Summary
COX-2 and its product PGE2 enhance carcinogenesis and tumor progression, which has been previously reported in melanoma. As most COX inhibitors cause much toxicity, the downstream microsomal PGE2 synthase-1 (mPGES1) is a consideration for targeting. Human melanoma TMAs were employed for testing mPGES1 protein staining intensity and percentage levels and both increased with clinical stage; employing a different Stage III TMA, mPGES1 intensity (not percentage) associated with reduced patient survival. Our results further show that iNOS was also highly expressed in melanoma tissues with high mPGES1 levels, and iNOS-mediated NO promoted mPGES1 expression and PGE2 production. An mPGES1specific inhibitor (CAY10526) as well as siRNA attenuated cell survival and increased apoptosis. CAY10526 significantly suppressed tumor growth and increased apoptosis in melanoma xenografts. Our findings support the value of a prognostic and predictive role for mPGES1, and suggest targeting this molecule in the PGE2 pathway as another avenue toward improving melanoma therapy.
Keywords: Prostaglandin E2, Microsomal PGE2 synthase-1, inducible nitric oxide synthase, melanoma, inflammation, cell survival
Introduction
Melanoma is the most aggressive form of skin cancer. Although melanoma constitutes a minority of skin cancers, it is considered to be the most deadly (Siegel et al., 2014). In the past three years, developments in targeted- and immuno-therapy have significantly impacted the lethality of melanoma (Shtivelman et al., 2014). Both the targeted- and immuno-therapy advances indicate that melanoma is responsive to systemic manipulation, but that research is urgently needed to address the molecular and biologic mechanisms that drive resistance. This realization also highlights the need for development of novel combination treatment strategies to overcome plasticity and resistance, such as intrinsic inflammatory ones. Melanoma progression is strongly associated with chronic inflammation because melanoma cells secrete high levels of proinflammatory cytokines and prostaglandins (PGs) (Hensler and Mueller, 2013; Richmond et al., 2009; Singh and Katiyar, 2011). PGE2, the most abundant PG in several types of human malignancies, is often associated with poor prognosis, and PGE2 has been shown to enhance carcinogenesis, angiogenesis, tumor growth, and metastasis (Brown and DuBois, 2005; Howe, 2007). Additionally, cyclooxygenase-2 (COX-2), one of the enzymes responsible for PGE2 production, has been correlated with the progression of melanoma (Johansson et al., 2009; Meyer et al., 2012). Thus, there is substantial interest in COX-2/PGE2 pathways as therapeutic targets for melanoma.
Nonsteroidal anti-inflammatory drugs (NSAIDs), such as aspirin and ibuprofen reduce the biosynthesis of PGE2 by inhibiting both COX-1 and COX-2 activity (Funk, 2001). Postmenopausal white women treated with aspirin had lower melanoma risk than those who did not take aspirin (Gamba et al., 2013). However, long-term use of NSAIDs can cause life-threatening side effects, such as gastrointestinal injury (Rainsford, 2007). COX-2–specific inhibitors were designed to minimize these side effects, but a recent clinical study indicated significantly increased risk for cardiovascular events such as sudden myocardial infarction and thrombosis owing to imbalance in the levels of prostacyclin (PGI2) and thromboxane A2, which are downstream products of COX-2 (Iyer et al., 2009). Because effective and safe alternatives to reduce PGE2 levels are needed, the focus of research has shifted to devising inhibitors for other downstream enzymes of COX-2, such as PGE2 synthase (PGES), as potential anti-inflammatory and anti-tumor therapies.
Microsomal PGES1 (mPGES1), mPGES2, and cytosolic PGES have been identified as potential inhibitors, but only mPGES1 is markedly induced by proinflammatory stimuli, often with the concomitant induction of COX-2. mPGES1 is functionally coupled with COX-2 in PGE2 production (Hara et al., 2010). mPGES1 upregulation has been identified in many cancers, including cancers of the lung and gastrointestinal tract (Yoshimatsu et al., 2001a; Yoshimatsu et al., 2001b). The genetic deletion of mpges1in mice reduced PGE2 and increased PGI2 in the circulation but had no effect on thromboxane biosynthesis, thrombogenesis, or blood pressure (Cheng et al., 2006). Thus, mPGES1 inhibition may retain anti-inflammatory and anti-tumor effects by suppressing PGE2, while avoiding the adverse cardiovascular consequences associated with COX-2 inhibitor–mediated PGI2 suppression. However, little is known about the expression and biological function of mPGES1 in melanoma. Here, we investigated the role of mPGES1 in melanoma progression and explored the therapeutic potential of regulating mPGES1 in melanoma.
Results
mPGES1 expression is increased in a human melanoma progression TMA
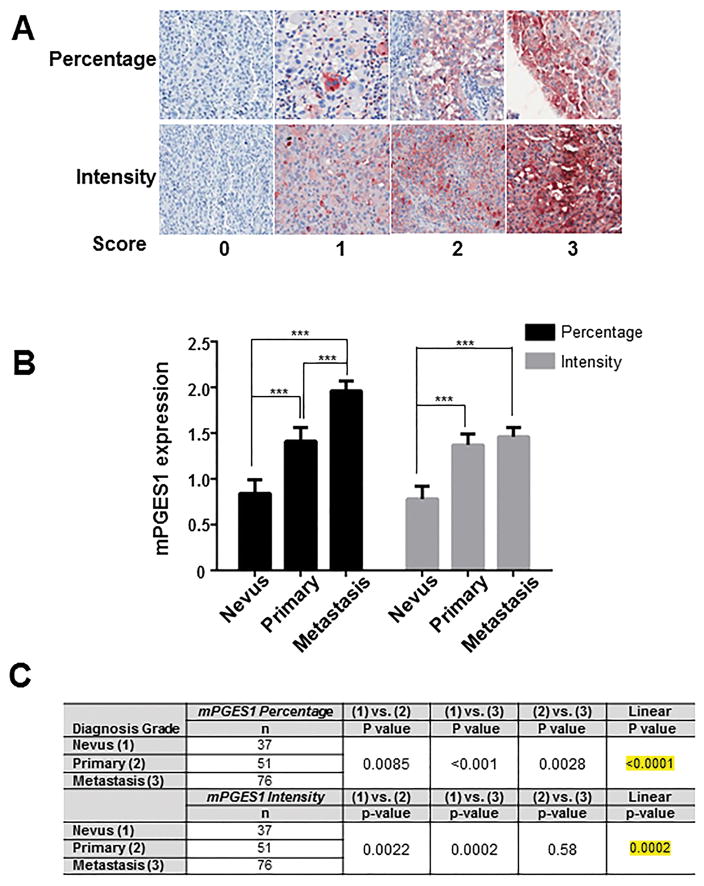
To determine whether mPGES1 is expressed in human melanoma tissue, we performed immunohistochemical (IHC) staining for mPGES1 protein on a melanoma progression tissue microarray (TMA) comprising 480 tissue cores from clinically stratified melanoma specimens from 164 patients (Table 1). We observed various levels of mPGES1 expression in tumor cells and scored for the percentage and intensity of positive staining (Figure 1A). Twenty-seven percent (44/164) of the samples had no mPGES1 expression, while 12%, 41%, and 20% had mPGES1 percentage expression levels 1, 2, and 3, respectively. For mPGES1 staining intensity, 20% of the samples had no expression, 43% had light intensity, 26% had medium intensity, and 11% had marked intensity. Table 1 presents frequencies of mPGES1 expression by percentage and intensity scores and by disease stage. The majority of samples with no expression were from patients with less advanced disease stage, while the majority of those with the highest expression were associated with a more advanced disease stage. The mean differences in mPGES1 expression measures were compared between patients with nevi, primary melanoma, and metastases; a significant difference in mean mPGES1 percentage expression was noted for all pairwise comparisons, and a significant linear trend was observed (P < 0.0001) (Figure 1B and C). Similarly, for mPGES1 intensity expression scores, significant pairwise differences were observed for nevi compared with primary melanoma as well as with metastases. However, mean mPGES1 intensity expression did not significantly differ between primary melanoma and metastatic samples (Figure 1B). Nonetheless, a significant linear trend was observed for mPGES1 intensity expression (P = 0.0002) (Figure 1C), suggesting that mPGES1 could be a progression marker for melanoma. Next, we assessed the prognostic effects (overall survival (OS) and recurrence-free survival (RFS)) of mPGES1 expression according to the percentage and intensity of staining in tumor cells. We performed IHC staining on samples from a TMA comprising lymph node metastases from 91 patients with stage IIIB or IIIC metastatic melanoma (Supplementary Table 1). Patients with increased intensity of mPGES1 expression had significantly increased risk of death compared with patients with no mPGES1 expression (hazards ratio [HR]=2.68; P = 0.014) (Table 2, top). As with OS, patients with high mPGES1 intensity expression in their tumor cells had significantly increased risk of recurrence and death compared with patients with no mPGES1 expression (HR=2.90; P = 0.006) (Table 2, bottom). There was no statistically significant association between mPGES1 percentage expression and OS and RFS (Table 2).
