SUMMARY
A critical issue in development is coordination of the activity of stem cell niches with differentiation of their progeny to ensure coherent organ growth. In the plant root, these processes take place at opposite ends of the meristem and must be coordinated with each other at a distance. Here we show that in Arabidopsis the gene SCR presides over this spatial coordination. In the organizing centre of the root stem cell niche, SCR directly represses the expression of the cytokinin-response transcription factor ARR1, which promotes cell differentiation, controlling auxin production via the ASB1 gene and sustaining stem cell activity. This allows SCR to regulate, via auxin, the level of ARR1 expression in the transition zone where the stem cell progeny leave the meristem, thus controlling the rate of differentiation. In this way, SCR simultaneously controls stem cell division and differentiation ensuring coherent root growth.
INTRODUCTION
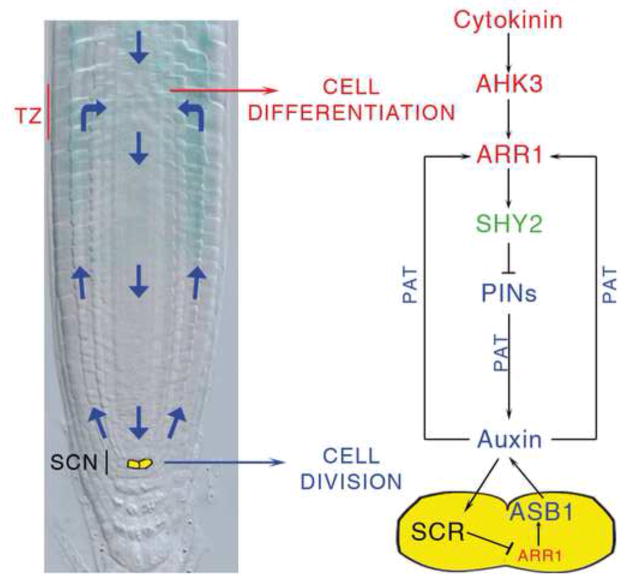
In multicellular organisms, stem cell division and differentiation of the progeny cells must be coordinated to ensure coherent growth. In the Arabidopsis root, stem cells reside in the apical region of the meristem where they surround a small group of organizer cells. Together, they form a stem cell niche (SCN) (van den Berg et al., 1997; Scheres, 2007) (Figure 1A). As in the animal stem cell niche, the organizer cells, called the quiescent center (QC), maintain the divisional activity of the surrounding stem cells by means of unknown short range signals (van den Berg et al., 1997; Scheres, 2007). Stem cells generate transit-amplifying cells, which undergo additional divisions then differentiate in the transition zone (TZ, Figure 1A) (Moubayidin et al., 2009; Perilli et al., 2010). For meristem maintenance, and therefore continuous root growth, the rate of differentiation of transit-amplifying cells must be equal to the rate of generation of new cells. We have shown that, in the TZ, this balance is ensured by the interaction between two hormones: cytokinin, which promotes cell differentiation (Dello Ioio et al., 2007), and auxin, which promotes cell division (Blilou et al., 2005). In particular, cytokinins are perceived at the TZ by the ARABIDOPSIS HISTIDINE KINASE 3 (AHK3) cytokinin receptor, which transfers the signal, via a phosphorelay, into the nucleus and activates two cytokinin primary response transcription factors, ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) and 12 (ARR12) (Hwang et al., 2012). These genes activate the transcription of the SHORT HYPOCOTYL 2 gene (SHY2), an inhibitor of auxin signalling (Tian et al., 2002; Dello Ioio et al., 2008). SHY2, in turn, negatively regulates the expression of several PIN-FORMED (PIN) genes encoding auxin transport facilitators (Friml, 2010), thus limiting auxin transport and distribution and allowing cell differentiation (Dello Ioio et al., 2008; Moubayidin et al., 2010).
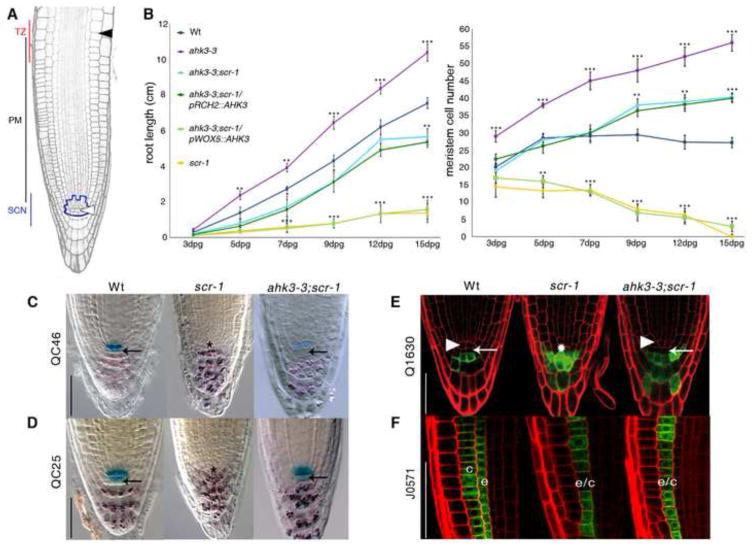
Figure 1. SCR maintains SCN activity by suppressing AHK3-mediated differentiation input in the QC cells.
(A) Longitudinal section of the Arabidopsis root meristem. SCN stem cell niche (highlight in blue), PM proximal meristem, TZ transition zone. The white and black arrowheads include meristematic cortex file, indicating QC and the last meristematic cortical cell, respectively.
(B) From left to right, root length and meristem cell number measured over time of wild-type (Wt), scr-1, ahk3-3, ahk3-3;scr-1, ahk3-3;scr-1/pRCH2::AHK3 and ahk3-3;scr-1/pWOX5::AHK3. dpg: days post germination. Error bars, SD; *p < 0.05, **p < 0.01, ***p < 0.001; Student’s t-test.
(C-E) Elimination of AHK3 activity rescues SCN defect of scr-1 mutant. Expression of QC46 and QC25 (C and D) and Q1630 markers
(E) in Wt, scr-1 and ahk3-3;scr-1 root tips. (C and D) Double labeling of QC and differentiated columella cells visualized by QC specific markers and amyloplast staining, respectively. Note that black arrows indicate restoration of columella stem cells activityin the ank3-3;scr-1 double mutant (C). Lack of Q1630 expression in the columella stem cells (white arrows) indicates that columella stem cell activity is restored in the ahk3-3;scr-1 double mutant. White arrowheads indicate QC position in Wt and ahk3-3;scr-1. Asterisk indicates the presumptive position of QC cells in scr-1 mutant.
(F) SCR-mediated cytokinin signalling suppression is not necessary for endodermis activity. J0571 expression in Wt, scr-1 and ahk3-3;scr-1. Note the lack of asymmetric cell division necessary to generate properly cortex (c) and endodermis (e) tissues in scr-1 (Di Laurenzio et al., 1996) and in the ahk3-3;scr-1 double mutant. The resulting monolayer is indicated by c/e. Scale bar represents 50 μm in all panels. See also Figure S1.
