Abstract
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts longer than 200 nucleotides. Many of these lncRNAs have regulatory functions and have recently emerged as major players in governing fundamental biological processes. Here we review the definition, distribution, identification, databases, analysis, classification and functions of lncRNAs. We also discuss the potential roles of lncRNAs in the etiological processes of psychiatric disorders and the implications for clinical diagnosis and treatment.
Keywords: LncRNAs, psychiatric disorders
Although only 1.2% of the mammalian genome encodes proteins, the genome is almost entirely transcribed, generating an enormous number of non-protein coding RNAs that include tens of thousands of long non-coding RNAs (lncRNAs) (> 200 nt) (Carninci et al., 2005; Mercer et al., 2009; Perkel, 2013). lncRNAs resemble protein-coding messenger RNAs (mRNAs) in sequence, but they can be distinguished based on multiple other features including expression levels, average number of exons, gene length, number of alternatively spliced isoforms, degree of tissue-specificity, signatures of conservation, presence of 5′ caps and polyA tails, rates of degradation/turn-over, etc. Expression of lncRNAs is tissue-, cell type- and developmental stage-specific (Amaral et al., 2008; Amaral and Mattick, 2008; Mercer et al., 2008). lncRNAs are expressed in various tissues (e.g., livers (Dong et al., 2014)) with over half of all lncRNAs expressed in the brain (Mercer et al., 2008). Most lncRNAs are permanently localized in the nucleus (Kapranov et al., 2007), with exceptions showing functionality in the cytoplasm (Carrieri et al., 2012; Kapranov et al., 2007; Mercer et al., 2009; Qureshi and Mehler, 2013; Taft et al., 2010). This specificity of tissue and subcellular distributions strongly suggests that the expression of lncRNAs is under precise regulatory control. This review summarizes the identification, databases, classification, analysis and functions of lncRNAs and their roles in various psychiatric disorders.
Identification of lncRNAs
Both RNA-Seq and microarray hybridization technologies can be used to identify lncRNAs, each with distinct advantages and limitations. Traditionally, lncRNAs can be identified by sequencing together with mRNAs and other RNAs using whole-transcriptome RNA-Seq technology. However, because lncRNAs tend to be expressed at much lower levels than mRNAs (Cabili et al., 2011; Cawley et al., 2004; Guttman et al., 2010; Kampa et al., 2004), to achieve adequate coverage of lncRNAs, the read depths of mRNAs will have to exceed around 10 times of the normal need in whole-transcriptome sequencing (Cabili et al., 2011; Derrien et al., 2012; Guttman et al., 2010; Yan et al., 2013). In addition, many lncRNAs are still undetectable by whole-transcriptome RNA-Seq even with the increase in overall read depth (Toung et al., 2011). In a word, this technique may not be the most effective to reliably and precisely quantify the low abundance lncRNA expression (Labaj et al., 2011). Instead, targeted RNA-Seq technology may better address these coverage issues, including lncRNA capture or rRNA+PolyA depletion to enrich lncRNAs before sequencing. Alternatively, some special microarrays can be used to identify low-abundance lncRNAs. For example, Arraystar Human LncRNA Microarray V3.0 (www.arraystar.com) has been designed to collect only lncRNAs and proximate mRNAs, so that users can enhance the expression signals of lncRNAs to a sufficient level by increasing the template input, and avoid cost on other unwanted sequences that whole-transcriptome RNA-Seq usually produces. However, the technical limitations of microarrays for the detection of low-abundant transcripts are well known, including low signal-to-noise ratios, and have in the past led to significant over-estimation of the extent and levels of intergenic transcription.
Many lncRNAs overlap with mRNAs, which brings a big challenge for researchers to distinguish between these two classes of transcripts if when the traditionally 3′-biased microarray probes are used, because these probes target the 3′UTRs that are shared by both types of transcripts. Transcript-specific probes that target only the exons or the splice junctions of each lncRNA transcript may enable more reliable and accurate detection of each individual transcript. Alternatively, directional RNA-Seq may also have significant advantages in addressing the issue of overlapping.
Many lncRNAs identified by RNA-Seq and microarray hybridization technologies fail to show conservation, and thus their functions are unclear. On this point, chromatin signature represents a distinct, third approach and may confer advantages (www.arraystar.com). On the basis of exploiting chromatin structure, this method identifies sets of functional large intergenic non-coding RNAs (lincRNAs) that show a high degree of evolutionary conservation. Mikkelsen et al. (2007) created a genome-wide chromatin-state map using chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-Seq) (Mikkelsen et al., 2007). This map marks the ‘K4–K36 domains’ from trimethylation of lysine 4 of histoneH3 (H3K4me3) at the promoters to trimethylation of lysine 36 of histone H3 (H3K36me3) along the length of the transcribed genomic regions. Guttman et al. (2009) searched for K4–K36 domains in this genome-wide chromatin-state map that reside outside known protein coding gene loci and do not overlap known miRNAs or endogenous short interfering RNAs (siRNAs), and then systematically revealed 1,675 K4–K36 (1,250 conservatively defined) domains (Guttman et al., 2009). Most of these K4–K36 domains encode functional lincRNAs that are highly conserved in nucleotide sequence and chromatin structure and are implicated in diverse biological processes including transcription of these sites (Guttman et al., 2009).
