zhao Y, Du LF, Yang SH, Li SC, Zhang YZ. (2001) Chloroplast composition and structure differences in a chlorophyll-reduced mutant of oilseed rape seedlings. Acta Bot Sin 43(8): 877–880. [Google Scholar]
- 8.Carol P, Stevenson D, Bisanz C, Breitenbach J, Sandmann G, Mache R, et al. (1999) Mutations in the Arabidopsis gene IMMUTANTS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terao T, Yamashita A, Katoh S. (1985) Chlorophyll b-deficient mutants of rice: I. Absorption and fluorescence spectra and chlorophyll a/b ratios. Plant Cell Physiol 26(7): 1361–1367. [Google Scholar]
- 10.Zhang HT, Li JJ, Yoo JH, Yoo SC, Cho SH, Koh HJ, et al. (2006) Rice chlorina-1 and chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol 62: 325–337. [DOI] [PubMed] [Google Scholar]
- 11.Yoo SC, Cho SH, Suqimoto H, Li JJ, Kusumi K, Koh HJ, et al. (2009) Rice Virescent3 and Stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150: 388–401. 10.1104/pp.109.136648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu QB, Jiang Y, Chong K, Yang ZN. (2009) AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. Plant J 59: 1011–1023. 10.1111/j.1365-313X.2009.03930.x [DOI] [PubMed] [Google Scholar]
- 13.Sawers RJH, Linley PJ, Gutierrez-Marcos JF, Delli-Bovi T, Farmer PR, Kohchi T, et al. (2004) The Elm1(ZmHy2) gene of maize encodes a phytochromobilin synthase. Plant Physiol 136(1): 2771–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi DY, Zheng X, Li L, Lin WH, Xie WJ, Yang JP, et al. (2013) Chlorophyll deficiency in the maize elongated mesocotyl2 mutant is caused by a defective heme oxygenase and delaying grana stacking. PLoS One 8(11): e80107 10.1371/journal.pone.0080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing AQ, Williams ME, Bourett TM, Hu WN, Hou ZL, Meeley RB, et al. (2014) A pair of homoeolog ClpP5 genes underlies a virescent yellow-like mutant and its modifier in maize. Plant J 79: 192–205. 10.1111/tpj.12568 [DOI] [PubMed] [Google Scholar]
- 16.Wu ZM, Zhang X, He B, Diao LP, Sheng SL, Wang JL, et al. (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Cheng ZJ, Ma XD, Wu H, Liu YL, Zhou KN, et al. (2013) A knockdown mutation of YELLOW-GREEN LEAF2 blocks chlorophyll biosynthesis in rice. Plant Cell Rep 32: 1855–1867. 10.1007/s00299-013-1498-y [DOI] [PubMed] [Google Scholar]
- 18.Tian XQ, Ling YH, Fang LK, Du P, Sang XC, Zhao FM, et al. (2013) Gene cloning and functional analysis of yellow green leaf3 (ygl3) gene during the whole-plant growth stage in rice. Genes Genom 35: 87–93. [Google Scholar]
- 19.Deng XJ, Zhang HQ, Wang Y, He F, Liu JL, Xiao X, et al. (2014) Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS One 9(6): e99564 10.1371/journal.pone.0099564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun XQ, Wang B, Xiao YH, Wan CM, Deng XJ, Wang PR. (2011) Genetic analysis and fine mapping of gene ygl98 for yellow-green leaf of rice. Acta Agronomica Sinica (Chinese Version) 37(6): 991–997. [Google Scholar]
- 21.Zhang FT, Luo XD, Hu BL, Wan Y, Xie JK. (2013) YGL138(t), encoding a putative signal recognition particle 54 kDa protein, is involved in chloroplast development of rice. Rice 6:7 10.1186/1939-8433-6-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klimyuk VI, Persello-Cartieaux F, Havaux M, Contard-David P, Schuenemann D, Meiherhoff K, et al. (1999) A chromodomain protein encoded by the Arabidopsis CAO gene is a plant-specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Bi YR, Li N. (2005) EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J 41(3): 364–375. [DOI] [PubMed] [Google Scholar]
- 24.Motohashi R, Ito T, Kobayashi M, Taji T, Nagata N, Asami T, et al. (2003) Functional analysis of the 37kDa inner envelope membrane polypeptide in chloroplast biogenesis using a Ds-tagged Arabidopsis pale-green mutant. Plant J 34(5): 719–731. [DOI] [PubMed] [Google Scholar]
