Abstract
Chronic obstructive pulmonary disease (COPD) is a possible risk factor for cardiovascular disease. The association of COPD with the pathogenicity of infection with Chlamydia pneumoniae and Mycoplasma pneumoniae is controversial. We conducted a cross-sectional study to clarify the association between atypical pneumoniae seropositivity and COPD in a general population. We also investigated genetic polymorphisms conferring susceptibility to a pneumonia titer.
The study included 9040 Japanese subjects (54 ± 13 years). COPD was defined as a ratio of forced expiratory volume in 1 second to forced vital capacity of less than 70%. Serum levels of IgA and IgG antibodies to C pneumoniae were determined using an enzyme-linked immunoassay, and M pneumoniae seropositivity was assessed by a particle agglutination test.
Subjects seropositive for C pneumoniae (26.1%) had a higher prevalence of COPD (seropositive, 5.8%; seronegative, 3.1%; P < 0.001) after adjustment for age, sex, height, weight, and smoking status. The association between M pneumoniae seropositivity (20.4%) and COPD was also significant in covariate-adjusted analysis (P < 0.001). A genome-wide association analysis of the C pneumoniae IgA index identified a susceptible genotype (rs17634369) near the IKZF1 gene, and the seropositive rate of C pneumoniae significantly differed among genotypes (AA, 22.5; AG, 25.3; GG, 29.7%, P < 0.001). On multiple regression analysis, seropositivity for both C pneumoniae (odds ratio = 1.41, P = 0.004) and M pneumoniae (odds ratio = 1.60, P = 0.002) was an independent determinant for COPD, while no direct association was found with the rs17634369 genotype.
Seropositivity for both C pneumoniae and M pneumoniae is an independent risk factor for COPD in the general population.
INTRODUCTION
Chronic obstructive pulmonary disease (COPD), as assessed by forced expiratory volume in 1 second (FEV1) and forced vital capacity (FVC), is an independent risk factor for the incidence of cardiovascular diseases, including stroke,1 myocardial infarction,2 and heart failure,3,4 as well as for dementia5 in the general population. COPD has also been associated with increased mortality.6 Although the most important risk factor for COPD is long-term smoking,7 only a small proportion of smokers develop airflow obstruction,8 and some lifelong nonsmokers develop COPD.9 These inconsistent results suggest that other risk factors are also causative in the pathogenesis of COPD.
Factors postulated to play a role in the pathogenesis of COPD to date include air pollution, respiratory infection in childhood, bronchial hyperresponsiveness, systemic inflammation, and genetic background.7,10,11 Infection with Chlamydia pneumoniae (C pneumoniae), the most common nonviral human respiratory pathogen, has also been proposed as a risk factor for COPD.12 Although the mechanism by which chronic infection with C pneumoniae might trigger the development of COPD is not precisely understood, 1 plausible mechanism may be the release of inflammatory cytokines and subsequent airway inflammation and tissue damage.12 Higher titers of C pneumoniae in blood specimens are commonly observed in patients with respiratory diseases, namely COPD,13 chronic bronchitis,14 and symptomatic respiratory disease,15 and in hospitalized patients with acute exacerbation of COPD.16 Further, approximately 50% of cases of exacerbated COPD are caused by bacterial infection, such as with C pneumoniae.17 Although the prevalence and mortality of COPD are increasing worldwide,18 the involvement of C pneumoniae in the development of COPD in the general population is largely unknown. Mycoplasma pneumoniae (M pneumoniae) is another atypical pathogen that causes community-acquired pneumonia. Infection or colonization of M pneumoniae in the lower airway is associated with the pathogenesis of bronchial asthma,19 suggesting that chronic infection with M pneumoniae may also be a risk factor for COPD.
Recent advances have enabled the analysis of millions of single nucleotide polymorphisms (SNPs) dispersed throughout the human genome. Genome-wide association studies (GWAS) conducted without a prior hypothesis have successfully identified susceptibility loci for various common diseases and quantitative traits. GWAS of lung function have identified multiple loci such as the hedgehog interacting protein (HHIP) and glutathione S-transferase C-terminal domain containing (GSTCD) gene region.20 Despite the possible existence of SNPs which might influence susceptibility to seropositivity for pneumoniae, and the relationship between C pneumoniae and M pneumoniae infection and pulmonary function, no study has yet explored SNPs associated with seropositivity for both strains.
Here, we conducted a cross-sectional study to investigate the association of C pneumoniae and M pneumoniae seropositivity with pulmonary function in a large general Japanese population. We also conducted a GWAS to explore SNPs for their association with seropositivity for both strains.
METHODS
Study Subjects
Study subjects were participants in the Nagahama Prospective Genome Cohort for Comprehensive Human Bioscience (the Nagahama Study). The Nagahama Study cohort was recruited between 2008 and 2010 from the general population (30–74 years old) living in Nagahama City, a largely suburban city of 125,000 inhabitants in Shiga Prefecture. Nagahama City residents aged 30 to 74 years at recruitment and without serious health problems who agreed to participate in the cohort study of their own accord were recruited via mass communications in the local community, such as public relations magazines and periodical newspapers.
Among a total of 9804 participants, we considered 9237 subjects as the total population in this study, consisting of 3246 subjects whose genome-wide SNP genotype data were available and 5991 other subjects as a subset of the remaining samples available for replication genotyping (Table S1). Among these, participants meeting any of the following conditions were excluded from subsequent association analysis for COPD: pregnancy (n = 40), history of lung cancer (n = 19), unsuccessful assessment of the Brinkman index (n = 23) or spirometric parameters (n = 24), and unavailability of serum M pneumoniae titer (n = 1) or rs17634369 genotype (n = 90). Eventually, a total of 9040 people were included as study subjects.