Databases of lncRNAs
The sequence information for lncRNAs are available in several public databases, including RefSeq release 60, UCSC hg19, GENCODE 17, RNAdb 2.0 (Pang et al., 2007), NRED (Dinger et al., 2009), Valadkhan Lab Functional lncRNA Database, LncRNADisease, lncRNAdb, NONCODE (Bu et al., 2012a) and Ensembl 37.59. Among them, RefSeq, GENCODE, Ensembl and UCSC are gene annotation databases that include varying numbers of annotated lncRNAs; Valadkhan Lab Functional lncRNAs, LncRNADisease and lncRNAdb are lncRNA databases; RNAdb 2.0 and NONCODE are RNA databases including lncRNAs; and NRED is a microarray expression database of lncRNAs. Some databases include unannotated RNAs that are not well defined and might have little or no expression data. To extract effective lncRNAs from these unannotated RNAs, one can screen these databases according to certain computational pipeline. For example, one can filter transcripts of known coding RNAs, structural RNAs (e.g., tRNAs, rRNAs), small ncRNAs, and highly similar sequences, examine whether each transcript contains a significant open reading frame (ORF), and, finally, retain only multiexonic transcripts > 200 nt.
Several ground-breaking landmark publications also reported lncRNAs 30,37–50. These lncRNAs (www.arraystar.com) include: (1) LincRNAs (1st set). Khalil et al. identified and characterized 3,289 lincRNAs by searching for intergenic K4-K36 domains in genome-wide chromatin-state maps (Khalil et al., 2009). (2) LincRNAs (2nd set). Cabili et al. defined a reference catalog of more than 8,000 lincRNAs using RNA-Seq data and public database information (Cabili et al., 2011). A total of 14,353 transcripts expressed from 4,662 stringently-defined lincRNAs were identified. (3) Transcribed Ultra-Conserved Regions encoding lncRNAs (T-UCRs). Ultra-conserved regions (UCRs) are intra- and intergenic sequences with >200nt that are 100% identical among humans, mice, and rats. 481 UCRs were identified by Bejerano et al. (Bejerano et al., 2004). 475 UCRs encode T-UCRs. (4) HOX LncRNAs. Rinn et al. identified 407 transcribed regions within the four HOX loci in humans (101 HOX exons, 75 introns and 231 intergenic ncRNA transcripts) (Rinn et al., 2007). (5) LncRNAs with Enhancer-like Function (LncRNA-a). Orom et al. identified about 3,000 lncRNAs with lncRNA-a using the GENCODE annotation (Harrow et al., 2006; Orom et al., 2010). (6) Finally, some lncRNAs with ORFs. Some sense-overlapping lncRNAs have an ORF that shares the same start codon as a protein-coding transcript; however, these short ORFs are unlikely to encode a protein for some reasons. About 709 sense-overlapping lncRNAs with such ORF types have been reported (www.arraystar.com). Actually, a large proportion of these lncRNAs have already been deposited into the above databases. Some of the putative lncRNAs lack expression data or have not been characterized in details either, and thus the effective lncRNAs should be extracted using the computational filtering pipelines as described above.