Root SCN maintenance requires the activity of SCR, a member of the GRAS family of transcription factors (Di Laurenzio et al., 1996; Pysh et al., 1999; Lee et al., 2008; Sabatini et al., 2003). SCR was originally identified as a key factor regulating the asymmetric cell division producing the cortex and endodermis (Di Laurenzio et al., 1996; Heidstra et al., 2004; Cui et al., 2007). More recently, it has also been shown to be involved in the specification of xylem cell types within the vascular tissue (Carlsbecker et al., 2010) and in controlling ground tissue stem cell asymmetric division (Sozzani et al., 2010; Cruz-Ramírez et al., 2012). In addition, SCR has been shown to be necessary and sufficient, when acting in the QC cells, to sustain QC functions and, as a consequence, the surrounding stem cells (Sabatini et al., 2003), but the molecular mechanism for this activity remains unknown.
While molecular mechanisms regulating stem cell division (Spradling et al., 2001; Scheres, 2007; Sablowski, 2011; Wolpert and Tickle, 2011) and transit-amplifying cell differentiation (Wolpert and Tickle, 2011; Dello Ioio et al., 2008; Tsukagoshi et al., 2010) have been described, little is known as to how these two events are coordinated (Mondal et al., 2011).
Here, we show that SCR acts in the QC to control, at the same time, stem cell activity and the differentiation of their progeny by cell-autonomously preventing expression of the cytokinin response regulator ARR1 in the QC, and non-cell autonomously controlling ARR1 expression in the TZ via auxin.
RESULTS
SCR sustains stem cell and meristem activity by suppressing cytokinin perception
We hypothesized that the coordination between stem cell activity in the SCN and cell differentiation in the TZ might be effected by a genetic interaction between key molecular components directly regulating each zone of the root meristem. We therefore asked whether a genetic interaction may exist between SCR, which is involved in QC and stem cell maintenance (Sabatini et al., 2003) and the cytokinin receptor AHK3, which mediates cell differentiation in the TZ (Dello Ioio et al., 2007). Mutation in the SCR gene results in meristem consumption and arrested root growth a few days after germination (Figure 1B), due to defective QC and stem cell activities (Figures 1C–1E) (Sabatini et al., 2003). Mutation in the AHK3 gene results in an enlarged meristem and a longer root, due to delayed cell differentiation in the TZ (Figure 1B) (Dello Ioio et al., 2007). Interestingly, in ahk3-3;scr-1 double mutants, meristem size and root growth are maintained over time (Figures 1B, S1A and S1D). In the scr-1 mutant, the QC markers, QC25 and QC46, are absent due to loss of QC identity, while cells immediately below the QC, at the position of columella stem cells, acquire differentiation markers such as amyloplasts and the Q1630 marker (Figures 1C–1E) (Sabatini et al., 2003). By contrast, in the ahk3-3;scr-1 double mutant, QC25 and QC46 expression is restored, and no amyloplasts or Q1630 expression are detected within the columella stem cell layer (Figures 1C–1E). In addition, root hairs and xylem strands, characteristic of fully differentiated epidermal and vascular cells, are present in scr-1 meristems (Figure S1D) (Sabatini et al., 2003) but are absent from the ahk3-3;scr-1 root meristem (Figure S1D). These data indicate that in ahk3-3;scr-1 roots stem cell maintenance is restored, leading to meristem maintenance and root growth. Thus, this suggests that SCR sustains stem cell and meristem activity by suppressing cytokinin perception. It is important to note that the ahk3-3;scr-1 double mutant still displays an undivided ground tissue layer (Figure 1F) (Di Laurenzio et al., 1996), and a lack of protoxylem cells characteristic of the scr-1 mutant (Figure S1B) (Carlsbecker et al., 2010), suggesting that its role in stem cell regulation is independent of its developmental role in these tissues.
SCR suppresses cytokinin activity in the QC
Comparison of the expression patterns of SCR (Wysocka-Diller et al., 2000) and AHK3 (Dello Ioio et al., 2007) (Figure S1C) suggested that these genes might genetically interact either in the QC, where SCR maintains QC identity (Sabatini et al., 2003), or in the endodermis of the TZ, where AHK3 mediates cell differentiation (Dello Ioio et al., 2007). To determine if and where the interaction occurs, we exploited a tissue-specific complementation approach to restore AHK3 activity (and thus cytokinin perception) either, specifically in the QC, or in the TZ of the ahk3-3;scr-1 double mutant. To accomplish this, we drove AHK3 expression with the WUS-RELATED HOMEOBOX 5 (WOX5) promoter, which confers expression in the QC, (Sakar et al., 2007) and the ROOT CLAVATA HOMOLOG 2 (RCH2) promoter, which confers expression in the TZ (Dello Ioio et al., 2007) (Figure S1C). In ahk3-3;scr-1/pRCH2::AHK3 plants, both meristem size and root growth are indistinguishable from that of ahk3-3;scr-1 (Figures 1B and S1A and S1D). By contrast, the meristem of ahk3-3;scr-1/pWOX5::AHK3 plants is consumed and root growth arrested as in the scr-1 mutant (Figures 1B and S1A and S1D). These results indicate that AHK3 and SCR interact in the QC, thus suggesting that SCR maintains the SCN and meristem activity by suppressing cytokinin signalling in these cells.
SCR prevents SCN differentiation and maintains meristem activity by suppressing ARR1
To understand how SCR represses cytokinin-mediated cell differentiation, we examined AHK3 expression in the scr-1 mutant by means of qRT-PCR analysis and a pAHK3::AHK3:GUS translational fusion (Dello Ioio et al., 2007). Interestingly, neither the expression nor the accumulation of AHK3 (Figures S2A and S2B) is altered in the scr-1 mutant. We have previously shown that AHK3 controls the rate of cell differentiation in the TZ via the activation of two primary cytokinin response transcription factors: ARR1 and ARR12 (Dello Ioio et al., 2007; Moubayidin et al., 2010). We therefore analysed the expression of ARR1 and ARR12 in scr-1 utilizing pARR1::ARR1:GUS and pARR12::ARR12:GUS translational fusions (Dello Ioio et al., 2007; Moubayidin et al., 2010), as well as by qRT-PCR. Neither ARR12 protein (Figure S2A) nor mRNA (Figure S2B) levels are altered, while the levels of both mRNA and protein of ARR1 are upregulated in both scr-1 and scr-4 mutant backgrounds (Figures 2A, 2B -scr-1 and 3A, 3B –scr-4). The ARR1 protein, which is normally expressed in all tissues of the TZ, was ectopically expressed in the proximal meristem, including the SCN, of scr plants (Figures 2B, 3A and 3B). This suggests that SCR maintains the activities of both QC and stem cells by suppressing ARR1 transcription, and therefore cytokinin signaling, in the QC. To further test this hypothesis, we analysed the root phenotype of plants carrying a double mutation in both SCR and ARR1 genes. Similar to the ahk3-3;scr-1 double mutant, the arr1-4;scr-1 double mutant displays sustained meristem and indeterminate root growth (Figures 2C, S2C and S2D) due to an active SCN, as visualized by the presence of QC markers absent in the scr-1 mutant (Figures 2D and S2E). In addition, amyloplasts were absent from the columella stem cells (Figures 2D and S2E). By contrast, the root phenotype of the arr12-1;scr-1 double mutant was indistinguishable from that of scr-1 (Figures 2C, S2C and S2D), consistent with the hypothesis that SCR maintains the stem cell niche and root growth by specifically suppressing ARR1 expression.