All study procedures were approved by the ethics committee of Kyoto University Graduate School of Medicine and the Nagahama Municipal Review Board. Written informed consent was obtained from all participants.
Basic Clinical Parameters
Clinical measurements and blood sampling were performed at enrollment. Medical history and smoking status were investigated using a structured questionnaire. The Brinkman index was calculated as the daily number of cigarettes smoked multiplied by the number of years spent smoking.
Evaluation of Pulmonary Function
Pulmonary function was measured by an FVC maneuver on a computed spirometer with automated quality checks (SP-350 COPD, Fukuda Denshi, Tokyo, Japan). Prebronchodilator spirometry was measured by certified medical technologists in accordance with a standardized protocol. COPD was defined by a ratio of FEV1 to FVC of less than 70%. Predicted normal values for FVC (FVC predicted) (L) and FEV1 (FEV1 predicted) (L) were calculated using the following equations, in accordance with guidelines developed by the Japanese Respiratory Society for the diagnosis and treatment of COPD (3rd edition; http://www.jrs.or.jp/uploads/uploads/files/photos/765.pdf): FVC male = 0.042 × height (cm) − 0.024 × age − 1.785; FVC female = 0.031 × height − 0.019 × age − 1.105; FEV1 male = 0.036 × height − 0.028 × age − 1.178; FEV1 female = 0.022 × height − 0.022 × age − 0.005.
Measurement of C pneumoniae IgA and IgG Indices
Levels of serum IgA and IgG antibodies to C pneumoniae were determined using a specific enzyme-linked immunoassay (ELISA) kit (HITAZYME C pneumoniae, Hitachi Chemical, Tokyo, Japan) that detects antibodies to the chlamydial outer membrane complex. IgA and IgG levels in each sample were expressed as the IgA or IgG index, respectively. Seropositivity to C pneumoniae was diagnosed as an IgA and IgG index value of more than 1.1 for both. Details of the ELISA assay and validity of the cut-off point have been described elsewhere.21 Mean intraassay coefficients of variation were 3.3% to 7.7% for the IgA index and 6.3% to 9.0% for the IgG index. Respective mean interassay coefficients of variation were 3.8% to 10.7% and 4.0% to 10.7%.
Evaluation of M pneumoniae Seropositivity
M pneumoniae seropositivity was assessed using a semiquantitative particle agglutination test kit (Serodia-Myco II, Fujirebio, Tokyo, Japan) consisting of gelatin particles coated with cell membrane components of M pneumoniae (Mac strain).22 Serum samples were serially diluted to give final dilutions of 1:40 to 1:20, 480. According to the manufacturer's instructions, antibody titers of 1:40 and higher were considered seropositive.23,24
Genome-Wide SNP Genotyping
DNA was extracted from peripheral blood samples by the phenol–chloroform method. Genome-wide SNP genotyping was performed on 3710 samples of participants who joined the Nagahama cohort from 2008 to 2009. A series of BeadChip DNA arrays were used for analysis, namely HumanHap610 quad (1828 samples), HumanOmni2.5-4 (1616 samples), HumanOmni2.5-8 (378 samples), HumanOmni2.5s (192 samples), and HumanExome (192 samples) (Illumina, San Diego, CA). Several samples were repeatedly genotyped using different arrays. As a reference panel, 192 samples were genotyped using HumanOmni2.5-8, HumanOmni2.5s, and HumanExome, and used in the following genotype imputation. Genotyping quality was controlled by excluding SNPs with a call rate below 99%, minor allele frequency below 0.01, or extreme deviation from Hardy–Weinberg equilibrium (P < 1.0 × 10−7). This threshold was defined to ensure a concordance ratio of SNPs genotyped by different SNP arrays greater than 99.99%.25 Genotype imputation was performed by a standard 2-step procedure using MACH ver. 1.0.16 software with 1792015 SNPs commonly genotyped in the 192 samples as a reference. Imputed SNPs with a minor allele frequency of less than 0.01 or R2 less than 0.5 were excluded from the following association analysis. Samples with a call rate less than 95% (n = 162) were excluded from analysis. Of the remaining 3548 subjects, 295 individuals were excluded as they showed high degrees of kinship (Pi-hat greater than 0.35, PLINK ver. 1.07), and 7 individuals were excluded as ancestry outliers, as identified by principal component analysis using the HapMap Phase 2 release 28 JPT dataset as reference (EIGENSTRAT ver. 2.0).
Replication Genotyping
Replication analysis was performed with a subset of the remaining samples from the Nagahama cohort (n = 5991). Genotypes were analyzed with a TaqMan probe assay using commercially available primer and probe sets purchased from Life Technologies Corporation (Carlsbad, CA). Fluorescence level of PCR products was measured using the 7900HT Fast Real-Time PCR System (Life Technologies).
Statistical Analysis
Genome-wide association analysis of the C pneumoniae IgA and IgG index values was performed by linear regression analysis under an additive genetic model adjusted for age, age-squared, sex, and body mass index, while analysis of M pneumoniae seropositivity was performed by a Chi-square test (PLINK). Rank-based inverse normal transformation was applied to the C pneumoniae IgA and IgG index values. Population stratification was adjusted using top principal components as covariates. Given an estimation of the appropriate testing burden in a GWAS in Europeans as a million, a P-value less than 5.0 × 10−8 was considered to have genome-wide significance.26 Based on findings that an additive model has reasonable power in detecting both additive and dominant effects,27 we applied only an additive model in the current GWAS.
