FLT3-internal-tandem-duplication (ITD)-mutated acute myeloid leukemia (AML) is difficult to treat and the majority of patients experience relapse within a short period of time after discontinuation of chemotherapy.1 Current therapeutic options include experimental trials using FLT3-tyrosine kinase-inhibitors (TKI) or allogeneic stem cell transplantation. Inhibitors that are currently investigated in advanced clinical trials with promising clinical responses include midostaurin (PKC412) and quizartinib (AC220). Most recently, Röllig and colleagues reported on a large multicenter trial where combination of chemotherapy with the FLT3-inhibitor sorafenib improved progression free survival in younger patients with AML (abstract #6, ASH annual meeting, 2014).2 Resistance-mediating mutations emerging upon long-term exposure to these inhibitors have confirmed mutated FLT3-kinase as a valid therapeutic target.3,4 Recently, we identified a subgroup of FLT3 length-mutations (~30% of patients that harbor an ITD mutation) with the ITD being located in the tyrosine kinase domain 1 (TKD1).5,6 Multivariate analysis of clinical data revealed that location of FLT3-ITDs within the β1-sheet of TKD1 is an independent unfavorable prognostic factor (that may even outweigh other prognostically relevant determinants such as allelic ratio)5 and predicts resistance to chemotherapy and inferior survival. A recent report by Schnittger et al.7 could not find a strict association with functional domains but confirmed that ITD-mutations localized more 5′ were associated with better outcome than more 3′ mutations.
In AML, it is of major clinical interest to identify patients that may benefit from targeted therapies in the future. So far, the significance of FLT3-kinase inhibitors in AML therapy remains a matter of current debate.
Therefore, we aimed to clarify whether distinct location of ITD-mutations may impact the biological phenotype. Depending on their location they may influence (i) transforming capacity, (ii) target gene expression and (iii) responsiveness to TKIs in vitro and in vivo.
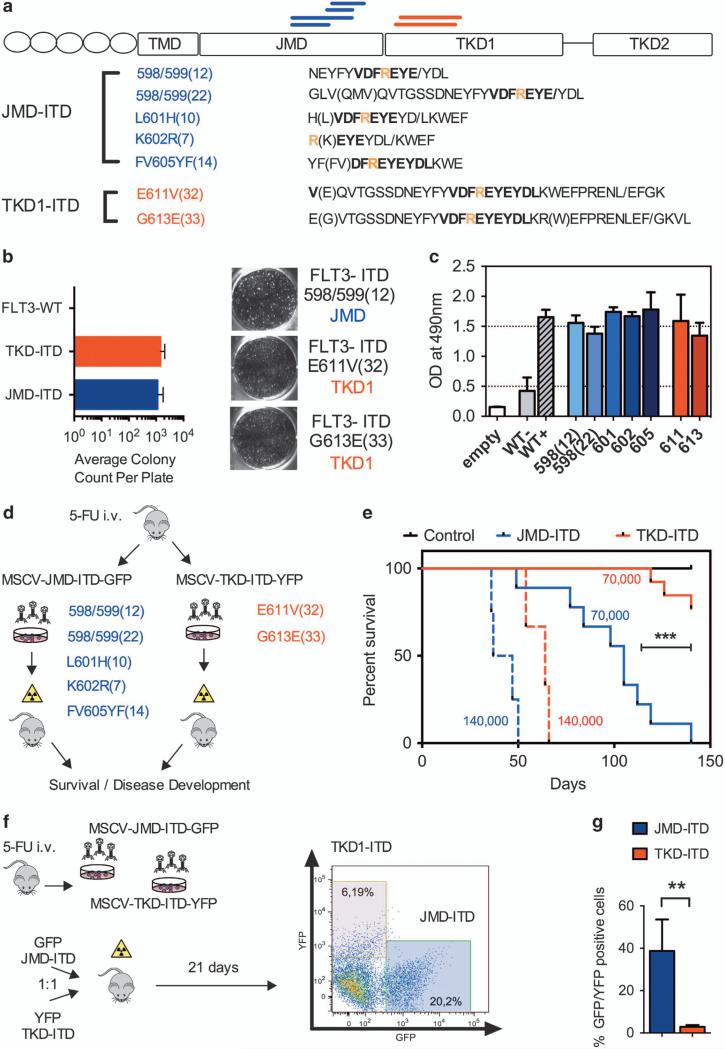
To assess for functional properties of differentially located FLT3-ITD-mutations, we amplified and sub-cloned a total of seven ITD-mutations from primary patient material. With limited data being available on functional properties of FLT3-ITD-mutations located in the TKD1, we sought to confirm that TKD1-ITDs show similar transforming capacity as juxtamembrane domain (JMD)-located ITD-mutations. BaF3- and 32D-cells were infected with each of five JMD-ITD-constructs and each of two TKD1-ITD-constructs (Figure 1a). As ITD-mutations are known to induce growth-factor independent colony formation in otherwise interleukin-3 (IL-3) dependent murine cell lines, we investigated all seven constructs for colony forming potential in methylcellulose. Colony-forming unit assays following IL3-withdrawal revealed a comparable degree of clonal growth formation irrespective of FLT3-ITD location (JMD- or TKD1-ITDs; Figure 1b). As expected, FLT3-wild-type controls did not generate any growth-factor independent colonies. To confirm these results, we assessed for metabolic activity by using MTS assays. Here, all transformed cell lines revealed similar growth-factor independent proliferative capacity (Figure 1c). FLT3-wild-type controls did not proliferate following IL-3 withdrawal. Western blotting confirmed constitutive auto-phosphorylation of the FLT3-receptor and comparable activation of downstream targets such as STAT5 (data not shown). Taken together, FLT3-ITDs exhibit comparable transforming capacity and activation of downstream signaling nodes irrespective of their location within the JM-domain or TKD1 in vitro.
