Abstract
Understanding how a single cell, the zygote, can divide and differentiate to produce the diverse animal cell types is a central goal of developmental biology research. The model organism Caenorhabditis elegans provides a system that enables a truly comprehensive understanding of this process across all cells. Its invariant cell lineage makes it possible to identify all of the cells in each individual and compare them across organisms. Recently developed methods automate the process of cell identification, allowing high-throughput gene expression characterization and phenotyping at single cell resolution. In this Review, we summarize the sequences of events that pattern the lineage including establishment of founder cell identity, the signaling pathways that diversify embryonic fate, and the regulators involved in patterning within these founder lineages before cells adopt their terminal fates. We focus on insights that have emerged from automated approaches to lineage tracking, including insights into mechanisms of robustness, context-specific regulation of gene expression, and temporal coordination of differentiation. We suggest a model by which lineage history produces a combinatorial code of transcription factors that act, often redundantly, to ensure terminal fate.
Keywords: Cell differentiation, quantitative microscopy, transcription factors, signal transduction, robustness
Background
Developmental control is a central function encoded in animal genomes. Decades of developmental genetics research has provided invaluable insights into signaling pathways and regulatory mechanisms. However due to the incredible complexity of animal development, most developmental decisions in most model organisms remain relatively unexplored. When Sydney Brenner set out on the search that ultimately led him to develop C. elegans as a model organism, one of his goals was to choose an animal simple enough to understand “completely” from genes to development and behavior (1987). One of the most striking features of C. elegans is its reproducible development and small cell count. Each adult hermaphrodite has the 959 somatic cells, and these cells are produced by an almost identical pattern of cell divisions in each animal, termed the “invariant lineage.” An additional ~2000 nuclei are present in the syncytial germ line of hermaphrodites, and these have no fixed lineage (Kimble and Hirsh 1979). In the 1970s and 1980s, Brenner’s original ambition inspired the Herculean efforts of others resulting in the complete cell lineage(Sulston and Horvitz 1977; Sulston et al. 1983), which has become a cornerstone for C. elegans research. At the time, tracing the embryonic lineage required laboriously following a small group of individual nuclei by eye while sitting at a microscope until an embryo hatched, then starting over with a new embryo and a new group of nuclei until each cell had been traced multiple times. In this way, Sulston et al both verified the invariance of the lineage, and defined the precise cell division patterns that produce each of the 558 terminal cells present when the embryo hatches and the 113 embryonic programmed cell deaths (Sulston et al. 1983).
Methods for lineage analysis in embryos
The long cell cycles lengths of the postembryonic lineage and the ease of identifying cells based on their position and morphology in larvae made this stage a practical tool for genetic screens for lineage regulators. Such screens for defects in postembryonic lineages led to major insights including the discoveries of miRNAs (Lee et al. 1993; Wightman et al. 1993), lateral inhibition between EGF and Notch signaling pathways (Kimble 1981; Seydoux and Greenwald 1989) and mechanisms controlling directed cell migration (Aroian et al. 1990; Han and Sternberg 1990) among many others.
In contrast, most of the embryonic lineage has proved more challenging to work with due to the difficulty of distinguishing cells that have very similar morphology and are rapidly dividing. While it was possible to identify maternal effect regulators of the very earliest cell fate distinctions acting prior to the 12-cell stage due to the larger cell sizes and the substantial changes in terminal cell types present in these mutants (Kemphues et al. 1986; Kemphues et al. 1988; Mello et al. 1992; Bowerman et al. 1993; Hutter and Schnabel 1994; Mello et al. 1994; Lin et al. 1995), characterizing defects in later decisions was more challenging. The visual lineage tracing methods used for postembryonic lineages and the initial map of the embryonic lineage were simply too cumbersome.
The development of 3D time-lapse microscopy and advent of efficient data storage systems allowed for the first time entire lineages of single embryos to be tracked by manually annotating each cell in each time point using custom software (Martinelli et al. 1997; Schnabel et al. 1997; Hench et al. 2009). Applying these approaches to mutants (e.g. (Hutter and Schnabel 1994; Kaletta et al. 1997; Lin et al. 1998)) or embryos where cells had been rearranged (Bischoff and Schnabel 2006) provided insight into signal transduction and lineage specification. However the methods were inefficient, requiring days or weeks of annotation for a single embryo, limiting the methods from widespread application.
Methods to automatically track nuclei from DIC embryos (Onami et al. 2001; Hamahashi et al. 2003) proved challenging due to low contrast, and were most useful for classifying RNAi-induced defects the first 2 embryonic divisions (Gunsalus et al. 2005; Sonnichsen et al. 2005). To get around the contrast problem, we and others (Bao et al. 2006; Dzyubachyk et al. 2009; Giurumescu et al. 2012; Mace et al. 2013) developed methods for cell identification and tracking in 4D images of embryos expressing fluorescently tagged histone proteins. These transgenes improve cell tracking because of the higher contrast of bright nuclei and dark background (cytoplasm), the retention of histones in chromatin during mitosis, and the ability with confocal microscopy to achieve sufficiently high resolution for identification of nuclei and divisions throughout the depth of a z-stack.
These efforts generated an integrated suite of methods and tools that dramatically increased efficiency in lineage tracing and allowed new applications. After collecting 4D fluorescent images using a wide variety of microscopy platforms (Bao et al. 2006; Murray and Bao 2012; Moore et al. 2013; Richards et al. 2013; Wu et al. 2013), the analysis software StarryNite identifies (Santella et al. 2010) and tracks (Santella et al. 2014) nuclei over time. The automated process is highly accurate (>99.6%) but the sheer number of cells being identified and tracked requires manual curation and editing of the automated output, which is facilitated by custom software (AceTree)(Boyle et al. 2006). Each step of this process has been refined over time to increase imaging throughput, reduce error rates in automated segmentation and tracking, and make curation more efficient (Aydin et al. 2010). Currently eight or more embryos can be imaged per day on resonance scanning or spinning disk imaging platforms and it is common to trace lineages up to the point where the embryo begins to move within the eggshell (Mace et al. 2013; Richards et al. 2013; Walton et al. 2015; Zacharias et al. 2015).
Automated lineage tracing allows large-scale mapping of gene expression and phenotypes
We adapted the automated lineage tracing approach to map gene expression at single cell resolution across the organism (Murray et al. 2008). The ubiquitous fluorescent histone images allow lineage tracing and a gene of interest can be tagged with a second color fluorescent reporter and imaged simultaneously to measure gene expression. The reporter for a gene of interest can be transcriptional or translational. A transcriptional reporter can contain as little as individual enhancer elements up to several kilobases of upstream intergenic sequence driving the expression of a stable nuclear localized fluorescent reporter, while a translational reporter incorporates a fluorescent tag on the protein of interest, either in the context of a fosmid or the endogenous locus (Murray et al. 2012; Sarov et al. 2012). Transcriptional reporters give more precise information about the location of relevant regulatory sequences and are easier to generate at scale but translational reporters with more genomic context better recapitulate the endogenous gene’s expression. Both types of reporters give qualitatively and quantitatively reproducible expression across embryos for the same reporter and allow identification of genes coexpressed in each cell. We used this approach to generate the EPIC database, which currently contains expression profiles for over 180 genes, mostly transcription factors (TFs). (Murray et al. 2012; Sarov et al. 2012; Mace et al. 2013; Araya et al. 2014). We and others have used this dataset to identify mechanisms and regulators controlling lineage identity and cell fate determination (see below).
Automated cell tracking is also a powerful tool for cellular resolution phenotyping. Each lineage is traced de novo without assuming particular cell division or cell migration patterns, allowing the quantitative comparison of wild-type and mutant embryos. We created a reference embryo model, and showed that features such as cell cycle length, cell position and division orientation are highly consistent across embryos under normal conditions, and become more variable after temperature stress (Bao et al. 2008; Richards et al. 2013). For example, the mean variability (Coefficient of Variation) of each cell cycle length is only ~4.4% at 22C. This high consistency between wild-type embryos simplifies identification of cell cycle and cell position defects in mutants or after RNAi (Ho et al. 2015; Kruger et al. 2015; Walton et al. 2015). Analyzing fluorescent reporters of terminal fate makes it possible to reliably characterize homeotic lineage transformations (Moore et al. 2013). This general approach has been applied to identify lineage transformations after RNAi for the majority of embryonic lethal genes (Du et al. 2014; Du et al. 2015).