Classification and functions of lncRNAs
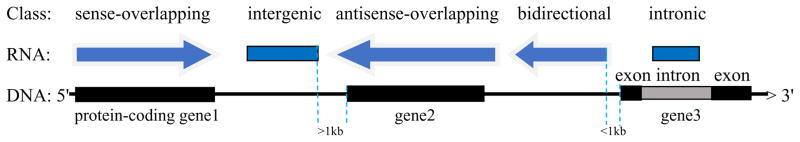
LncRNAs have recently emerged as major players in governing fundamental biological processes. Recent evidence suggests that lncRNAs are involved in a wide variety of cellular functions, including epigenetic silencing, transcriptional regulation, RNA processing and modification (Amaral et al., 2008; Mercer et al., 2009; Wang et al., 2008), and implicated in neural plasticity (Sartor et al., 2012), neuropathological process (Bu et al., 2012b), neurotransmission (Qureshi et al., 2010), and stress response (Sartor et al., 2012). A large proportion of lncRNAs may cis-regulate their neighboring protein-coding genes, so analyzing the genomic context of lncRNAs can help predict their functional roles. According to the positional relationship between lncRNAs and their associated protein-coding genes, lncRNAs can be classified as intergenic, intronic, antisense, sense overlapping, and bidirectional lncRNAs (www.arraystar.com) (Figure 1). 1. Intergenic lncRNAs (lincRNAs) are located between protein-coding genes and are at least 1 kb away from the nearest protein-coding genes. For example, a sense lincRNA, i.e., proliferating cell nuclear antigen pseudogene 1 (PCNAP1; 1,055 nt), is located between ADH4 and ADH6. LincRNAs are implicated in diverse biological processes, including embryonic stem cell pluripotency, cell-cycle regulation and immune surveillance. They usually interact with chromatin modifying proteins (PRC2, SCMX and CoREST) to regulate the expression of proximate genes (Khalil et al., 2009). 2. Intronic lncRNAs are located within the intron of annotated protein coding genes. Most of them show the same tissue expression patterns as the host genes, and may stabilize the host transcripts or regulate their alternative splicing (Nakaya et al., 2007). 3. Sense-overlapping lncRNAs can be considered transcript isoforms of protein-coding mRNAs, because they overlap with the host gene on the same genomic strand. The majority of these IncRNAs lack substantial open reading frames (ORFs) for protein translation. Some others contain ORFs that share the same start codons as the host transcripts but are unlikely to encode a protein because of non-sense mediated decay (NMD) that limits the translation of mRNAs with premature termination stop codons and triggers NMD-mediated destruction of the mRNA, or because of an upstream alternative ORF which inhibits the translation of the predicted ORF. Some sense-overlapping lncRNAs share stop codons with host mRNAs on the same genomic strand. For example, IPO11-LRRC70 read-through overlaps IPO11 mRNA and shares a stop codon on the same genomic strand. This lncRNA is close to the peak association marker rs7445832 (p=6.2×10−9) for alcohol and nicotine codependence in European-Americans and Australians as identified in a GWAS (Zuo et al., 2013). 4. Antisense-overlapping lncRNAs: Natural antisense transcripts (NATs) are RNA molecules that are transcribed from the opposite strand of many protein coding (sense) genes and overlap in part with well-defined spliced sense or intronless sense mRNAs. NATs bind to sense RNA and/or proteins to regulate transcription and translation. For example, a large antisense-overlapping lncRNA, i.e., LOC100507053 (213kb), covers ADHs 5, 4, 6 and 1A genes that form a risk genomic region for alcoholism demonstrated by numerous GWASs and candidate gene studies (Gelernter et al., 2009; Li et al., 2011, 2012). This class of lncRNA frequently uses diverse transcriptional and post-transcriptional regulatory mechanisms to fulfill a wide variety of biological roles (Sartor et al., 2012). These IncRNAs usually have a tendency to undergo fewer splicing events and typically show lower abundance than sense transcripts (He et al., 2008). The basal expression levels of antisense-overlapping lncRNAs and sense mRNAs in different tissues and cell lines can be either positively or negatively regulated (Katayama et al., 2005; Okada et al., 2008). 5. Bidirectional lncRNAs are oriented head to head with a protein-coding gene within 1kb. A bidirectional lncRNA transcript exhibits an expression pattern similar to its host gene, suggesting that they may be subject to shared regulatory pressures. However, discordant expression relationships between bidirectional lncRNAs and protein coding gene pairs have also been identified, challenging the assertion that lncRNA transcription occurs solely to “open” chromatin to promote the expression of neighboring coding genes (Chakalova et al., 2005; Mercer et al., 2008; Struhl, 2007).
Figure 1. Classification of LncRNAs.
[Sense, LncRNAs are transcribed from the same genomic strand as the protein-coding mRNAs; Intergenic, located between two protein-coding genes and at least 1 kb away from these genes; Antisense, transcribed from the antisense strand; Intronic, located within the intron of protein coding genes; Bidirectional, oriented head to head with a protein-coding gene within 1kb. Arrow direction: 5′ → 3′]
Additionally, some lncRNAs may trans-regulate distant protein-coding genes. RNA Immunoprecipitation sequencing (RIP-Seq) or microarray (RIP-Chip) technology has identified many lncRNAs that interact with specific RNA binding proteins (RBPs) (Zhao et al., 2010). LncRNAs may function by interacting with these RBPs.
Roles of lncRNAs in psychiatric disorders
Recent findings that suggest a functional role of lncRNAs in various aspects of cell biology have increased awareness of their potential to contributing towards diseases. To determine which lncRNAs are related to various diseases becomes the logical and necessary next step in identifying the missing regulatory pathways following a long history of attention to the coding regions and small ncRNAs like miRNAs. For instance, by analyzing the differential expression, fold change of expression, classification, and regulatory effects of lncRNAs, many association studies have identified lncRNAs in association with Alzheimer’s disease, substance dependence, schizophrenia, bipolar disorder, depression, autism spectrum disorder (ASD) and panic disorder.