Table 1.
Frequency of mPGES1 expression levels by disease stage
| Diagnosis stage | mPGES1 percentage
|
|||
|---|---|---|---|---|
| 0 (n=44) n (%) |
1 (n=20) n (%) |
2 (n=68) n (%) |
3 (n=32) n (%) |
|
|
| ||||
| Nevus | ||||
| Thin (n=17) | 8 (47) | 2 (12) | 7 (41) | 0 |
| Thick (n=20) | 10 (50) | 6 (30) | 3 (15) | 1 (5) |
| Both (n=37) | 18 (49) | 8 (22) | 10 (27) | 1 (3) |
|
| ||||
| Primary | ||||
| Thin (n=31) | 12 (39) | 4 (13) | 14 (45) | 1 (3) |
| Thick (n=20) | 3 (15) | 2 (10) | 10 (50) | 5 (25) |
| Both (n=51) | 15 (29) | 6 (12) | 24 (47) | 6 (12) |
|
| ||||
| Metastasis | ||||
| LN (n=30) | 4 (13) | 1 (3) | 10 (33) | 15 (50) |
| Viscera (n=46) | 7 (15) | 5 (11) | 24 (52) | 10 (22) |
| Both (n=76) | 11 (14) | 6 (8) | 34 (45) | 25 (33) |
|
| ||||
| Disease stage |
mPGES1 intensity
|
|||
|
0 (n=33) n (%) |
1 (n=70) n (%) |
2 (n=43) n (%) |
3 (n=18) n (%) |
|
|
| ||||
| Nevus | ||||
| Thin (n=17) | 8 (47) | 3 (18) | 6 (35) | 0 |
| Thick (n=20) | 10 (50) | 6 (30) | 4 (20) | 0 |
| Both (n=37) | 18 (49) | 9 (24) | 10 (27) | 0 |
|
| ||||
| Primary | ||||
| Thin (n=31) | 6 (19) | 11 (35) | 12 (39) | 2 (6) |
| Thick (n=20) | 2 (10) | 9 (45) | 7 (35) | 2 (10) |
| Both (n=51) | 8 (16) | 20 (39) | 19 (37) | 4 (8) |
|
| ||||
| Metastasis | ||||
| LN (n=30) | 2 (7) | 19 (63) | 6 (20) | 3 (10) |
| Viscera (n=46) | 5 (11) | 22 (48) | 8 (17) | 11 (24) |
| Both (n=76) | 7 (9) | 41 (54) | 14 (18) | 14 (18) |
Abbreviations: LN, lymph node.
Figure 1. mPGES1 elevation according to melanoma progression.
(A) Representative immunohistochemical (IHC) staining for mPGES1 in serial human melanoma tissue cores. Scoring (0, 1, 2, or 3) for percentage (top) or intensity (bottom) of mPGES1 staining. (B) Mean intensity scores and percentage scores of mPGES1 staining by disease stage in the Melanoma Progression Tissue Microarray. Columns, mean; error bars, SD. P >0.001 (***) was considered statistically significant. (C) Pairwise differences in mPGES1 expression by disease stage were assessed by analysis of variance.
Table 2.
Overall survival and recurrence-free survival analysis of mPGES1 expression in patients with stage III
| Overall survival | ||||||
|---|---|---|---|---|---|---|
| Expression | Group | N | Death (n) | HR | 95% CI | P value |
| mPGES1 number (N=91) | 0 | 32 | 26 | - | - | - |
| 1 | 22 | 12 | 0.71 | 0.36, 1.40 | 0.32 | |
| 2 | 29 | 19 | 0.81 | 0.45, 1.47 | 0.49 | |
| 3 | 8 | 6 | 1.24 | 0.51, 3.02 | 0.64 | |
| mPGES1 intensity (N=91) | 0 | 27 | 22 | - | - | - |
| 1 | 33 | 18 | 0.68 | 0.36, 1.27 | 0.22 | |
| 2 | 21 | 14 | 0.78 | 0.40, 1.53 | 0.48 | |
| 3 | 10 | 9 | 2.68 | 1.22, 5.87 | 0.0141 | |
| Recurrence-free survival | ||||||
|---|---|---|---|---|---|---|
| Expression | Group | N | Deaths (n) | HR | 95% CI | P value |
| mPGES1 number (N=91) | 0 | 32 | 30 | - | - | - |
| 1 | 22 | 20 | 1.16 | 0.66, 2.05 | 0.61 | |
| 2 | 29 | 23 | 0.95 | 0.55, 1.63 | 0.84 | |
| 3 | 8 | 7 | 1.22 | 0.54, 2.80 | 0.63 | |
| mPGES1 intensity (N=91) | 0 | 27 | 26 | - | - | - |
| 1 | 33 | 28 | 1.17 | 0.68, 2.00 | 0.57 | |
| 2 | 21 | 16 | 0.84 | 0.45, 1.58 | 0.59 | |
| 3 | 10 | 10 | 2.9 | 1.37, 6.16 | 0.0055 | |
Abbreviations: HR, hazards ratio; CI, confidence interval.
Human melanoma cells with mPGES1 expression have higher PGE2 levels
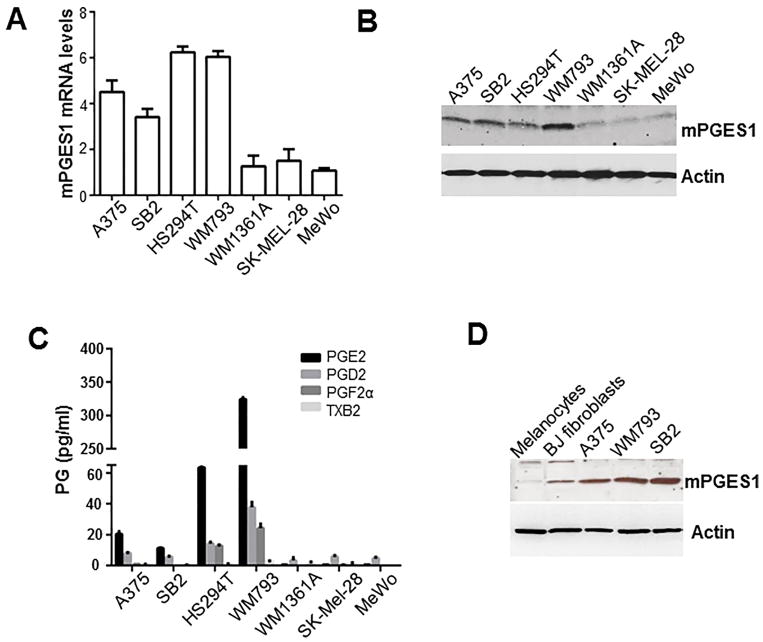
To determine if human melanoma cells express mPGES1, we investigated mPGES1 mRNA and mPGES1 protein expression in seven melanoma cell lines. Our RT-qPCR and Western blotting results demonstrated that expression of mPGES1 mRNA and mPGES1 protein was high in A375, SB2, HS294T, and WM793 cells but low in WM1361A, SK-MEL-28, and MeWo cells (Figure 2A and B). We have analyzed the mPGES1 expression in 61 human melanoma cell lines which is from the Cancer Cell Line Encyclopedia (CCLE) database and found that a subset of human melanoma cells has high mPGES1 mRNA expression (Supplementary Figure 1A). In addition, our lipid mass spectrometry analysis showed that the four cell lines (A375, SB2, HS294T, and WM793) that had upregulation of mPGES1 had higher PGE2 levels than the three cell lines that did not have mPGES1 upregulation (WM1361A, SK-MEL-28, and MeWo (Figure 2C).
Figure 2. mPGES1 and PGE2 levels in human melanoma cells.
We used Western blotting and real-time quantitative polymerase chain reaction to determine the mRNA (A) and protein (B) levels of mPGES1, respectively, in seven human melanoma cell lines (C) Prostaglandin (PG) levels were measured in the culture media by lipid mass spectrometry. (D) mPGES1 protein was detected in normal human epidermal primary melanocytes and BJ fibroblasts and melanoma cells (A375, SB2, and WM793). Abbreviation: TXB2, thromboxane B2.