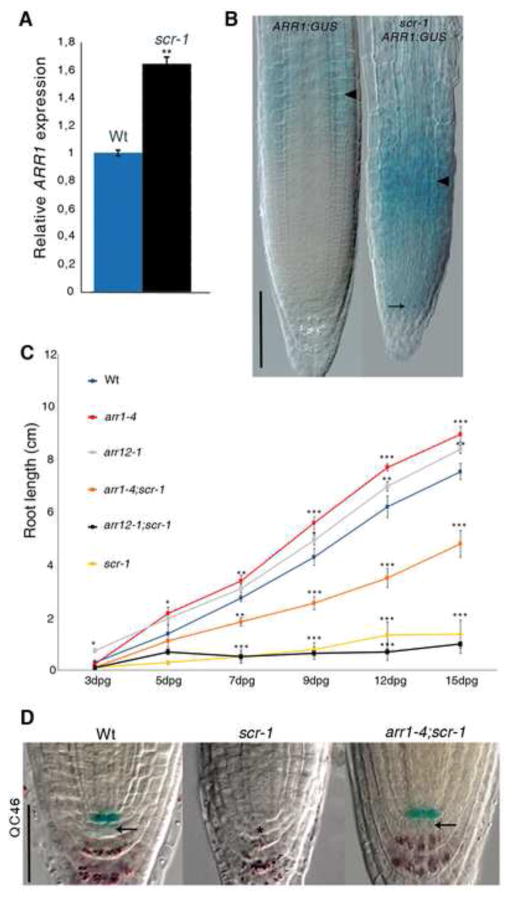
Figure 2. SCR maintains SCN activity by suppressing ARR1.
(A) qRT-PCR showing low level of ARR1 transcript in Wt compared to scr-1 roots. Relative expression is normalized to ACTIN2. Error bars, SD; **p < 0.01 Student’s t-test.
(B) Expression of pARR1::ARR1:GUS construct in Wt roots and in scr-1 mutant. Black arrowheads indicate the position of transition zone; arrow indicates ARR1 ectopic expression in the scr-1 QC. Scale bar represents 100 μm.
(C) Root length measured over time for Wt, scr-1, arr1-4, arr12-1, arr1-4;scr-1, arr12-1;scr-1. dpg: days post germination. Error bars, SD; *p < 0.05, **p < 0.01, ***p < 0.001; Student’s t-test.
(D) QC46 expression in, from left to right, Wt, scr-1 and arr1-4;scr-1. Double labeling of QC and differentiated columella cells visualized by QC46 and amyloplast staining, respectively. Note that black arrows indicate restoration of columella stem cells activity in the arr1-4;scr-1 double mutant. Asterisk indicates position of QC cells in the scr-1 mutant. The same results have been obtained with the arr1-3;scr-1, arr1-3;scr-4 and arr1-4;scr-4 double mutant combinations (data not shown). Scale bar represents 50 μm. See also Figure S2.
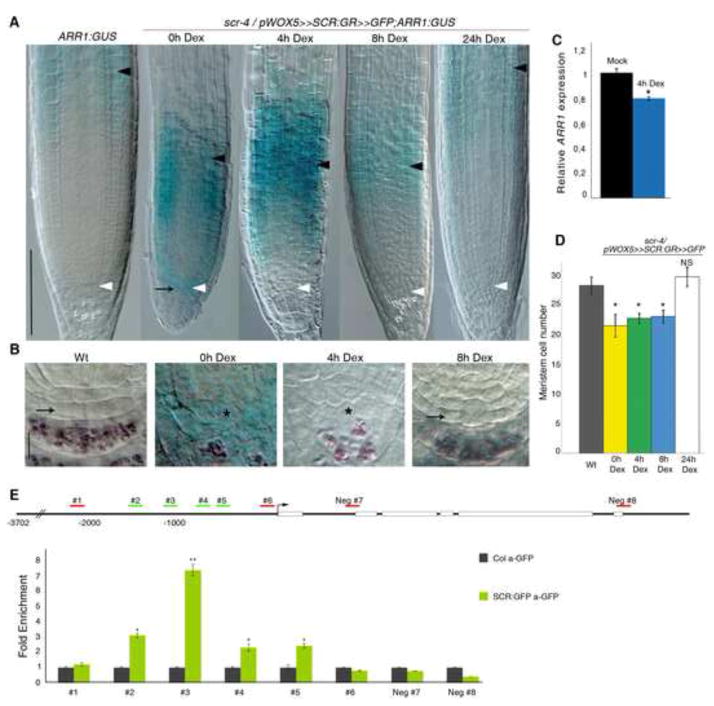
Figure 3. SCR controls stem cell activity by suppressing directly ARR1 in the QC.
(A) Expression analysis of the ARR1:GUS translational fusion in, from left to right, Wt and scr-4/pWOX5≫SCR:GR≫GFP roots untreated (0h Dex) or treated with Dex for 4, 8 and 24 hours. Black arrow indicates ectopic ARR1 expression in the QC. Black and white arrowheads indicate, respectively, the cortex TZ and the QC. Scale bar represents 100 μm.
(B) Stem cell niche of, from left to right, ARR1:GUS and scr-4/pWOX5≫SCR:GR≫GFP;ARR1:GUS untreated (0h Dex) or treated with Dex for 4 and 8 hours. Black arrow indicates active stem cells in Wt and scr-4 roots after 8 hours of Dex treatment. Asterisk indicates the presumptive position of QC cells in scr-4 untreated and treated with Dex for 4 hours. Scale bar represents 10 μm.
(C) qRT-PCR performed on sorted QC cells of scr-4 roots carrying the pWOX5≫SCR:GR≫GFP construct, showing significant down regulation of ARR1 transcription upon 4 hours of Dex treatment. Relative expression is normalized to ACTIN2. Error bars, SD; *p < 0.05; Student’s t-test.
(D) Meristem cell number of, from left to right, Wt and scr-4/pWOX5≫SCR:GR≫GFP roots untreated (0h Dex) or treated with Dex for 4, 8 and 24 hours. Error bars, SD; *p < 0.05, NS, not significant; Student’s t-test.
(E) ChIP-RT qPCR of the ARR1 promoter using 5 days old Col-0 (grey bars) and pSCR::SCR:GFP scr-4 (green bars) plants. ChIP samples were prepared from QC-enriched material. RT-qPCR results are shown as fold enrichment compared to Col-0. We found significant enrichment for the several fragments (#2, #3, #4 and #5) bound by SCR by scanning the 2186 Kb of sequence upstream of ARR1. The data shown are representative of three independent biological experiments with similar results. Error bars show the standard deviations of the ChIP-qPCR reactions performed in triplicate. *p < 0.05, **p < 0.01; Student’s t-test. See also Figure S3.
SCR directly suppresses ARR1 in the QC and acts non-cell autonomously to control ARR1 at the TZ
The fact that SCR is expressed exclusively in the QC and endodermal lineage implies that SCR regulates ARR1 cell autonomously in the QC but that outside the QC its regulation is non-cell autonomous.