Alzheimer’s disease
Alzheimer’s disease (AD) is the main cause of dementia in the elderly population worldwide. Adult neurogenesis appears to be upregulated very early in AD pathogenesis in response to some specific aggregates of beta-amyloid (Aβ) peptides, exhausting the neuronal stem cell pools in the brain. LncRNAs exhibit aberrant expression in AD. Recently, it has been demonstrated that an antisense lncRNA, BACEAS, exhibited elevated expression in several brain regions in individuals with AD. BACEAS regulates the expression of the sense beta-secretase-1 (BACE1) gene, a crucial enzyme in AD etiology (Faghihi et al., 2008; Modarresi et al., 2011). Upon exposure to various cell stressors including beta-amyloid 1-42 (Aβ 1-42), expression of BACE1-AS becomes elevated, increasing BACE1 mRNA stability and generating additional Aβ 1-42 through a post-transcriptional feed-forward mechanism. Alternatively, alteration of the expression for BACE1-AS may also mediate changes at an epigenetic level to effect gene expression and contribute to disease etiology, suggesting that this lncRNA may serve as an attractive drug target candidate for AD (Faghihi et al., 2008) (www.arraystar.com).
Recent studies indicated that sortilin-related receptor 1 (SORL1) is a risk gene for late-onset AD. An antisense-overlapping lncRNA, 51A, maps in intron 1 of SORL1 gene and is frequently upregulated in expression in the cerebral cortices of individuals with AD. 51A expression drives a splicing shift of SORL1 from the synthesis of the canonical long protein variant A to an alternatively spliced protein form. This process, resulting in a decreased synthesis of SORL1 variant A, is associated with impaired processing of amyloid precursor protein (APP), leading to increased Aβ formation that is implicated in neurodegeneration. (Ciarlo et al., 2013)
Another lncRNA, Brain Cytoplasmic RNA 200-Alpha (BC200), a human analog of brain cytoplasmic RNA 1 (BC1), is a translational regulator that is selectively targeted to somatodendritic domains of neurons. It modulates local protein synthesis in postsynaptic dendritic microdomains, contributing to the maintenance of long-term synaptic plasticity. Dysfunctional plasticity has been posited as a starting point for the neurodegenerative changes as observed in AD. BC200 was significantly up-regulated in AD brains, specifically in Brodmann’s area 9 and the hippocampus, regions that frequently are involved in the disease. Relative BC200 levels in these brain areas are correlated with the disease severity. In more advanced stages of the disease, BC200 is mis-localized and clustered in the perikaryon. These observations suggest that deregulation of these synaptic lncRNAs is involved in the synaptic and neural network dysfunction in both early and later stages of AD. (Mus et al., 2007)
Other lncRNAs implicated in the etiology of AD include GDNF-AS1 (Airavaara et al., 2011), CDKN2B-AS1 (Zuchner et al., 2008), HAR1A (Harries, 2012), HAR1B (Harries, 2012), SNHG3 and SOX2-OT (Arisi et al., 2011).
Substance dependence
Dysregulation of many lncRNAs has been reported to contribute to substance use disorders including alcohol, nicotine, heroin and cocaine dependence. For example, nuclear enriched abundant transcript 2 (NEAT2), an lncRNA regulating synapse formation (Bernard et al., 2010), was up-regulated in alcoholics’ brain (Kryger et al., 2012); NEAT2, NEAT1, myocardial infarction associated transcript (MIAT) and maternally expressed 3 transcript (MEG3) were up-regulated in the nucleus accumbens (NAc) of heroin abusers (Michelhaugh et al., 2011); and NEAT2, MIAT, MEG3 and empty spiracles homeobox 2 opposite strand transcript (EMX2OS) were elevated in the NAc of cocaine abusers (Michelhaugh et al., 2011). Smokers had dramatically elevated imprinted maternally expressed transcript (H19) expression in airway epithelium (Kaplan et al., 2003); demethylation of H19 was correlated to chronic alcohol use in males (Ouko et al., 2009); and many lncRNAs were reported to be involved in cocaine-induced neural plasticity in the NAc and in risk for cocaine dependence (Bu et al., 2012b).
