Abstract
STUDY QUESTION
Can we make the comet assay (single-cell gel electrophoresis) for human sperm a more accurate and informative high throughput assay?
SUMMARY ANSWER
We developed a standardized automated high throughput comet (HT-COMET) assay for human sperm that improves its accuracy and efficiency, and could be of prognostic value to patients in the fertility clinic.
WHAT IS KNOWN ALREADY
The comet assay involves the collection of data on sperm DNA damage at the level of the single cell, allowing the use of samples from severe oligozoospermic patients. However, this makes comet scoring a low throughput procedure that renders large cohort analyses tedious. Furthermore, the comet assay comes with an inherent vulnerability to variability. Our objective is to develop an automated high throughput comet assay for human sperm that will increase both its accuracy and efficiency.
STUDY DESIGN, SIZE, DURATION
The study comprised two distinct components: a HT-COMET technical optimization section based on control versus DNAse treatment analyses (n = 3–5), and a cross-sectional study on 123 men presenting to a reproductive center with sperm concentrations categorized as severe oligozoospermia, oligozoospermia or normozoospermia.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Sperm chromatin quality was measured using the comet assay: on classic 2-well slides for software comparison; on 96-well slides for HT-COMET optimization; after exposure to various concentrations of a damage-inducing agent, DNAse, using HT-COMET; on 123 subjects with different sperm concentrations using HT-COMET. Data from the 123 subjects were correlated to classic semen quality parameters and plotted as single-cell data in individual DNA damage profiles.
MAIN RESULTS AND THE ROLE OF CHANCE
We have developed a standard automated HT-COMET procedure for human sperm. It includes automated scoring of comets by a fully integrated high content screening setup that compares well with the most commonly used semi-manual analysis software. Using this method, a cross-sectional study on 123 men showed no significant correlation between sperm concentration and sperm DNA damage, confirming the existence of hidden chromatin damage in men with apparently normal semen characteristics, and a significant correlation between percentage DNA in the tail and percentage of progressively motile spermatozoa. Finally, the use of DNA damage profiles helped to distinguish subjects between and within sperm concentration categories, and allowed a determination of the proportion of highly damaged cells.
LIMITATIONS, REASONS FOR CAUTION
The main limitations of the HT-COMET are the high, yet indispensable, investment in an automated liquid handling system and heating block to ensure accuracy, and the availability of an automated plate reading microscope and analysis software.
WIDER IMPLICATIONS OF THE FINDINGS
This standardized HT-COMET assay offers many advantages, including higher accuracy and evenness due to automation of sensitive steps, a 14.4-fold increase in sample analysis capacity, and an imaging and scoring time of 1 min/well. Overall, HT-COMET offers a decrease in total experimental time of more than 90%. Hence, this assay constitutes a more efficient option to assess sperm chromatin quality, paves the way to using this assay to screen large cohorts, and holds prognostic value for infertile patients.
STUDY FUNDING/COMPETING INTEREST(S)
Funded by the CIHR Institute of Human Development, Child and Youth Health (IHDCYH; RHF 100625). O.A. is a fellow supported by the Fonds de la Recherche du Québec - Santé (FRQS) and the CIHR Training Program in Reproduction, Early Development, and the Impact on Health (REDIH). B.R. is a James McGill Professor. The authors declare no conflicts of interest.
Keywords: sperm chromatin quality, sperm DNA damage, high throughput comet assay, fertility, DNA damage profiling, fertility prognosis
Introduction
Since 1980, the World Health Organization (WHO) has developed and updated standard semen reference values and operating procedures to assess sperm quality based on concentration, morphology and motility (World Health Organization, 2010). While fundamental to establish an initial diagnosis, these criteria have long been shown to be poor predictors of fertility, in part due to the inherent geographical, seasonal and intra-individual heterogeneity of human semen (Lewis, 2007). More importantly, these parameters do not assess the extent of any damage in the sperm nuclear material. In the era of assisted reproductive technologies, several studies have shown an association between sperm DNA integrity and fertility, success of IVF, pregnancy rates and outcomes (Zini et al., 2014). Recently, increased sperm DNA damage in men has been shown to adversely affect embryo quality starting at Day 2 of early embryonic development and continuing after embryo transfer, resulting in reduced implantation rates and pregnancy outcomes (Simon et al., 2014), further suggesting a prognostic value of sperm chromatin quality assays.
A number of sperm chromatin quality and DNA integrity assays allow for the detection of chromosomal aberrations, DNA fragmentation and compaction and protamination defects (Delbes et al., 2009; Shamsi et al., 2011). Among those, the single-cell gel electrophoresis, or comet assay, gives a comprehensive measure of sperm DNA integrity by revealing, under specific treatment conditions, single and double strand DNA breaks and alkali-labile sites (Singh et al., 1988). This technique involves the embedding of spermatozoa in low melting point agarose followed by a number of treatments designed to induce cell lysis and DNA unwinding. After a brief electrophoresis, each cell is stained with a fluorescent dye and produces a structure resembling a comet, consisting of a round shaped head made of high molecular weight undamaged DNA, and a tail containing the leading ends and lower heterogeneous molecular weight fragments of damaged DNA. Each comet is given a score called the tail extent moment (TEM), a product of the percentage DNA in the tail (accounting for the proportion of damaged DNA) and the tail length (determined by the relative size of DNA fragments); however, while TEM is theoretically more informative, it is poorly standardized across laboratories because it has no generally accepted units. Hence, for the purpose of this study, both the percentage DNA in the tail and the TEM will be given.