Figure 1.
(a) Location and amino acid sequence of seven FLT3-ITD-mutations investigated, five located in the JMD (JMD-ITD, blue) and two located in the TKD1-domain (TKD1-ITD, red, all located within the β1-sheet). The conserved amino acid residue R595 is indicated as a reference in yellow. FLT3-ITD-mutations are named regarding to their insertion site followed by its amino acid length: FLT3-ITD-mutations include (1) integration between two codons, for example, ITD598/599 (ref. 13) and (2) integration into a codon leads to a point mutation with an amino acid exchange: here, the origin amino acid is placed before the integration site and is followed by the point mutation, for example, ITDK602R.8 All mutations were amplified by PCR from primary patient samples after informed consent. Murine cell lines (BaF3 and 32D) and primary murine bone marrow cells were stably transduced with the respective retroviral constructs as previously described14 (b) Average colony count of FLT3-ITD transduced BaF3 cells in methylcellulose confirms transformation capacity of FLT3-ITD-mutations. FLT3-ITD and FLT3-WT cells were seeded at a fixed density (1×103 cells/ml) in methylcellulose (Stem Cell Technologies, Cologne, Germany; #3231). FLT3-ITD-mutations induce high numbers of colonies in methylcellulose irrespective of the location (JMD or TKD1), whereas cells transduced with FLT3-WT do not induce any colony formation. Each data point represents mean ± s.d. Each experiment was performed >3 times. (c) Location of FLT3-ITD within the TKD1 does not impact proliferative capacity of FLT3-ITD transduced BaF3 cells as assessed by MTS assays (Promega, Mannheim, Germany). 5 × 103 cells were seeded per well and optical density (OD) at 490 nm was measured after 72 h. Each data point represents mean ± s.d. Each experiment was performed >3 times. (d) Experimental scheme: Balb/C donor mice were injected with 5-FU (150 mg/kg) and harvested bone marrow (BM) cells were infected with the respective ITD-constructs (murine stem cell virus (MSCV)-JMD-GFP or MSCV-TKD1-GFP). Equal amounts of GFP-positive cells were injected at two different dilutions: 140 000 GFP+ cells (n = 5 for JMD- and TKD1-ITD-constructs, respectively) and 70 000 GFP+ cells (n = 19 for JMD and n = 13 for TKD1-ITDs). (e) Survival of lethally irradiated (13 Gy) 6–8-week-old female Balb/C recipients of FLT3-ITD-transduced BM cells. TKD1-ITD-transduced BMC (red) show delayed onset of disease and reduced penetrance at lower dilutions compared with JMD-ITDs (blue). Survival differences of the two groups (JMD- versus TKD-ITD) were calculated using Fisher's exact test: using a group size of >10 animals per construct we would be able to determine a significant difference in survival with a power >80% given a death rate of >75% in one of group and <15% in the other group at a specific time point. (f) Competitive transplantation of cells harboring different FLT3-ITD-mutations (distinct ITD location). 20 000 GFP and 20 000 yellow fluorescent protein (YFP)-positive sorted cells (FLT3-ITD-JMD or FLT3-ITD-TKD1, respectively) were transplanted together into lethally irradiated recipient mice. Outgrowth of GFP-positive cells confirms competitive advantage of JMD-ITD infected cells in vivo (**P<0.01; n = 5 in three independent experiments). ***P<0.001.