Next, we investigated mPGES1 expression levels in normal epidermal melanocytes and normal BJ fibroblasts. As shown in Figure 2D and supplementary Figure 1B, BJ cells and melanocytes had low mPGES1 expression compared with A375, WM793, and SB2 cells. These results suggest that some but not all melanoma cell lines, and none of the normal cells, express high levels of mPGES1 with PGE2 production.
iNOS signaling positively regulates mPGES1
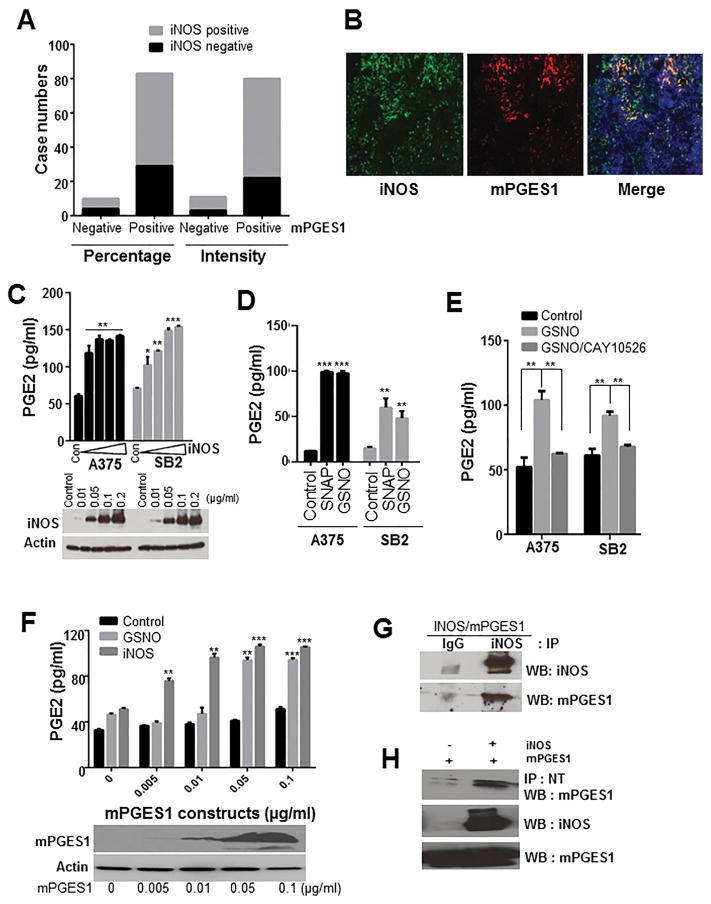
It has been shown that COX-2 and inducible nitric oxide synthase (iNOS) can be produced simultaneously in the same tissues in several inflammatory models (Cuzzocrea and Salvemini, 2007). Our previous studies showed that iNOS expression was elevated during melanoma progression and that high iNOS levels were strongly correlated with poor patient survival (Ekmekcioglu et al., 2006). In the current study, we first investigated whether mPGES1 is co-expressed with iNOS in melanoma patient tissues. Interestingly, we found that about 90% of mPGES1–positive samples also stained positive for the iNOS antibody (Figure 3A and Supplementary Table 2). Additionally, we performed co-immunofluorescent staining in three human melanoma patient specimens and analysis of the Manders’ Coefficients (value is between 0 and 1, 1 is high colocalization) using the Mander’s Calculator ImageJ plugin. The Manders’ Coefficients for localization between iNOS and mPGES2 staining in specimens tested were 0.865, 0.939 and 0.946, suggesting the co-localized expression between mPGES1 and iNOS in melanoma specimens (Figure 3B). Therefore, we hypothesized that cross-talk between mPGES1 and iNOS might regulate the respective activities of these proteins. We specifically tested the effect of PGE2 on NO production and also the effect of iNOS and NO on PGE2 production in A375 and SB2 melanoma cells. We observed that PGE2 treatment did not affect iNOS or NO levels in these cells (Supplementary Figure 2). However, transient iNOS expression, which uses the gene-induced expression of iNOS to mimic the microenvironment driven up regulation, significantly enhanced PGE2 production (Figure 3C). Moreover, NO donors, GSNO and SNAP, significantly enhanced PGE2 production (Figure 3D). These results suggest that iNOS and NO are upstream of PGE2 biosynthesis in melanoma cells.
Figure 3. iNOS expression and NO donors regulate mPGES1 activity and PGE2 production in melanoma.
(A) Columns demonstrate N numbers of negative or positive staining of mPGES1 and iNOS. (B) A representative images of the staining of mPGES1 (red), iNOS (green) and DAPI (blue) in human melanoma specimens. (C) A375 and SB2 cells were transiently transfected with vector or iNOS construct. These cells were incubated in fresh medium for 2 days, and then iNOS expression, total nitrite, and PGE2 levels were determined. P <0.05 (*), P <0.001 (**), and P >0.001 (***) was considered statistically significant. (D) Cells were exposed to SNAP or GSNO (100 μM) for 24 h, and we performed an enzyme immunoassay to measure PGE2 levels. (E) Cells were pretreated with CAY10526 (5 μM) for 1 h, and then PGE2 levels were determined in cells exposed to GSNO (100 μM). (F) HEK293 cells were transfected with the indicated concentrations of mPGES1 construct and exposed to GSNO for 24 h or co-transfected with iNOS construct, and the PGE2 levels were measured. Actin and mPGES1 levels were determined using Western blotting. (G) HEK293T cells were co-transfected with iNOS and mPGES1 constructs. Then cell extracts were subjected to immunoprecipitation (IP) with immunoglobulin G (IgG) or anti-iNOS antibody, and the precipitates were analyzed by Western blot using the indicated antibodies (H) The extracts of HEK293T cells transfected with mPGES1 or mPGES1/iNOS constructs were subjected to IP with anti-nitrotyrosine antibody (NT), and mPGES1 levels were determined in the precipitates. Abbreviations: WB, Western blotting; IP, immunoprecipitation.
Next, to determine the role of mPGES1 in this pathway, we examined the effect of mPGES1 inhibition on NO–induced PGE2 production. We subjected cells to CAY10526, which inhibits PGE2 production through the selective modulation of mPGES1 expression but does not affect COX-2 (Guerrero et al., 2007). CAY10526 suppressed PGE2 production by GSNO (Figure 3E). Consistent with these results, knockdown of mPGES1 dramatically blocked GSNO–mediated PGE2 production (Supplementary Figure 3). Further, iNOS expression and GSNO–mediated PGE2 production depended on mPGES1 expression in HEK293 cells (Figure 3F). Taken together, these data indicate that mPGES1 plays a key role in the crosstalk between NO and PGE2.
Although several reports have shown that NO increases PGE2 levels through upregulating COX-2 expression in various cells, including macrophages and colonic fibroblasts (Salvemini et al., 1993; Zhu et al., 2012), we found that NO donors and iNOS expression did not affect the expression of COX-2 or mPGES1 in A375 and SB2 cells (Supplementary Figure 4A and B). One study reported that iNOS binds, S-nitrosylates, and activates COX-2 (Kim et al., 2005). S-nitrosylation is a reversible and specific posttranslational modification on cysteine caused by NO and regulates the activity of many targets, including metabolic, structural, cytoskeletal, and signaling proteins (Gould et al., 2013). Therefore, co-immunoprecipitation and biotin-switch assays were performed to test the interaction of mPGES1 with iNOS and the S-nitrosylation of mPGES1. mPGES1 formed a complex with iNOS in HEK293 cells expressing both mPGES1 and iNOS (Figure 3G). However, our biotin-switch assay results demonstrated that GSNO treatment did not increase S-nitrosylation of mPGES1, whereas GSNO induced S-nitrosylation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which was used as a positive control (Supplementary Figure 4C). Because nitration at tyrosine residues in proteins is a prominent posttranslational modification caused by NO (Yakovlev and Mikkelsen, 2010), we investigated whether iNOS caused tyrosine nitration of mPGES1. All proteins with tyrosine nitration were precipitated with a specific anti-nitrotyrosine antibody, and mPGES1 was detected. We found that mPGES1 was tyrosine-nitrated as a function of iNOS expression (Figure 3H). Our data imply that iNOS regulates mPGES1 activity through a posttranslational nitration modification, which ultimately enhances PGE2 production.