Of all sperm DNA integrity test procedures, the comet assay offers the unique feature of collecting data at the level of the single cell: the assessment of 50 individual comets is enough to deliver accurate information on sperm quality (Olive and Banáth, 2006), opening the possibility of analysis of severely oligozoospermic patients. However, this feature, paired with the 2-well output of the classic comet assay, makes comet assessment a low throughput procedure that renders analyses of large cohorts very challenging. Furthermore, due to the high sensitivity of unwound DNA and of the multiple technical steps of the assay, the comet assay comes with an inherent vulnerability to variability that calls for standardization (Speit et al., 2009; Zini et al., 2014). The objective of this study is to develop a standardized high throughput comet assay for human sperm with improved accuracy and reproducibility that will provide additional information for the diagnosis of DNA integrity.
Materials and Methods
Subject recruitment
Approval to conduct this study has been obtained from the McGill University Hospital Centre Institutional Review Board (A03-M27-10B). Informed consent was obtained from a series of men presenting to the McGill University Hospital Reproductive Center.
Sperm samples
Fresh ejaculated semen samples from subjects aged 18–54 years old (average ± SEM: 29.80 ± 0.68) were obtained and analyzed according to the WHO guidelines (2010). All semen samples were collected and frozen without freezing media in 500 µl aliquots by immediate storage at −80°C. Detailed information on patients is given in Supplementary data, Table SI.
Sample preparation
Frozen semen samples were thawed at room temperature, spun at 500 × g for 5 min at 4°C in a fixed-angle rotor centrifuge, and seminal plasma was removed and replaced by an equivalent volume of filtered 1× phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl; 10 mM Na2HPO4; 2 mM KH2PO4; pH 7.4). Each sample was diluted to a concentration of 1.5 × 105 cells/ml in PBS, aliquoted and stored at −80°C until further use. This concentration was determined to be optimal for comet density and the avoidance of abundant overlapping objects. For a positive control, samples were spun at 500 × g for 5 min at 4°C, all of the seminal plasma was removed and replaced by an equivalent volume of DNAse solution [30 mM Tris–HCl; 4 mM MgCl2; 1 mM dithiothreitol (DTT; Roche Diagnostics, Basel, Switzerland); 100 Kunitz units (KU)/ml DNAse (Sigma Aldrich®, Saint Louis, MO, USA)]. Tubes were placed at 37°C for 20 min, spun at 500 × g for 5 min and washed with 1× PBS. The washing step was repeated three times. The number of sperm in each sample was counted using a hemocytometer; the sample was diluted to a concentration of 1.5 × 105 cells/ml in PBS, aliquoted and stored until further use at −80°C. Sperm from a normozoospermic patient with both sperm concentration and percentage progressive motility above the 75th percentile of the WHO reference ranges of fertile men was used as a control across experiments to ensure inter-experimental reproducibility (Patient #1; Supplementary data, Table SI).
2-Well. comet assay
Low melting point agarose (0.5%) (Sigma Aldrich®) was prepared in PBS using a 85°C water bath for 1 h and cooled down to 37°C in an incubator overnight. Fifty microliters of each sample were diluted in 500 µl of 37°C warm low melting point agarose, and 50 µl of this mix were immediately spread on a 2-well CometSlide™ (Trevigen®, Gaithersburg, MD, USA). The slides were left on ice for 20 min and subsequently immersed in the following solutions: (i) a lysis solution (2.5 M NaCl; 0.5 M EDTA; 10 mM Tris; pH 10) containing 10% dimethylsulfoxide (DMSO; Fisher Scientific, Pittsburgh, PA, USA), 1 mM DTT and 1% Triton X-100 (Sigma Aldrich®) for 60 min at 4°C; (ii) a 4 mM lithium 3,5-diiodosalicylate (LIS; Sigma Aldrich®) solution for 90 min at room temperature; (iii) an alkaline solution (1 mM EDTA; pH 12.1) for 45 min at room temperature. For the neutral comet assay, the alkaline solution was replaced with 1× Tris Borate EDTA buffer (TBE; 89 mM Tris; 89 mM boric acid; 2 mM EDTA; pH 7.4). Electrophoresis was then carried out using the CometAssay® horizontal electrophoresis system (Trevigen®) at 4°C at 0.7 V/cm for 5 min in 1× TBE (pH 8). Slides were fixed in chilled 70% ethanol for 5 min, air-dried at room temperature, and protected from light for 48–72 h prior to scoring to bring all comets to the same focal plane. Due to light sensitivity, all steps from lysis onward were executed in a dark room.
High throughput comet assay (HT-COMET)
Low melting point agarose was prepared as described above and 500 μl were distributed in each well of a Costar™ 96-deep-well plate (Fisher Scientific, Pittsburgh, PA, USA). The agarose was kept warm with an INHECO CPAC heating block (Inheco, Martinsried, Germany). Diluted sperm samples were randomly distributed in a Costar™ 96-well cell culture plate (Fisher Scientific) and kept on ice until mixing. The Janus® automated workstation (PerkinElmer®, Waltham, MA, USA) was programmed to perform the following steps: (i) pipet 50 μl of diluted sperm from the 96-well culture plate; (ii) dispense these 50 μl into 37°C warm low melting point agarose in the 96-deep-well plate; (iii) pipet five times up and down to ensure proper mixing; (iv) dispense 80 μl of mix per well on a 96-well CometSlide™ (Trevigen®) to ensure the covering of the whole well; (v) pipet back 60 μl to leave 20 μl of evenly spread mix on each well. From that point on, the 96-well CometSlides™ underwent the same treatment as the 2-well slides.
Imaging
Comets were stained using 50 or 20 μl of 1:10 000 SYBR® Gold nucleic acid gel stain (Life Technologies, Carlsbad, CA, USA) in TBE for 2- and 96-well slides, respectively. Comets were imaged using the fully automated Operetta® high content imaging system (PerkinElmer®). Images were acquired using a 10×, 0.3NA air objective. About 100 and 15 pictures/well were taken for 2- and 96-well slides, respectively.