In the remainder of this Review, we summarize previous knowledge about pathways controlling lineage fate, and then provide examples of insights gleaned from these automated lineaging resources and methods
Specification of founder lineage identities
A major problem in C. elegans developmental biology is to determine how cell fate is encoded in the lineage. While spatial cues are important for organization of the body axes and for specific signals (especially through the Notch pathway), many cell fate decisions are determined by lineage history as opposed to spatial patterning; isolated early embryonic blastomeres often divide to produce the same terminally differentiated fates as they would have in the embryo (e.g. (Cowing and Kenyon 1996; Hashimshony et al. 2015)). Over the last three decades geneticists have worked out many of the major pathways involved. Our current understanding of this process can be summarized as follows. First, maternally provided regulators are sorted to early blastomeres, and their activity leads to initial establishment of lineage identity for the major founder lineages (AB, E, MS, C/D, P4). Following this establishment phase, each founder cell divides in a series of synchronous cleavage divisions. Despite the synchrony of these divisions, they are not fate-symmetric. Instead, the action of signaling pathways, especially the Wnt and Notch pathways, ensures that the daughters of nearly every division have unique molecular identities. In most cases the relevant targets of these pathways are not known, probably due to both the historical difficulty of identifying cells in mid-embryogenesis and the fact that many of these early zygotic regulators are fully or partially redundant (see below). Finally, cells begin to adopt their terminal tissue identity, a process controlled primarily by the appropriate expression of conserved metazoan tissue-specific master regulators.
Lineage tracing methods have been informative for each of these steps, but the details of how lineage identity is translated into fate remains elusive for most cells. Three major pathways have been implicated in these processes: PAR polarity, and the Notch and Wnt signaling pathways. As excellent reviews describe each of these pathways’ roles in C. elegans embryogenesis (Nance 2005; Priess 2005; Sawa 2012; Hoege and Hyman 2013; Sawa and Korswagen 2013), we focus here on describing how these pathways are integrated to regulate lineage and tissue identity.
Establishment of the A-P axis by sperm entry and PAR-mediated polarity
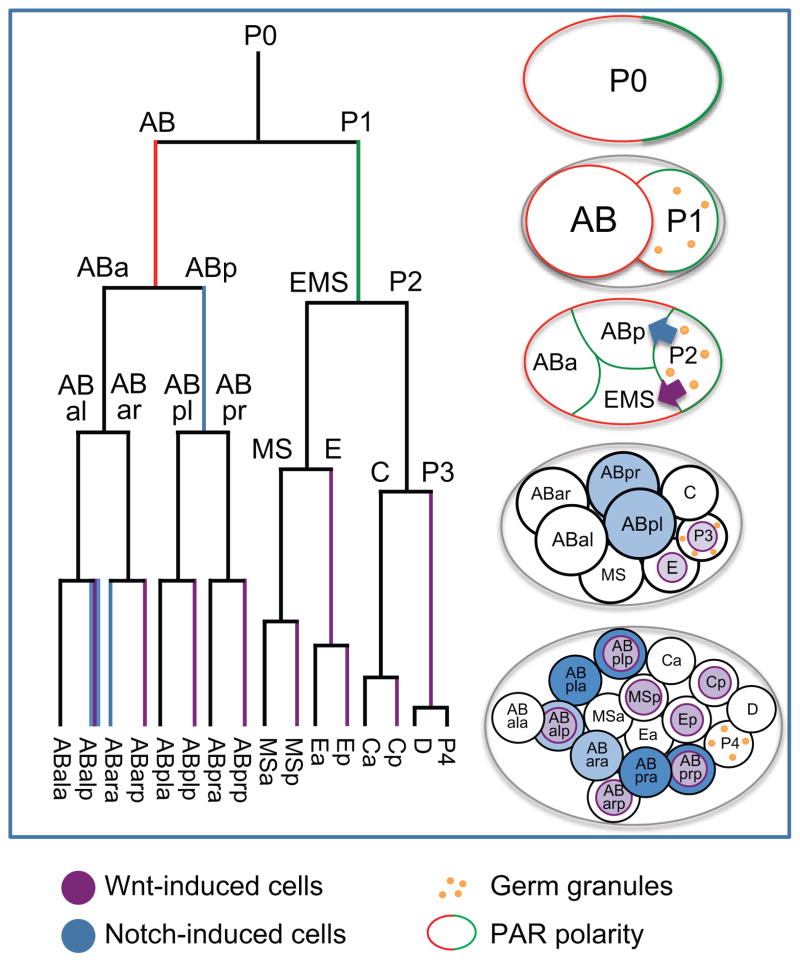
The location of sperm entry during fertilization determines the initial anterior-posterior axis of the zygote by regulating microtubule dynamics and asymmetric inheritance of maternal factors through the PAR protein polarity system (Kemphues et al. 1986; Kemphues et al. 1988; Boyd et al. 1996; Cowan and Hyman 2004; Tsai and Ahringer 2007; Hoege and Hyman 2013). As a result, the two cells resulting from first cleavage (AB and P1) inherit distinct combinations of cytoplasmic factors. In the next division AB divides symmetrically to generate ABa and ABp, while P1 divides asymmetrically using the PAR polarity system to generate EMS (specified by the activity of the transcription factor SKN-1 (Bowerman et al. 1993)) and P2 (which contains global transcriptional inhibitors such as oma-1 and pie-1 that prevent expression of SKN-1 targets (Mello et al. 1996; Guven-Ozkan et al. 2008)), giving the embryo four cells.
Notch induction of ABp fate
The symmetry of the AB daughters is broken by signaling through the Notch pathway. The Notch receptor protein GLP-1 is present in both equivalent AB daughters (Mango et al. 1994; Mello et al. 1994), and the Notch ligand APX-1 is expressed only in P2 (Priess and Thomson 1987; Mickey et al. 1996). The diamond-shaped orientation of the four cells (Figure 1) ensures that ABp contacts P2 but ABa does not. This allows specific induction of Notch target genes in ABp; these targets include the Hes family of bHLH transcriptional repressors ref-1 and hlh-25/26/27/28/29. These factors repress the transcription of tbx-37 and tbx-38, limiting their expression to the ABa lineage (Neves and Priess 2005) where they redundantly promote ABa fate (Good et al. 2004).
Figure 1.
Establishment of cell fate in the early embryo. Red = PAR-3/PAR-6/PKC-3 localization, Green = PAR-1/PAR-2 localization, Purple = Wnt signaling/signaled cells, Blue = Notch signaling/signaled cells. Yellow dots = P granules
Reiterative PAR polarity and PAL-1 in C lineage and germline fates
In addition to the P1 division, PAR polarity recursively polarizes the daughters of P2 (C and P3) and P3 (D and P4) (Guo and Kemphues 1995; Boyd et al. 1996). As a result, the maternally expressed homeodomain transcription factor PAL-1 is translated in the EMS, C and D cells. In EMS, SKN-1 antagonizes the C-fate inducing activity of PAL-1 activity (Hunter and Kenyon 1996, ), but SKN-1 is degraded by the time C and D are born (see below). Thus in the C and D lineages cells PAL-1 can act as a master regulator of lineage identity, allowing induction of skin and muscle fates, in part through a positive feedback loop in which maternal PAL-1 induces zygotic PAL-1 expression (Baugh et al. 2005; Fukushige and Krause 2005). P4 expresses none of these maternal transcription factors and inherits global silencers of transcription (in the P-granules) and regulators that allow it to adopt a germline identity (reviewed in (Strome 2005)).
Wnt induction of EMS fate
In EMS, the presence of SKN-1 in the absence of PIE-1 activates zygotic expression of transcription factors important for EMS fate, including med-1 and med-2 (Maduro et al. 2001; Maduro et al. 2015). A Wnt signal from P2 then orients the EMS division (Goldstein 1995) (Thorpe et al. 1997) and cooperates with SKN-1, MED-1/2 and PAL-1 to induce the daughter in contact with P2 (E) to express the GATA factors end-1 and end-3 (Maduro et al. 2005b), which are necessary and sufficient for intestinal fate specification(Zhu et al. 1997; Zhu et al. 1998; Maduro et al. 2005a). The other daughter (MS) instead expresses the T-box TF TBX-35 and the homeodomain TF CEH-51, which together regulate the fate of that lineage (Broitman-Maduro et al. 2006; Broitman-Maduro et al. 2009).
Recursive activation and repression by the TCF/β-catenin asymmetry pathway are a lineal cue to diversify cell fates
Genetic screens identified two genes, pop-1 and lit-1, with mutant phenotypes that suggested they play broad roles in symmetry breaking between sister cells throughout the embryo (Kaletta et al. 1997; Lin et al. 1998). pop-1 encodes a TCF family transcription factor (Lin et al. 1995) and lit-1 encodes a Nemo-like Kinase homolog (Rocheleau et al. 1999). These factors act downstream of Wnt and a parallel MAPK signaling pathway (Thorpe et al. 1997; Park et al. 2004) to distinguish the fates of most sister cell pairs. The mechanism for this was worked out primarily in the EMS division described above but appears to work similarly in later divisions. Posterior-produced Wnt ligand orients the spindle of each dividing cell along the anterior-posterior axis, in part by localizing the Frizzled receptor MOM-5 to the posterior of the cell (Thorpe et al. 1997; Park et al. 2004; Goldstein et al. 2006). A non-transcriptional role of Wnt signaling through mom-5 in parallel with a Src-related signaling pathway are required for normal division orientation (Bei et al. 2002).