mPGES1 plays a critical role in cell survival
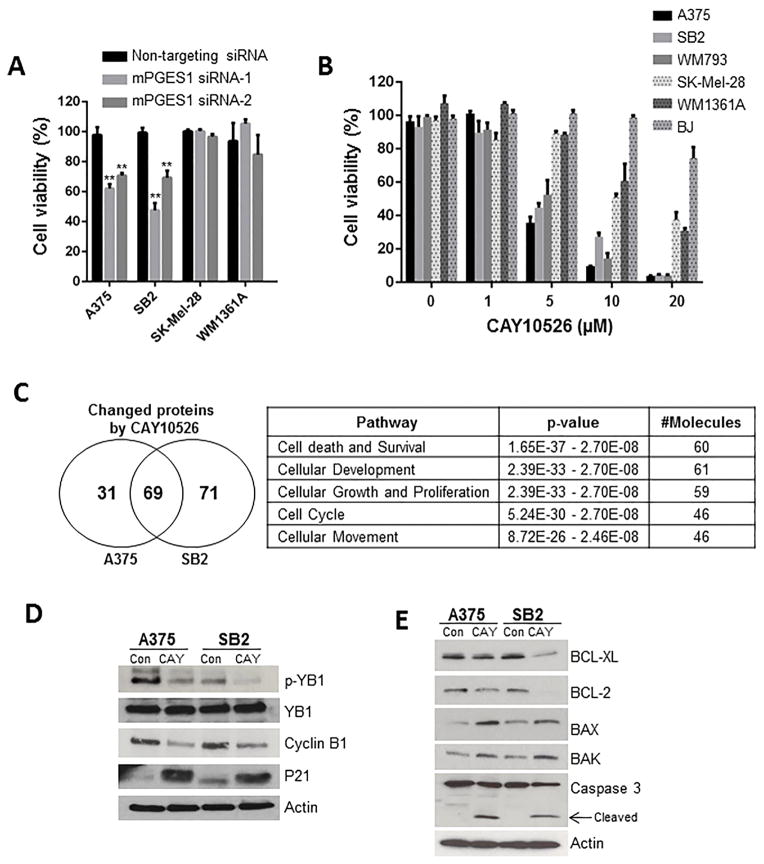
Because PGE2 has been reported to have a survival and proliferative effect in several types of cancer (Wang and Dubois, 2010), we examined the contribution of mPGES1 in human melanoma cell survival. Knockdown of mPGES1 dramatically suppressed cell viability in A375 and SB2 melanoma cells (high mPGES1) but not SK-MEL-28 and WM1361A (low mPGES1) (Figure. 4A). Also, CAY10526 inhibited cell viability (median inhibitory concentration [IC50] < 5 μM) in three melanoma cell lines expressing mPGES1 (A375, SB2, and WM793). This effect was significantly lower (IC50 > 10 μM) in other melanoma cells (SK-MEL-28 and WM1361A) as well as in normal fibroblasts (BJ) that had low expression of mPGES1 (Figure 4B). These results were confirmed with second mPGES1 inhibitor, MF63 (Supplementary Figure 5).
Figure 4. mPGES1 inhibition suppresses cell survival in human melanoma cells.
(A) A375, SB2, SK-Mel-28, and WM1361A cells were transfected with non-targeting and two mPGES1 siRNAs, respectively. After 72 h, viability of transfected cells was determined using PrestoBlue cell viability assay. Cell lysates were subjected to Western blotting for mPGES1 and actin (B) BJ fibroblast and melanoma cells were treated with the indicated concentrations of CAY10526 for 72 h and were subjected to a cell viability assay. The solid and dotted columns show cell survival in cells with high expression (A375, SB2, and WM793) or low expression (SK-MEL-28, WM1361A, and BJ) of mPGES1, respectively. (C) A375 and SB2 cells were treated with CAY10526 (5 μM) for 48 h and then were subjected to reverse phase protein array analysis. A total of 69 proteins were identified as significantly changed by CAY10526 treatment in both A375 and SB2 cells (P < 0.005). (D and E) Western blot analysis was performed in A375 and SB2 cells exposed to CAY10526 (5 μM) for 48 h to determine the levels of phosphorylated Y-box binding protein 1 (p-YB-1), YB-1, cyclin B1, p21 (D) BCL-XL, BCL2, BAX, BAK, and cleaved caspase 3 (E).
To explore the potential target proteins’ response to mPGES1 inhibition, we used reverse phase protein array (RPPA) to quantify the expression or phosphorylation of 217 proteins involved in the cell cycle, apoptosis, angiogenesis, migration, and adhesion. Target proteins changed by CAY10526 were selected according to statistical significance by Student t test (P < 0.05). We found that CAY10526 changed the expression or phosphorylation of 69 proteins in two mPGES1–dependent cell lines A375 and SB2 (Figure 4C and Supplementary Table 3). In addition, ingenuity pathway analysis (IPA) demonstrated that 60 of these 69 (87%) proteins are involved in cell death and survival (Figure 4C).
RPPA and Western blotting results showed that CAY10526 suppressed the phosphorylation of Y-box binding protein 1 (YB-1), an oncogenic translation factor that regulates proliferation, survival, and migration; the phosphorylation of YB-1 is important for nuclear translocation and oncogenic function (Sinnberg et al., 2012). Additionally, CAY10526 attenuated the expression of cyclin B1 (a positive target of YB-1) and increased the expression of p21 (a negative target of YB-1) (Figure 4D). It reported that YB-1 inhibition decreased proliferation and induced apoptosis in multiple myeloma (Chatterjee et al., 2008). As shown in Figure 4 E, CAY10526 reduced BCL-2 and BCL-XL (anti-apoptotic) protein levels and increased BAX and BAK (pro-apoptotic) as well as cleaved caspase 3 levels. These findings suggest that CAY10526-mediated inhibition of the YB-1 pathway may be one of the mechanisms by which mPGES1 inhibitor promotes apoptosis. Taken together, these results suggest that mPGES1 production regulates melanoma cell growth via enhancing cell survival.
mPGES1 inhibition suppresses melanoma growth in mouse xenograft model
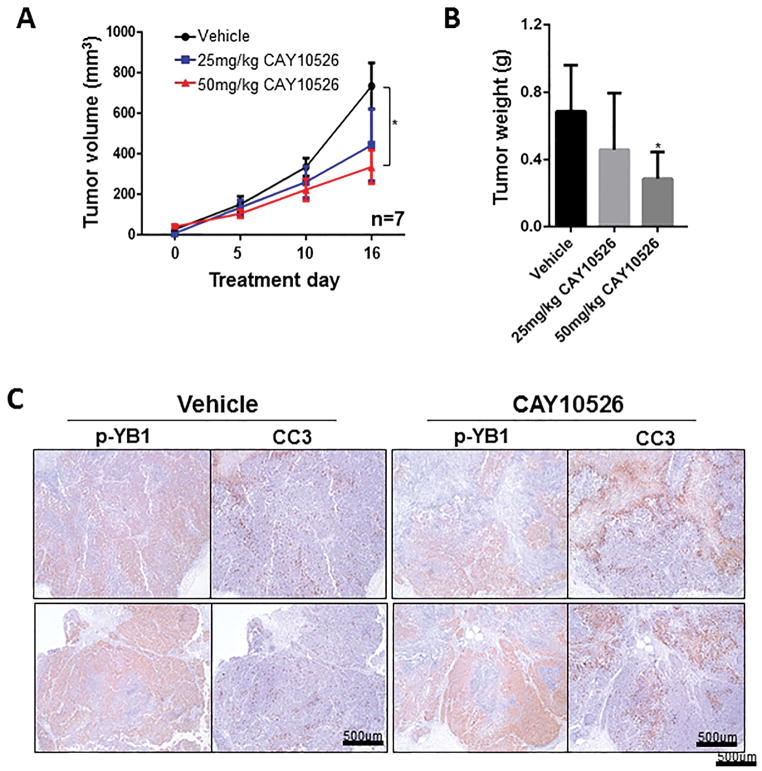
To validate our in vitro cell survival results, we tested mPGES1 inhibition in a subcutaneous xenograft mouse model by injecting A375 cells into the flanks of nude mice and treating the resulting tumors with various concentrations of CAY10526. Consistent with the in vitro results, CAY10526 (50 mg/kg) significantly reduced tumor volume and weight, by 43% and 48%, respectively (Figure 5A and B). Next, to confirm the molecular regulation by CAY10526 in vivo, we performed IHC on tumor samples for phosphorylated YB-1 and cleaved caspase 3. This analysis demonstrated that YB-1 phosphorylation was dramatically decreased in tissues treated with CAY10526. In contrast, CAY10526 treatment resulted in increased levels of cleaved caspase 3 (Figure 5C).
Figure 5. mPGES1 inhibition suppressed human melanoma cell survival.
(A) A375 cells were injected subcutaneously into the flanks of nude mice. Mice were randomized into the following treatment groups (n=7): Vehicle, CAY10526 (25 mg/kg), or CAY10526 (50 mg/kg). Mice were treated from day 7 after injection to day 15 and tumor size was measured with an external caliper. (B) Tumor weight at the end of the experiment. (C) Tumor tissues from mice treated with vehicle and CAY10526 (50 mg/kg) were immunostained for phosphorylated Y-box binding protein 1 (p-YB-1) and cleaved caspase 3 (CC3).
Discussion
Our study is the first to demonstrate the unique association between mPGES1 expression and melanoma progression. We found that the mPGES1/PGE2 pathway is downstream of iNOS signaling, which is one of the biomarkers and potential targets in melanoma. Furthermore, mPGES1 inhibition suppressed melanoma cell survival. Our results suggest that mPGES1 could be a predictive marker and therapeutic target in melanoma. Furthermore, inflammatory mediators should be considered not only in cancer prevention but also as targets for cancer treatment.