Scoring
Images were analyzed with the Columbus™ software (PerkinElmer®). The analytical part of the software consists of analysis building blocks that can be arranged in sequence to generate a complete analysis. Building blocks for object identification can be customized by choosing between different algorithms and adjusting thresholds and expected object size parameters. To identify and analyze comets, nuclei were first identified as primary objects. Intensity and morphological properties were then calculated to be able to filter out debris and other false-positive objects. Comet tails were identified as secondary objects using the find cytoplasm building block. This building block searches for areas with above-background signal around each identified nucleus. Morphological properties of the tail were then calculated and used to filter out false-positive tail identifications. Incomplete comets at the image boundaries were also eliminated. To calculate the raw tail length in each identified comet the length of the nucleus was subtracted from the ‘cytoplasmic’ length. The percentage of nuclear DNA was calculated by dividing the sum of pixel-intensities in the nucleus by the combined sum of pixel-intensities in nucleus and tail. The proportion of signal in the tail was calculated accordingly. TEM was calculated as the proportion of cytoplasmic (tail) intensity multiplied by tail length. The detailed sequential analysis is available in Supplementary data, Table SII; a Columbus™ compatible file is available upon request. For a given analysis, the inter- and intra-assay coefficients of variation of the Columbus™ software are equal to 0%. For software comparison (Fig. 1), all comets that were imaged with the Operetta® high content imaging system and scored with Columbus™ were also scored using the Komet™ 6 (Andor Technology, Belfast, UK) acquisition and analysis software.
Figure 1.
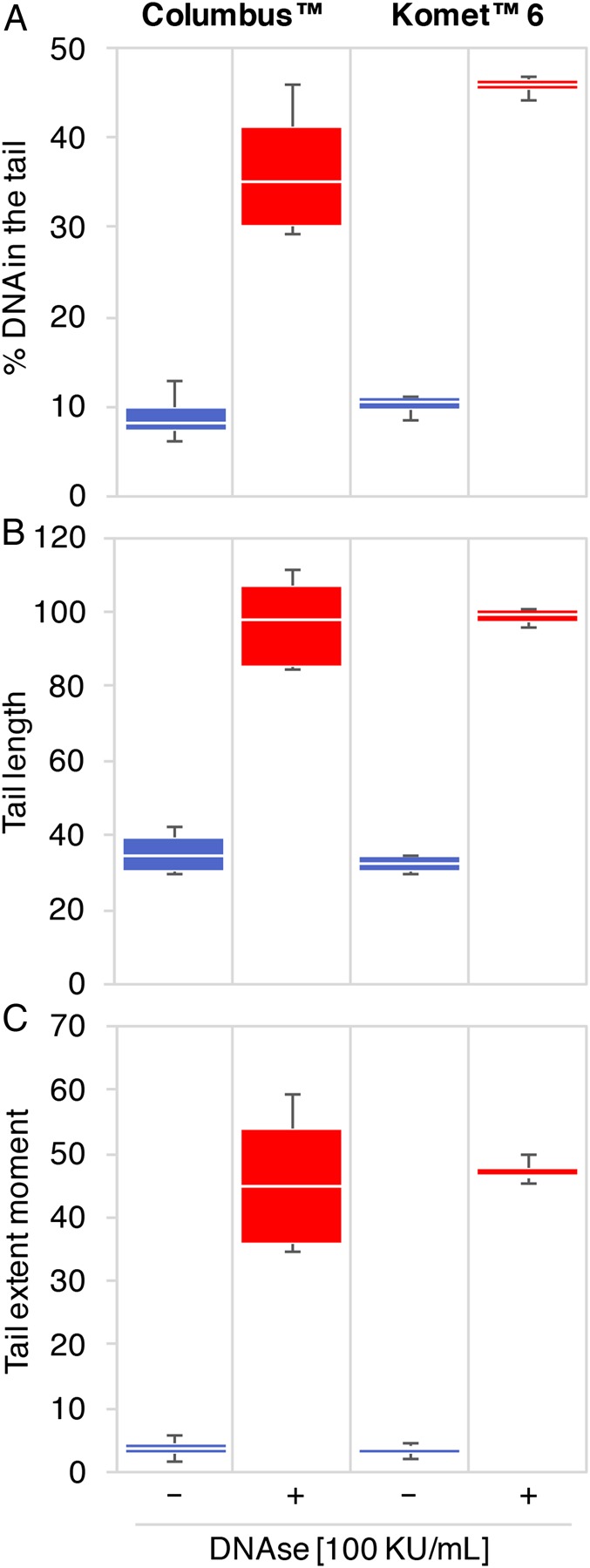
Software comparison for comet scoring. Percentage DNA in the tail (A), tail length (B) and TEM (C) of comets scored with the software program Columbus™ (left panel) or Komet™ 6 (right panel). Spermatozoa from patient #1 were either left untreated (blue bars) or treated with 100 KU/ml DNAse (red bars) (n = 4). Comets from 2-well slides were imaged using the Operetta® system and scored with both software programs. No significant difference was detected between software programs within treatment groups using the non-parametric Mann–Whitney test.