Brain-derived neurotrophic factor (BDNF) gene, a gene known to be involved in substance dependence, e.g., cocaine dependence (Ghitza et al., 2010), is controlled by a conserved antisense lncRNA, i.e., BDNF-AS (Modarresi et al., 2012). BDNF-AS suppresses BDNF mRNA expression by altering chromatin structure at the BDNF gene locus. Inhibiting BDNF-AS by siRNA or other methods robustly increased BDNF mRNA and protein expression and enhanced neuronal outgrowth. BDNF-AS is dysregulated in response to chronic drug use and contributes to drug-seeking behaviors. (Sartor et al., 2012)
Schizophrenia
Alternative splicing of some mRNAs is associated with the pathology of schizophrenia (SZ). Many disease-associated genes displayed aberrant splicing patterns. Gomafu is an lncRNA highly regulated by neural activity. It binds directly to splicing factors, and is significantly downregulated in the cortex of SZ patients. Modulation of Gomafu expression alters splicing patterns of at least two SZ-associated genes. Knockdown of Gomafu resulted in the upregulation of SZ pathology-related splice variants of DISC1 and ERBB4, consistent with the observation that overexpression of these same splice variants are associated with SZ. In contrast, Gomafu overexpression produced significant downregulation of the same disease-associated splice variants of both genes (Barry et al., 2014). This suggests that the lncRNA Gomafu may contribute to the pathogenic splicing pattern of these key SZ genes. (Guennewig and Cooper, 2014)
DLG2AS, aka PSZA11q14, is an antisense-overlapping lncRNA to DLG-2 gene, located within the first intron of DLG-2. It acts as an antisense regulator of DLG-2, which controls the assembly of functional N-methyl-D-aspartate (NMDA) receptors. Its expression was reduced in the brains of SZ patients, specifically in Brodmann’s areas 9, 21 and 22 and in the hippocampus, indicating that it may be involved in at least some cases of SZ (Polesskaya et al., 2003). Schizophrenia spectrum disorders have also been linked to the reelin (RELN) gene and its antisense transcript HAR1 (Tamura et al., 2007). Other lncRNAs have been reported to be implicated in SZ, including C6UAS (Morelli et al., 2000) and LINC00271 (Amann-Zalcenstein et al., 2006).
Autism spectrum disorders
Autism spectrum disorders (ASDs) include various developmental disorders, including autism, pervasive developmental disorder not otherwise specified (PDD-NOS), Rett syndrome, Fragile X syndrome and the Asperger syndrome. Common symptoms of the various ASDs include problems of reciprocal social interactions, verbal and non-verbal communication, and rigid and stereotyped behaviors. ASD is a clinically and etiologically heterogeneous disorder with a complex genetic architecture. In the last decade, several studies reported aberrant expression of lncRNAs, suggesting that lncRNAs contributed to ASD risk. Recently, Ziats and Rennert (2013) showed that over 200 lncRNAs were differentially expressed in a microarray of postmortem prefrontal cortex and cerebellum tissue of ASD patients (Ziats and Rennert, 2013).
(1) Autism
Vincent et al. (2002) identified a novel autism locus, which includes the gene RAY1/ST7 (Vincent et al., 2002). This locus contains at least four lncRNAs, i.e., ST7OT1-4, both on the sense and antisense strands that potentially regulate RAY1/ST7. Additionally, moesin regulates neuronal architecture and the lncRNA MSNP1AS, transcribed in antisense to a moesin pseudogene, is 94% identical and antisense to moesin and can bind moesin mRNA (Kerin et al., 2012; Le Meur et al., 2005). Overexpression of MSNP1AS in cultured cells led to decreased moesin levels while MSNP1AS transcript levels were 12-fold higher in postmortem brain samples from autism cases (Kerin et al., 2012). High levels of the MSNP1AS transcript were associated with the presence of an autism risk SNP and thus MSNP1AS is strongly positioned to be an lncRNA risk factor for autism (Kerin et al., 2012). Finally, mutations in the X-chromosome PTCHD1 gene have been reported to involve X-linked intellectual disability (ID) and autism (Filges et al., 2011; Noor et al., 2010). Several lines of evidence suggest that PTCHD1 might have a causative role in a subset of ID and/or autism patients (Filges et al., 2011). On the antisense strand of the PTCHD1 gene, several overlapping lncRNAs (PTCHD1AS1, PTCHD1AS2 and PTCHD1AS3) were detected, which may serve as regulators for PTCHD1.
(2) Rett syndrome
Rett syndrome is a rare, severe, “girls only” form of autism, usually identified in the first two years of life. It is characterized by arrested development between 6 and 18 months of age, regression of acquired skills, loss of speech, stereotypical movements, seizures, and ID. Mutations in the methyl CpG binding protein 2 (MECP2), which binds methylated CpGs and can both activate and repress transcription, were first described to be the cause of the disorder (Amir et al., 1999). While assessing the transcriptome of male Mecp2 hemizygous knockout mouse brains (Petazzi et al., 2013), it was revealed that the lncRNAs AK081227 and AK087060 were both significantly upregulated as compared to wild-type littermates. Importantly, overexpression of AK08127 was associated with the downregulation of its host coding protein gene, the GABA receptor subunit Rho 2. This suggests that transcriptional dysregulation of lncRNAs may have the capacity to contribute to the etiology of Rett syndrome.