Melanoma is among the most highly immunogenic cancers, often associated with massive immune cell infiltration and sometimes exhibiting spontaneous regression (Krelin et al., 2013; Tikoo and Haass, 2015). Consequently, inflammatory factors represent important biomarkers in melanoma (Karagiannis et al., 2014). There is also a strong correlation between PGE2 mediated chronic inflammation and worse long-term outcomes associated with cancer (Wang and DuBois, 2013). Despite having this knowledge base, very little is known about the role of the COX-2-mPGES1-PGE2 axis in melanoma biology. Several studies have supported that COX-2 is a biomarker that is often upregulated in melanoma. Becker et al. showed a correlation between COX-2 staining intensity and Breslow thickness in melanoma (Becker et al., 2009). Furthermore, Chwirot and Kuzbicki reported a higher COX-2 staining intensity in melanoma lesions than in benign nevi (Chwirot and Kuzbicki, 2007). Similar to COX-2, mPGES1 is an inducible factor regulated by proinflammotory mediators and is upregulated in many cancers including lung and gastrointestinal cancers (Yoshimatsu et al., 2001a; Yoshimatsu et al., 2001b). Therefore, we assessed the associations between mPGES1 expression measure and clinical characteristics in patients with stage III melanoma. There were no statistically significant associations between mPGES1 expression measure and stage III diagnosis, ulceration, Breslow thickness, or BRAF mutations status (Data not shown). However, our present findings show that mPGES1 expression was lower in nevi than in primary melanoma and metastatic samples (Figure 1B), and patients with thick primary melanomas had a significantly higher mPGES1 percentage expression than patients with thin primary melanomas. These findings suggest that mPGES1, in concert with COX-2 may be progression markers for melanoma.
Cross-talk between the NO and PG pathways has been reported (Uno et al., 2004; Weinberg, 2000). The NO and PG pathways share several similarities, and the two molecules were produced simultaneously in the same tissues in several inflammation models (Cuzzocrea and Salvemini, 2007). Several studies found that iNOS and COX-2 were induced in macrophages and colonic fibroblasts treated with lipopolysaccharide to produce both NO and PGE2; the iNOS and COX-2 activity was suppressed by an iNOS inhibitor (Salvemini et al., 1993; Zhu et al., 2012). Another study found that COX-2 inhibitors attenuated interleukin-1β–stimulated iNOS expression and NO production in osteoarthritic chondrocytes (Fioravanti et al., 2012). Hence, the interaction between NO and PG is not unidirectional and differs according to cell type and conditions. Our findings in human melanoma cells showed that iNOS signaling was an upstream activator of the PG pathway. Several mechanisms have been proposed to explain how NO mediates PG production, including both transcriptional and posttranslational regulation. We found that NO increased tyrosine nitration of mPGES1. Peroxynitrite (ONOO-), a secondary oxide of NO, stimulates nitration of tyrosine residues, which alters the structure and function of each target protein, and mPGES1 has 6 tyrosine residues (Y28, 58, 80, 89, 117, and 130). Our site-directed mutagenesis approach showed that mPGES1 activity was partially attenuated by Y89F or Y130F mutation (Supplementary Figure 6). Three-dimensional protein structure homology modeling was performed using SWISS-MODEL and AstexViewer (Bordoli et al., 2009; Hartshorn, 2002). Tyrosine 89 is positioned in the second transmembrane domain (TM2) near the luminal side of the membrane and does not engage GSH but when converted to hydrophobic F may cause a structural closure that decreases substrate access. Tyrosine 130 is a key side chain that contributes to forming the binding pocket for glutathione (GSH), which coordinates AA substrate entry into the enzyme. Therefore, these residues are located in the putative active site of mPGES1, implying that NO may affect the ligand binding affinity of mPGES1. However, we need to further characterize the contribution of the individual tyrosine residues of mPGES1 to NO-induced tyrosine nitration and mPGES1activation. Our previous studies showed elevated expression of iNOS in 60% of human metastatic melanoma samples, and iNOS was strongly correlated with poor patient survival (Ekmekcioglu et al., 2006). Our current findings showed that the iNOS expression pattern was similar to that of mPGES1 in human melanoma tissues (Figure 3). Therefore, iNOS activates the mPGES1 pathway and their cross-talk likely boosts inflammation, favoring melanoma tumor progression.
PGE2 is one of the major proinflammatory and carcinogenic PGs, as has been confirmed by several animal studies (Isono et al., 2011; Zang et al., 2013). One study found that mPGES1 knockdown suppressed hepatocellular carcinoma growth (Zang et al., 2013); another study found that mPGES1−/− mice exhibited reduced tumor growth in bone marrow compared with wild-type mice (Isono et al., 2011). These results, together with the findings of the current study, indicate that mPGES1 is an important target for cancer prevention and treatment. Several studies have shown that CAY10526 inhibited mPGES1 expression and PGE2 production (Cavar et al., 2010; Yu et al., 2011) and also suppressed cholangiocarcinoma cell proliferation and migration, which are regulated by PGE2 (Jongthawin et al., 2014). Our results support these findings: we found that CAY10526 attenuated melanoma cell viability via enhancing the apoptotic pathway (Figure 4). Our findings suggest that YB-1 signaling is one of the mechanisms through which CAY10526 promotes melanoma cell apoptosis. YB-1 can function as an oncoprotein and is correlated with progression in several malignancies, including melanoma (Bargou et al., 1997; Gimenez-Bonafe et al., 2004). YB-1controls apoptosis and cell-cycle arrest by transcriptionally repressing the gene encoding p53 (p21) and inhibiting p53-dependent apoptosis by direct protein interaction. YB-1 activates transcription of E2F1 growth-associated gene targets (cyclin B1), thus enhancing cell proliferation and survival (Kosnopfel et al., 2014). Another mechanism of CAY10526 could be PGE2 independent; our study showed that the inhibitory effect of CAY10526 on melanoma cell survival was partially recovered by exogenous PGE2 treatment (Supplementary Figure 7), indicating iNOS–independent regulation. Like PGE2, PGD2 is also a predominant product of PGH2. One study showed via enzymatic analysis that mPGES1 increased PGE2 but decreased PGD2 levels in vitro (Yu et al., 2011). PGD2, unlike PGE2, has been demonstrated to have anti-tumor effects on cancer cells (Yoshida et al., 1998). We postulate that CAY10526’s antitumor effects are due to the downregulation of the PGE2 pathway and the upregulation of PGD2. However, further investigation is needed to test this hypothesis.
In conclusion, our study is the first to show that the mPGES1 is associated with melanoma progression. Additionally, we demonstrated that iNOS signaling enhances the mPGES1/PGE2 pathway, which indicates cross-talk between these two inflammatory pathways. Our findings showed that mPGES1 inhibition suppressed melanoma cell survival. Therefore, mPGES1 could be a useful therapeutic target for treating melanoma. mPGES1’s function and cross-talk with inflammatory mediators may further our understanding of how inflammation promotes cancer progression.
Materials and Methods
Reagents
Antibodies for iNOS, actin, p21, and cyclin B1 were purchased from Santa Cruz Biotechnology. Antibodies for COX-2, mPGES1, and PGE2were purchased from Cayman Chemical. Antibodies for phosphorylated YB-1, BCL-XL, BCL2, BAX, BAK, and cleaved caspase 3 were purchased from Cell Signaling Technology. S-nitrosoglutathione [GSNO] or soluble N-ethylmaleimide-sensitive factor attachment proteins [SNAP] were purchased from Enzo Life Sciences. Anti-nitrotyrosine antibody was obtained from Abcam. PGE2 inhibitor CAY10526 was provided by Cayman Chemical.
Cell culture
Normal BJ fibroblasts; melanoma cell lines A375, HS294T, WM793, SK-MEL-28, and MeWo; and human embryonic kidney HEK293 cells were purchased from the American Type Culture Collection. SB2 melanoma cells were provided by Dr. Michael Davies at The University of Texas MD Anderson Cancer Center (Houston, TX). WM1361A melanoma cells were obtained from Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA). Cells were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum in a 5% carbon dioxide atmosphere.
Co-immunofluorescent staining
Paraffin-embedded specimens were treated with xylene and ethanol to remove the paraffin. The slides were immersed in Borg Decloaker solution (Biocare Medical, Inc.) and boiled in a pressure cooker at 125°C for 5 min for antigen retrieval. Endogenous peroxidase activity was blocked by incubating the slides for 10 min in 3% hydrogen peroxide containing phosphate-buffered saline solution. The slides were blocked with 5% normal goat serum and were incubated with primary antibodies (iNOS monoclonal antibody; BD Biosciences and mPGES1 polyclonal antibody; Novus Biologicals) overnight at 4°C. HRP-conjuated secondary antibodies were then applied to the slides. Thereafter, Alexa Flour® 488 labeled tyramide for iNOS and Alexa Flouro® 594 for mPGES1 were used to detect the specific signals. The nuclei were stained with DAPI. Their localization and then have calculated the Manders’ Coefficients using the Mander’s Calculator ImageJ plugin.