Statistical analyses
Statistical analyses were conducted using GraphPad Prism® 6 (GraphPad Software, Inc., La Jolla, CA, USA). Software equivalence was assessed by comparing comet tail length, percentage of DNA in the tail and TEM using the Mann–Whitney non-parametric test; HT-COMET score homogeneity across the slide was assessed using the Kruskal–Wallis non-parametric test; effects of exposure to increasing concentrations of DNAse were assessed using the Kruskal–Wallis non-parametric test and post hoc Dunn's multiple comparison test on selected comparisons; correlations between sperm DNA damage and classic semen parameters were assessed using the Spearman's rank correlation coefficient. In Figs 1–3, data are presented as box and whisker plots and tests were performed on medians. Data were expressed as mean and SEM; in Fig. 4, data are expressed as average relative to control; in Fig. 5, single-cell data are represented; in Supplementary data, Fig. S1, data are expressed as mean ± SEM. A P value <0.05 was considered as significant.
Figure 3.
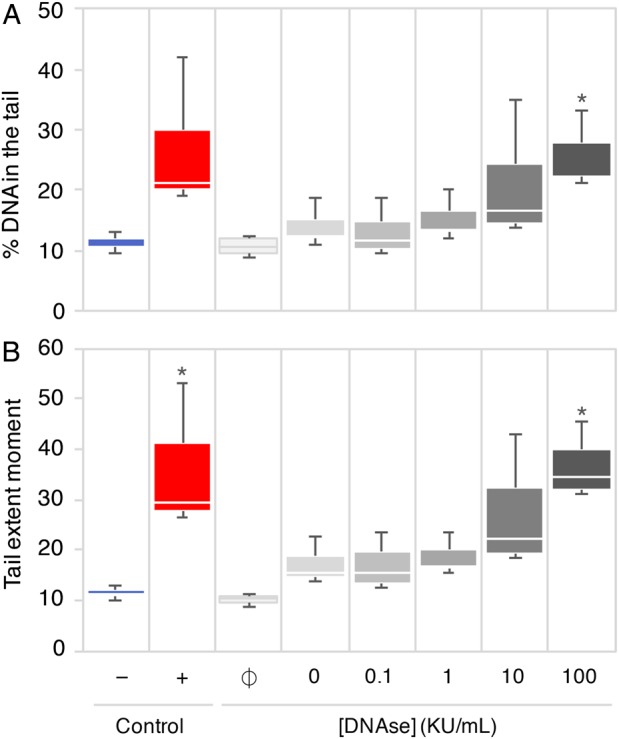
Detection of DNAse-induced damage to human sperm DNA with the high throughput comet assay. Percentage DNA in the tail (A) and TEM (B) of comets after exposure to increasing concentrations of DNAse. The negative (−) and positive (+) control conditions correspond to the sperm of patient #1 untreated or treated with 100 KU/ml DNAse, respectively. Spermatozoa from patient #2 were either untreated (ϕ) or treated with 0, 0.1, 1, 10 or 100 KU/ml DNAse (n = 3). Sperm from patients #1 and #2 treated with 100 KU/ml DNAse displayed significantly higher DNA damage compared with their untreated equivalent using the non-parametric Kruskal–Wallis test (*P < 0.05).
Figure 4.
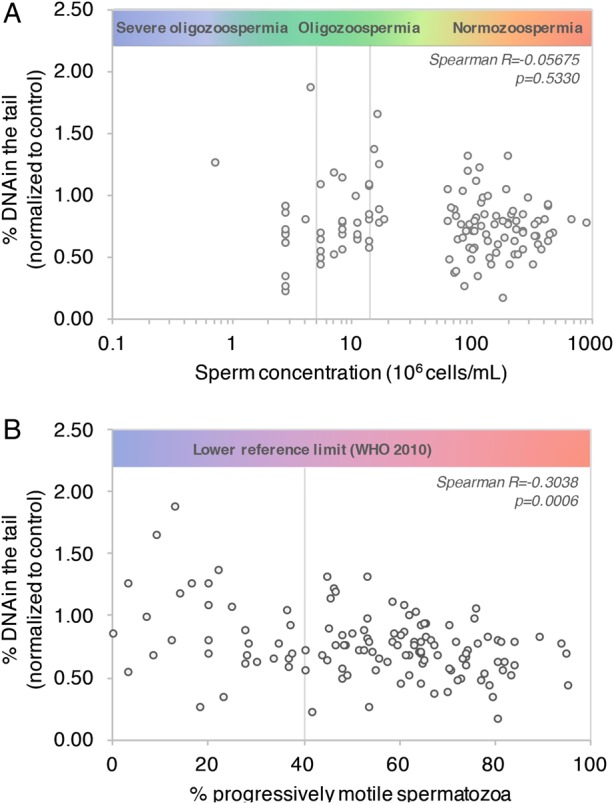
Correlation between DNA damage and World Health Organization recommended semen characteristics. Percentage DNA in the tail (normalized to control patient #1) was plotted as a function of sperm concentration (A) and percentage of progressively motile spermatozoa (B). Percentage progressively motile sperm was calculated as the sum of rapid and slow progressive motility. n = 123 patients. Spearman's rank test correlation coefficient and P value are indicated in italic.
Figure 5.