To analyse SCR activity in the QC, we employed the GAL4VP16/UAS transactivation system (Brand and Perrimon, 1993; Sabatini et al., 2003). This allowed us to express an inducible version of the SCR protein fused to the glucocorticoid receptor (GR) in the scr-4 mutant background and under control of the WOX5 promoter, and simultaneously mark the SCR:GR expressing cells with GFP (scr-4/pWOX5≫SCR:GR≫GFP plants) (Figure S3A). Four hours of SCR:GR activation with dexamethasone (Dex) were sufficient to downregulate ARR1 mRNA specifically in the QC region, as shown by qRT-PCR experiments on sorted QC cells (using fluorescence activated cell sorting) (Figure 3C). However, there was no change in expression in the rest of the root meristem, as visualized by pARR1::ARR1:GUS expression (Figures 3A and 3B, compare 0h Dex with 4h Dex). ARR1 down-regulation was followed by recovery of stem cell activity but not meristem size, after 8 hours of Dex treatment (Figures 3A, 3B and 3D, compare 0h Dex with 8h Dex). This was coincident with the time when ARR1 expression was repositioned at the TZ (Figure 3A, 8h Dex). Root meristem size recovery occurred after 24 hours of Dex treatment (Figures 3A and 3D, 24h Dex). These results support the hypothesis that SCR suppresses ARR1 expression in the QC to maintain stem cell activity. Importantly, these results also provide evidence that SCR acts on ARR1 non-cell autonomously to position it at the TZ thereby controlling meristem size.
To determine if SCR binds directly to the ARR1 promoter, we performed chromatin immunoprecipitation (ChIP) followed by quantitative PCR (ChIP-qPCR) on plants containing a complementing SCR:GFP chimeric protein under the control of its own promoter in the scr-4 background. Initially, we used whole root meristems but found no enrichment for binding to the ARR1 promoter region with the exception of region #3, where enrichment was not statistically significant (Figure S3B). If SCR binds the ARR1 promoter specifically in the QC, we reasoned that we may have been diluting the ChIP signal when we used the entire meristem. Therefore, we next performed ChIP on extracts from dissected longitudinal sections (3–5 cell lengths) that encompassed the QC (Brady et al., 2007). In this QC-enriched material we found that binding to region #3 in the ARR1 promoter was significantly enriched (Figure 3E), providing evidence that SCR directly binds to the ARR1 promoter in the QC.
Suppression of ARR1 expression in the QC titrates auxin production
We have previously shown that ARR1 promotes cell differentiation in the TZ by directly activating the SHY2 gene (Dello Ioio et al., 2008). Therefore, we hypothesized that in the QC of scr-1 roots, SHY2 is activated by ARR1, which triggers the onset of differentiation. However, we did not detect ectopic expression of SHY2 in the scr-1 QC (data not shown), and elimination of SHY2 activity in the scr-1 mutant did not restore root growth and meristem size (Figures S2C and D). This suggests that ectopic expression of ARR1 in the scr QC induces stem cell differentiation via a different mechanism.
It was recently shown that cytokinin can induce auxin biosynthesis in the Arabidopsis root apex (Jones et al., 2010) and that high levels of auxin in the SCN result in stem cell differentiation (Ding and Friml, 2010). We observed abnormally high levels of auxin in scr-1 root meristems, as visualized by DR5::GFP expression (Sabatini et al., 1999; Ottenschläger et al., 2003) and as measured by mass spectrometry (Figures 4A and 4B). DR5::GFP localization reverted to normal in the arr1-4;scr-1 double mutant (Figure 4A), suggesting that excess auxin biosynthesis in scr-1 is induced by ectopic expression of ARR1 in the QC. Among the auxin biosynthesis genes induced by cytokinin (Jones et al., 2010) and expressed in the QC (Sun et al., 2008; Bartel and Fink, 1994; http://www.arexdb.org), NITRILASE 3 (NIT3), NITRILASE 4(NIT4) and YUCCA6 (YUC6) are over-expressed in the scr-1 mutant and revert to wild type level in the arr1-4;scr-1 (Figure S4A). Since these genes control different branches of the tryptophan-dependent auxin biosynthesis pathway (Stepanova et al., 2011), we decided to prevent enzymatic redundant activities by focusing on the ANTHRANILATE SYNTHASE BETA SUBUNIT 1 (ASB1) gene, which is expressed in the SCN and catalyzes a rate-limiting step of tryptophan biosynthesis (Stepanova et al., 2005). ASB1 is induced by cytokinin, as visualized by the pASB1::GUS transcriptional fusion and confirmed by qRT-PCR (Figures S4B and S4C). Moreover, its induction is dependent on ARR1 function, since cytokinin treatment of the arr1-4 mutant had no effect on ASB1 levels (Figures S4B and S4C). In addition, a very brief staining of this transcriptional fusion shows that the expression of ASB1 is highly upregulated in the QC of the scr-1 mutant and reverted to wild-type levels in the arr1-4;scr-1 double mutant (Figures 4C and S4D). This suggests that overexpression of ASB1 in the scr-1 mutant background depends on ARR1. To exclude the possibility that ASB1 transcriptional activation is under the direct controls of SCR, we performed ChIP-qPCR analysis as described above for ARR1. Our ChIP-qPCR results suggest that SCR does not directly regulate ASB1 (Figure S3C). Thus, in the QC of scr-1, loss of repression of ARR1 leads to over expression of ASB1 and consequently over-accumulation of auxin in the root meristem.
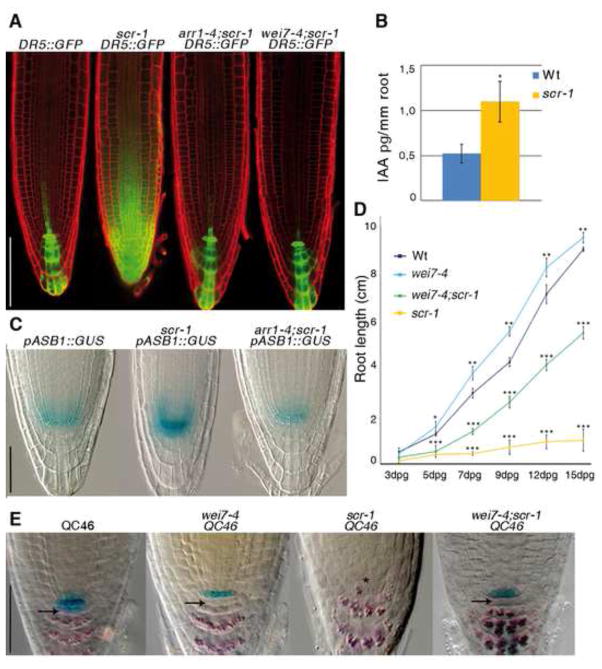
Figure 4. SCR controls auxin biosynthesis in the QC via ARR1.
(A) DR5::GFP expression in, from left to right, Wt, scr-1, arr1-4;scr-1 and wei7-4;scr-1 5-day old root meristems. Scale bar represents 100 μm.
(B) Concentration of free IAA in root tips of Wt and scr-1. Error bars, SD; *p < 0.05; Student’s t-test.