In other association analyses, differences in numeric variables among subgroups were assessed by analysis of variance, while differences in frequency were assessed by a Chi-square test. Factors independently associated with COPD were identified by multiple logistic regression analysis. All statistical analyses were performed using JMP 9.0.2 software (SAS Institute, Cary, NC), with a conventional P-value less than 0.05 considered to indicate statistical significance.
RESULTS
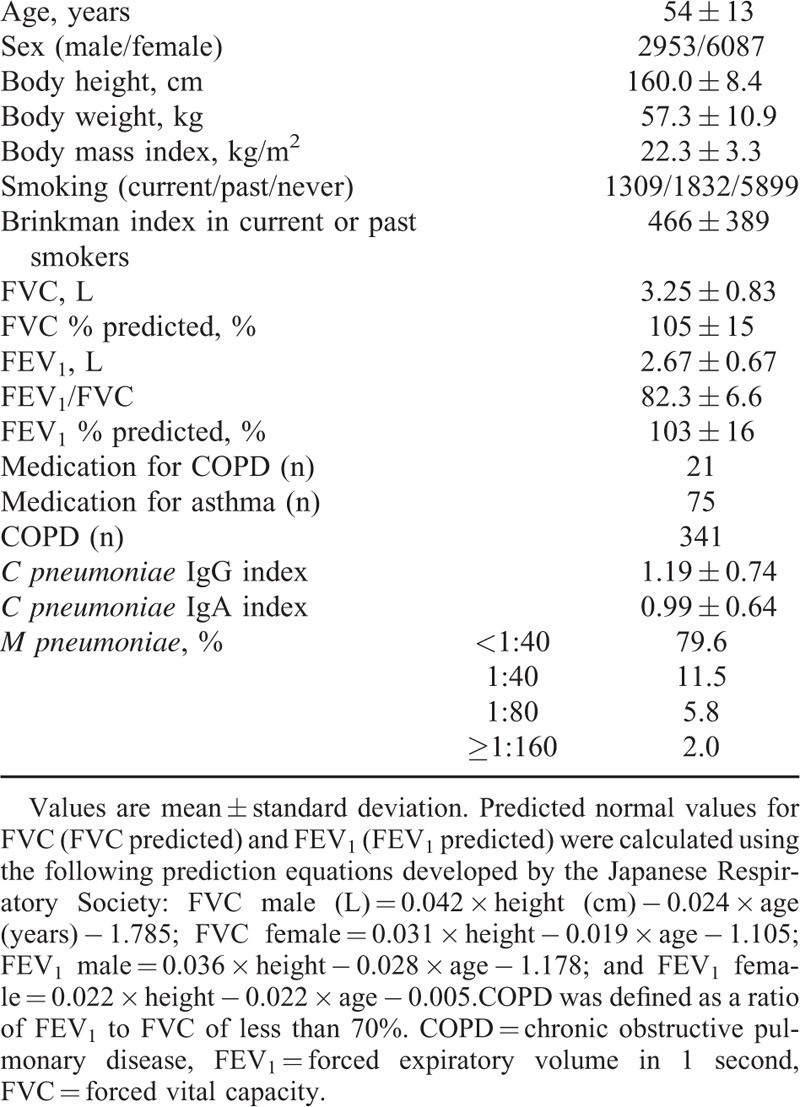
Table 1 shows the clinical characteristics of the study subjects. A total of 341 (3.8%) subjects were diagnosed with COPD. Subjects with COPD were significantly older (COPD 61 ± 12 vs control 53 ± 13 years, P < 0.001), and mostly male (61.0% vs 31.6%, P < 0.001) and current or past smokers (58.7% vs 33.8%, P < 0.001). Differences in major clinical parameters by severity of pulmonary function are detailed in Table S2.
TABLE 1.
Clinical Characteristics of Study Subjects (n = 9040)
C pneumoniae and M pneumoniae Seropositivity With Pulmonary Function
The frequency of C pneumoniae and M pneumoniae seropositivity was 26.1% and 20.4%, respectively. The number of subjects seropositive for C pneumoniae increased with age, whereas that for M pneumoniae showed the opposite relationship (Figure S1). Associations of seropositivity for both strains with pulmonary function are summarized in Table 2 and Figure S2. Subjects seropositive for C pneumoniae exhibited significantly lower pulmonary function and a higher frequency of COPD. Although the values of the C pneumonia e IgA and IgG indices were linearly associated with smoking intensity as assessed by the Brinkman index (Figure 1), significantly lower pulmonary function in seropositive subjects remained after adjustment for the interrelationships (Table 2). Conversely, the pulmonary function of subjects seropositive for M pneumoniae was slightly better than that of seronegative subjects (Table 2), but these associations were lost after adjustment for covariates, including age, presumably due to the markedly younger age of seropositive subjects. In contrast, the association between M pneumoniae seropositivity and COPD became significant in the covariate-adjusted analysis (odds ratio = 1.63). Similar results were observed in an analysis which excluded subjects receiving treatment for asthma or COPD (n = 90) (Table S3).
TABLE 2.