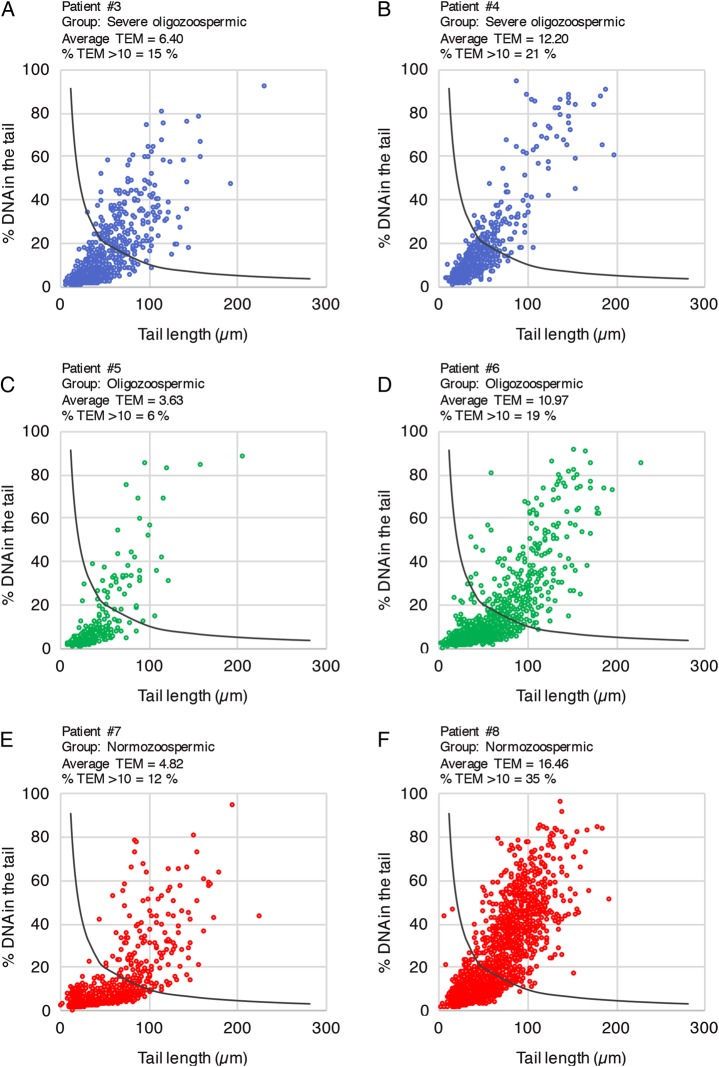
DNA damage profiling using the automated alkaline HT-COMET. Comet data from individual spermatozoa from selected severe oligozoospermic (A and B), oligozoospermic (C and D) and normozoospermic (E and F) patients were used to produce individual DNA damage profiles as a proposed tool for fertility prognosis. n = 6 wells from one 96-well slide were plotted for each patient. The dark curve identifies the limit of a TEM equal to 10 as a threshold for high damage: every dot above the curve represents a highly damaged cell. The patient number, fertility group, average TEM and percentage of highly damaged cells within the total population of comets (% TEM >10) are indicated above each profile.
Results
Automated scoring with Columbus™ compared with semi-manual scoring with Komet™ 6
To evaluate the accuracy of comet automated scoring, spermatozoa from patient #1 were either left untreated or treated with 100 KU/ml DNAse to create artificial DNA breaks, and run on classic 2-well comet slides. Comets were either scored automatically, using our custom Columbus™ analysis, or semi-manually, using the widely used Komet™ 6 software. We scored an average ± SEM of 326 ± 49 comets and 396 ± 71 comets per well with Komet™ 6 and Columbus™, respectively, in untreated conditions. In DNAse-treated samples, we scored an average ± SEM of 163 ± 6 comets using Komet™ 6 and of 149 ± 11 comets using Columbus™. Median values for comet percentage DNA in the tail (Fig. 1A), tail length (Fig. 1B) and TEM (Fig. 1C) were not significantly different between the two software analyses.
Automated HT-COMET assay
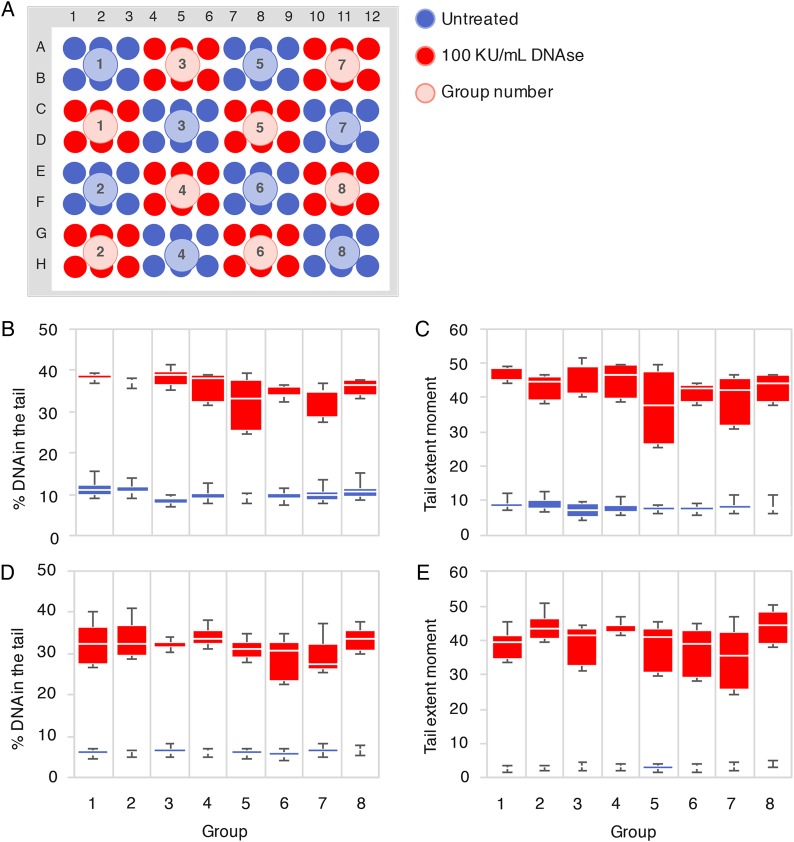
The transition from 2- to 96-well slides was done using a patterned plate map to test for the consistency of DNA damage measurements across the slide (Fig. 2A). Group wise manual dispensation of the mix of sperm and low melting point agarose led to artifactual trends in DNA damage as we progressed along the slide (Supplementary data, Fig. S1). Multiple attempts and observation led us to hypothesize that this phenomenon was due to a technical limiting factor: by the time the experimenter had dispensed the mix in the last well, the agarose in the first well had shrunk and polymerized, creating a gradient of agarose texture across the slide. The use of a multichannel pipet or of a heating block during distribution did not prevent these artifacts. As a consequence, we robotized the mixing and dispensation steps of the experiment so that all 96 wells were managed at the same time. This produced even results across the slide in both alkaline (Fig. 2B and C) and neutral (Fig. 2D and E) conditions. Compared with neutral treatment, alkaline processing of the same samples produced comets that had 2.42 ± 0.18 times higher average TEM in the untreated group, and only 1.10 ± 0.04 times in the DNAse-treated group. The generation of longer comets in alkaline conditions is well documented in the literature and is due to the unravelling of alkali-labile sites (such as apurinic and apyrimidinic sites which arise from the loss of a damaged base) and further detachment of DNA strand breaks from the nuclear matrix (Karlsson, 2010). However, after exposure to 100 KU/ml of DNAse, the generation of innumerable small fragments of DNA detached from the nuclear matrix by nature produces comparable comets in both neutral and alkaline conditions.