The activity of Frizzled receptors (MOM-5 in the early embryo (Park et al. 2004)) and other Wnt pathway components also cause preferential nuclear import of the β-catenin protein SYS-1 (Huang et al. 2007; Phillips et al. 2007) and the β-catenin-related protein WRM-1 (Nakamura et al. 2005) in the posterior daughter, which is closer to the source of Wnt. WRM-1 then recruits LIT-1 to phosphorylate the TF POP-1, inducing its partial nuclear export (Rocheleau et al. 1999; Lo et al. 2004). POP-1 is thought to bind to targets in both cells (Maduro et al. 2002) and the posterior Wnt signaled daughter expresses target genes activated by the TCF-β-catenin complex, while in the unsignaled anterior cell, TCF represses these target genes, possibly by recruiting Groucho and other co-repressors (Calvo et al. 2001; Shetty et al. 2005). Manual lineage tracing of mutants for pop-1 or the nuclear export regulator lit-1 indicated that these mutants exhibit widespread anterior-to-posterior or posterior-to-anterior fate transformations (Kaletta et al. 1997; Lin et al. 1998), and most embryonic divisions are polarized along the A-P axis (Sulston et al. 1983; Richards et al. 2013) indicating a broad role for this pathway in fate specification, but until recently only a few embryonic POP-1 targets had been identified (Maduro et al. 2002; Streit et al. 2002; Maduro et al. 2005b; Shetty et al. 2005; Bertrand and Hobert 2009). As described later in this review, the EPIC single cell expression database has allowed identification of many additional TFs that require POP-1 for their expression patterns (Murray et al. 2012; Zacharias et al. 2015).
Later Notch inductions
While the Wnt pathway is notable for its broad activity in distinguishing daughters of nearly every embryonic division, the Notch signaling pathway also plays a major role for a more restricted set of cells. The C. elegans genome encodes two Notch receptor genes: lin-12 and glp-1 (reviewed in (Greenwald 2012; Greenwald and Kovall 2013)). In addition to the signal through GLP-1 that distinguishes ABp from ABa at the four-cell stage, about half a dozen additional later Notch inductions specify the anterior pharynx and break left-right symmetry in a small number of cells derived from the ABp and E lineages (reviewed in (Moskowitz and Rothman 1996; Priess 2005; Rasmussen et al. 2008). Unlike the Wnt pathway, it appears that Notch regulates a common set of targets, the ref-1/Hes family of bHLH-1 TFs in many of the cells in which the pathway is activated (Neves and Priess 2005). We used automated lineage tracing of embryos expressing a LIN-12::GFP fusion to identify additional cells where this Notch receptor may be active, as judged by the accumulation of the GFP-tagged C-terminal intracellular domain in the nucleus (Sarov et al. 2012). These findings show the utility of applying lineage analysis to the study of signaling pathways—it can identify divisions in which the pathway is active that may be too late in development or too downstream of initial defects to be efficiently identified by mutant phenotypes.
The TCF/β-catenin asymmetry pathway regulates diverse fates across the lineage
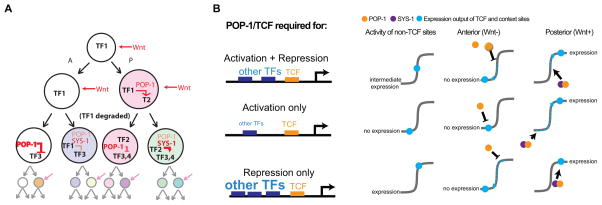
The successive rounds of POP-1/β-catenin asymmetry in sister cells are a major contributor to patterning the diverse fates of the 558 terminal embryonic cells. Several groups have proposed that this pathway patterns the lineage by iterative binary differentiation of sister cells (e.g. (Huang et al. 2007; Bertrand and Hobert 2010)). In this model, the expression of POP-1 target genes in a posterior daughter cell can cause its daughters to express different target genes in response in the next cell division (Figure 2). Bertrand and Hobart defined these as successive transient regulatory states in which a particular combination of transcription factors exists in the nucleus of a given cell for a short period of time, perhaps as short as a single cell cycle. These transient regulatory states represent a sequential combinatorial code that ultimately defines each cell during development. This model requires POP-1 to regulate different targets in different cells, and so a key question is how input from POP-1 is integrated with other cues to ensure expression only in the appropriate context.
Figure 2.
Model for quantitative regulation by the TCF/β-catenin asymmetry pathway. A) SYS-1β-catenin levels are higher in posterior cells where it complexes with low levels of POP-1/TCF to activate TFs that can partner with POP-1/SYS-1 in later cells to regulate new targets. Quantitative differences in POP-1 and SYS-1 levels due to memory of signaling states from previous mitoses further influences target selection. B) Proposed model explaining the three categories of TCF targets: targets that require TCF for repression and activation have intermediate activity provided by additional “context TFs,” targets that require TCF only for activation have low levels of context TF activity, and targets that require TCF only for repression have high levels of context TF activity.
Synthetic enhancers containing a large enough number of POP-1 binding sites drive embryonic expression in reproducible subset of posterior daughter cells after the 100 cell stage, with activity correlated with nuclear β-catenin concentrations (see below) (Zacharias et al. 2015). Surprisingly, even reporters with seven POP-1 binding sites do not drive substantial expression in the E cell, an early cell where multiple likely direct POP-1 targets have been identified (Maduro et al. 2002; Maduro et al. 2005b; Shetty et al. 2005). The explanation for this is likely that these direct targets contain binding sites for additional TFs (including SKN-1, PAL-1, MED-1 and MED-2) that are co-expressed only in the E cell and its close relatives (EMS and MS) (Maduro et al. 2005b; Maduro et al. 2015). The E cell may rely on these co-regulators to help activate direct POP-1 targets because the levels of the POP-1 co-activator SYS-1 are relatively low compared to those observed later development (Zacharias et al. 2015). Together these factors induce a moderate level of expression of direct target genes like end-1 and end-3 in both E and MS even in the absence of POP-1. (Maduro et al. 2002; Maduro et al. 2005b; Shetty et al. 2005; Maduro et al. 2015; Zacharias et al. 2015) The additional influence of POP-1 binding upstream of these direct targets boosts expression in the posterior sister E and represses expression in the anterior sister MS. Thus competence for POP-1-dependent expression is defined by other TFs bound to an enhancer and POP-1 converts these enhancers into A-P switches (Maduro et al. 2002; Shetty et al. 2005). Similar results have been seen for other targets including the myogenic factor hlh-1 in the C lineage muscle (Lei et al. 2009), and the homeobox TF ceh-10 in the AIY neuron (Bertrand and Hobert 2009).
Chromatin states may also play a role in the ability of Wnt signals to activate different targets in cells with different lineage histories. In the specification of the E and MS sister lineages, the histone acetyltransferase, CBP-1, counters the repressive activity of the histone deacetylase, HDA-1, in the E lineage to allow activation of end-1 in response to Wnt pathway activation (Shi and Mello 1998; Calvo et al. 2001). In the MS lineage, POP-1 binding in the absence of SYS-1 recruits HDA-1 and another repressor, UNC-37/groucho, to repress expression of end-1 (Calvo et al. 2001). It remains unclear whether the activity of CBP-1 in the MS lineage is offset by the higher concentration of POP-1 in MS or if another mechanism drives its differential activity in E and MS.
Quantitative differences in nuclear TCF and β-catenin encode a memory of previous states
The POP-1/SYS-1 system is described above as a binary switch that acts at each division across the lineage to promote expression in the posterior daughter and repress expression in the anterior daughter. We tested whether quantitative variation in this pathway’s activity might influence target selection by using automated lineage tracing to measure nuclear levels of fluorescently tagged SYS-1 and POP-1 (Zacharias et al. 2015). We found that not all posterior sister cells are equivalent; instead they show substantial and reproducible quantitative differences in nuclear SYS-1. One major source of this variation is lineage history; “double-posterior” cells (posterior daughters of mothers that were themselves posterior sister cells and thus had high nuclear SYS-1) have substantially more nuclear SYS-1 than single-posterior cells whose mother had low nuclear SYS-1. The reciprocal pattern exists for POP-1, with higher levels in double-anterior cells (unsignaled for two consecutive generations). The net effect is that the stoichiometric ratio of POP-1 to SYS-1 varies depending multiple generations of lineage history. Since this stoichiometry is thought to determine the balance between POP-1 mediated repression and POP-1:SYS-1 mediated activation, we predicted that POP-1 should be a more potent activator in double-posterior cells. We tested this by using synthetic enhancers containing different numbers of POP-1 binding sites. Synthetic enhancers containing seven POP-1 binding sites were expressed in many posterior lineages with a bias for double-posterior lineages. As predicted by the quantitative model, this bias was stronger for enhancers containing fewer POP-1 binding sites; a three-site enhancer is expressed only in triple-posterior cells. Thus the POP-1:SYS-1 is able to activate targets with weaker binding sites more strongly in these cells with history of multiple consecutive signaling events. A similar logic suggests that POP-1 may be a more potent repressor in double-anterior cells, although this was not yet experimentally tested.