Differences in Clinical Parameters of C pneumoniae and M pneumoniae Seropositive Patients
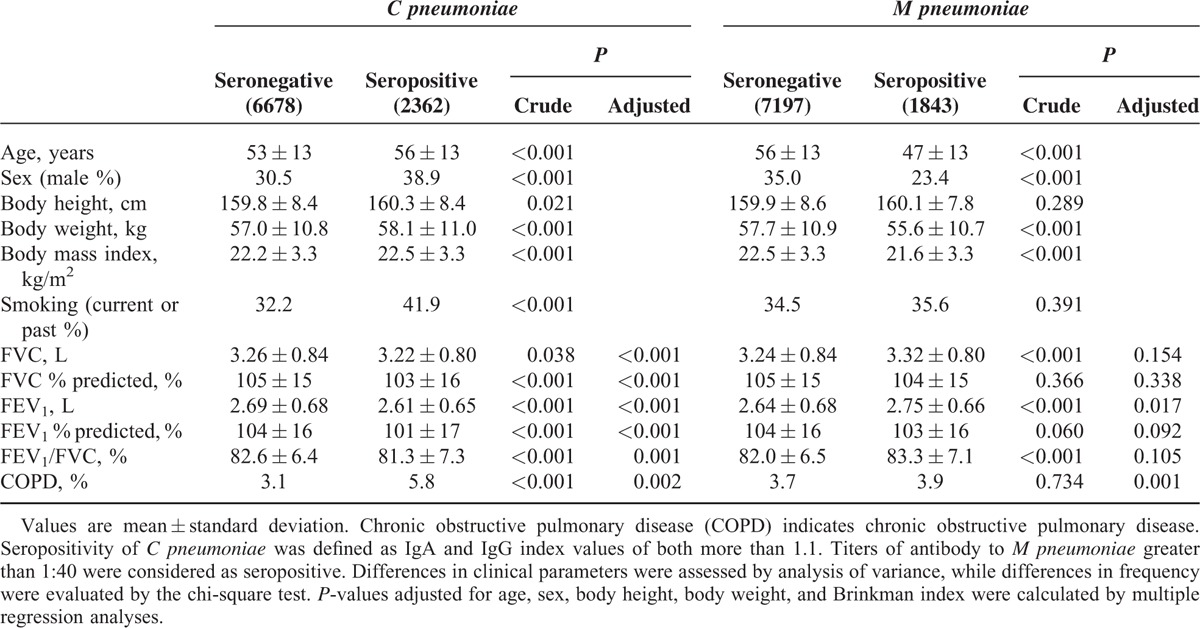
FIGURE 1.
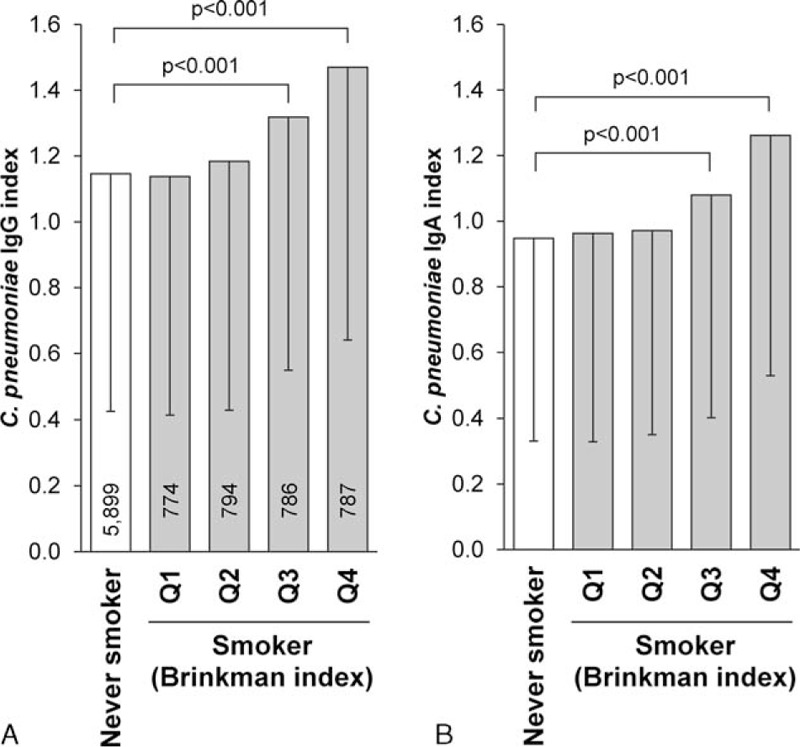
Mean IgA and IgG indices by smoking status and quartile of Brinkman index. Values are mean ± standard deviation of IgG (A) and IgA (B) indices. Smokers were divided into quartiles according to Brinkman index (Q1: <159.7, Q2: 160.0–374.0, Q3: 375.0–684.8, Q4: 685.0–2784). Numbers of subjects in each subgroup are shown in the column. Statistical significance was assessed by analysis of covariance and post-hoc analysis was performed by Dunnett test.
GWAS of C pneumoniae IgA and IgG Indices
GWAS of the C pneumoniae IgA index values identified strong association signals at 6p22.3 (rs9460391, β = 0.163, P = 6.9 × 10−8), 7p12 (rs17634369, β = 0.134, P = 3.0 × 10−8), and 13q33 (rs942102, β = −0.134, P = 7.7 × 10−8) (Figure S3, Table S4). Additional genome-wide significant signals were found in the human leukocyte antigen gene region, but these were lost after adjustment for population stratification (Figures S4 and S5). No significant signals were found in GWAS of the C pneumoniae IgG index value (Figures S3 and S4), or in analysis of M pneumoniae seropositivity (Figure S6). Statistical power of the GWAS is shown in Figure S8. Additional susceptible SNPs with a lower frequency or smaller effect size might be detectable with a greater number of subjects.
Replication Analysis for the Candidate SNPs
Replication analysis of the remaining Nagahama samples confirmed the positive association of rs17634369 (β = 0.080, P = 1.4 × 10−5), but not rs9460391 (P = 0.414) or rs942102 (P = 0.621) (Table S4). Per-allele effect size of rs17634369 on the C pneumoniae IgA index value calculated from the combined datasets used in the GWAS, and replication analysis was approximately 0.1 (P = 1.3 × 10−11). SNP rs17634369 was located approximately 22 kb upstream of the IKZF1 gene, where no other genes were mapped (Figure S7), and was in strong linkage disequilibrium with the SNP rs4917014 (D′ = 0.921, r2 = 0.773), which was previously identified as conferring susceptibility to systemic lupus erythematosus (SLE).28 There were no significant associations between previously validated SNPs for SLE and C pneumoniae IgA index value (Table S5).