(C) Expression of the pASB1::GUS construct in, from left to right, Wt, scr-1 and arr1-4;scr-1. Note that plants were stained for only 30 minutes. Scale bar represents 50 μm.
(D) Root length measured over time of Wt, scr-1, wei7-4, and wei7-4;scr-1. dpg: days post germination. Error bars, SD; *p < 0.05, **p < 0.01, ***p < 0.001; Student’s t-test.
(E) QC46 expression and lugol staining of Wt, wei7-4, scr-1 and wei7-4;scr-1. Black arrows indicate stem cells activity in Wt, wei7-4 and wei7-4;scr-1. Asterisk indicate the presumptive position of QC cells in scr-1. The same result has been obtained with the wei7-1;scr-1 double mutant combination. Scale bar represents 50 μm. See also Figure S4.
To determine if over-accumulation of auxin is responsible for stem cell differentiation in scr-1, we introduced the ASB1 loss-of-function mutation wei7-4 (WEAK ETHYLENE INSENSITIVE 7) (Stepanova et al., 2005) into the scr-1 background. Consistent with our hypothesis, the root of wei7-4;scr-1 displays a stable root meristem and indeterminate growth (Figures 4D and S4E) due to a functional SCN (Figures 4E and S4F). This phenotype is similar to the one of arr1-4;scr-1 previously described (Figures 2C and S2D). In addition, DR5::GFP expression in the root tip of wei7-4;scr-1 reverted to lower levels compared to scr-1 (Figure 4A), further supporting the hypothesis that ASB1 upregulation in the scr-1 QC is responsible for the abnormal accumulation of auxin in this mutant.
Therefore, our working model is that in scr-1, lack of ARR1 repression in the QC cells leads to ASB1-dependent auxin over-accumulation responsible for stem cell inactivation, meristem consumption and determinate root growth (Figure S4G). At the same time, we are showing that SCR can be induced by auxin (Figure S5A), thus generating a local feedback loop involved in homeostasis of auxin levels responsible for SCN activity.
SCR regulates ARR1 non-cell autonomously through auxin
We next asked if auxin could mediate the non-cell autonomous effect of SCR on ARR1 expression at the TZ (Figure 3A). We found that exogenous auxin application induces ARR1 expression in the TZ (Figures S5B and S5C). To further test this hypothesis, we employed a construct expressing the IAAH gene, the bacterial auxin biosynthetic enzyme, driven by the WOX5 promoter (pWOX5::IAAH) (Figure S5D) (Blilou et al., 2005). In pWOX5::IAAH plants auxin biosynthesis can be induced specifically in the QC after indole-3-acetamide (IAM) auxin precursor treatment (Blilou et al., 2005). We observed that, as early as at 3 hours of IAM treatment, a QC-specific increase of auxin levels was sufficient to induce ARR1 expression in the TZ but not in the QC (Figures 5A and 5B). To verify that auxin produced in the QC was transported in the TZ and that this resulted in ARR1 transcriptional activation, we treated plants in the presence of N-naphthylphthalamic acid (NPA), an inhibitor of auxin transport (Jacobs and Rubery, 1988). Production of auxin in the QC, via the pWOX5::IAAH/IAM system, resulted in accumulation of the auxin sensor DR5::GFP at the TZ. We could show that this accumulation is prevented via NPA treatment (Blilou et al 2005) (Figure S5D). Interestingly lack of DR5::GFP accumulation at the TZ upon NPA treatment was accompanied by lack of ARR1 induction at the TZ (Figure 5A), strongly suggesting that auxin is the non-cell autonomous factor that controls ARR1 activity in the TZ from the QC.
Figure 5. SCR controls ARR1 activity at the TZ via auxin.
(A) Expression of the ARR1:GUS translational fusion in, from left to right, mock-treated pWOX5::IAAH root, pWOX5::IAAH root treated with IAM, pWOX5::IAAH root treated with IAM and NPA, wei7-4 roots untreated or treated with IAA. Black and white arrowheads indicate, respectively, the cortex TZ and the QC. Roots were analyzed 5dpg. Scale bar represents 100 μm.
(B) qRT-PCR showing upregulation of ARR1 transcription in pWOX5::IAAH roots upon 3 hours of IAM treatment. Error bars, SD; **p < 0.01; Student’s t-test.
(C) qRT-PCR showing low ARR1 transcript levels in wei7-4. Error bars, SD; *p < 0.05; Student’s t-test.
(D) Root meristem cell number in, from left to right, Wt, wei7-4, Wt upon 16 hours of IAA treatment, wei7-4 upon 16 hours of IAA treatment. Error bars, SD; *p < 0.05, NS, not significant; Student’s t-test. Roots were analyzed 5dpg.
(E) Root meristem cell number in, from left to right, mock-treated pWOX5::IAAH root, pWOX5::IAAH treated 24 hours with IAM, mock-treated arr1-4;pWOX5::IAAH and arr1-4;pWOX5::IAAH treated 24 hours with IAM. Error bars, SD; *p < 0.05, **p < 0.01; Student’s t-test. Roots were analyzed 5dpg. See also Figure S5.
To determine if ASB1-dependent auxin production in the QC could control ARR1 activity in the TZ, we analyzed ARR1 expression in the wei7-4 mutant. We observed lower ARR1 mRNA and protein levels and an enlarged root meristem (Figures 5A, 5C and 5D). Interestingly, ARR1 protein levels reverted to wild type after 3 hours of exogenous auxin application (Figure 5A), followed by root meristem size complementation after 16 hours of auxin application (Figure 5D). We then asked if auxin produced in the QC is sufficient to induce cell differentiation in the TZ and if so, if it acts through ARR1. Consistent with this hypothesis, we observed that root meristem size of wild-type plants carrying a pWOX5::IAAH construct was reduced after 24 hours of IAM supplying, and thus of auxin production, while no meristem size reduction was observed when the same construct was introduced into the arr1-4 background (Figure 5E). Taken together, these results indicate that auxin is the non-cell autonomous signal, which originates in the QC and controls ARR1 activity in the TZ (Figure 6). We hypothesize that heightened levels of auxin induce cell differentiation in the TZ via the cytokinin signalling pathway.
Figure 6. Model for the spatial coordination between SCN and TZ.
Model showing how SCR presides over the spatial coordination between stem cell niche (SCN) and transition zone (TZ) activity. In the QC (yellow), SCR represses ARR1, which in turn controls auxin production via ASB1, thus enabling cell division along the trans-amplifying zone. SCR also exerts, via polar auxin transport (PAT), a long distance control on ARR1 at the TZ, enabling cytokinin to sustain cell differentiation via SHY2 (Dello Ioio et al., 2008).