The mechanisms underlying this memory of cellular signaling history are not yet fully clear. The enrichment of SYS-1 in double-posterior cells requires a functional proteasome (Zacharias et al. 2015), suggesting that the lack of activity of the conserved β-catenin destruction complex in the signaled posterior daughters could allow SYS-1 to accumulate in the absence of transcriptional or translational compensation (Phillips et al. 2007; Baldwin and Phillips 2014). The accumulation of POP-1 in double-anterior cells is independent of the proteasome, indicating it uses a separate mechanism. One possible mode of regulation is via the pathway controlling POP-1 nuclear export in signaled cells. For example, the lit-1 kinase that phosphorylates POP-1 and triggers its export (Takeshita and Sawa 2005) could be differentially inherited between unsignaled and signaled daughter cells. Future imaging and genetic studies will be needed to test these mechanisms.
Posterior lineally repetitive TFs are context-specific targets of POP-1
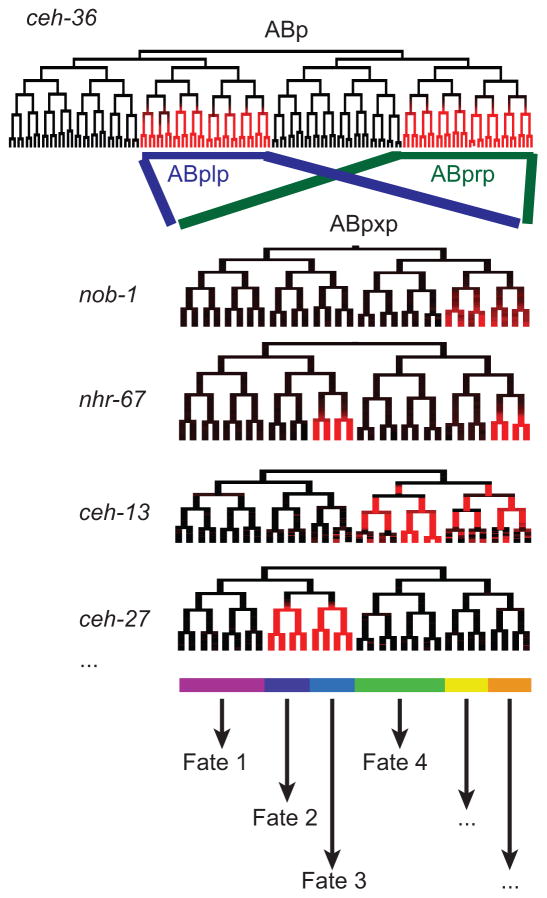
To identify candidate regulators of lineage identity that may act downstream of Wnt to specify cell fates, we traced the expression patterns for reporters of hundreds of embryonic regulators through the 350-cell stage by lineage tracing (Murray et al. 2012; Sarov et al. 2012; Mace et al. 2013; Araya et al. 2014). We identified the expected pattern for known maternally controlled regulators of early blastomere identity, with expression limited to a single lineage. Surprisingly, we did not identify any additional TFs expressed in single lineages later in development. Instead, many are expressed in a type of pattern that we refer to as “lineally repetitive” (Figure 3). This class of expression is defined by expression in multiple lineages comprised of either related posterior or related anterior daughter cells, but not both, typically in progenitors of multiple tissue types (Murray et al, 2012). This type of expression had been previously identified for the POU homeodomain TF unc-86 (Baumeister et al. 1996) and the Hox TF ceh-13 (Streit et al. 2002); our work suggests it is a widespread mode of regulation, with at least 30 TFs (of 127 examined in the initial study) showed this type of expression pattern (Murray et al. 2012).
Figure 3.
Model for combinatorial control by lineally repetitive TFs, using the ABplp/prp lineage as an example. ceh-36 and unc-30 are expressed throughout the ABplp and ABprp lineages (which are symmetric for all TFs included in this figure). Later TFs including nob-1, nhr-67, ceh-13, ets-7 and ceh-27 are expressed in distinct subsets of this pattern. We propose that the combinations of these and other lineally repetitive TFs then go on to specify terminal fates combinatorially. Redundancy between these genes may explain the frequent low penetrance phenotypes seen for mutants.
About half of these TFs with lineally repetitive expression are specifically in multiple related lineages derived from posterior daughter cells where the Wnt pathway canonically activates targets through POP-1 (Figure 2). Nearly all of these TFs’ expression is altered when pop-1, sys-1 or other pathway components are depleted by RNAi (Zacharias et al. 2015), and each carries predicted POP-1 binding sites in its regulatory sequences, consistent with them being targets of POP-1. Intriguingly, for some of these targets pop-1 RNAi causes gain of expression in anterior sister lineages of the normally expressing cells, while for others it causes loss of expression in posterior lineages. This is consistent with the lineage specificity of each target being defined by binding sites for other transcription factors (“context sites”), with POP-1 acting as a switch to confer posterior-specific expression. Such enhancers could fall into two categories. In one, the context sites alone are sufficient to drive expression throughout the relevant lineage; in this case POP-1-mediated repression in the anterior sister cell leads to posterior-specific expression. In the second, the context sites are insufficient to drive robust expression; in this case the additional activity of the POP-1:SYS-1 complex pushes the target above the threshold required for expression (Figure 2B).
This provides a way evolution could exploit the quantitative differences in nuclear POP-1 and SYS-1 described above to help ensure context specific expression. To drive expression specifically in double-posterior lineages, where POP-1:SYS-1 activation is strongest, an enhancer with very weak activity in the absence of POP-1 and/or with low affinity POP-1 binding sites could be used. Consistent with this, genes expressed in double-posterior lineages are more likely to require pop-1 for activation as opposed to repression (Zacharias et al. 2015). Detailed identification of the binding sites for POP-1 and context TFs in multiple target enhancers will be needed in order to determine the relative balance of these mechanisms in regulating the lineally repetitive TFs.
Overall, it appears that Wnt is not acting as a traditional morphogen during C. elegans embryonic development. The secretion of Wnt ligand from cells at the posterior of the embryo does not cause cells at the posterior to respond more strongly, its primary role seems to be to orient divisions, ensuring cell fates are coupled appropriately to embryonic position (Bischoff and Schnabel 2006; Goldstein et al. 2006; Zacharias et al. 2015). Furthermore, Wnt is not acting strictly as a temporal morphogen: levels of SYS-1 and POP-1 rise quickly after mitosis and plateau until the next division, even for cells with very different cell cycle lengths (Zacharias et al. 2015). Cells’ responses to Wnt occur during particular phases of mitosis (Mizumoto and Sawa 2007; Sugioka et al. 2011). This suggests that the Wnt response may be gated on mitosis—a cell cannot respond to an additional Wnt signal until it re-enters mitosis. Therefore, we propose that Wnt behaves as a transmitotic morphogen; it has different effects when a cell responds a signal in consecutive rounds of mitosis. Thus, the number of consecutive rounds of mitosis in which a cell has high SYS-1 and low POP-1 has the potential to trigger distinct fates.
Possible mechanisms controlling expression in anterior sister lineages
These results provide a framework for understanding how lineage specification may work in posterior sister cells, but how genes become expressed in anterior sister-specific patterns is less well understood. One mechanism is for Wnt and POP-1 to activate expression of repressors in posterior sister lineages, and these repressors then prevent expression of their targets in those lineages, leading to anterior-specific expression. This has been shown for example in the C lineage, where the lineage-specific TF PAL-1 combines with POP-1 to regulate expression of the myogenic TF hlh-1/MyoD exclusively in the posterior granddaughters of the C founder cell (Lei et al. 2009). hlh-1 then represses the skin specification TF elt-1/GATA, leading to its anterior-specific expression (Yanai et al. 2008).
However several lines of evidence suggest that Wnt pathway components may directly regulate some anterior sister-specific genes. Similar numbers of genes are expressed in “anterior” lineally repetitive patterns as in posterior-specific patterns (Murray and Bao 2012). The time between commitment (birth of the anterior lineage) and first detection of fluorescent reporters is similar between the two classes, and not delayed for the anterior-specific genes as would be predicted from the indirect repression model. The t-box TF tbx-35, which specifies the fate of the anterior-specific MS cell, is misexpressed in E (the posterior sister of MS) when pop-1 is depleted but not when the two Wnt-regulated GATA TFs (end-1 and end-3) required for E fate are lost. This suggests that tbx-35 is not subject to POP-1 dependent indirect repression by end-1 and end-3 and is instead either repressed by other POP-1 targets or directly repressed by POP-1 (Broitman-Maduro et al. 2006; Broitman-Maduro et al. 2009). Finally, a recent study suggested that the homeodomain TF ttx-3, which specifies the fate of the “AIY mother cell” (an anterior sister cell), is directly regulated by POP-1 in the absence of SYS-1 in an “opposite” fashion to canonical targets (Murgan et al. 2015). The model proposed is that POP-1 is recruited to a ttx-3 enhancer not through direct DNA binding to its motif, but indirectly through the lineage-specific TF REF-2 (itself an anterior lineage-specific TF), with the REF-2:POP-1 complex activating ttx-3 expression directly in the anterior cell through an unknown mechanism. In the posterior sister, SYS-1 interferes with the formation or activity of this complex, thus preventing expression. Several examples of this type of opposite regulation have now been identified in different model systems (Piepenburg et al. 2000; Theisen et al. 2007; Blauwkamp et al. 2008; Zhang et al. 2014). The proposed mechanisms include not just indirect recruitment of the TCF factor but also binding of TCF to noncanonical motifs or adjacent to cofactors, allosterically affecting TCF function, or at sites overlapping a separate regulator, preventing its binding. The relative importance of these different mechanisms across the diversity of anterior-specific genes remains to be determined.