SNP rs17634369 and C pneumoniae IgA Index
Figure 2A shows the differences in the C pneumoniae IgA index values for the rs17634369 genotype. Subjects with the G allele showed a significantly higher IgA index value. The seropositive rate of C pneumoniae defined by the IgA index (Figure 2B) and the IgA and IgG indices (Figure 2C) linearly increased with the number of G alleles. Associations between the rs17634369 genotype and C pneumoniae IgA index value (P < 0.001), as well as the seropositive rate defined by IgA and IgG indices (P < 0.001), remained significant after adjustment for age, sex, and smoking status.
FIGURE 2.
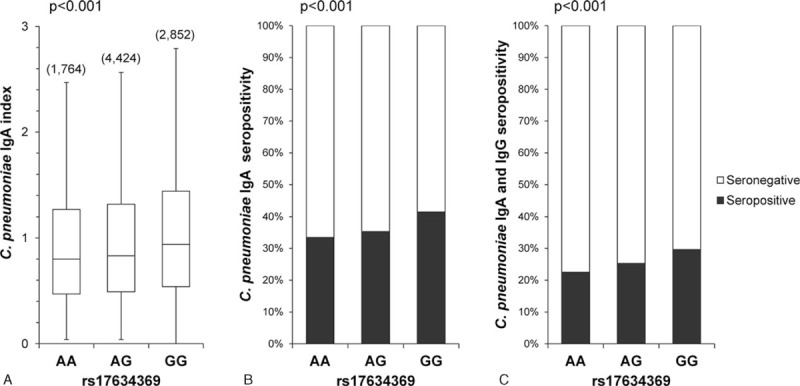
Association of rs17634369 genotype with C pneumoniae IgA index value and seropositivity. (A) Box plot of C pneumonia e IgA index by rs17634369 genotype. Number of subjects for each genotype is shown in parentheses. Statistical significance was assessed by analysis of variance. (B) Seropositivity of C pneumoniae defined by the IgA index. An IgA index value of more than 1.1 was considered as seropositive. Statistical significance was assessed by the Chi-square test. (C) Seropositivity of C pneumonia e defined by the IgA or IgG index. Seropositivity was defined as IgA and IgG index values of more than 1.1 for both.
Multivariate Analysis for COPD
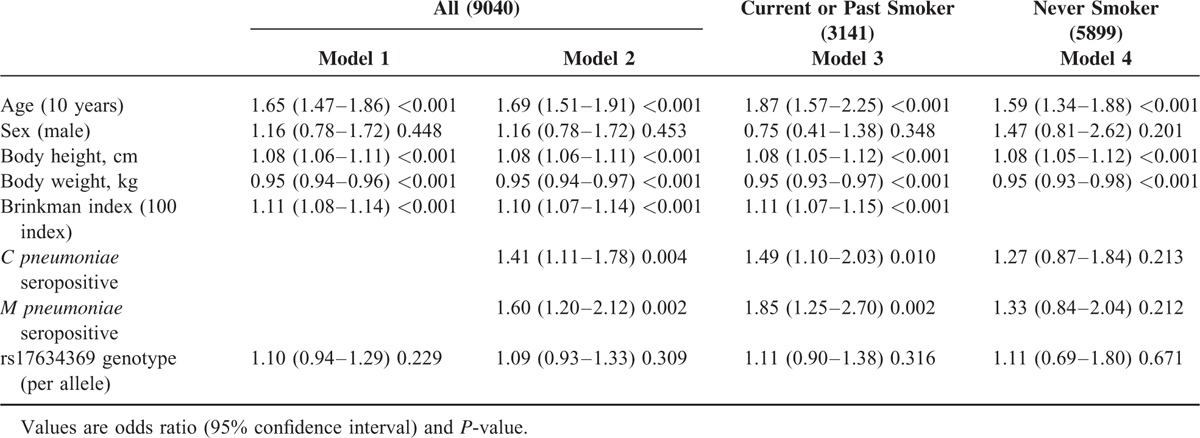
A multiple logistic regression analysis was used to further identify factors independently associated with COPD (Table 3). Results showed that both C pneumoniae and M pneumoniae seropositivity were independently associated with COPD (Model 1), even when individuals taking medication for asthma and COPD were excluded (Table S6), while the rs17634369 genotype was not directly associated with COPD (Model 1), even following adjustment for C pneumoniae and M pneumoniae seropositivity (Model 2). In a separate analysis by smoking status, positive associations between pneumoniae seropositivity and COPD were observed only in current or past smokers (Model 3), and not in never-smokers (Model 4). No interaction was observed between the rs17634369 genotype and C pneumoniae seropositivity (P = 0.846).
TABLE 3.
Multiple Logistic Regression Analysis for Chronic Obstructive Pulmonary Disease (COPD)
DISCUSSION
In this study, we demonstrated that seropositivity for C pneumoniae and M pneumoniae was positively associated with COPD. To our knowledge, this is the 1st study to clearly show the risk of C pneumoniae and M pneumoniae seropositivity for COPD in a general population, particularly in smokers. Further, we identified SNPs conferring susceptibility for C pneumoniae seropositivity, although no direct association with COPD was observed.