(3) Fragile X Syndrome
Fragile X syndrome (FXS) is the most common known single gene cause of ASD. It is inherited via an X-linked dominant pattern and characterized by moderate to severe mental retardation, macroorchidism, and distinct facial features. The disorder is caused by an unstable expansion of a CGG repeat in the fragile X mental retardation 1 gene (FMR1), that leads to the silencing of the gene by methylation of the repeat and the promoter (Sutcliffe et al., 1992), resulting in decreased fragile X mental retardation protein (FMRP) levels in the brain (Devys et al., 1993). Accumulating evidence suggests that the etiology of the disorder is influenced by lncRNAs. FMRP, the protein encoded by FMR1, acts as a translational repressor of specific mRNAs at the synapse and is associated with the dendritic lncRNA and BC1 (Zalfa et al., 2003). BC1 enables the interaction of FMRP with the target mRNAs; and FMRP can directly bind to BC1 and its human analog BC200 via its N-terminus. Of note, the 5′ stem loop of BC1 is involved in FMRP recognition and this region is complementary to FMRP target mRNAs (Zalfa et al., 2005). Taken together, the studies suggested that BC1 is a lncRNA that is essential for the repression of mRNAs via FMRP and loss of this repression in FXS patients could result in synaptic dysfunction.
The promoter of FMR1 is bidirectional and can also give rise to the lncRNA FMR4 (FMR1AS1 or ASFMR1), a gene transcribed in the antisense orientation and overlaps the CGG repeat region. FMR4 is similar to FRM1 in being silenced in FXS patients. Alternative splicing of FMR4 seems to exhibit premutation specific profiles and is upregulated in premutation carriers (Khalil et al., 2008; Ladd et al., 2007). Following siRNA knockdown of FMR4, alterations in cell cycle and apoptosis were reported. Conversely, overexpression of FMR4 resulted in increased cell proliferation. Additionally, knockdown of FMR4 did not influence FMR1 expression and vice versa, suggesting an independent mechanism from FMR1 (Khalil et al., 2008). Together, these findings point toward a contribution of FMR4 in the pathology of FXS.
Recently, Pastori et al. (2014) discovered two new lncRNAs in the FMR1 gene locus: FMR5 and FMR6. FMR5 was similarly expressed in brain regions from unaffected and premutation individuals and full mutation patients, whereas FMR6 was silenced in full mutation and premutation carriers. According to the authors, this might suggest an abnormal transcription or chromatin remodeling prior to transition to the full mutation. In addition to the finding that both FMR5 and FMR6 are expressed in blood leukocytes, these lncRNAs are potentially useful as biomarkers in FXS. (Pastori et al., 2014)
Multiple mental illnesses
Some lncRNAs have been suggested to be involved in multiple mental illnesses. Disruption of the ‘disrupted in schizophrenia-1’ (DISC1) locus has been linked to the development of schizophrenia, schizoaffective disorder, bipolar disorder, major depression and autistic spectrum disorders (Brandon et al., 2009; Chubb et al., 2008). DISC1 is regulated by lncRNA DISC2 (Millar et al., 2000), which may also represent an excellent candidate for susceptibility to these disorders (Taylor et al., 2003). Additionally, lncRNA DAOA-AS1 (G72/G30) has been associated with schizophrenia (Ma et al., 2006; Yue et al., 2006), bipolar disorders (Hattori et al., 2003) and panic disorders (Schumacher et al., 2005).
In summary, lncRNAs are a diverse group of non-coding RNAs that play critical roles in many cellular processes. Accumulating evidence suggests that they are involved in many psychiatric disorders. Having a better understanding of lncRNAs’ roles in psychiatric disorders will not only enrich functional annotation of the non-coding regions of human genome, but also have tremendous potential to advance our understanding of specific regulatory pathways for the risk DNA variants to affect the development of psychiatric disorders. These studies have the potential to discover novel biomarkers and drug targets that can be used to facilitate the diagnosis, treatment and prognosis of psychiatric disorders.
Acknowledgments
This work was supported in part by NIH grants K01 DA029643, R21 AA021380, R21 AA020319 and R21AA023237, the National Alliance for Research on Schizophrenia and Depression (NARSAD) Award 17616 and the ABMRF/The Foundation for Alcohol Research grant award.
Footnotes
Conflict of interest
Authors declare no conflict of interest related to this review.