- 25.Naested H, Holm A, Jenkins T, Nielsen HB, Harris CA, Beale MH, et al. (2004) Arabidopsis VARIEGATED 3 eneodes a chloroplast targeted, zinc-finger Protein required for chloroplast and palisade cell development. J Cell Sci 117: 4807–4818. [DOI] [PubMed] [Google Scholar]
- 26.Nagata N, Tanaka R, Satoh S, Tanaka A. (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of prochlorococcus species. Plant Cell 17: 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beale SI. (2005) Green genes gleaned. Trends Plant Sci 10(7): 309–312. [DOI] [PubMed] [Google Scholar]
- 28.Comparot-Moss S, Denyer K. (2009) The evolution of the starch biosynthetic pathway in cereals and other grasses. J Exp Bot 60(9): 2481–2492. 10.1093/jxb/erp141 [DOI] [PubMed] [Google Scholar]
- 29.Kumar AM, Soll D. (2000) Antisense HEMA1 RNA expression inhibits heme and chlorophyll biosynthesis in Arabidopsis. Plant Physiol 122: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing C, Wang GX, Huang JL, Wu JZ. (2008) Research on Chlorophyll mutation of plants and molecular mechanism. Biotechnology Bulletin 5: 10–12. [Google Scholar]
- 31.Sawers RJH, Linley PJ, Farmer PR, Hanley NP, Costich DE, Terry MJ, et al. (2002) elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol 130(1): 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawers RJH, Viney J, Farmer PR, Bussey RR, Olsefski G, Anufrikova K, et al. (2006) The maize Oil Yellow1 (Oy1) gene encodes the I subunit of magnesium chelatase. Plant Mol Biol 60: 95–106. [DOI] [PubMed] [Google Scholar]
- 33.Schultes NP, Sawers RJH, Brutnell TP, Krueger RW. (2000) Maize high chlorophyll fluorescent 60 mutation is caused by an Ac disruption of the gene encoding the chloroplast ribosomal small subunit protein 17. Plant J 21(4): 317–327. [DOI] [PubMed] [Google Scholar]
- 34.Kirst H, Melis A. (2014) The chloroplast signal recognition particle (CpSRP) pathway as a tool to minimize chlorophyll antenna size and maximize photosynthetic productivity. Biotechnol Adv 32: 66–72. 10.1016/j.biotechadv.2013.08.018 [DOI] [PubMed] [Google Scholar]
- 35.Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, Yano S, et al. (2004) Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16(1): 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aldridge C, Cain P, Robinson C. (2009) Protein transport in organelles: Protein transport into and across the thylakoid membrane. Febs J 276(5): 1177–1186. 10.1111/j.1742-4658.2009.06875.x [DOI] [PubMed] [Google Scholar]
- 37.Arnon DI. (1949) Copper enzymes in chloroplasts polyphenoloxidase in beta vulgaris. Plant Physiol 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A 81(24): 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J, Liu L, Xu YB, Chen CR, Rong TZ, Ali FH, et al. (2013) Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res 20: 497–509. 10.1093/dnares/dst026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyozuka J, McElroy D, Hayakawa T, Xie Y, Wu R, Shimamoto K. (1993) Light-regulated and cell-specific expression of tomato rbcS-gusA and rice rbcS-gusA fusion genes in transgenic rice. Plant Physiol 102(3): 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bedbrook JR, Link G, Coen DM, Bogorad L. (1978) Maize plastid gene expressed during photoregulated development. Proc Natl Acad Sci U S A 75(7): 3060–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crossland LD, Rodermel SR, Bogorad L. (1984) Single gene for the large subunit of ribulosebisphosphate carboxylase in maize yields two differentially regulated mRNAs. Proc Natl Acad Sci U S A 81(13): 4060–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olinares PD, Kim J, van Wijk KJ. (2011) The Clp protease system: A central component of the chloroplast protease network. Biochim Biophys Acta 1807(8): 999–1011. 10.1016/j.bbabio.2010.12.003 [DOI] [PubMed] [Google Scholar]
- 44.Voelker R, Berkan A. (1995) Nuclear genes required for post-translational steps in the biogenesis of the chloroplast cytochrome b6f complex in maize. Mol Gen Genet 249: 507–514. [DOI] [PubMed] [Google Scholar]