Figure 2.
Automated 96-well high throughput comet assay (HT-COMET). (A) Each well was filled with a mix of low melting point agarose and either untreated or DNAse-treated (100 KU/ml) spermatozoa from patient #1 in eight groups of six wells. Median values of percentage DNA in the tail (B) and TEM (C) per well per group after an alkaline comet assay (n = 5) are shown. Median values of percentage DNA in the tail (D) and TEM (E) per well per group after a neutral comet assay (n = 3) are shown. No significant difference between groups was detected using the non-parametric Kruskal–Wallis test.
HT-COMET assay detects a wide range of DNA damage
To ensure that HT-COMET would allow for the detection of a wide range of DNA damage, we induced increasing amounts of DNA damage by treating spermatozoa from patient #2 (Supplementary data, Table SI) with progressively higher concentrations of DNAse (Fig. 3A and B). Untreated and DNAse-treated sperm from patient #1 were used as negative and positive controls, respectively. The automated HT-COMET accurately detected increasing DNA damage.
DNA damage does not correlate with classical sperm quality parameters
We conducted a study on 123 patients with different sperm concentrations and motility. Samples were run in two replicate wells of 96-well slides. Our analysis allowed for the detection of an average (±SEM) of 104 ± 9 comets per well across fertility groups; there was no correlation between the number of comets detected and sperm concentration (data not shown). No significant correlation was found between the percentage DNA in the tail (standardized to untreated control patient #1) and sperm concentration (Fig. 4A; Spearman R=−0.05675; P = 0.5330). A significant correlation between percentage DNA in the tail and percentage of progressively motile spermatozoa was found (Fig. 4B; Spearman R=−0.3038; P = 0.0006).
DNA damage profiling
By using single-comet data obtained with our automated analysis, we plotted a DNA damage profile for two severe oligozoospermic (Fig. 5A and B), two oligozoospermic (Fig. 5C and D) and two normozoospermic patients (Fig. 5E and F). This allowed us to visualize the extent and heterogeneity of DNA damage in the whole spermatozoa population and evaluate the proportions of highly damaged cells. For that purpose, we arbitrarily chose a TEM of 10. We plotted this threshold as a black curve on each profile (y = 10/x): every dot above the curve represents a highly damaged cell. For a clinical use of the DNA damage profiling, a threshold would have to be defined from a large cohort of proven biological fathers.
DNA damage profiling allowed us to do three levels of analysis: across fertility groups, within a fertility group and within a patient's spermatozoa population. For instance, comparison between fertility groups revealed that severe oligozoospermic patient #3 (panel A) displayed 15% of highly damaged cells versus 35% for normozoospermic patient #8 (panel F), exemplifying the absence of correlation between sperm count and sperm chromatin quality.
Comparison within fertility groups revealed highly heterogeneous profiles. Hence, oligozoospermic patients #5 (panel C) and #6 (panel D) displayed a percentage of cells above threshold of 6 and 19%, respectively, with corresponding average TEMs of 3.63 and 10.97. Similarly, normozoospermic patients #7 (panel E) and #8 (panel F) produced 12 and 35% of highly damaged cells, with corresponding average TEM of 4.82 and 16.46, respectively. Interestingly, the DNA damage profile of severely oligozoospermic patient #4 (panel B) revealed that a small subpopulation of highly damaged cells accounted for the elevated TEM, while severely oligozoospermic patient #3 (panel A) displayed a more continuous pattern. These three examples highlight the value of HT-COMET DNA damage profiling as a visual tool to discriminate patients with theoretically comparable sperm quality parameters.
Discussion
Since the early 2000s, multiple attempts have been made to circumvent the inherent low throughput and laboriousness of the comet assay, mostly within the frame of genotoxicity screening of drug compounds in somatic cells. The concept of a multigel assay arose in 2007 (Witte et al., 2007). The first high throughput comet protocol for somatic cells was meant to avoid cell trypsinization and involved cell culture within multichamber plates with removable walls followed by agarose coating (Stang and Witte, 2009). The use of manually dispensed minigels on GelBond® sheets was also proposed (Gutzkow et al., 2013). More recently, homemade micropatterned arrays allowing single-cell trapping were developed and elaborated into CometChip arrays (Wood, 2010; Weingeist et al., 2013; Ge et al., 2015). The CometChip array technique involved the automatic detection and scoring of comets using a suite of custom analyses coded in MATLAB®. However, all of the above-mentioned methods require intensive manual handling of both the cells and the agarose film, which can be a compounding factor for variability. More importantly, none of these methods were specifically developed for the chromatin of spermatozoa, the condensed and insoluble nature of which requires a specific treatment.