First, we aimed to functionally characterize ITD-mutations depending on their location in vivo. Currently, various mouse models are available to investigate function of FLT3-ITD transformed cells, including genetically engineered mouse models. As we aimed to compare all different ITDs derived and sub-cloned from primary patient cells (indicated in Figure 1a), we chose a retroviral transduction model, in which hematopoietic progenitors are being transformed in vitro and transplanted into irradiated recipient mice (Figure 1d). Injection of primary murine bone marrow cells harboring the respective ITDs led to induction of a lethal myeloproliferation (Figure 1e) as previously described.8 At a cell dose of 140 000 green fluorescent protein (GFP)-positive cells per recipient animal, all constructs used for cellular transformation showed 100% disease penetrance with a slight and non-significant disease delay for the TKD1-ITDs. Unexpectedly, ITDs located in the TKD1 led to delayed onset of disease and decreased penetrance when injected at higher dilutions of 70 000 GFP-positive cells per recipient animal (Figure 1e). This may indicate differential proliferative capacity of primary murine (Balb/C) bone marrow cells transformed by FLT3-ITD-mutations, depending on their location. To confirm our findings and to exclude inter-individual differences between recipient hosts, we aimed to compare proliferative capacity of differentially located ITDs within the same recipient animal (Figure 1f). Indeed, competitive transplantation of JMD- and TKD1-ITDs labeled with different fluorochromes (GFP or YFP) revealed competitive advantage for JMD-ITDs over TKD1-ITDs within the same recipient mouse (Figure 1g). For the first time these data confirm distinct functional properties of FLT3-ITD-mutations depending on their location in vivo.
Proliferative capacity, however, does not necessarily correlate with drug resistance. Several other types of leukemia harbor (partially pre-existing) resistance-mediating mutations that are selected upon chemotherapy or kinase-inhibitor treatment.3,9 In AML, multiple FLT3-ITD-mutations can be present within the same patient. Along these lines, less sensitive FLT3-ITD-mutations could be selected upon FLT3-TKI-treatment. Treatment with FLT3-TKI is a promising therapeutic approach. FLT3-kinase inhibitors midostaurin (PKC412) and quizartinib (AC220) have already proven significant clinical activity in advanced clinical trials.3,4,10 To assess for differential sensitivity in respect to ITD-location, we performed drug treatment with either compound at clinically relevant doses in vitro.
Although nanomolar concentrations (10–100 nm) of midostaurin (PKC412) induced apoptosis in 32D-cells harboring JMD-ITDs, ITDs located within the TKD1 revealed significantly decreased sensitivity to TKI treatment in vitro (Figure 2a). These effects were detectable in two different cellular backgrounds (BaF3- and 32D-cells) after 24 h TKI exposure, respectively (BaF3, Figure 2a and 32D, data not shown). This phenotype could be re-capitulated using quizartinib as a more selective and potent FLT3-inhibitor. Upon 10–100 nm of quizartinib (AC220) a similar degree of resistance became evident for ITDs located in the TKD1 (Figure 2b). In line with previously published data,5,6 these findings indicate differential sensitivity of FLT3-ITDs to TKI-therapy depending on the location of the FLT3-ITD in vitro.