- 45.Craig E, Kang PJ, Boorstein W. (1990) A review of the role of 70 kDa heat shock proteins in protein translocations across membranes. Antonie Van Leeuwenhoek 58: 137–146. [DOI] [PubMed] [Google Scholar]
- 46.Tu CJ, Schuenemann D, Hoffman NE. (1999) Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J Biol Chem 274(38): 27219–27224. [DOI] [PubMed] [Google Scholar]
- 47.Jonas-Straube E, Hutin C, Hoffman NE, Schuenemann D. (2001) Functional Analysis of the protein-interacting domains of chloroplast SRP43. J Biol Chem 76(27): 24654–24660. [DOI] [PubMed] [Google Scholar]
- 48.Amin P, Sy DAC, Pilgrim ML, Parry DH, Nussaume L, Hoffman NE. (1999) Arabidopsis mutants lacking the 43- and 54-kilodalton subunits of the chloroplast signal recognition particle have distinct phenotypes. Plant Physiol 121(1): 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klimyuk VI, Nussaume L, Harrison K, Jones JDG. (1995) Novel GUS expression patterns following transposition of an enhancer trap Ds element in Arabidopsis. Mol Gen Genet 249(4): 357–365. [DOI] [PubMed] [Google Scholar]
- 50.Hutin C, Havaux M, Carde JP, Kloppstech K, Hoffman N, Nussaume L. (2002) Double mutation cpSRP43—/cpSRP54— is necessary to abolish the cpSRP pathway required for thylakoid targeting of the light-harvesting chlorophyll proteins. Plant J 29(5): 531–543. [DOI] [PubMed] [Google Scholar]
- 51.Kirst H, Garcia-Cerdan JG, Zurbriggen A, Ruehle T, Melis A. (2012) Truncated phytosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol 160: 2251–2260. 10.1104/pp.112.206672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pilgrim ML, van Wijk KJ, Parry DH, Sy DA, Hoffman NE. (1998) Expression of a dominant negative form of cpSRP54 inhibits chloroplast biogenesis in Arabidopsis. Plant J 13: 177–186. [DOI] [PubMed] [Google Scholar]
- 53.Nussaume L. (2008) Chloroplast SRP takes another road. Nat Chem Biol 4(9): 529–531. 10.1038/nchembio0908-529 [DOI] [PubMed] [Google Scholar]
- 54.Falk S, Sinning I. (2010) cpSRP43 is a novel chaperone specific for light-harvesting chlorophyll a, b-binding proteins. J Biol Chem 285(28): 21655–21661. 10.1074/jbc.C110.132746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stengel KF, Holdermann I, Cain P, Robinson C, Wild K, Sinning I. (2008) Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321(5886): 253–256. 10.1126/science.1158640 [DOI] [PubMed] [Google Scholar]
- 56.Tzvetkova-Chevolleau T, Hutin C, Noel LD, Goforth R, Carde JP, Caffarri S, et al. (2007) Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Settles AM, Yonetani A, Baron A, Bush DR, Cline K, Martienssen R. (1997) Sec-independent protein translocation by the maize Hcf106 protein. Science 278: 1467–1470. [DOI] [PubMed] [Google Scholar]
- 58.Cline K, Henry R. (1996) Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol 12: 1–26. [DOI] [PubMed] [Google Scholar]
- 59.Schnell DJ. (1998) Protein targeting to the thylakoid membrane. Annu Rev Plant Physiol Plant Mol Biol 49: 97–126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNAMAN 5.0 software was used for sequence alignment. The light blue and red lines represent ANK_2 and ANK region, respectively, and the dark blue line represents CHROMO region.
(TIF)
The light blue and red lines represent ANK_2 and ANK region in YGL-1, respectively, and the dark blue line represents CHROMO region in YGL-1. Accession numbers of the protein sequence were as follows: Zea mays [YGL-1, DAA43190.1]; Sorghum bicolor [XP_002465920.1]; Setaria italica [XP_004985832.1]; Oryza sativa [NP_001048866.1]; Brachypodium distachyon [XP_003562136.1]; Arabidopsis thaliana [NP_566101]; Physcomitrella patens [XP_001771412.1]; Selaginella moellendorffii [XP_002989974.1]; Auxenochlorella protothecoides [XP_011399198.1]; Chlamydomonas reinhardtii [AGC59877.1]. DNAMAN software was used for the sequence alignment.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.