Lineally repetitive transcription factors act redundantly to regulate robust lineage identity
Each individual lineally repetitive TF is expressed in many lineages, but combinations of these TFs uniquely mark each lineage (Figure 3). Thus these TFs represent a combinatorial encoding of lineage history that could be used to translate this history into terminal fates. Support for the importance of these TFs comes from the fact that deletion mutants for many of these are reported to be lethal either singly or in combination with deletion of a redundant homolog (The C. elegans knockout consortium 2012; Howe et al. 2015). Mammalian orthologs of many of these are important for regulating cell fate in progenitor cells (e.g. (Lufkin et al. 1991; Acampora et al. 1995; Acampora et al. 1996; Monaghan et al. 1997; Rossel and Capecchi 1999; Pabst et al. 2000; Martinez-Morales et al. 2001; Wellik et al. 2002; Levantini et al. 2003; Wellik and Capecchi 2003; Kmita et al. 2005; Tischfield et al. 2005; Zhang et al. 2006)), suggesting the possibility that this regulation could be conserved. Together, this indicates that Wnt pathway activity distinguishes lineages by generating a combinatorial code of lineally repetitive TFs that then act to ensure appropriate expression of the “terminal selector” TFs responsible for establishing and maintaining terminal fates.
Why then have genetic screens identified so many maternally expressed genes involved in early lineage specification but so few zygotic genes involved in progenitor identity? The answer may be in the redundancy built into the network to ensure robustness. Most well characterized early zygotic regulators have one or more redundant paralogs. In some cases, these were identified fortuitously; for example the redundant E lineage master regulators end-1 and end-3 (Zhu et al. 1997; Maduro et al. 2005a) and ABa-lineage regulators tbx-37 and tbx-38 (Good et al. 2004) are each in close proximity to their paralog on the same chromosome, allowing them to be identified based on large deletions that remove both genes. In other cases (Andachi 2004; Neves and Priess 2005), the redundant paralogs were predicted from sequence homology and tested directly for redundancy by simultaneously disrupting both genes. Genome-wide expression time courses of early embryos has identified many TFs and other genes expressed in early embryos whose functions are unknown; many of these fall into groups of paralogs suggesting they may play similarly redundant developmental roles (Robertson et al. 2004; Baugh et al. 2005; Levin et al. 2012; Hashimshony et al. 2015).
This redundancy could result from the constraints associated with rapid cell division and simultaneous transcription. Expression in the context of rapid cell division is likely be noisy. One approach to buffer against this transcriptional noise is by averaging. For example in the syncytial early Drosophila embryo, noise in transcription of the critical zygotic regulator Hunchback in individual nuclei is buffered by spatial averaging as molecules diffuse between adjacent cells (Little et al. 2013). Both spatial and temporal averaging likely also occurs in the C. elegans oogenesis, since each oocyte contains mRNA and protein produced by hundreds of nuclei in the syncytial germline (Kimble and Hirsh 1979; Wolke et al. 2007). These mechanisms are not feasible in the rapidly dividing and fully cellularized early C. elegans embryo; instead the embryo benefits from averaging across multiple paralogous gene copies. Consistent with this, a maternal effect mutation in the skn-1 gene results in partial penetrance loss of gut specification (Bowerman et al. 1992). This phenotypic variability was shown to result from increased expression variability of the skn-1 target end-1 coupled to the loss of expression of its redundant paralog end-3 (Raj et al. 2010). Similarly, robust end-1 and end-3 expression depends on the binding of the upstream pair of redundant GATA TFs med-1 and med-2 (Maduro et al. 2015). Together this suggests that one role of seemingly redundant paralogs expressed in early embryogenesis is to increase the robustness of differentiation.
Genetic redundancy does not have to occur between paralogs. Indeed several examples of non-paralogous parallelism have been identified. We showed that the homeodomain TFs ceh-36/Otx and unc-30/Pitx are redundantly required for robust specification of two early embryonic lineages (Walton et al. 2015). These TFs are coexpressed early in the ABplp and ABprp lineages and have distinct expression in other lineages. We used automated lineage tracing to test for defects in cell cycle length, cell migration and division orientation in embryos carrying mutations in these genes. ceh-36 mutant embryos have low penetrance defects in a wide range of cells that normally express that TF and all hatch, with ~60% larvae arresting later in development, while unc-30 mutants have no early embryonic defects. However the double mutant is 100% lethal and has both higher-penetrance and higher-frequency (across cells) defects in the ABplp and ABprp lineages. Both ceh-36 and unc-30 had been previously identified in genetic screens for defects in specification of specific terminal neurons. unc-30 was particularly well-known as a master-regulator of GABAergic neuronal identity (Jin et al. 1994), but our results show that in ABplp and ABprp it regulates other non-neuronal fates. Other embryonic TFs also play distinct roles postembryonically (Bishop and Guarente 2007) suggesting this may be a common principle and emphasizing the importance of context in determining TF function.
Even in cases where the individual TF mutants are lethal, defects in cell fate specification may be low-penetrance or absent. For example, embryos lacking the conserved myogenic TF hlh-1/MyoD, make a fairly normal quantity of muscle cells, and die as larvae due to defects in muscle function (Krause 1995). It is only when the parallel regulators unc-120/SRF and hnd-1/HAND are removed as well that muscle differentiation is completely lost (Fukushige et al. 2006). Similarly, in unc-30;ceh-36 double mutants, we observed partially penetrant defects in cell division and position as well as expression of the downstream lineage-specific TF mls-2/Hmx (Walton et al. 2015). This suggests that even essential TFs or TF combinations may have additional redundant partners in any given cellular differentiation decision, thus lethality in mutant animals is a function of the number and identity of cells impacted along with the penetrance of the individual defects.
Temporal coordination of proliferation and differentiation
Redundancy may also provide a buffer for the rapid pace of early embryonic development. As noted above, early C. elegans embryonic cells have both rapid rates of cell division and simultaneous expression of genes important for lineage specification. This provides major challenges to the organism. In early zygotic cells interphase can be as short as 10 minutes (Sulston et al. 1983; Schnabel et al. 1997; Bao et al. 2008; Richards et al. 2013). Assuming a transcription rate of ~1–1.5 kb/minute as has been seen in other organisms (Garcia et al. 2013), production of a gene with a 10kb primary transcript would be severely limited in these cell cycles, and even shorter transcripts are limited in how many messages can be produced. Cells needs to transcribe and translate critical regulatory TFs rapidly enough to allow them to act as early as 15 minutes later in daughter cells to regulate additional fate decisions. Due to this so-called “intron delay,” early zygotic genes have short primary transcripts in Drosophila, zebrafish and mouse (Heyn et al. 2014) as well as worms. In C. elegans, early zygotic primary transcripts are on average half the length of those for maternally expressed genes. The presence of redundant paralogs provides another way to boost total production of important regulators.
An additional remaining question is how the timing of differentiation is coordinated with the cell cycle. Different lineally repetitive TFs are often expressed in subsequent cell cycles within the same lineage, with distinct regulatory decisions separated by as little as 15 minutes (Figure 3). Yet, the pattern of fate specification remains robust over a 2-fold range of cell cycle rates between 15°C and 25°C and in delayed mutants such as clk-1 (Wong et al. 1995; Nair et al. 2013). An attractive possibility would be coupling of expression to a temporal cue such as mitosis. Pioneering work showed that this is not true for genes in clonal lineages such as the E (intestine) lineage, which goes on to express terminal differentiation markers even if cell division is blocked pharmacologically at the 8-cell stage (Edgar and McGhee 1988). We showed that when the cell cycle is specifically perturbed using cell cycle mutants, in both the clonal E lineage and the C lineage, which gives rise to multiple tissues, genes are expressed at the normal time, even if an inappropriate number of cells are present (Nair et al. 2013). This appears to rule out the cell cycle lock as a dominant mechanism, although it remains possible that other genes are regulated by the cell cycle more directly.