DISCUSSION
In both plants and animals, organ morphogenesis depends on the coordination between the activities of cells belonging to different tissues and at different developmental stages. In particular, SCN activity and differentiation of their progeny must be coordinated to ensure coherent growth. As observed in animal organogenesis (e.g. growth of mammalian bones or the gut crypt) (Spradling et al., 2001; Wolpert and Tickle, 2011), stem cell division and terminal differentiation of the progeny in the plant root take place in physically distinct zones separated by a group of transit-amplifying cells (Scheres, 2007; Sablowski, 2011). Here we show that in Arabidopsis, the SCR gene acts from the QC to coordinate stem cell division and cell differentiation (model in Figure 6). It had been previously shown that SCR is necessary and sufficient to maintain the SCN (Sabatini et al., 2003) but the molecular mechanisms through which it sustains QC and stem cell activity are unknown. We show that SCR directly represses ARR1 specifically in the QC thereby repressing cytokinin dependent cell differentiation and sustaining SCN activity (model in Figure 6). This is in line with what is already observed in animal systems where master regulatory transcription factors mediate stem cell maintenance through direct transcriptional repression of factors that in turn would promote differentiation (Bernstein et al., 2006; Mikkelsen et al., 2007). Ectopic expression of ARR1 in the scr-1 QC induces stem cell differentiation via a mechanism different from the one acting in the TZ, which relies on the transcriptional activation of the SHY2 gene (Dello Ioio et al., 2008). In the QC, ARR1 regulates auxin production by modulating expression of the auxin biosynthesis gene ASB1. ASB1 controls a rate-limiting step in auxin biosynthesis and is specifically expressed in the SCN, being most active in the QC. Interestingly, other genes involved in auxin biosynthesis and acting downstream of the ASB1 pathway, such as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) (Stepanova et al., 2008), NIT3 (Sun et al., 2009), CYTOCHROME P450 FAMILY 79 SUBFAMILY B POLYPEPTIDE 2 (CYP79B2) and CYP79B3 (Ljung et al., 2005) and YUCCA2 (YUC2) (Sun et al., 2009), are specifically expressed in the QC, thus suggesting that the QC is the primary source of auxin in the root meristem. A future challenge will be to understand how auxin produced in the QC controls the activity of the surrounding stem cells. Recently, it has been shown that SCR in the cortex/endodermis stem cell is part of a complex specifically involved in ground tissue stem cell asymmetric division (Cruz-Ramírez et al., 2012). The activity of this complex depends on auxin levels (Cruz-Ramírez et al., 2012) leading to the intriguing hypothesis that SCR, by controlling auxin biosynthesis in the QC, controls division of the other stem cell types modulating the activities of similar complexes. On the other hand, it is well known that titration of auxin levels in the SCN is critical to the maintenance of stem cell activity since it has been shown that high levels of auxin promote stem cell differentiation (Ding and Friml, 2010).
By controlling the level of auxin production in the QC, SCR also exerts a long-distance control on ARR1 in the TZ. Auxin produced by ASB1 in the QC, distributed in the meristem via the auxin transport facilitators (PIN, PGP genes etc.) (Friml, 2010), is sufficient to activate expression of ARR1 in the TZ, where ARR1 controls SHY2 expression. SHY2 negatively controls PINs activities and auxin transport enabling cytokinin to trigger differentiation of transit-amplifying cells (model in Figure 6) (Dello Ioio et al., 2008). It is worth mentioning that SCR, by controlling activity of the PHABULOSA gene (Carlsbecker et al., 2010) also controls ARR1 protein activity by regulating cytokinin biosynthesis in the vascular tissues (Dello Ioio et al., 2012).
In conclusion, we propose a model in which the SCN organizer not only controls division of the surrounding stem cells (van den Berg et al., 1997), but also regulates the differentiation of the transit-amplifying cells in the TZ. QC activity depends on SCR (Sabatini et al., 2003), which suppresses ARR1 thus titrating auxin production via the ASB1 gene. Auxin produced in the QC not only controls stem cell division activity by an as yet unknown mechanism, but at the same time acts as a long distance signal to fine tune the level of ARR1 transcription in the TZ, thus controlling the rate of differentiation. In this way a single gene, SCR, regulates the spatial coordination between stem cell division and differentiation ensuring coherent root growth.
EXPERIMENTAL PROCEDURES
Plant materials
The Arabidopsis thaliana ecotypes Columbia (Col-0), Wassilewskija (Ws) and Landsberg erecta (Ler) were used. ahk3-3, arr1-4, arr1-3, arr12-1, wei7-4 and wei7-1 mutants are in Col-0 background (Dello Ioio et al., 2007; Dello Ioio et al., 2008; Stepanova et al., 2005), shy2-31 mutant is in Ler background (Dello Ioio et al., 2008), scr-1 and scr-4 mutants are in Ws background (Fukaki et al., 1998; Sabatini et al., 2003). pAHK3::AHK3:GUS, pARR12::ARR12:GUS, pARR1::ARR1:GUS, pWOX5::GFP, QC25, QC46, DR5::GFP, pSCR::SCR:GFP, pWOX5:IAAH/DR5::GFP and pASB1::GUS transgenic plants have been described previously (Dello Ioio et al., 2007; Dello Ioio et al., 2008; Blilou et al., 2005; Sabatini et al., 2003; Stepanova et al., 2005; Ottenschläger et al., 2003). Q1630 and J0571 lines belong to Haseloff collection (http://www.plantsci.cam.ac.uk/Haseloff/assembly/page167/index.html).
Plant growth conditions
Seeds were surface sterilized using 50% bleach for 10 minutes and then rinsed four times with sterile water. After 5 days of cold treatment, A. thaliana seeds were plated and grown, in a near vertical position, at 22°C in long-day conditions (16 hours light/8 hours dark cycle) on 1/2 MS (Murashige & Skoog - Duchefa) supplemented with sucrose as previously described (Perilli and Sabatini, 2010). Plants for IAA measurement were grown as previously described (Ljung et al., 2005).
Root length and meristem size analysis
For root length measurements, plates were photographed and the resulting images were analyzed using the ImageJ software available on-line (http://rsbweb.nih.gov/ij/). For meristem size analysis, roots were prepared in a glass slide with chloralhydrate as described in Perilli and Sabatini, 2010, then observed using an Axio Imager.A2 (Zeiss) light microscopy. To measure root meristem size, the number of cortical cells along a cell file (from the QC to the first elongated cell) were counted. To measure root development over time, root length and meristem size were analyzed at different days post germination (dpg): 3 (dpg), 5dpg, 7dpg, 9dpg, 12dpg and 15dpg. For each experiment, at least 90 roots were analyzed, mean and standard deviation were calculated.
Hormonal treatments
5-day old seedlings were transferred to solid ½ MS medium containing mock conditions or a suitable concentration of hormone. For auxin treatment, we used indole-3-acetic acid (IAA, Duchefa) at a final concentration of 5 μM, prepared from a 10 mM stock in ethanol. Plants were treated for 2, 3 and 16 hours. For cytokinin treatment, we used trans-zeatin (tZ, Duchefa) at a final concentration of 5 μM, prepared from a 30 mM stock in 1N NaOH. Plants were treated for 4 hours. For auxin induction specifically within QC cells, plants carrying the pWOX5::IAAH construct, in wild type and in arr1-4 mutant background, were treated for 3 hours with 10 μM indole-3-acetamide (IAM, Sigma-Aldrich), a precursor of IAA, prepared from a 10 mM stock in DMSO. To inhibit auxin transport, plants were pretreated 1 hour with 50 μM N-naphthylphthalamic acid (NPA, Duchefa), and then treated for 3 hours with both 10 μM IAM and 50 μM NPA together. For each experiment, at least 90 roots were analyzed.