Melanoma tissue microarray
The Melanoma Progression TMA (Nazarian et al., 2010) was developed as a collaborative effort of the Skin Specialized Programs of Research Excellence at Harvard University (Cambridge, MA), MD Anderson Cancer Center, and University of Pennsylvania (Philadelphia, PA). This TMA was designed to provide samples of tumors from each stage in melanocytic tumor progression, according to the American Joint Committee on Cancer staging system. This TMA contains 480 tissue cores from 170 patients, consisting of benign nevi (132 cores from 36 patients), primary cutaneous melanomas (196 cores from 59 patients), melanoma metastases to lymph nodes (60 cores from 29 patients), and melanoma metastases to visceral organs (92 cores from 46 patients). Six samples of 170 patients were nonevaluable and were excluded from this analysis. The Melanoma Stage III TMA consisting of tumor samples from 118 patients with stage III melanoma, was also used. Of these 118 patients, 27 patients were excluded from the outcome analysis due to nonevaluable mPGES1 expression measurements or lack of clinical data. Most of the patients represented in this TMA were male, with a median age of 56 years at both initial and stage III diagnoses. Seventy percent of the patients had died (63/91) while eleven (12%) patients were alive with no progression of disease at the last evaluation date. The median follow-up time for all patients was 2.3 years (range: 0.2 – 14.9) and for patients still alive was 5.6 years (range: 2.2 – 14.9).
Immunostaining was scored separately for two variables, first according to the percentage of melanoma cells with positive staining (<5% = 0, 5%–25% = 1, 26%–75% = 2, and >75% = 3) and then according to the overall intensity of the immunoreactivity of positive cells (no staining = 0, light staining = 1, moderate staining = 2, and marked staining = 3). Cases were considered to be positive if either the percentage or intensity score was 1–3. The slides were manually scored independently by two researchers without prior knowledge of tumor stage or data.
RT-qPCR
Total RNA was isolated with an RNeasy Mini kit (QIAGEN). We synthesized cDNA from total RNA (2 μg) by using High-Capacity cDNA Reverse Transcription kits (Applied Biosystems) and then mixed the cDNA with SYBR Green PCR Master Mix (Applied Biosystems), sterile water, and primers for mPGES1 (Sense 5′-CACAGCCTGGTGATGAGC-3′ and anti-sense 5′-CCGCTTCCCAGAGGATCT-3′) or GAPDH (Sense 5′-TGCACCACCAACTGCTTAGC-3′ and antisense 5′-GGCATGGACTGTGGTCATGAG-3′). Real-time PCR was performed using MasterCycler RealPlex (Eppendorf), and the mRNA expression of target genes relative to the mRNA of GAPDH was calculated.
Western blotting
Total proteins were separated by loading total cell lysate (20 μg) on a 10% sodium dodecyl sulfate polyacrylamide gel and were transferred to a nitrocellulose membrane. Membranes were blocked with 5% non-fat dry milk and were incubated with primary antibodies. Secondary antibody conjugated to horseradish peroxidase (1:2,000; Vector Laboratories, Inc.) was used to detect primary antibodies, and enzymatic signals were visualized by chemiluminescence.
Prostaglandin measurement
PGs were extracted and analyzed by lipid mass spectrometry, as previously described (Yang et al., 2002). Also, a PGE2 Express EIA kit (Cayman Chemical) was used to measure PGE2 concentration.
DNA construction
iNOS and mPGES1 cDNA (OriGene) were amplified with pfuUltra Hotstart DNA polymerase (Stratagene). The PCR products were digested with KpnI and XhoI for iNOS and with EcoRI and XhoI for mPGES1 and were cloned into the pCMV-Sport 6 vector (Addgene). Cells were transfected with plasmids using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s specifications.
RNA interference
Two siRNAs targeting mPGES1 were purchased from Sigma-Aldrich. Cells were transfected with 20 nmol/L of mPGES1 siRNA or non-targeting siRNA using Lipofectamine RNAiMAX (Invitrogen), according to the manufacturer’s specifications. The efficacy of knockdown was confirmed by Western blot analysis.
Co-immunoprecipitation
Cell lysate (1 mg) was incubated with antibodies (2 μg) overnight. Protein A/G magnetic beads (Pierce) were added to each immunoprecipitation reaction. After the immunoprecipitation reaction, sodium dodecyl sulfate sample buffer (2×) was added, and the samples were boiled and subjected to Western blotting as described above.
Cell viability assay
Cell viability was determined using PrestoBlue Cell Viability Reagent (Invitrogen). Cells were seeded into 96-well plates at a density of 3,000 cells per well and were treated with CAY10526for 3 days. Cells were incubated with PrestoBlue reagent for 30 min, and fluorescence was read (excitation 560 nm and emission 615 nm).
Reverse phase protein array
A375 and SB2 cells were treated with CAY10526 (5 μM) for 2 days. Samples were probed with 217 validated primary antibodies by RPPA at the MD Anderson Cancer Center RPPA Core Facility. Expression or phosphorylation of proteins changed by CAY10526, were selected according to statistical significance by Student t test (P < 0.05). The Ingenuity Pathway Analysis program (Ingenuity Systems Inc., Redwood City, CA, USA) was used to identify functional pathways associated with the differentially expressed or phosphorylated proteins by CAY10526.
Xenograft mouse study
All mice were housed and treated in accordance with protocols approved by the Institutional Animal Care and Use Committee at MD Anderson. 6–8-week-old male athymic nu/nu mice were used for xenograft mouse study. A375 cells (1 × 106) were subcutaneously injected into the flanks of nude mice. Seven days after injection, mice were randomized and started on daily intraperitoneal treatment with CAY10526 (25 mg/kg or 50 mg/kg, dissolved in 1% Tween 80) or vehicle for 15 days. Tumor growth was monitored for 16 days, and then the mice were euthanized using carbon dioxide asphyxiation. Tumors were excised and weighed and measured.
Statistical analysis
Patient characteristics were summarized with descriptive statistics. Pairwise differences in means for PGE2 were assessed using Student’s t test. For the assessment of mPGES1 in the melanoma TMA, analysis of variance was used for pairwise differences and to test for linear trends. OS was computed from the date of stage III diagnosis to the last known vital status report. Patients alive at the last follow-up date were censored. RFS was computed from the date of stage III diagnosis to the first date of a subsequent disease event (i.e., local/regional, distant, or both) or death (if the patient died without disease progression) or to the last date of follow-up for which the patient showed no evidence of disease. Patients with no evidence of disease at the last follow-up date were censored. Associations between mPGES1 and OS and RFS were evaluated using univariate Cox proportional hazards regression models.
Supplementary Material
Supplementary Figure 1. mPGES1 expression in human melanoma cells and normal human epidermal primary melanocytes
(A) PTGES mRNA expressions of melanoma cell lines were downloaded from the Cancer Cell Line Encyclopedia (CCLE) (http://www.broadinstitute.org/ccle/home). Gene expressions were normalized by the CCLE using RMA normalization, and then the normalized data were log2 transformed.
(B) mPGES1 mRNA was detected in two normal human epidermal primary melanocytes, A375 and HS294T cells.
Supplementary Figure 2. Effect of PGE2 on iNOS expression and NO production
(A) A375 and SB2 cells were treated with 1 μM PGE2 for 12 h and then iNOS mRNA was determined.
(B) NO levels was measured in A375 cells treated 1 μM PGE2 for 24 h using DAF-FM assay.
Supplementary Figure 3. Effect of mPGES1 knockdown on GSNO-enhanced PGE2 production
(A) A375 and SB2 cells transfected with non-targeting siRNA or mPGES1 siRNA were treated with GSNO (100 μM) for 24 h, and then PGE2 levels were determined. (B) Cell lysates were subjected to Western blotting for mPGES1 and actin.
Supplementary Figure 4. Effects of iNOS/NO on the expression and S-nitrosylation of mPGES1
(A) A375 and SB2 cells were treated with SNAP or GNSO (100 μM) for 24 h and were subjected to Western blotting to detect the expression of cyclooxygenase-2 (COX-2) and mPGES1. (B) A375 and SB2 cells were transfected with iNOS construct and iNOS, and then mPGES1 and COX-2 expression was determined. (C) HEK293 cells were transfected with mPGES1 and treated with GSNO. S-nitrosylation of mPGES1 was analyzed with a biotin-switch assay. Abbreviations: CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Supplementary Figure 5. Effects of MF63 on cell viability in human melanoma cells.
BJ fibroblast and melanoma cells were treated with the indicated concentrations of MF63 for 72 h and were subjected to a cell viability assay. The solid and dotted columns show cell survival in cells with high expression (A375, SB2, and WM793) or low expression (SK-MEL-28, WM1361A, and BJ) of mPGES1, respectively.