Dynamic turnover of transcription factors is likely to play a role in temporal control; one striking result of our survey of translational reporters by lineage tracing is how common such dynamics are. Many TFs are initially expressed in a broad lineage and but rapidly degraded in some or all cells, leading to a dynamic temporal expression domain. The most striking examples of this were several factors that were present in either lineage-specific patterns or in all embryonic cells for brief temporal pulses, typically 2–3 cell cycles in length (Sarov et al. 2012; Walton et al. 2015). This turnover is sometimes important for developmental progression. A survey of embryonic lethal RNAi treatments by automated lineage tracing found that genes involved in ubiquitin-mediated degradation of SKN-1 are important for progression in the EMS lineage (Du et al. 2014; Du et al. 2015). SKN-1 protein in EMS specifies that lineage’s fate, and SKN-1 is degraded when EMS divides to generate an anterior mesodermal daughter MS and a posterior endodermal daughter E. Blocking this degradation leads to a reiteration of the EMS fate, where the cell that should be MS divides to produce an MS-like daughter and an E-like daughter. Similar fate reiterations occur in other lineages after loss of proteolysis regulators, suggesting that degradation of progenitor fate regulators may be a general mechanism for directional differentiation.
Prospects for the future of C. elegans lineage analysis
There is still much to learn from the C. elegans lineage. While the latest approaches allow lineage tracing through the onset of movement, extending automated lineage tracing through the second half of embryogenesis (or postembryonically) is more challenging because the embryo begins to twitch (and later roll) within the eggshell. One approach is to use even faster, less phototoxic imaging approaches such as inverted Selective Plane Illumination Microscopy (iSPIM) or Bessel Beam microscopy, which allows collection of 4D image series at 2-second resolution over the full 14-hour time period from first cleavage to hatch (Wu et al. 2013; Santella et al. 2015). Combining this greater temporal resolution with approaches to correct for the movements that do occur in between 2-second intervals, may allow specific subsets of cells, or even full lineages, to be followed during the period when the embryo is moving, allowing late-onset expression to be accurately annotated and allowing detailed description of cell morphology changes over time. This could be especially useful for neurons, for which the complex shape changes occurring during this phase of development are a powerful phenotype (e.g. (Heiman and Shaham 2009)).
Extending the resource of expression patterns to the full set of embryonically expressed transcription factors, not to mention other genes, remains a major challenge. With improved methods for single-cell transcriptomics (Macosko et al. 2015), it may be possible to define expression dynamics across the lineage genome-wide. However, answering many of the remaining questions about fate specification will require detailed dissection of individual enhancers and their constituent TF binding sites, where imaging based methods are currently the only practical method. Additional improvements to image quality and cell tracking algorithms could further reduce the error rate and the amount of hands-on time needed for curation, allowing higher throughput. Together, the next ten years should see a clear model emerge of how the remarkable C. elegans lineage is regulated.
Acknowledgments
We thank Meera Sundaram and current and former members of the Murray laboratory, especially Travis Walton, for many helpful discussions. We also thank Zhirong Bao for suggestions and comments on the manuscript. This work was funded in part by funding from the NIH to JM (GM105676) and ALZ (GM111825).
Literature Cited
- Acampora D, Mazan S, Avantaggiato V, Barone P, Tuorto F, Lallemand Y, Brulet P, Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nature genetics. 1996;14(2):218–222. doi: 10.1038/ng1096-218. [DOI] [PubMed] [Google Scholar]
- Acampora D, Mazan S, Lallemand Y, Avantaggiato V, Maury M, Simeone A, Brulet P. Forebrain and midbrain regions are deleted in Otx2−/− mutants due to a defective anterior neuroectoderm specification during gastrulation. Development. 1995;121(10):3279–3290. doi: 10.1242/dev.121.10.3279. [DOI] [PubMed] [Google Scholar]
- Andachi Y. Caenorhabditis elegans T-box genes tbx-9 and tbx-8 are required for formation of hypodermis and body-wall muscle in embryogenesis. Genes Cells. 2004;9(4):331–344. doi: 10.1111/j.1356-9597.2004.00725.x. [DOI] [PubMed] [Google Scholar]
- Araya CL, Kawli T, Kundaje A, Jiang L, Wu B, Vafeados D, Terrell R, Weissdepp P, Gevirtzman L, Mace D, et al. Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature. 2014;512(7515):400–405. doi: 10.1038/nature13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroian RV, Koga M, Mendel JE, Ohshima Y, Sternberg PW. The let-23 gene necessary for Caenorhabditis elegans vulval induction encodes a tyrosine kinase of the EGF receptor subfamily. Nature. 1990;348(6303):693–699. doi: 10.1038/348693a0. [DOI] [PubMed] [Google Scholar]
- Aydin Z, Murray JI, Waterston RH, Noble WS. Using machine learning to speed up manual image annotation: application to a 3D imaging protocol for measuring single cell gene expression in the developing C. elegans embryo. BMC Bioinformatics. 2010;11:84. doi: 10.1186/1471-2105-11-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AT, Phillips BT. The tumor suppressor APC differentially regulates multiple beta-catenins through the function of axin and CKIalpha during C. elegans asymmetric stem cell divisions. Journal of cell science. 2014;127(Pt 12):2771–2781. doi: 10.1242/jcs.146514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Murray JI, Boyle T, Ooi SL, Sandel MJ, Waterston RH. Automated cell lineage tracing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Zhao Z, Boyle TJ, Murray JI, Waterston RH. Control of cell cycle timing during C. elegans embryogenesis. Dev Biol. 2008;318(1):65–72. doi: 10.1016/j.ydbio.2008.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development. 2005;132(8):1843–1854. doi: 10.1242/dev.01782. [DOI] [PubMed] [Google Scholar]
- Baumeister R, Liu Y, Ruvkun G. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes & Development. 1996;10(11):1395–1410. doi: 10.1101/gad.10.11.1395. [DOI] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3(1):113–125. doi: 10.1016/s1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hobert O. Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Developmental Cell. 2009;16(4):563–575. doi: 10.1016/j.devcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand V, Hobert O. Lineage programming: navigating through transient regulatory states via binary decisions. Current Opinion in Genetics & Development. 2010;20(4):362–368. doi: 10.1016/j.gde.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M, Schnabel R. A posterior centre establishes and maintains polarity of the Caenorhabditis elegans embryo by a Wnt-dependent relay mechanism. PLoS biology. 2006;4(12):e396. doi: 10.1371/journal.pbio.0040396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447(7144):545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Blauwkamp TA, Chang MV, Cadigan KM. Novel TCF-binding sites specify transcriptional repression by Wnt signalling. EMBO J. 2008;27(10):1436–1446. doi: 10.1038/emboj.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman B, Draper BW, Mello CC, Priess JR. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell. 1993;74(3):443–452. doi: 10.1016/0092-8674(93)80046-h. [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68(6):1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122(10):3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Boyle TJ, Bao Z, Murray JI, Araya CL, Waterston RH. AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinformatics. 2006;7(1):275. doi: 10.1186/1471-2105-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro G, Lin KT, Hung WW, Maduro MF. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development. 2006;133(16):3097–3106. doi: 10.1242/dev.02475. [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development. 2009;136(16):2735–2746. doi: 10.1242/dev.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo D, Victor M, Gay F, Sui G, Luke MP, Dufourcq P, Wen G, Maduro M, Rothman J, Shi Y. A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegans embryogenesis. EMBO J. 2001;20(24):7197–7208. doi: 10.1093/emboj/20.24.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004;431(7004):92–96. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- Cowing D, Kenyon C. Correct Hox gene expression established independently of position in Caenorhabditis elegans. Nature. 1996;382(6589):353–356. doi: 10.1038/382353a0. [DOI] [PubMed] [Google Scholar]
- Du Z, Santella A, He F, Shah PK, Kamikawa Y, Bao Z. The Regulatory Landscape of Lineage Differentiation in a Metazoan Embryo. Dev Cell. 2015;34(5):592–607. doi: 10.1016/j.devcel.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Santella A, He F, Tiongson M, Bao Z. De novo inference of systems-level mechanistic models of development from live-imaging-based phenotype analysis. Cell. 2014;156(1–2):359–372. doi: 10.1016/j.cell.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzyubachyk O, Jelier R, Lehner B, Niessen W, Meijering E. Model-based approach for tracking embryogenesis in Caenorhabditis elegans fluorescence microscopy data. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference. 2009;2009:5356–5359. doi: 10.1109/IEMBS.2009.5334046. [DOI] [PubMed] [Google Scholar]