GUS histochemical assay
To visualize QC25, QC46, pASB1::GUS, pAHK3::AHK3:GUS pARR12::ARR12:GUS and pARR1::ARR1:GUS lines, GUS histochemical assay was performed using the ß-glucoronidase substrate X-gluc (5-bromo-4-chloro-3-indolyl glucuronide, Duchefa) dissolved in N-N-dimethyl-formamide. X-gluc solution, 100 mM Na2HPO4, 100 mM NaH2PO4, 0.5 mM K3 Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X100 and 1 mg/ml X-gluc, was prepared as previously described (Perilli and Sabatini, 2010). 5-day old seedlings were incubated for 16 hours at 37°C in the dark and imaged using the Axio Imager.A2 (Zeiss) microscopy. pASB1::GUS in Figure 4C was stained for 30 minutes only. For each experiment, at least 90 roots were analyzed.
Lugol staining
Starch granules in the root tips were stained with Lugol’s solution (Carlo Erba Reagenti) for few seconds and then immediately observed and photographed at microscopy as previously described (van den Berg et al., 1997; Sabatini et al., 2003).
Confocal image processing
Confocal images of median longitudinal sections of 5-day old roots were taken using a ZEISS LSM 780 microscope. A 10 μg/ml Propidium Iodide (Sigma) solution was used to visualize the cell wall.
Analysis of ARR1 expression in vivo using GUS histochemical assay
In vivo analysis of ARR1 expression in SCR:GR plants after Dex treatment were performed as follows: scr-4/pWOX5≫SCR:GR≫GFP;pARR1::ARR1:GUS were treated for 4, 8 and 24 hours with 2 μM Dex or an equivalent amount of DMSO as mock treatment. After Dex treatment, plants were stained 16 hours for the ß-glucoronidase activity of pARR1::ARR1:GUS transgene and visualized as described above. Starch granules were visualized and meristem cell number was counted as described above (Dello Ioio et al., 2007; Sabatini et al., 2003; Perilli and Sabatini, 2010). To analyze long distance effect of both auxin and polar auxin transport on ARR1, 5-day old pWOX5::IAAH;pARR1::ARR1:GUS plants were treated for 3 hours with 10 μM IAM and with both 10 μM IAM and 50 μM NPA as described above. After treatment, plants were stained for 16 hours for the ß-glucoronidase activity of pARR1::ARR1:GUS transgene and visualized as described previously (Dello Ioio et al., 2007; Perilli and Sabatini, 2010). Meristem cell numbers were counted as described above. For exogenous auxin application plants were treated for 3 hours with 5 μM IAA and stained as described above. For each experiment, at least 90 roots were analyzed.
FACS and qRT-PCR experiments
scr-4/pWOX5≫SCR:GR≫GFP seeds were plated, on top of nylon mesh (Nitex 03-100/44, Sefar America), on½ MS agar (+ 1% sucrose) plates at a density of approximately 500 seeds per row 5-day old seedlings were transferred onto plates containing 10 μM dexamethasone (Dex, Sigma-Aldrich), prepared from a 10 mM stock in dimethysulfoxid (DMSO), or an equivalent amount of DMSO, as mock treatment. Root tips (3–4 mm) were collected 4 hours after Dex treatment and protoplasted. QC cells expressing GFP were isolated on a fluorescence activated cell sorter (Becton Dickinson FACSAriaII) as described (Nawy et al., 2005). Sorted cells were collected directly into a RNA lysis buffer (Qiagen RLT buffer, RNeasy Micro kit), mixed and immediately frozen at −80°C. Experiments were carried out in two consecutive days, obtaining two biological replicates for each treatment (Mock and Dex). mRNA was later isolated and treated with DNase using TURBO DNA-free™ Kit. Each RNA sample was reverse transcribed using the SuperScript™ III first-strand Synthesis system (Invitrogen) according to the manufacturer’s instructions. qRT-PCR was performed in triplicates from each RNA sample and repeated twice using SYBR Green PCR Master mix (Applied Biosystems) with ViiA™ 7 Real-Time PCR system (Applied Biosystems). Expression levels were calculated relative to ACTIN2 using the 2−ΔΔct method. Statistical analysis was done in MS Exel (ANOVA: Single Factor) using p<0.05. Primers were designed according to the recommendations of Applied Biosystems. Quantitative RT-PCR (qRT-PCR) analysis was conducted using the gene-specific primers listed below:
ARR1 FW: TTGAAGAAACCGCGTGTCGTCT and ARR1 RV: CCTTCTCAACGCCGAGCTGATTAA for ARR1
ACT FW: CCTTCTCAACGCCGAGCTGATTAA and ACT RV: GTGGATTCCAGCAGCTTCCAT for ACTIN2
Chromatin immunoprecipitation followed by gene-specific quantitative real-time PCR (ChIP-qRT-PCR)
ChIP was conducted as in Sozzani et al., 2010 on either Col-0, as control, or pSCR::SCR:GFP/scr-4 5 days old dissected roots which included longitudinal sections comprised of 3–5 cell lengths from the QC as described in Brady et al., 2007. Immunoprecipitation was done using a rabbit polyclonal antibody to GFP (ab290; Abcam Ltd., Cambridge, United Kingdom). DNA from the ChIP, for both Col-0 and pSCR::SCR:GFP experiments, was individually amplified using a random-primer based genome amplification method according to the Agilent Mammalian ChIP-on-chip protocol with minor modifications. Briefly, chromatin from the Col-0 and pSCR::SCR:GFP was blunted using T4 DNA polymerase, then linkers were added using T4 DNA ligase (oligo JW102 5′-GCGGTGACCCGGGAGATCTGAATTC-3′ and oligo JW103 5′-GAATTCAGATC-3′). The IP was then amplified with 15 PCR cycles (1st LM-PCR) and then the reaction was diluted and used as template for a second round of 20 cycles (2nd LM-PCR). The DNA was cleaned up each time using the MiniElute Reaction Cleanup kit (Qiagen). Enrichment of the ARR1 putative target promoter-region DNA was determined using RT-qPCR. A qPCR efficiency of 2-fold amplifications per cycle was assumed, and sequences from ubiquitin 10 (UBQ10-F: GGCCTTGTATAATCCCTGATGAATAAG; UBQ10-R: AAAGAGATAACAGGAACGGAAACATAGT) were used to normalize the results between samples. Tiling along the At3G16857 ARR1 was done using the following sets of adjacent specific amplified regions along the putative 2186 kb region of the ARR1 promoter. In ascending order upstream from the ARR1 ATG:
100 - 0 bp: ARR1#6-F: TCGAATAAAACTCGCGTCAA; ARR1#6-R: ACCTCTCTCTATGTAGCTCGAACC
578 – 396 bp: ARR1#5-F: GCAACATCCAAAATGTTCTTCT; ARR1#5-R: GCTGACATGTCCTTTATTTTCATC
687 - 580 bp: ARR1#4-F: CCCACTTTCTTCCCAATTAT; ARR1#4-R: TGAGCTCAAATTCTGGGTTA
1120 - 1049 bp: ARR1#3-F: GTAGGTGATGATGGGAAATG; ARR1#3-R: TACGTCTACGCACATTCAAG
1568 - 1497 bp: ARR1#2-F: CATTCAGATGAACCATTTGC; ARR1#2-R: TGTGCGCTTCTTCTCTTAAA
2186 - 2100 bp: ARR1#1-F: GATAGATGGAGAGGTCGATG; ARR1#1-R: GCTGAGAAGCTTTCCCTATT
As negative control we used the following primers:
(+) 595 – 696 bp: ARR1#7-F: TCATGAGGTCCTTGTTGTCAG; ARR1#7-R: TGCATTTCGTTACTGCATGTC
(+) 2990 - 3061 bp: ARR1#8-F: CTTTGACGCATACTCCATCG; ARR1#8-R: GTAGTCCCTCCGGTTTATCG
Detailed descriptions of the transgenic lines construction and analysis, IAA measurement, ChIP techniques and transcript level analysis are provided in the Supplementary materials.