References
- Airavaara M, Pletnikova O, Doyle ME, Zhang YE, Troncoso JC, Liu QR. Identification of novel GDNF isoforms and cis-antisense GDNFOS gene and their regulation in human middle temporal gyrus of Alzheimer disease. J Biol Chem. 2011;286:45093–45102. doi: 10.1074/jbc.M111.310250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann-Zalcenstein D, Avidan N, Kanyas K, Ebstein RP, Kohn Y, Hamdan A, et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur J Hum Genet. 2006;14:1111–1119. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Arisi I, D’Onofrio M, Brandi R, Felsani A, Capsoni S, Drovandi G, et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J Alzheimers Dis. 2011;24:721–738. doi: 10.3233/JAD-2011-101881. [DOI] [PubMed] [Google Scholar]
- Barry G, Briggs JA, Vanichkina DP, Poth EM, Beveridge NJ, Ratnu VS, et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry. 2014;19:486–494. doi: 10.1038/mp.2013.45. [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon NJ, Millar JK, Korth C, Sive H, Singh KK, Sawa A. Understanding the role of DISC1 in psychiatric disease and during normal development. J Neurosci. 2009;29:12768–12775. doi: 10.1523/JNEUROSCI.3355-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012a;40:D210–215. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Hu Z, Chen F, Zhu R, Deng Y, Shao X, et al. Transcriptome analysis of long non-coding RNAs of the nucleus accumbens in cocaine-conditioned mice. J Neurochem. 2012b;123:790–799. doi: 10.1111/jnc.12006. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Replication and transcription: shaping the landscape of the genome. Nat Rev Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. The DISC locus in psychiatric illness. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Ciarlo E, Massone S, Penna I, Nizzari M, Gigoni A, Dieci G, et al. An intronic ncRNA-dependent regulation of SORL1 expression affecting Abeta formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis Model Mech. 2013;6:424–433. doi: 10.1242/dmm.009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devys D, Lutz Y, Rouyer N, Bellocq JP, Mandel JL. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Jia D, Xue P, Cui X, Li K, Zheng S, et al. Genome-wide analysis of long noncoding RNA (lncRNA) expression in hepatoblastoma tissues. PLoS One. 2014;9:e85599. doi: 10.1371/journal.pone.0085599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filges I, Rothlisberger B, Blattner A, Boesch N, Demougin P, Wenzel F, et al. Deletion in Xp22.11: PTCHD1 is a candidate gene for X-linked intellectual disability with or without autism. Clin Genet. 2011;79:79–85. doi: 10.1111/j.1399-0004.2010.01590.x. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler HR, Panhuysen C, Weiss RD, Brady K, Poling J, et al. Dense genomewide linkage scan for alcohol dependence in African Americans: significant linkage on chromosome 10. Biol Psychiatry. 2009;65:111–115. doi: 10.1016/j.biopsych.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghitza UE, Zhai H, Wu P, Airavaara M, Shaham Y, Lu L. Role of BDNF and GDNF in drug reward and relapse: a review. Neurosci Biobehav Rev. 2010;35:157–171. doi: 10.1016/j.neubiorev.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guennewig B, Cooper AA. The central role of noncoding RNA in the brain. Int Rev Neurobiol. 2014;116:153–194. doi: 10.1016/B978-0-12-801105-8.00007-2. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans. 2012;40:902–906. doi: 10.1042/BST20120020. [DOI] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, et al. Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet. 2003;72:1131–1140. doi: 10.1086/374822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R, Luettich K, Heguy A, Hackett NR, Harvey BG, Crystal RG. Monoallelic up-regulation of the imprinted H19 gene in airway epithelium of phenotypically normal cigarette smokers. Cancer Res. 2003;63:1475–1482. [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kerin T, Ramanathan A, Rivas K, Grepo N, Coetzee GA, Campbell DB. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci Transl Med. 2012;4:128ra140. doi: 10.1126/scitranslmed.3003479. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryger R, Fan L, Wilce PA, Jaquet V. MALAT-1, a non protein-coding RNA is upregulated in the cerebellum, hippocampus and brain stem of human alcoholics. Alcohol. 2012;46:629–634. doi: 10.1016/j.alcohol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Labaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of RNA-Seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27:i383–391. doi: 10.1093/bioinformatics/btr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol Psychiatry. 2011;70:504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhao H, Gelernter J. Further clarification of the contribution of the ADH1C gene to vulnerability of alcoholism and selected liver diseases. Hum Genet. 