Although several studies reported that patients with COPD had a significantly higher titer of serum C pneumoniae IgA or IgG,13–15 the East London COPD study in 110 patients with stable COPD reported an inconsistent result.29 A plausible reason for this discrepancy is insufficient statistical power due to the relatively small sample size. For example, von Hertzen et al13 compared IgA and IgG titers in 54 COPD patients and 321 healthy controls, and Brandén et al15 investigated the association of chronic C pneumoniae infection with longstanding airway symptoms in 199 patients, of whom 30 were diagnosed with COPD. In a general population, 1 study investigated the association of C pneumoniae infection with pulmonary function in 1773 men30 and failed to find any association with outcome measures, including baseline spirometric parameters and FEV1 decline during the 5-year follow-up period. However, the frequency of current or past smokers in the previous study (>80%) was more than double that in our present study, and more than double that in the National Survey on Circulatory Disorders of Japan,31 which reported that the frequency of current or past smokers in Japan is approximately 40%. The harmful effects of smoking might therefore render the potential pathogenicity of C pneumoniae infection undetectable. Given that our present study was based on a large population with a ratio of smokers to nonsmokers equivalent to that of the national survey, our observations may provide strong evidence that C pneumoniae seropositivity is a risk factor for reduced pulmonary function in general populations.
M pneumoniae is another atypical pathogen of community-acquired pneumonia. Although several small studies reported a possible risk of serologically identified M pneumoniae for acute exacerbation of COPD,32,33 other studies denied this etiological relationship.34,35 Clinical and epidemiological data on the pulmonary risk of M pneumoniae infection is thus limited and conflicting even in COPD patients. In addition, no data are available on the association between M pneumoniae infection and spirometric parameters. Our present findings are therefore important in helping to elucidate the pulmonary risk of M pneumoniae infection in a general population. Further, the risk of M pneumoniae infection was equal to or somewhat greater than that of C pneumoniae. Of note, however, subjects infected with M pneumoniae first appeared to have better pulmonary function, presumably due to their younger age.
In general populations, even mild to moderate airflow limitation and COPD have been suggested to confer a risk for the incidence of cardiovascular diseases.2–5 One potential factor in explaining the possible relationship between COPD and cardiovascular outcomes is atherosclerosis.36 Given that the present and previous reports showed a positive association between C pneumoniae seropositivity and atherosclerotic vascular change,37,38 as well as cardiovascular and all-cause mortality,39C pneumoniae infection might be a factor underlying the relationship between COPD and cardiovascular outcomes.
The rs17634369 genotype was significantly associated with the seropositive prevalence of C pneumoniae. According to the Encyclopedia of DNA Elements (ENCODE) project datasets (UCSC genome browser: http://genome.ucsc.edu/), this polymorphism lies approximately 22 kb upstream of the IKZF1 gene. A recent systemic expression quantitative trait locus (eQTL) analysis40 found that the T allele of the rs4917014 genotype, a proxy of the G allele of the rs17634369 genotype, was associated with increased expression of the IKZF1 gene (cis-effect). Further, very recently, the rs4917014 genotype was identified as susceptible for cold medicine-related Stevens–Johnson syndrome via a mechanism involving a change in the quantitative ratio of the IKZF1 alternative splicing isoforms Ik1 and Ik2.41 Although we did not perform functional analysis for the relationship between rs17634369 or rs4917014 genotype and C pneumoniae Ig A index, these previous results are sufficient to consider IKZF1 as a factor in C pneumoniae seropositivity.
IKZF1 encodes the transcription factor “Ikaros.” Ikaros was reported to be associated with transcriptional regulation of human STAT4,42 which is involved in cell-mediated immune responses via Th1 cell development.43 The STAT4 pathway may therefore help explain the association between IKZF1 genotype and C pneumoniae seropositivity. The eQTL analysis also reported a significant association between the rs4917014 genotype and increased expression of genes involved in the type 1 interferon response (trans-effect).40 Given that increased expression of interferon-α response genes is a distinct expression pattern in SLE patients,44 and that the rs4917014 genotype confers susceptibility to SLE,28 the increased C pneumoniae IgA levels and higher risk of SLE in individuals with the risk allele might partially share a common pathophysiological pathway.
The microimmunofluorescence (MIF) test is currently the objective standard for the serodiagnosis of C pneumoniae, but interlaboratory variability is an unanswered issue. An ELISA is another convenient method of serodiagnosis. However, discrepancies in the detection ratio between indirect (MIF, ELISA) and direct (PCR) methods remain to be resolved. Our present findings may suggest that the discrepancy in detection ratio among populations, which plays at least some role in the controversy over C pneumoniae infection in chronic inflammatory diseases, may in part be due to genetic heritability.45
Several limitations of our study warrant mention. First, we could not distinguish between persistent and past infection of pneumoniae by measuring seropositivity. Although the presence of pneumoniae antibodies may reflect the presence of prior infection, this may not be a valid measure of persistent and chronic active reinfection. A guideline from a workshop on the standardization of C pneumoniae diagnostic methods46 suggested that no valid serological marker specific for chronic C pneumoniae infection is available and that any interpretation of infection status on the basis of single titer readings should be done with care. Second, we might not have entirely excluded subjects with bronchial asthma due to a lack of postbronchodilator spirometry values, or subjects with other pulmonary diseases such as diffuse bronchiectasis and bronchiolitis obliterans. Given the positive association between atypical bacterial infection and asthma,47 our findings might have been confounded by interaction with the reversible airflow obstruction. However, our observed positive association between C pneumoniae and M pneumoniae seropositivity and COPD remained significant after the exclusion of subjects with self-reported medication for bronchial asthma or COPD, or with pulmonary disease. Third, as our study subjects were Japanese, the results of this study might not be simply extrapolatable to other populations with different environmental backgrounds. Further, the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/) shows ethnic differences in the rs17634369 genotype, with the highest frequency of the G allele in Africans, followed by Asians and Caucasians.