Spermatozoa were initially described to exhibit higher baseline levels of DNA migration than somatic cells with the comet assay (Singh et al., 1989). This, combined with the extreme sensitivity of the assay to the approach used to decondense sperm chromatin (Speit et al., 2009), gave the comet assay an inherent high degree of variability. One of the most critical and debated parameters in sperm chromatin decondensation is pH: erroneous attempts to associate the nature of DNA breaks with pH conditions are often made (Collins et al., 2008), and there are nearly as many different protocols as there are laboratories. For the purpose of this study, denaturation was carried out at pH 12.1 for alkaline and pH 7.4 for neutral conditions, respectively; the alkaline condition has been shown to be more sensitive than the neutral one (Collins, 2004). Due to the highly compacted nature of sperm chromatin, another fundamental aspect of the sperm comet assay is the introduction of additional treatment steps to initiate decondensation of the chromatin and allow the migration of DNA from the nucleus. While many protocols initially used exposure to proteinase K at either room temperature or 37°C for 1–18 h (Speit et al., 2009), more recent studies use a 90-min exposure to the chaotropic agent lithium 3,5-diiodosalicylate (LIS) at room temperature right after lysis. LIS appeared to us to be the better option as it shortens and simplifies the process, prevents agarose softening as it is active at room temperature, and was proved to give lower background levels of DNA migration in alkaline conditions (Donnelly et al., 1999).
Although treatment with different enzymes or at different pH is often identified as a major factor in comet assay variability, another major variable is often overlooked. Indeed, agarose evenness across and between the slides is a sine qua non-condition to reach reproducibility and accuracy. Heterogeneous agarose levels and textures can lead to artifactual trends in DNA damage. Homogeneous distribution of agarose is ensured by a strict control of agarose temperature when mixed with sperm and pipetted onto the slide, and a quick dispensation of the mix across conditions. While this is easily achieved on 2-well slides, it proved to be impossible for HT-COMET without the use of a deep-well plate temperature control unit and a robotic dispensation of the mix. Hence, we used an automated liquid handling workstation to perform a controlled mixing of sperm and low melting point agarose by programming five gentle up-and-down aspiration/dispensation cycles at exactly 37°C, circumventing experimenter variability and avoiding the creation of additional damage to the sperm chromatin by harsh pipetting. The automated workstation then dispensed the mix in all wells simultaneously. The automation of the process proved to be both efficient and time-saving, cutting down mixing and dispensation time to 75 s. However, it requires a major and yet unavoidable investment in equipment and software, which may limit the accessibility of HT-COMET.
The analysis of traditional comets is a tedious process involving manual selection, focusing, imaging and semi-automated scoring. This substantial task accounts for a significant part of the classical comet low throughput. In this study, we use a conventional, fully integrated high content screening setup for automatic detection and scoring of comets and demonstrate its comparability with the widely used Komet™ software. It is interesting to note the recent publication of the open-source software OpenComet (Gyori et al., 2014), which can be used for comet analysis as a plug-in in Image J; however, the limitation of this configuration is that the software only allows the analysis of a few images at a time which makes the scoring of a complete 96-well plate time consuming and laborious.
Using 123 subjects, we found no significant correlation between sperm chromatin quality and sperm concentration. We therefore confirmed the existence of hidden abnormalities in the semen of men with apparently normal semen parameters (Saleh et al., 2002), further stressing the poor discriminative power of classical semen analysis and the need to include sperm chromatin quality assays as independent predictors of fertility (Giwercman et al., 2010). Interestingly, sperm motility displayed a significant inverse correlation with sperm chromatin quality. If this corroborates studies pointing at sperm motility as an accurate criterion in the evaluation of infertility (Nallella et al., 2006), the predictive value of sperm motility and other WHO parameters is still very much debated (Lewis, 2007).
We used the automated HT-COMET assay developed here to illustrate the usefulness of DNA damage profiling as a potential prognostic tool for infertility patients. By plotting the percentage of DNA in the tail against tail length, we can visualize a patient's sperm chromatin damage profile, determine the proportions of highly damaged cells and ultimately get a sense of the nature and extent of the chromatin damage. Indeed, the shape of each scatter plot can ultimately inform on the overall nature of DNA damage within a sample: a trend towards a horizontal cloud suggests longer comets, i.e. multiple smaller pieces of DNA migrating further, while a convergence towards the vertical axis reveals the presence of short DNA rich comet tails, i.e. of longer fragments of DNA. Because sperm chromatin damage has been shown to affect embryo quality, development, transfer and implantation as well as pregnancy rates in humans (Simon et al., 2014), we propose that automated HT-COMET DNA damage profiling could be of use in guiding patients' options when considering assisted reproductive technologies.
By circumventing the major disadvantages of the classical comet assay, the high throughput comet assay pushes the comet method to another level of public health interest. One of the main drawbacks of the classic comet assay is its low throughput, attributable to both technical (single and 2-well slides) and data extraction (manual or semi-automated scoring of comets) requirements. With HT-COMET, the use of 96-well slides, paired with the three-slots tray of the horizontal electrophoresis system, allows an increase in sample size by 14.4-fold if using singlet testing. The automation of the most sensitive steps of the assay limits experimenter variability and ensures reproducibility across cohorts. More importantly, the automation of image acquisition, comet detection and scoring considerably reduces total experimental timing, with 1440 images being taken and scored in <100 min, cutting down the total analysis time to 1.0 min/well versus 20–30 min on a classical manual 2-well setting. Hence, when used at its full capacity of 144 samples in duplicate and compared with the classical 2-well comet assay, HT-COMET reduces agarose handling, mixing and distribution time by 79%, treatment procedure time by 93% and imaging and scoring time by 95%, reducing overall experimental time by more than 90%. Further optimization should allow us to bring HT-COMET to its full potential by extending the analysis to any cell type in any species.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
O.A. and B.R.: designed research and wrote the manuscript; O.A.: performed research and analyzed the data; W.E.R.: designed the automated liquid handling and analysis; P.C.: obtained ethics approval, designed recruitment program and obtained all human samples.