2012;131:1361–1374. doi: 10.1007/s00439-012-1163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Qin W, Wang XY, Guo TW, Bian L, Duan SW, et al. Further evidence for the association between G72/G30 genes and schizophrenia in two ethnically distinct populations. Mol Psychiatry. 2006;11:479–487. doi: 10.1038/sj.mp.4001788. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Patel NS, Sahagan BG, Wahlestedt C, Lopez-Toledano MA. Knockdown of BACE1-AS Nonprotein-Coding Transcript Modulates Beta-Amyloid-Related Hippocampal Neurogenesis. Int J Alzheimers Dis. 2011;2011:929042. doi: 10.4061/2011/929042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli C, Magnanini C, Mungall AJ, Negrini M, Barbanti-Brodano G. Cloning and characterization of two overlapping genes in a subregion at 6q21 involved in replicative senescence and schizophrenia. Gene. 2000;252:217–225. doi: 10.1016/s0378-1119(00)00231-6. [DOI] [PubMed] [Google Scholar]
- Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor A, Whibley A, Marshall CR, Gianakopoulos PJ, Piton A, Carson AR, et al. Disruption at the PTCHD1 Locus on Xp22.11 in Autism spectrum disorder and intellectual disability. Sci Transl Med. 2010;2:49ra68. doi: 10.1126/scitranslmed.3001267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, et al. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum Mol Genet. 2008;17:1631–1640. doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0--an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori C, Peschansky VJ, Barbouth D, Mehta A, Silva JP, Wahlestedt C. Comprehensive analysis of the transcriptional landscape of the human FMR1 gene reveals two new long noncoding RNAs differentially expressed in Fragile X syndrome and Fragile X-associated tremor/ataxia syndrome. Hum Genet. 2014;133:59–67. doi: 10.1007/s00439-013-1356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel JM. Visiting “noncodarnia”. Biotechniques. 2013;54:301, 303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- Petazzi P, Sandoval J, Szczesna K, Jorge OC, Roa L, Sayols S, et al. Dysregulation of the long non-coding RNA transcriptome in a Rett syndrome mouse model. RNA Biol. 2013;10:1197–1203. doi: 10.4161/rna.24286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya OO, Haroutunian V, Davis KL, Hernandez I, Sokolov BP. Novel putative nonprotein-coding RNA gene from 11q14 displays decreased expression in brains of patients with schizophrenia. J Neurosci Res. 2003;74:111–122. doi: 10.1002/jnr.10752. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Long Non-coding RNAs: Novel Targets for Nervous System Disease Diagnosis and Therapy. Neurotherapeutics. 2013;10:632–646. doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, St Laurent G, 3rd, Wahlestedt C. The Emerging Role of Non-Coding RNAs in Drug Addiction. Front Genet. 2012;3:106. doi: 10.3389/fgene.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Abou Jamra R, Becker T, Klopp N, Franke P, Jacob C, et al. Investigation of the DAOA/G30 locus in panic disorder. Mol Psychiatry. 2005;10:428–429. doi: 10.1038/sj.mp.4001598. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nelson DL, Zhang F, Pieretti M, Caskey CT, Saxe D, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol Psychiatry. 2007;12:519, 593–600. doi: 10.1038/sj.mp.4002014. [DOI] [PubMed] [Google Scholar]
- Taylor MS, Devon RS, Millar JK, Porteous DJ. Evolutionary constraints on the Disrupted in Schizophrenia locus. Genomics. 2003;81:67–77. doi: 10.1016/s0888-7543(02)00026-5. [DOI] [PubMed] [Google Scholar]
- Toung JM, Morley M, Li M, Cheung VG. RNA-sequence analysis of human B-cells. Genome Res. 2011;21:991–998. doi: 10.1101/gr.116335.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JB, Petek E, Thevarkunnel S, Kolozsvari D, Cheung J, Patel M, et al. The RAY1/ST7 tumor-suppressor locus on chromosome 7q31 represents a complex multi-transcript system. Genomics. 2002;80:283–294. doi: 10.1006/geno.2002.6835. [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- Yue W, Liu Z, Kang G, Yan J, Tang F, Ruan Y, et al. Association of G72/G30 polymorphisms with early-onset and male schizophrenia. Neuroreport. 2006;17:1899–1902. doi: 10.1097/WNR.0b013e3280102ed4. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Adinolfi S, Napoli I, Kuhn-Holsken E, Urlaub H, Achsel T, et al. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J Mol Neurosci. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Gilbert JR, Martin ER, Leon-Guerrero CR, Xu PT, Browning C, et al. Linkage and association study of late-onset Alzheimer disease families linked to 9p21.3. Ann Hum Genet. 2008;72:725–731. doi: 10.1111/j.1469-1809.2008.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang XY, Wang F, Li CS, Lu L, Ye L, et al. Genome-wide significant association signals in IPO11-HTR1A region specific for alcohol and nicotine codependence. Alcohol Clin Exp Res. 2013;37:730–739. doi: 10.1111/acer.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]