In summary, our study clarified that dual C pneumoniae and M pneumoniae seropositivity is an independent risk factor for pulmonary function. Further investigations are needed to clarify mechanisms regarding the disease pathogenicity of pneumoniae infection.
Supplementary Material
Acknowledgments
The authors thank University Grant and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science & Technology in Japan, and a research grant from the Takeda Science Foundation for the support. The authors also thank Dr Yoshihiko Kotoura for his help in clinical measurements, and the Nagahama City Office and nonprofit organization Zeroji Club for their help in conducting the Nagahama Study; and the editors of DMC Corporation for their help in the preparation of this manuscript.
Footnotes
Abbreviations: COPD = chronic obstructive pulmonary disease, FEV1 = forced expiratory volume in 1 second, FVC = forced vital capacity, GWAS = genome-wide association study, SLE = systemic lupus erythematosus, SNP = single nucleotide polymorphism.
This study was supported by a University Grant and Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science & Technology in Japan, and a research grant from the Takeda Science Foundation.
The authors have no conflicts of interest to disclose
REFERENCES
- 1.Hozawa A, Billings JL, Shahar E, et al. Lung function and ischemic stroke incidence: the Atherosclerosis Risk in Communities study. Chest 2006; 130:1642–1649. [DOI] [PubMed] [Google Scholar]
- 2.Engström G, Lind P, Hedblad B, et al. Lung function and cardiovascular risk: relationship with inflammation-sensitive plasma proteins. Circulation 2002; 106:2555–2560. [DOI] [PubMed] [Google Scholar]
- 3.Agarwal SK, Heiss G, Barr RG, et al. Airflow obstruction, lung function, and risk of incident heart failure: the Atherosclerosis Risk in Communities (ARIC) study. Eur J Heart Fail 2012; 14:414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Georgiopoulou VV, Kalogeropoulos AP, Psaty BM, et al. Lung function and risk for heart failure among older adults: the Health ABC Study. Am J Med 2011; 124:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao KM, Ho CH, Ko SC, et al. Increased risk of dementia in patients with chronic obstructive pulmonary disease. Medicine 2015; 94:e930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miniati M, Monti S, Pavlickova I, et al. Survival in COPD: impact of lung dysfunction and comorbidities. Medicine 2014; 93:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Marco R, Accordini S, Marcon A, et al. Risk factors for chronic obstructive pulmonary disease in a European cohort of young adults. Am J Respir Crit Care Med 2011; 183:891–897. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher C, Peto R. The natural history of chronic airflow obstruction. Br Med J 1977; 1:1645–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stang P, Lydick E, Silberman C, et al. The prevalence of COPD: using smoking rates to estimate disease frequency in the general population. Chest 2000; 117:354S–359S. [DOI] [PubMed] [Google Scholar]
- 10.Brutsche MH, Downs SH, Schindler C, et al. Bronchial hyperresponsiveness and the development of asthma and COPD in asymptomatic individuals: SAPALDIA cohort study. Thorax 2006; 61:671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D, Wu Y, Tian P, et al. Adipokine CTRP-5 as a potential novel inflammatory biomarker in chronic obstructive pulmonary disease. Medicine 2015; 94:e1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papaetis GS, Anastasakou E, Orphanidou D. Chlamydophila pneumoniae infection and COPD: more evidence for lack of evidence? Eur J Intern Med 2009; 20:579–585. [DOI] [PubMed] [Google Scholar]
- 13.von Hertzen L, Isoaho R, Leinonen M, et al. Chlamydia pneumoniae antibodies in chronic obstructive pulmonary disease. Int J Epidemiol 1996; 25:658–664. [DOI] [PubMed] [Google Scholar]
- 14.Blasi F, Damato S, Cosentini R, et al. Chlamydia pneumoniae and chronic bronchitis: association with severity and bacterial clearance following treatment. Thorax 2002; 57:672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandén E, Koyi H, Gnarpe J, et al. Chronic Chlamydia pneumoniae infection is a risk factor for the development of COPD. Respir Med 2005; 99:20–26. [DOI] [PubMed] [Google Scholar]
- 16.Lieberman D, Ben-Yaakov M, Lazarovich Z, et al. Chlamydia pneumoniae infection in acute exacerbations of chronic obstructive pulmonary disease: analysis of 250 hospitalizations. Eur J Clin Microbiol Infect Dis 2001; 20:698–704. [DOI] [PubMed] [Google Scholar]
- 17.Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med 2008; 359:2355–2365. [DOI] [PubMed] [Google Scholar]
- 18.National Heart, Lung and Blood Institute. Morbidity and Mortality: 2012 Chart Book on Cardiovascular, Lung and Blood Diseases. Bethesda, Maryland: National Institutes of Health; 2012. [Google Scholar]
- 19.Kraft M, Cassell GH, Henson JE, et al. Detection of Mycoplasma pneumoniae in the airways of adults with chronic asthma. Am J Respir Crit Care Med 1998; 158:998–1001. [DOI] [PubMed] [Google Scholar]
- 20.Repapi E, Sayers I, Wain LV, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet 2010; 42:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohara K, Tabara Y, Yamamoto Y, et al. Chlamydia pneumoniae seropositivity is associated with increased plasma levels of soluble cellular adhesion molecules in community-dwelling subjects: the Shimanami Health Promoting Program (J-SHIPP) study. Stroke 2002; 33:1474–1479. [DOI] [PubMed] [Google Scholar]