Supplementary Figure 6. Effects of tyrosine residue mutation on mPGES1 activity
HEK293 cells were transfected with the mutated or wild-type (WT) microsomal PGE2 synthase-1 (mPGES1) plasmids with the inducible nitric oxide synthase (iNOS) construct, and then cell lysates and conditioned medium were collected to determine PGE2, mPGES1, and iNOS levels. Abbreviations: vec, vector control
Supplementary Figure 7. PGE2 partially rescued cells from CAY10526-induced cell death.
A375 cells were treated with PGE2 (1 μM) and CAY10526 (5 μM) for 72 h and cell viability determined.
Supplementary Table 1. Characteristics of patients from the stage III melanoma tissue microarray (TMA)
Supplementary Table 2. iNOS expression in mPGES1–positive tissue
Supplementary Table 3. 69 proteins regulated by CAY10526
Significance.
Chronic inflammation is strongly associated with the progression of human melanoma, and a clearer understanding of the molecular basis of this inflammation is greatly needed. Our data provides the first evidence for the COX-2-linked PGE2 synthase (mPGES1) as a novel marker and possible target for consideration of regulation. Cross-talk of mPGES1 with other inflammatory mediators, suggests a functional network, and the elucidation of such networks in cancer is needed to increase our understanding of this poorly defined system. Understanding inflammation’s role in melanoma progression and elucidating novel translational insights is likely to provide new approaches for targeting melanoma vulnerabilities.
Acknowledgments
This work was supported by the UT MD Anderson Cancer Center SPORE in Melanoma (P50 CA093459) funded from the NCI, Aim at Melanoma Foundation and the Miriam & Jim Mulva Research Funds, and the MD Anderson CCSG grant (P30 CA016672) funded from the NIH National Cancer Institute (SK, YH, SE, and EAG), Outrun the Sun, Inc. as part of the Outrun the Sun Melanoma Research Scholar program (SK), the Mike Hogg Fund (2014–005) (SK), and the philanthropic funds provided to the melanoma moonshot (EAG). We would like to thank the Reverse Phase Protein Array Core Facility at MD Anderson Cancer Center (funded by the National Cancer Institute Cancer Center Support Grant P30 CA016672) and Sandra Kinney for technical assistance.
References
- Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dorken B, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature medicine. 1997;3:447–50. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- Becker MR, Siegelin MD, Rompel R, Enk AH, Gaiser T. COX-2 expression in malignant melanoma: a novel prognostic marker? Melanoma research. 2009;19:8–16. doi: 10.1097/CMR.0b013e32831d7f52. [DOI] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nature protocols. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- Brown JR, Dubois RN. COX-2: a molecular target for colorectal cancer prevention. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:2840–55. doi: 10.1200/JCO.2005.09.051. [DOI] [PubMed] [Google Scholar]
- Cavar I, Kelava T, Vukojevic K, Saraga-Babic M, Culo F. The role of prostaglandin E2 in acute acetaminophen hepatotoxicity in mice. Histology and histopathology. 2010;25:819–30. doi: 10.14670/HH-25.819. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Rancso C, Stuhmer T, Eckstein N, Andrulis M, Gerecke C, Lorentz H, Royer HD, Bargou RC. The Y-box binding protein YB-1 is associated with progressive disease and mediates survival and drug resistance in multiple myeloma. Blood. 2008;111:3714–22. doi: 10.1182/blood-2007-05-089151. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wang M, Yu Y, Lawson J, Funk CD, Fitzgerald GA. Cyclooxygenases, microsomal prostaglandin E synthase-1, and cardiovascular function. The Journal of clinical investigation. 2006;116:1391–9. doi: 10.1172/JCI27540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwirot BW, Kuzbicki L. Cyclooxygenase-2 (COX-2): first immunohistochemical marker distinguishing early cutaneous melanomas from benign melanocytic skin tumours. Melanoma research. 2007;17:139–45. doi: 10.1097/CMR.0b013e3280dec6ac. [DOI] [PubMed] [Google Scholar]
- Cuzzocrea S, Salvemini D. Molecular mechanisms involved in the reciprocal regulation of cyclooxygenase and nitric oxide synthase enzymes. Kidney international. 2007;71:290–7. doi: 10.1038/sj.ki.5002058. [DOI] [PubMed] [Google Scholar]
- Ekmekcioglu S, Ellerhorst JA, Prieto VG, Johnson MM, Broemeling LD, Grimm EA. Tumor iNOS predicts poor survival for stage III melanoma patients. International journal of cancer Journal international du cancer. 2006;119:861–6. doi: 10.1002/ijc.21767. [DOI] [PubMed] [Google Scholar]
- Fioravanti A, Tinti L, Pascarelli NA, Di Capua A, Lamboglia A, Cappelli A, Biava M, Giordani A, Niccolini S, Galeazzi M, et al. In vitro effects of VA441, a new selective cyclooxygenase-2 inhibitor, on human osteoarthritic chondrocytes exposed to IL-1beta. Journal of pharmacological sciences. 2012;120:6–14. doi: 10.1254/jphs.12016fp. [DOI] [PubMed] [Google Scholar]
- Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–5. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Gamba CA, Swetter SM, Stefanick ML, Kubo J, Desai M, Spaunhurst KM, Sinha AA, Asgari MM, Sturgeon S, Tang JY. Aspirin is associated with lower melanoma risk among postmenopausal Caucasian women: the Women’s Health Initiative. Cancer. 2013;119:1562–9. doi: 10.1002/cncr.27817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Bonafe P, Fedoruk MN, Whitmore TG, Akbari M, Ralph JL, Ettinger S, Gleave ME, Nelson CC. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. The Prostate. 2004;59:337–49. doi: 10.1002/pros.20023. [DOI] [PubMed] [Google Scholar]
- Gould N, Doulias PT, Tenopoulou M, Raju K, Ischiropoulos H. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. The Journal of biological chemistry. 2013;288:26473–9. doi: 10.1074/jbc.R113.460261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero MD, Aquino M, Bruno I, Terencio MC, Paya M, Riccio R, Gomez-Paloma L. Synthesis and pharmacological evaluation of a selected library of new potential anti-inflammatory agents bearing the gamma-hydroxybutenolide scaffold: a new class of inhibitors of prostanoid production through the selective modulation of microsomal prostaglandin E synthase-1 expression. Journal of medicinal chemistry. 2007;50:2176–84. doi: 10.1021/jm0700823. [DOI] [PubMed] [Google Scholar]
- Hara S, Kamei D, Sasaki Y, Tanemoto A, Nakatani Y, Murakami M. Prostaglandin E synthases: Understanding their pathophysiological roles through mouse genetic models. Biochimie. 2010;92:651–9. doi: 10.1016/j.biochi.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. Journal of computer-aided molecular design. 2002;16:871–81. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- Hensler S, Mueller MM. Inflammation and skin cancer: old pals telling new stories. Cancer journal. 2013;19:517–24. doi: 10.1097/PPO.0000000000000010. [DOI] [PubMed] [Google Scholar]
- Howe LR. Inflammation and breast cancer. Cyclooxygenase/prostaglandin signaling and breast cancer. Breast cancer research : BCR. 2007;9:210. doi: 10.1186/bcr1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono M, Suzuki T, Hosono K, Hayashi I, Sakagami H, Uematsu S, Akira S, Declerck YA, Okamoto H, Majima M. Microsomal prostaglandin E synthase-1 enhances bone cancer growth and bone cancer-related pain behaviors in mice. Life sciences. 2011;88:693–700. doi: 10.1016/j.lfs.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Iyer JP, Srivastava PK, Dev R, Dastidar SG, Ray A. Prostaglandin E(2) synthase inhibition as a therapeutic target. Expert opinion on therapeutic targets. 2009;13:849–65. doi: 10.1517/14728220903018932. [DOI] [PubMed] [Google Scholar]
- Johansson CC, Egyhazi S, Masucci G, Harlin H, Mougiakakos D, Poschke I, Nilsson B, Garberg L, Tuominen R, Linden D, et al. Prognostic significance of tumor iNOS and COX-2 in stage III malignant cutaneous melanoma. Cancer immunology, immunotherapy : CII. 2009;58:1085–94. doi: 10.1007/s00262-008-0631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongthawin J, Chusorn P, Techasen A, Loilome W, Boonmars T, Thanan R, Puapairoj A, Khuntikeo N, Tassaneeyakul W, Yongvanit P, et al. PGE signaling and its biosynthesis-related enzymes in cholangiocarcinoma progression. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014 doi: 10.1007/s13277-014-2021-y. [DOI] [PubMed] [Google Scholar]