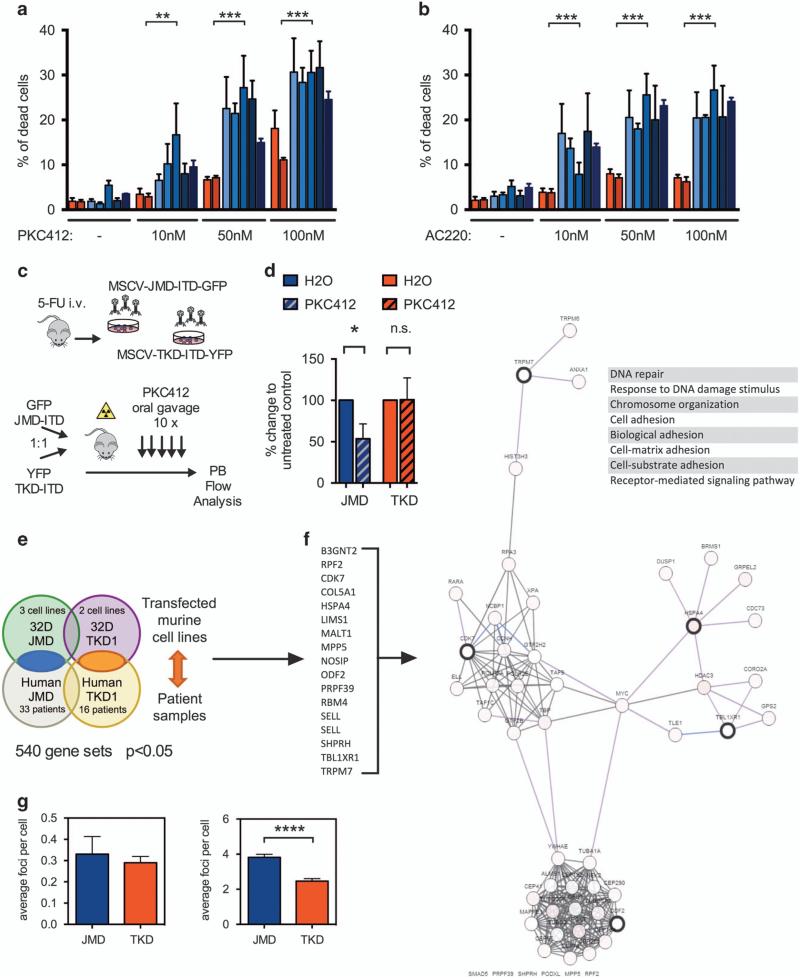
Figure 2.
(a, b) Induction of apoptosis in FLT3-ITD transduced BaF3 cells treated for 24 h with PKC412 (a) and AC220 (b) at low nanomolar concentrations (10–100 nm). The percentage of apoptotic cells was determined by propidium iodide (PI) staining and confirmed by SytoxBlue (Invitrogen, Darmstadt, Germany) and AnnexinV staining (Biolegend, San Diego, CA, USA). Cells transduced with TKD1-ITD-constructs (red) reveal decreased sensitivity to TKI treatment compared with cells harboring JMD-ITDs (blue) in vitro (***P<0.001). Each data point represents mean ± s.d. Experiments were performed at least in triplicate and a Mann–Whitney-test was applied for statistical comparison using GraphPad Prism (GraphPad Inc., La Jolla, CA, USA). (c) Experimental scheme: In vivo TKI treatment of competitively transplanted FLT3-ITD-mutations, depending on ITD location. 20 000 GFP and 20 000 YFP-positive sorted cells (FLT3-ITD-JMD or FLT3-ITD-TKD1, respectively) were transplanted competitively into lethally irradiated recipients and peripheral blood was monitored by flow cytometry. (d) Treatment with PKC412 by oral gavage (50 mg/kg/day for 10 days) leads to reduction of JMD-ITD- but not TKD1-ITD-clones within the same recipient host, indicating resistance of the respective TKD1-clones to TKI treatment in vivo (*P<0.05; n>5/cohort in two independent experiments). Experiments were performed at least in triplicate and a Mann–Whitney-test was applied for statistical comparison using GraphPad Prism (GraphPad Inc.). (e) Global gene-expression profiling (GEP) was performed on murine 32D-cells transfected with either FLT3-ITD mutation (JMD or TKD1) using Affymetrix GeneChip Mouse Genome 430 2.0 Arrays as previously reported (data available at http://www.ncbi.nlm.nih.gov/projects/geo; GSE56241), and data was compared with human FLT3-ITD signatures.15 Primary patient samples were analyzed according to the FLT3-ITD location: 33 patients with JMD-ITDs and 16 patients with TKD1-ITDs. 540 probe sets were differentially expressed between JMD- and TKD1-ITDs after stringent selection (P<0.05). (f) Network view of selected genes (=seed nodes with thick black borders) identified by the comparison of JMD-ITDs and TKD1-ITDs in murine 32D cells and primary patient samples in the context of biological interactions derived from public pathway databases (pathway and interaction data is from Human Protein Reference Database, Reactome, NCI-Nature Pathway Interaction Database and the MSKCC Cancer Call Map, as derived from Pathway Commons). Each gene in the network view is color coded with multi-dimensional genomic data derived from the TCGA AML data (each node is color coded along a white to red color gradient, indicating the frequency of alteration across AML; deeper red indicates higher frequency of alteration).16 Network visualization was