- Edgar LG, McGhee JD. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 1988;53(4):589–599. doi: 10.1016/0092-8674(88)90575-2. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20(24):3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;132(8):1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23(21):2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurumescu CA, Kang S, Planchon TA, Betzig E, Bloomekatz J, Yelon D, Cosman P, Chisholm AD. Quantitative semi-automated analysis of morphogenesis with single-cell resolution in complex embryos. Development. 2012 doi: 10.1242/dev.086256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. Cell contacts orient some cell division axes in the Caenorhabditis elegans embryo. The Journal of cell biology. 1995;129(4):1071–1080. doi: 10.1083/jcb.129.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Takeshita H, Mizumoto K, Sawa H. Wnt signals can function as positional cues in establishing cell polarity. Dev Cell. 2006;10(3):391–396. doi: 10.1016/j.devcel.2005.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development. 2004;131(9):1967–1978. doi: 10.1242/dev.01088. [DOI] [PubMed] [Google Scholar]
- Greenwald I. Notch and the awesome power of genetics. Genetics. 2012;191(3):655–669. doi: 10.1534/genetics.112.141812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I, Kovall R. Notch signaling: genetics and structure. WormBook. 2013:1–28. doi: 10.1895/wormbook.1.10.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JD, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436(7052):861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81(4):611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Guven-Ozkan T, Nishi Y, Robertson SM, Lin R. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell. 2008;135(1):149–160. doi: 10.1016/j.cell.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamahashi S, Urai M, Onami S. Automatics measurement system for early embryonic cell lineage. 14th international C elegans meeting abstracts.2003. [Google Scholar]
- Han M, Sternberg PW. let-60, a gene that specifies cell fates during C. elegans vulval induction, encodes a ras protein. Cell. 1990;63(5):921–931. doi: 10.1016/0092-8674(90)90495-z. [DOI] [PubMed] [Google Scholar]
- Hashimshony T, Feder M, Levin M, Hall BK, Yanai I. Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature. 2015;519(7542):219–222. doi: 10.1038/nature13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman MG, Shaham S. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell. 2009;137(2):344–355. doi: 10.1016/j.cell.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hench J, Henriksson J, Luppert M, Burglin TR. Spatio-temporal reference model of Caenorhabditis elegans embryogenesis with cell contact maps. Developmental biology. 2009;333(1):1–13. doi: 10.1016/j.ydbio.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 2014;6(2):285–292. doi: 10.1016/j.celrep.2013.12.030. [DOI] [PubMed] [Google Scholar]
- Ho VW, Wong MK, An X, Guan D, Shao J, Ng HC, Ren X, He K, Liao J, Ang Y, et al. Systems-level quantification of division timing reveals a common genetic architecture controlling asynchrony and fate asymmetry. Mol Syst Biol. 2015;11(6):814. doi: 10.15252/msb.20145857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Hyman AA. Principles of PAR polarity in Caenorhabditis elegans embryos. Nat Rev Mol Cell Biol. 2013;14(5):315–322. doi: 10.1038/nrm3558. [DOI] [PubMed] [Google Scholar]
- Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, Done J, Down T, Gao S, Grove C, et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCFPOP-1 and its coactivator beta-catenin SYS-1. Development. 2007;134(14):2685–2695. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell. 1996;87(2):217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- Hutter H, Schnabel R. glp-1 and inductions establishing embryonic axes in C. elegans. Development. 1994;120(7):2051–2064. doi: 10.1242/dev.120.7.2051. [DOI] [PubMed] [Google Scholar]
- Jin Y, Hoskins R, Horvitz HR. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature. 1994;372(6508):780–783. doi: 10.1038/372780a0. [DOI] [PubMed] [Google Scholar]
- Kaletta T, Schnabel H, Schnabel R. Binary specification of the embryonic lineage in Caenorhabditis elegans. Nature. 1997;390(6657):294–298. doi: 10.1038/36869. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52(3):311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Kemphues KJ, Wolf N, Wood WB, Hirsh D. Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans. Dev Biol. 1986;113(2):449–460. doi: 10.1016/0012-1606(86)90180-6. [DOI] [PubMed] [Google Scholar]
- Kimble J. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol. 1981;87(2):286–300. doi: 10.1016/0012-1606(81)90152-4. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70(2):396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kmita M, Tarchini B, Zakany J, Logan M, Tabin CJ, Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435(7045):1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- Krause M. MyoD and myogenesis in C. elegans. Bioessays. 1995;17(3):219–228. doi: 10.1002/bies.950170308. [DOI] [PubMed] [Google Scholar]
- Kruger AV, Jelier R, Dzyubachyk O, Zimmerman T, Meijering E, Lehner B. Comprehensive single cell-resolution analysis of the role of chromatin regulators in early C. elegans embryogenesis. Dev Biol. 2015;398(2):153–162. doi: 10.1016/j.ydbio.2014.10.014. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lei H, Liu J, Fukushige T, Fire A, Krause M. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development. 2009;136(8):1241–1249. doi: 10.1242/dev.030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levantini E, Giorgetti A, Cerisoli F, Traggiai E, Guidi A, Martin R, Acampora D, Aplan PD, Keller G, Simeone A, et al. Unsuspected role of the brain morphogenetic gene Otx1 in hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(18):10299–10303. doi: 10.1073/pnas.1734071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Developmental cell. 2012;22(5):1101–1108. doi: 10.1016/j.devcel.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92(2):229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83(4):599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154(4):789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117(1):95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66(6):1105–1119. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- Mace DL, Weisdepp P, Gevirtzman L, Boyle T, Waterston RH. A High-Fidelity Cell Lineage Tracing Method for Obtaining Systematic Spatiotemporal Gene Expression Patterns in Caenorhabditis elegans. G3 (Bethesda) 2013;3(5):851–863. doi: 10.1534/g3.113.005918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Broitman-Maduro G, Choi H, Carranza F, Wu AC, Rifkin SA. MED GATA factors promote robust development of the C. elegans endoderm. Dev Biol. 2015;404(1):66–79. doi: 10.1016/j.ydbio.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Hill RJ, Heid PJ, Newman-Smith ED, Zhu J, Priess JR, Rothman JH. Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev Biol. 2005a;284(2):509–522. doi: 10.1016/j.ydbio.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Kasmir JJ, Zhu J, Rothman JH. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol. 2005b;285(2):510–523. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Lin R, Rothman JH. Dynamics of a developmental switch: recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Developmental biology. 2002;248(1):128–142. doi: 10.1006/dbio.2002.0721. [DOI] [PubMed] [Google Scholar]
- Maduro MF, Meneghini MD, Bowerman B, Broitman-Maduro G, Rothman JH. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Molecular cell. 2001;7(3):475–485. doi: 10.1016/s1097-2765(01)00195-2. [DOI] [PubMed] [Google Scholar]
- Mango SE, Thorpe CJ, Martin PR, Chamberlain SH, Bowerman B. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development. 1994;120(8):2305–2315. doi: 10.1242/dev.120.8.2305. [DOI] [PubMed] [Google Scholar]
- Martinelli SD, Brown CG, Durbin R. Gene expression and development databases for C. elegans. Seminars in Cell and Developmental Biology. 1997;8(5):459–467. doi: 10.1006/scdb.1997.0171. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales JR, Signore M, Acampora D, Simeone A, Bovolenta P. Otx genes are required for tissue specification in the developing eye. Development. 2001;128(11):2019–2030. doi: 10.1242/dev.128.11.2019. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Krause M, Weintraub H, Priess JR. The pie-1 and mex-1 genes and maternal control of blastomere identity in early C. elegans embryos. Cell. 1992;70(1):163–176. doi: 10.1016/0092-8674(92)90542-k. [DOI] [PubMed] [Google Scholar]
- Mello CC, Draper BW, Priess JR. The maternal genes apx-1 and glp-1 and establishment of dorsal-ventral polarity in the early C. elegans embryo. Cell. 1994;77(1):95–106. doi: 10.1016/0092-8674(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Schubert C, Draper B, Zhang W, Lobel R, Priess JR. The PIE-1 protein and germline specification in C. elegans embryos. Nature. 1996;382(6593):710–712. doi: 10.1038/382710a0. [DOI] [PubMed] [Google Scholar]
- Mickey KM, Mello CC, Montgomery MK, Fire A, Priess JR. An inductive interaction in 4-cell stage C. elegans embryos involves APX-1 expression in the signalling cell. Development. 1996;122(6):1791–1798. doi: 10.1242/dev.122.6.1791. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H. Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev Cell. 2007;12(2):287–299. doi: 10.1016/j.devcel.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Bock D, Gass P, Schwager A, Wolfer DP, Lipp HP, Schutz G. Defective limbic system in mice lacking the tailless gene. Nature. 1997;390(6659):515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- Moore JL, Du Z, Bao Z. Systematic quantification of developmental phenotypes at single-cell resolution during embryogenesis. Development. 2013;140(15):3266–3274. doi: 10.1242/dev.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz IP, Rothman JH. lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are regulated by maternal GLP-1 signaling in Caenorhabditis elegans. Development. 1996;122(12):4105–4117. doi: 10.1242/dev.122.12.4105. [DOI] [PubMed] [Google Scholar]