Funding
Funded by the CIHR Institute of Human Development, Child and Youth Health (IHDCYH; RHF 100625). O.A. is a fellow supported by the Fonds de la Recherche du Québec - Santé (FRQS) and the CIHR Training Program in Reproduction, Early Development, and the Impact on Health (REDIH). B.R. is a James McGill Professor.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Robert G. Berger and Dr Barbara F. Hales for their insight and help with the project.
References
- Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol 2004;26:249–261. [DOI] [PubMed] [Google Scholar]
- Collins AR, Oscoz AA, Brunborg G, Gaivão I, Giovannelli L, Kruszewski M, Smith CC, Stetina R. The comet assay: topical issues. Mutagenesis 2008;23:143–151. [DOI] [PubMed] [Google Scholar]
- Delbes G, Hales BF, Robaire B. Toxicants and human sperm chromatin integrity. Mol Hum Reprod 2009;16:14–22. [DOI] [PubMed] [Google Scholar]
- Donnelly ET, McClure N, Lewis SE. The effect of ascorbate and alpha-tocopherol supplementation in vitro on DNA integrity and hydrogen peroxide-induced DNA damage in human spermatozoa. Mutagenesis 1999;14:505–512. [DOI] [PubMed] [Google Scholar]
- Ge J, Chow DN, Fessler JL, Weingeist DM, Wood DK, Engelward BP. Micropatterned comet assay enables high throughput and sensitive DNA damage quantification. Mutagenesis 2015;30:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giwercman A, Lindstedt L, Larsson M, Bungum M, Spano M, Levine RJ, Rylander L. Sperm chromatin structure assay as an independent predictor of fertility in vivo: a case-control study. Int J Androl 2010;33:e221–e227. [DOI] [PubMed] [Google Scholar]
- Gutzkow KB, Langleite TM, Meier S, Graupner A, Collins AR, Brunborg G. High-throughput comet assay using 96 minigels. Mutagenesis 2013;28:333–340. [DOI] [PubMed] [Google Scholar]
- Gyori BM, Venkatachalam G, Thiagarajan PS, Hsu D, Clement M-V. OpenComet: an automated tool for comet assay image analysis. Redox Biol 2014;2:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson HL. The comet assay in nanotoxicology research. Anal Bioanal Chem 2010;398:651–666. [DOI] [PubMed] [Google Scholar]
- Lewis SEM. Is sperm evaluation useful in predicting human fertility? Reproduction 2007;134:31–40. [DOI] [PubMed] [Google Scholar]
- Nallella KP, Sharma RK, Aziz N, Agarwal A. Significance of sperm characteristics in the evaluation of male infertility. Fertility and Sterility 2006;85:629–634. [DOI] [PubMed] [Google Scholar]
- Olive PL, Banáth JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 2006;1:23–29. [DOI] [PubMed] [Google Scholar]
- Saleh RA, Agarwal A, Nelson DR, Nada EA, El-Tonsy MH, Alvarez JG, Thomas AJ, Sharma RK. Increased sperm nuclear DNA damage in normozoospermic infertile men: a prospective study. Fertility Sterility 2002;78:313–318. [DOI] [PubMed] [Google Scholar]
- Shamsi MB, Imam SN, Dada R. Sperm DNA integrity assays: diagnostic and prognostic challenges and implications in management of infertility. J Assist Reprod Genet 2011;28:1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon L, Murphy K, Shamsi MB, Liu L, Emery B, Aston KI, Hotaling J, Carrell DT. Paternal influence of sperm DNA integrity on early embryonic development. Hum Reprod 2014;29:2402–2412. [DOI] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res 1988;175:184–191. [DOI] [PubMed] [Google Scholar]
- Singh NP, Danner DB, Tice RR, McCoy MT, Collins GD, Schneider EL. Abundant alkali-sensitive sites in DNA of human and mouse sperm. Exp Cell Res 1989;184:461–470. [DOI] [PubMed] [Google Scholar]
- Speit G, Vasquez M, Hartmann A. The comet assay as an indicator test for germ cell genotoxicity. Mutat Res 2009;681:3–12. [DOI] [PubMed] [Google Scholar]
- Stang A, Witte I. Performance of the comet assay in a high-throughput version. Mutat Res 2009;675:5–10. [DOI] [PubMed] [Google Scholar]
- Weingeist DM, Ge J, Wood DK, Mutamba JT, Huang Q, Rowland EA, Yaffe MB, Floyd S, Engelward BP. Single-cell microarray enables high-throughput evaluation of DNA double-strand breaks and DNA repair inhibitors. Cell Cycle 2013;12:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte I, Plappert U, de Wall H, Hartmann A. Genetic toxicity assessment: employing the best science for human safety evaluation part III: the comet assay as an alternative to in vitro clastogenicity tests for early drug candidate selection. Toxicol Sci 2007;97:21–26. [DOI] [PubMed] [Google Scholar]
- Wood DK. Single cell trapping and DNA damage analysis using microwell arrays. PNAS 2010;107:10008–100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Examination and Processing of Human Semen. Geneva: World Health Organization, 2010. [Google Scholar]
- Zini A, Albert O, Robaire B. Assessing sperm chromatin and DNA damage: clinical importance and development of standards. Andrology 2014;2:322–325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.