- Karagiannis P, Fittall M, Karagiannis SN. Evaluating biomarkers in melanoma. Frontiers in oncology. 2014;4:383. doi: 10.3389/fonc.2014.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–70. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- Kosnopfel C, Sinnberg T, Schittek B. Y-box binding protein 1--a prognostic marker and target in tumour therapy. European journal of cell biology. 2014;93:61–70. doi: 10.1016/j.ejcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Krelin Y, Berkovich L, Amit M, Gil Z. Association between tumorigenic potential and the fate of cancer cells in a syngeneic melanoma model. PloS one. 2013;8:e62124. doi: 10.1371/journal.pone.0062124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Fuchs TJ, Bosserhoff AK, Hofstadter F, Pauer A, Roth V, Buhmann JM, Moll I, Anagnostou N, Brandner JM, et al. A seven-marker signature and clinical outcome in malignant melanoma: a large-scale tissue-microarray study with two independent patient cohorts. PloS one. 2012;7:e38222. doi: 10.1371/journal.pone.0038222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian RM, Prieto VG, Elder DE, Duncan LM. Melanoma biomarker expression in melanocytic tumor progression: a tissue microarray study. Journal of cutaneous pathology. 2010;37(Suppl 1):41–7. doi: 10.1111/j.1600-0560.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- Rainsford KD. Anti-inflammatory drugs in the 21st century. Sub-cellular biochemistry. 2007;42:3–27. doi: 10.1007/1-4020-5688-5_1. [DOI] [PubMed] [Google Scholar]
- Richmond A, Yang J, Su Y. The good and the bad of chemokines/chemokine receptors in melanoma. Pigment cell & melanoma research. 2009;22:175–86. doi: 10.1111/j.1755-148X.2009.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvemini D, Misko TP, Masferrer JL, Seibert K, Currie MG, Needleman P. Nitric oxide activates cyclooxygenase enzymes. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7240–4. doi: 10.1073/pnas.90.15.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E, Davies MQ, Hwu P, Yang J, Lotem M, Oren M, Flaherty KT, Fisher DE. Pathways and therapeutic targets in melanoma. Oncotarget. 2014;5:1701–52. doi: 10.18632/oncotarget.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- Singh T, Katiyar SK. Green tea catechins reduce invasive potential of human melanoma cells by targeting COX-2, PGE2 receptors and epithelial-to-mesenchymal transition. PloS one. 2011;6:e25224. doi: 10.1371/journal.pone.0025224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sinnberg T, Sauer B, Holm P, Spangler B, Kuphal S, Bosserhoff A, Schittek B. MAPK and PI3K/AKT mediated YB-1 activation promotes melanoma cell proliferation which is counteracted by an autoregulatory loop. Experimental dermatology. 2012;21:265–70. doi: 10.1111/j.1600-0625.2012.01448.x. [DOI] [PubMed] [Google Scholar]
- Tikoo S, Haass NK. Friends or Foes: IL-10 and TGF-beta in melanoma. Experimental dermatology. 2015 doi: 10.1111/exd.12661. [DOI] [PubMed] [Google Scholar]
- Uno K, Iuchi Y, Fujii J, Sugata H, Iijima K, Kato K, Shimosegawa T, Yoshimura T. In vivo study on cross talk between inducible nitric-oxide synthase and cyclooxygenase in rat gastric mucosa: effect of cyclooxygenase activity on nitric oxide production. The Journal of pharmacology and experimental therapeutics. 2004;309:995–1002. doi: 10.1124/jpet.103.061283. [DOI] [PubMed] [Google Scholar]
- Wang D, Dubois RN. Eicosanoids and cancer. Nature reviews Cancer. 2010;10:181–93. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Dubois RN. An inflammatory mediator, prostaglandin E2, in colorectal cancer. Cancer journal. 2013;19:502–10. doi: 10.1097/PPO.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JB. Nitric oxide synthase 2 and cyclooxygenase 2 interactions in inflammation. Immunologic research. 2000;22:319–41. doi: 10.1385/IR:22:2-3:319. [DOI] [PubMed] [Google Scholar]
- Yakovlev VA, Mikkelsen RB. Protein tyrosine nitration in cellular signal transduction pathways. Journal of receptor and signal transduction research. 2010;30:420–9. doi: 10.3109/10799893.2010.513991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Felix E, Madden T, Fischer SM, Newman RA. Quantitative high-performance liquid chromatography/electrospray ionization tandem mass spectrometric analysis of 2- and 3-series prostaglandins in cultured tumor cells. Analytical biochemistry. 2002;308:168–77. doi: 10.1016/s0003-2697(02)00218-x. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Ohki S, Kanazawa M, Mizunuma H, Kikuchi Y, Satoh H, Andoh Y, Tsuchiya A, Abe R. Inhibitory effects of prostaglandin D2 against the proliferation of human colon cancer cell lines and hepatic metastasis from colorectal cancer. Surgery today. 1998;28:740–5. doi: 10.1007/BF02484622. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu K, Altorki NK, Golijanin D, Zhang F, Jakobsson PJ, Dannenberg AJ, Subbaramaiah K. Inducible prostaglandin E synthase is overexpressed in non-small cell lung cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001a;7:2669–74. [PubMed] [Google Scholar]
- Yoshimatsu K, Golijanin D, Paty PB, Soslow RA, Jakobsson PJ, Delellis RA, Subbaramaiah K, Dannenberg AJ. Inducible microsomal prostaglandin E synthase is overexpressed in colorectal adenomas and cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001b;7:3971–6. [PubMed] [Google Scholar]
- Yu R, Xiao L, Zhao G, Christman JW, Van Breemen RB. Competitive enzymatic interactions determine the relative amounts of prostaglandins E2 and D2. The Journal of pharmacology and experimental therapeutics. 2011;339:716–25. doi: 10.1124/jpet.111.185405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang S, Ni M, Lian Y, Zhang Y, Liu J, Huang A. Expression of microsomal prostaglandin E2 synthase-1 and its role in human hepatocellular carcinoma. Human pathology. 2013;44:1681–7. doi: 10.1016/j.humpath.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Zhu M, Lance P. iNOS signaling interacts with COX-2 pathway in colonic fibroblasts. Experimental cell research. 2012;318:2116–27. doi: 10.1016/j.yexcr.2012.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. mPGES1 expression in human melanoma cells and normal human epidermal primary melanocytes
(A) PTGES mRNA expressions of melanoma cell lines were downloaded from the Cancer Cell Line Encyclopedia (CCLE) (http://www.broadinstitute.org/ccle/home). Gene expressions were normalized by the CCLE using RMA normalization, and then the normalized data were log2 transformed.
(B) mPGES1 mRNA was detected in two normal human epidermal primary melanocytes, A375 and HS294T cells.
Supplementary Figure 2. Effect of PGE2 on iNOS expression and NO production
(A) A375 and SB2 cells were treated with 1 μM PGE2 for 12 h and then iNOS mRNA was determined.
(B) NO levels was measured in A375 cells treated 1 μM PGE2 for 24 h using DAF-FM assay.
Supplementary Figure 3. Effect of mPGES1 knockdown on GSNO-enhanced PGE2 production
(A) A375 and SB2 cells transfected with non-targeting siRNA or mPGES1 siRNA were treated with GSNO (100 μM) for 24 h, and then PGE2 levels were determined. (B) Cell lysates were subjected to Western blotting for mPGES1 and actin.
Supplementary Figure 4. Effects of iNOS/NO on the expression and S-nitrosylation of mPGES1
(A) A375 and SB2 cells were treated with SNAP or GNSO (100 μM) for 24 h and were subjected to Western blotting to detect the expression of cyclooxygenase-2 (COX-2) and mPGES1. (B) A375 and SB2 cells were transfected with iNOS construct and iNOS, and then mPGES1 and COX-2 expression was determined. (C) HEK293 cells were transfected with mPGES1 and treated with GSNO. S-nitrosylation of mPGES1 was analyzed with a biotin-switch assay. Abbreviations: CTL, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Supplementary Figure 5. Effects of MF63 on cell viability in human melanoma cells.
BJ fibroblast and melanoma cells were treated with the indicated concentrations of MF63 for 72 h and were subjected to a cell viability assay. The solid and dotted columns show cell survival in cells with high expression (A375, SB2, and WM793) or low expression (SK-MEL-28, WM1361A, and BJ) of mPGES1, respectively.
Supplementary Figure 6. Effects of tyrosine residue mutation on mPGES1 activity
HEK293 cells were transfected with the mutated or wild-type (WT) microsomal PGE2 synthase-1 (mPGES1) plasmids with the inducible nitric oxide synthase (iNOS) construct, and then cell lysates and conditioned medium were collected to determine PGE2, mPGES1, and iNOS levels. Abbreviations: vec, vector control
Supplementary Figure 7. PGE2 partially rescued cells from CAY10526-induced cell death.
A375 cells were treated with PGE2 (1 μM) and CAY10526 (5 μM) for 72 h and cell viability determined.
Supplementary Table 1. Characteristics of patients from the stage III melanoma tissue microarray (TMA)
Supplementary Table 2. iNOS expression in mPGES1–positive tissue
Supplementary Table 3. 69 proteins regulated by CAY10526