- Murgan S, Kari W, Rothbacher U, Iche-Torres M, Melenec P, Hobert O, Bertrand V. Atypical Transcriptional Activation by TCF via a Zic Transcription Factor in C. elegans Neuronal Precursors. Dev Cell. 2015;33(6):737–745. doi: 10.1016/j.devcel.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Bao Z. Automated Lineage and Expression Profiling in Live Caenorhabditis elegans Embryos. Cold Spring Harb Protoc. 2012;2012(8) doi: 10.1101/pdb.prot070615. [DOI] [PubMed] [Google Scholar]
- Murray JI, Bao Z, Boyle TJ, Boeck ME, Mericle BL, Nicholas TJ, Zhao Z, Sandel MJ, Waterston RH. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5(8):703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JI, Boyle TJ, Preston E, Vafeados D, Mericle B, Weisdepp P, Zhao Z, Bao Z, Boeck ME, Waterston R. Multidimensional regulation of gene expression in the C. elegans embryo. Genome research. 2012 doi: 10.1101/gr.131920.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G, Walton T, Murray JI, Raj A. Gene transcription is coordinated with, but not dependent on, cell divisions during C. elegans embryonic fate specification. Development. 2013;140(16):3385–3394. doi: 10.1242/dev.098012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kim S, Ishidate T, Bei Y, Pang K, Shirayama M, Trzepacz C, Brownell DR, Mello CC. Wnt signaling drives WRM-1/beta-catenin asymmetries in early C. elegans embryos. Genes & development. 2005;19(15):1749–1754. doi: 10.1101/gad.1323705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J. PAR proteins and the establishment of cell polarity during C. elegans development. Bioessays. 2005;27(2):126–135. doi: 10.1002/bies.20175. [DOI] [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8(6):867–879. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Onami S, Hamahashi S, Nagasaki M, Miyano S, Kitano H. Automatic acquisition of cell lineage through 4D microscopy and analysis of early C. elegans embryogenesis. Foundations in Systems Biology. 2001;1:39–55. [Google Scholar]
- Pabst O, Herbrand H, Takuma N, Arnold HH. NKX2 gene expression in neuroectoderm but not in mesendodermally derived structures depends on sonic hedgehog in mouse embryos. Dev Genes Evol. 2000;210(1):47–50. doi: 10.1007/pl00008188. [DOI] [PubMed] [Google Scholar]
- Park FD, Tenlen JR, Priess JR. C. elegans MOM-5/frizzled functions in MOM-2/Wnt-independent cell polarity and is localized asymmetrically prior to cell division. Curr Biol. 2004;14(24):2252–2258. doi: 10.1016/j.cub.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kidd AR, 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104(9):3231–3236. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Vorbruggen G, Jackle H. Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol Cell. 2000;6(1):203–209. [PubMed] [Google Scholar]
- Priess JR. Notch signaling in the C. elegans embryo. WormBook. 2005:1–16. doi: 10.1895/wormbook.1.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell. 1987;48(2):241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463(7283):913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JP, English K, Tenlen JR, Priess JR. Notch signaling and morphogenesis of single-cell tubes in the C. elegans digestive tract. Dev Cell. 2008;14(4):559–569. doi: 10.1016/j.devcel.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JL, Zacharias AL, Walton T, Burdick JT, Murray JI. A quantitative model of normal Caenorhabditis elegans embryogenesis and its disruption after stress. Developmental biology. 2013;374(1):12–23. doi: 10.1016/j.ydbio.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SM, Shetty P, Lin R. Identification of lineage-specific zygotic transcripts in early Caenorhabditis elegans embryos. Developmental biology. 2004;276(2):493–507. doi: 10.1016/j.ydbio.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, Priess JR, Davis RJ, Mello CC. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97(6):717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Rossel M, Capecchi MR. Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development. 1999;126(22):5027–5040. doi: 10.1242/dev.126.22.5027. [DOI] [PubMed] [Google Scholar]
- Santella A, Catena R, Kovacevic I, Shah P, Yu Z, Marquina-Solis J, Kumar A, Wu Y, Schaff J, Colon-Ramos D, et al. WormGUIDES: an interactive single cell developmental atlas and tool for collaborative multidimensional data exploration. BMC Bioinformatics. 2015;16:189. doi: 10.1186/s12859-015-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella A, Du Z, Bao Z. A semi-local neighborhood-based framework for probabilistic cell lineage tracing. BMC Bioinformatics. 2014;15:217. doi: 10.1186/1471-2105-15-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santella A, Du Z, Nowotschin S, Hadjantonakis AK, Bao Z. A hybrid blob-slice model for accurate and efficient detection of fluorescence labeled nuclei in 3D. BMC Bioinformatics. 2010;11:580. doi: 10.1186/1471-2105-11-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, Murray JI, Schanze K, Pozniakovski A, Niu W, Angermann K, Hasse S, Rupprecht M, Vinis E, Tinney M, et al. A genome-scale resource for in vivo tag-based protein function exploration in C. elegans. Cell. 2012;150(4):855–866. doi: 10.1016/j.cell.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H. Control of cell polarity and asymmetric division in C. elegans. Curr Top Dev Biol. 2012;101:55–76. doi: 10.1016/B978-0-12-394592-1.00003-X. [DOI] [PubMed] [Google Scholar]
- Sawa H, Korswagen HC. Wnt signaling in C. elegans. WormBook. 2013:1–30. doi: 10.1895/wormbook.1.7.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Developmental Biology. 1997;184(2):234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Greenwald I. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell. 1989;57(7):1237–1245. doi: 10.1016/0092-8674(89)90060-3. [DOI] [PubMed] [Google Scholar]
- Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Developmental biology. 2005;285(2):584–592. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Shi Y, Mello C. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 1998;12(7):943–955. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434(7032):462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Streit A, Kohler R, Marty T, Belfiore M, Takacs-Vellai K, Vigano MA, Schnabel R, Affolter M, Muller F. Conserved regulation of the Caenorhabditis elegans labial/Hox1 gene ceh-13. Developmental biology. 2002;242(2):96–108. doi: 10.1006/dbio.2001.0544. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. WormBook. 2005:1–10. doi: 10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka K, Mizumoto K, Sawa H. Wnt regulates spindle asymmetry to generate asymmetric nuclear beta-catenin in C. elegans. Cell. 2011;146(6):942–954. doi: 10.1016/j.cell.2011.07.043. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology. 1983;100(1):64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Takeshita H, Sawa H. Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes & development. 2005;19(15):1743–1748. doi: 10.1101/gad.1322805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans knockout consortium. large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2012;2(11):1415–1425. doi: 10.1534/g3.112.003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen H, Syed A, Nguyen BT, Lukacsovich T, Purcell J, Srivastava GP, Iron D, Gaudenz K, Nie Q, Wan FY, et al. Wingless directly represses DPP morphogen expression via an armadillo/TCF/Brinker complex. PLoS One. 2007;2(1):e142. doi: 10.1371/journal.pone.0000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90(4):695–705. doi: 10.1016/s0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, et al. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nature genetics. 2005;37(10):1035–1037. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Ahringer J. Microtubules are involved in anterior-posterior axis formation in C. elegans embryos. J Cell Biol. 2007;179(3):397–402. doi: 10.1083/jcb.200708101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T, Preston E, Nair G, Zacharias AL, Raj A, Murray JI. The Bicoid class homeodomain factors ceh-36/OTX and unc-30/PITX cooperate in C. elegans embryonic progenitor cells to regulate robust development. PLoS Genet. 2015;11(3):e1005003. doi: 10.1371/journal.pgen.1005003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301(5631):363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Hawkes PJ, Capecchi MR. Hox11 paralogous genes are essential for metanephric kidney induction. Genes & development. 2002;16(11):1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wolke U, Jezuit EA, Priess JR. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 2007;134(12):2227–2236. doi: 10.1242/dev.004952. [DOI] [PubMed] [Google Scholar]
- Wong A, Boutis P, Hekimi S. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics. 1995;139(3):1247–1259. doi: 10.1093/genetics/139.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1987. [Google Scholar]
- Wu Y, Wawrzusin P, Senseney J, Fischer RS, Christensen R, Santella A, York AG, Winter PW, Waterman CM, Bao Z, et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol. 2013;31(11):1032–1038. doi: 10.1038/nbt.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Baugh LR, Smith JJ, Roehrig C, Shen-Orr SS, Claggett JM, Hill AA, Slonim DK, Hunter CP. Pairing of competitive and topologically distinct regulatory modules enhances patterned gene expression. Mol Syst Biol. 2008;4:163. doi: 10.1038/msb.2008.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias AL, Walton T, Preston E, Murray JI. Quantitative Differences in Nuclear beta-catenin and TCF Pattern Embryonic Cells in C. elegans. PLoS Genet. 2015;11(10):e1005585. doi: 10.1371/journal.pgen.1005585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes & development. 2006;20(10):1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CU, Blauwkamp TA, Burby PE, Cadigan KM. Wnt-mediated repression via bipartite DNA recognition by TCF in the Drosophila hematopoietic system. PLoS Genet. 2014;10(8):e1004509. doi: 10.1371/journal.pgen.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12(24):3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11(21):2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]