Supplementary Material
Acknowledgments
We thank Ben Scheres and Marta Del Bianco for reading the manuscript, José M. Alonso for providing material and Leonardo Giustini for technical support. This work was supported by The European Research Council (to S.S., L.M. and S.P.), by Grant BB/E024858 (Plant Stem Cell Network) within the European Research Area in Plant Genomics framework (I.T.), a Marie Curie European Reintegration Grant (B.R.), Giovanni Armenise-Harvard Foundation career development grant (R.S). Netherlands Genomics Initiative/Netherlands Organization for Scientific Research grants from the Centre for BioSystems Genomics (D.B.) and Horizon program (R.H.), and by Istituto Pasteur-Fondazione Cenci Bolognetti, MIUR PRIN and MIUR FIRB ERA-PG (to P.C.). The authors declare no competing financial interests.
Footnotes
Supplementary Information contains: Supplementary Figures and Legends 1-5; Supplementary Methods and additional References.
Supplementary Figure 1 provides additional data on the genetic relationship between, and the tissue specificity of, SCR and the cytokinin signaling pathway. It is related to main Figure 1 because it provides further evidence that SCR suppresses AHK3-mediated differentiation input in the QC cells.
Supplementary Figure 2 provides additional data on the genetic relationship between SCR and the cytokinin-response transcription factor ARR1. It is related to main Figure 2 because it provides further evidence that SCR specifically suppresses ARR1-mediated differentiation input indicating that ectopic expression of ARR1 in the scr QC induces cell differentiation in the SCN via a mechanism different from that in the TZ.
Supplementary Figure 3 provides additional data on the molecular relationship between SCR and ARR1. It is related to main Figure 3 because it provides further evidence that SCR directly suppresses ARR1 in the QC cells.
Supplementary Figure 4 shows how SCR maintains SCN activity. It is related to main Figure 4 since it further confirms that SCN and meristem defects of scr mutant are dependent on the ARR1-mediated control of ASB1 expression.
Supplementary Figure 5 shows that both SCR and ARR1 are auxin inducible. It is related to main Figure 5 because it provides further evidence that SCR controls ARR1 at the TZ from the QC, via auxin.
References
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433:39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci USA. 1994;91:6649–6653. doi: 10.1073/pnas.91.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vatén A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell. 2012;150:1002–1015. doi: 10.1016/j.cell.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Linhares FS, Scacchi E, Casamitjana-Martinez E, Heidstra R, Costantino P, Sabatini S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr Biol. 2007;17:678–682. doi: 10.1016/j.cub.2007.02.047. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Galinha C, Fletcher AG, Grigg SP, Molnar A, Willemsen V, Scheres B, Sabatini S, Baulcombe D, Maini PK, et al. A PHABULOSA/cytokinin feedback loop controls root growth in Arabidopsis. Curr Biol. 2012;22:1699–1704. doi: 10.1016/j.cub.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Ding Z, Friml J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci USA. 2010;107:12046–12051. doi: 10.1073/pnas.1000672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J. Subcellular trafficking of PIN auxin efflux carriers in auxin transport. Eur J Cell Biol. 2010;89:231–235. doi: 10.1016/j.ejcb.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Fukaki H, Wysocka-Diller J, Kato T, Fujisawa H, Benfey PN, Tasaka M. Genetic evidence that the endodermis is essential for shoot gravitropism in Arabidopsis thaliana. Plant J. 1998;14:425–430. doi: 10.1046/j.1365-313x.1998.00137.x. [DOI] [PubMed] [Google Scholar]
- Heidstra R, Welch D, Scheres B. Mosaic analyses using marked activation and deletion clones dissect Arabidopsis SCARECROW action in asymmetric cell division. Genes Dev. 2004;18:1964–1969. doi: 10.1101/gad.305504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–80. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988;15:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- Jones B, Gunnerås SA, Petersson SV, Tarkowski P, Graham N, May S, Dolezal K, Sandberg G, Ljung K. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell. 2010;22:2956–2969. doi: 10.1105/tpc.110.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Kim B, Song SK, Heo JO, Yu NI, Lee SA, Kim M, Kim DG, Sohn SO, Lim CE, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:659–670. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell. 2005;17:1090–1104. doi: 10.1105/tpc.104.029272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal BC, Mukherjee T, Mandal L, Evans CJ, Sinenko SA, Martinez-Agosto JA, Banerjee U. Interaction between differentiating cell- and niche-derived signals in hematopoietic progenitor maintenance. Cell. 2011;147:1589–1600. doi: 10.1016/j.cell.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Dello Ioio R, Di Mambro R, Costantino P, Sabatini S. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. CurrBiol. 2010;20:1138–1143. doi: 10.1016/j.cub.2010.05.035. [DOI] [PubMed] [Google Scholar]
- Nawy T, Lee JY, Colinas J, Wang JY, Thongrod SC, Malamy JE, Birnbaum K, Benfey PN. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell. 2005;17:1908–1925. doi: 10.1105/tpc.105.031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L, Di Mambro R, Sabatini S. Cytokinin-auxin crosstalk. Trends Plant Sci. 2009;14:557–562. doi: 10.1016/j.tplants.2009.06.010. [DOI] [PubMed] [Google Scholar]
- Ottenschläger I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, Ishikawa H, Evans M, Palme K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci USA. 2003;100:2987–2991. doi: 10.1073/pnas.0437936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S, Moubayidin L, Sabatini S. The molecular basis of cytokinin function. Curr Opin Plant Biol. 2010;13:21–26. doi: 10.1016/j.pbi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–119. doi: 10.1046/j.1365-313x.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey PN, Leyser O, Bechtold N, Weisbeek P, et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell. 1999;99:463–72. doi: 10.1016/s0092-8674(00)81535-4. [DOI] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003;17:354–358. doi: 10.1101/gad.252503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R. Plant stem cell niches: from signalling to execution. Curr Opin Plant Biol. 2010;14:4–9. doi: 10.1016/j.pbi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- Scheres Stem-cell niches: nursery rhymes across kingdoms. Nature Reviews Molecular Cell Biology. 2007;8:345–354. doi: 10.1038/nrm2164. [DOI] [PubMed] [Google Scholar]
- Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jürgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–91. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 2009;21:1495–1511. doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- Wolpert L, Tickle C. Principles of development. New York: Oxford University Press; 2011. [Google Scholar]
- Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.