- 22.Barker CE, Sillis M, Wreghitt TG. Evaluation of Serodia Myco II particle agglutination test for detecting Mycoplasma pneumoniae antibody: comparison with mu-capture ELISA and indirect immunofluorescence. J Clin Pathol 1990; 43:163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shankar EM, Kumarasamy N, Balakrishnan P, et al. Seroprevalence of Mycoplasma pneumoniae in HIV-infected patients using a microparticle agglutination test. J Med Microbiol 2006; 55:759–763. [DOI] [PubMed] [Google Scholar]
- 24.Kung CM, Wang HL. Seroprevalence of Mycobacterium pneumoniae in healthy adolescents in Taiwan. Jpn J Infect Dis 2007; 60:352–354. [PubMed] [Google Scholar]
- 25.Finner H, Strassburger K, Heid IM, et al. How to link call rate and p-values for Hardy-Weinberg equilibrium as measures of genome-wide SNP data quality. Stat Med 2010; 29:2347–2358. [DOI] [PubMed] [Google Scholar]
- 26.Pe’er I, Yelensky R, Altshuler D, et al. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol 2008; 32:381–385. [DOI] [PubMed] [Google Scholar]
- 27.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol 2007; 31:358–362. [DOI] [PubMed] [Google Scholar]
- 28.Han JW, Zheng HF, Cui Y, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009; 41:1234–1237. [DOI] [PubMed] [Google Scholar]
- 29.Seemungal TA, Wedzicha JA, MacCallum PK, et al. Chlamydia pneumoniae and COPD exacerbation. Thorax 2002; 57:1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strachan DP, Carrington D, Mendall M, et al. Chlamydia pneumoniae serology, lung function decline, and treatment for respiratory disease. Am J Respir Crit Care Med 2000; 161:493–497. [DOI] [PubMed] [Google Scholar]
- 31.Takashima N, Miura K, Hozawa A, et al. Population attributable fraction of smoking and metabolic syndrome on cardiovascular disease mortality in Japan: a 15-year follow up of NIPPON DATA90. BMC Public Health 2010; 10:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varma-Basil M, Dwivedi SK, Kumar K, et al. Role of Mycoplasma pneumoniae infection in acute exacerbations of chronic obstructive pulmonary disease. J Med Microbiol 2009; 58:322–326. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman D, Lieberman D, Ben-Yaakov M, et al. Serological evidence of Mycoplasma pneumonia infection in acute exacerbation of COPD. Diagn Microbiol Infect Dis 2002; 44:1–6. [DOI] [PubMed] [Google Scholar]
- 34.Diederen BM, van der Valk PD, Kluytmans JA, et al. The role atypical respiratory pathogens in exacerbations of chronic obstructive pulmonary disease. Eur Respir J 2007; 30:240–244. [DOI] [PubMed] [Google Scholar]
- 35.Goh SK, Johan A, Cheong TH, et al. A prospective study of infections with atypical pneumonia organisms in acute exacerbations of chronic bronchitis. Ann Acad Med Singapore 1999; 28:476–480. [PubMed] [Google Scholar]
- 36.Iwamoto H, Yokoyama A, Kitahara Y, et al. Airflow limitation in smokers is associated with subclinical atherosclerosis. Am J Respir Crit Care Med 2009; 179:35–40. [DOI] [PubMed] [Google Scholar]
- 37.Markus HS, Sitzer M, Carrington D, et al. Chlamydia pneumoniae infection and early asymptomatic carotid atherosclerosis. Circulation 1999; 100:832–837. [DOI] [PubMed] [Google Scholar]
- 38.Player MS, Mainous AG, 3rd, Everett CJ, et al. Chlamydia pneumoniae and progression of subclinical atherosclerosis. Eur J Prev Cardiol 2014; 21:559–565. [DOI] [PubMed] [Google Scholar]
- 39.Strachan DP, Carrington D, Mendall MA, et al. Relation of Chlamydia pneumoniae serology to mortality and incidence of ischaemic heart disease over 13 years in the caerphilly prospective heart disease study. BMJ 1999; 318:1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013; 45:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueta M, Sawai H, Sotozono C, et al. IKZF1, a new susceptibility gene for cold medicine-related Stevens-Johnson syndrome/toxic epidermal necrolysis with severe mucosal involvement. J Allergy Clin Immunol 2015; 135:1538–1545. [DOI] [PubMed] [Google Scholar]
- 42.Yap WH, Yeoh E, Tay A, et al. STAT4 is a target of the hematopoietic zinc-finger transcription factor Ikaros in T cells. FEBS Lett 2005; 579:4470–4478. [DOI] [PubMed] [Google Scholar]
- 43.Kaplan MH, Sun YL, Hoey T, et al. Impaired IL-12 responses and enhance development of Th2 cells in Stat4-deficient mice. Nature 1996; 382:174–177. [DOI] [PubMed] [Google Scholar]
- 44.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 2003; 100:2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villegas E, Sorlózano A, Gutiérrez J. Serological diagnosis of Chlamydia pneumoniae infection: limitations and perspectives. J Med Microbiol 2010; 59:1267–1274. [DOI] [PubMed] [Google Scholar]
- 46.Dowell SF, Peeling RW, Boman J, et al. Standardizing Chlamydia pneumoniae assays: recommendations from the Centers for Disease Control and Prevention (USA) and the Laboratory Centre for Disease Control (Canada). Clin Infect Dis 2001; 33:492–503. [DOI] [PubMed] [Google Scholar]
- 47.Sutherland ER, Martin RJ. Asthma and atypical bacterial infection. Chest 2007; 132:1962–1966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