powered by Cytoscape Web via the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). Gene expression changes predict abnormal regulation of DNA repair, chromatin organization, adhesion and cellular signaling in TKD1-ITDs compared with JMD-ITDs. (g) Upon cellular stress (3 Gy irradiation), 32D-cells harboring TKD-ITDs reveal less H2AX foci when compared with JMD-ITD-mutations (right panel; experiments were performed at least in triplicate and a Mann–Whitney-test was applied for statistical comparison using GraphPad Prism, GraphPad Inc.). After 120 min in culture, cells were collected and spun onto microscope slides by centrifugation (2500 r.p.m. for 5 min, followed by 4000 r.p.m. for 5 min). Fixation and permeabilization was performed with methanol and acetone, followed by 1 h blocking with 5% bovine serum albumin in phosphate buffered saline. Cells were stained with phospho-H2AX (Ser139; Cell Signaling Technology, Danvers, MA, USA) and incubated with a secondary Cy3-conjugated antibody (Sigma-Aldrich, Taufkirchen, Germany). Finally, cells were counterstained with DAPI (4′,6-Diamidine-2′-phenylindole dihydrochloride) containing mounting medium (Dako, Denmark). Immunofluorescence was measured by using a Zeiss confocal microscope (Zeiss × 40 objective, Zeiss, Germany). For non-irradiated 32D cells (left panel) similar amount of H2AX foci were detectable, with a trend towards lower foci number in the TKD-ITD group. **P<0.01, ****P<0.0001.
To assess for sensitivity of different ITD-mutations depending on their location in vivo we transplanted equal numbers of different ITDs within the same recipient host and performed TKI-treatment per gavage (Figure 2c). In this setting, treatment with pi midostaurin (PKC412) led to significant reduction of JMD-ITD-transformed cells in vivo while leaving TKD1-ITD transformed cells largely unaffected (Figure 2d). These findings confirmed differential sensitivity of FLT3-ITDs in vivo, depending on their location. Moreover, reduced sensitivity of TKD-ITDs to TKI treatment could explain selection of TKD1-located ITDs in vivo and is consistent with our findings in vitro.
Differential sensitivity may arise from altered expression of transcriptional target genes. o gain insights into the mechanisms underlying the susceptibilities of JMD- and TKD1-ITD-mutations in vitro, we performed whole-genome transcriptional profiling of 32D-cells infected with ITD-mutations of different locations (three JMD-ITDs and two TKD1-ITDs) and primary patient cells harboring ITDs in either location (JMD or TKD1; 49 patient samples, 33 and 16, respectively; Figure 2e). 540 probe sets were differentially expressed between JMD- and TKD1-ITDs after stringent selection (P<0.05). These expression changes predicted abnormal regulation of DNA repair, chromatin organization and adhesion in TKD1-ITD-mutations compared with JMD-ITDs (Figure 2f).
Abnormal regulation of DNA repair may explain the observed phenotype as it may result in increased apoptosis of cells upon induction of cellular stress, for example, by targeted therapies, chemotherapy or irradiation. To confirm differential DNA-damage response in our cellular models, we studied the spatial and temporal distribution of foci of the phosphorylated form of the histone protein H2AX (gamma-H2AX), which are sensitive markers for double-strand breaks. Although non-irradiated FLT3-ITD transformed cells showed comparable levels of H2AX foci (Figure 2g, left panel), TKD1-ITD transformed cells revealed significantly less H2AX foci following irradiation when compared with JMD-ITDs (Figure 2g, right panel). Increased levels of DNA-damage in the JMD-ITDs may promote induction of apoptosis, whereas effective DNA-damage repair in TKD-ITD transformed cells may protect from targeted and cytotoxic treatment.
Taken together, our findings confirm that FLT3-ITD-location influences disease biology and leads to changes in global gene expression. In our model, ITD-location alters proliferative capacity and sensitivity to FLT3-TKI-treatment in vivo. All ITD-mutations investigated were located within the β1-sheet of TKD1 which is more 5′ than previously characterized ITD-variants that also revealed a resistance phenotype.6 Patients harboring TKD1-ITD may not benefit from concomitant or sequential treatment with TKI. This finding may help to stratify patients towards allogeneic stem cell transplantation versus chemotherapy in combination with kinase-inhibitor treatment in future clinical trials. Potential caveats of our study include impact of ITD-size on the phenotype. TKD1-ITDs have already been described to be longer5,7,11 and ITD-size has been controversially discussed for many years as a potential prognostic factor.1,7,11–13 Variability in ITD-size is—in part—reflected by the ITD-constructs used in our study. However, we did not find gross differences in sensitivity to TKI within the respective location (JMD or TKD1) depending on ITD-size.
As an outlook, ITD-mediated differences in DNA repair may facilitate therapeutic strategies targeting DNA-damage response pathways to sensitize TKD1-ITDs to chemotherapy or targeted therapy, however, these pathways need to be investigated in depth in further pre-clinical studies.
ACKNOWLEDGEMENTS
This work was supported by a grant from the German Research Council (DFG) FI405/5-1 to TF and FHH, and BU 1339/5-1 to LB and KD; the DFG graduate school GRK 1167 (P17) to TF and a Heisenberg-Stipend to LB (BU 1339/3-1). Part of this work was also funded by the German Cancer Aid (Deutsche Krebshilfe e.V.; grants #110058 and #110062 to TF). SAA was supported by NCI grant CA66996 and the Leukemia and Lymphoma Society. FHH was supported by the EHA/ASH TRTH program 2012.
Footnotes
AUTHOR CONTRIBUTIONS
PAT, TSM, AP, TMS, AB, ZW, AJD and SW. Designed research, performed research and collected data. LB, SAA, KD, FHH and TF. Analyzed and interpreted data and performed statistical analysis. FHH wrote the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Gale RE, Green C, Allen C, Mead AJ, Burnett AK, Hills RK, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with 5 NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 2.Röllig C, Müller-Tidow C, Hüttmann A, Noppeney R, Kunzmann V, Baldus CD, et al. Sorafenib Versus Placebo in Addition to Standard Therapy in Younger Patients with Newly Diagnosed Acute Myeloid Leukemia: Results from 267 Patients Treated in the Randomized Placebo-Controlled SAL-Soraml Trial. Blood. 2014;124:6. [Google Scholar]
- 3.Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- 4.Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kayser S, Schlenk RF, Correa Londono M, Breitenbuecher F, Wittke K, Du J, et al. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood. 2009;114:2386–2392. doi: 10.1182/blood-2009-03-209999. [DOI] [PubMed] [Google Scholar]
- 6.Breitenbuecher F, Schnittger S, Grundler R, Markova B, Carius B, Brecht, et al. Identification of a novel type of ITD mutations located in nonjuxtamembrane domains of the FLT3 tyrosine kinase receptor. Blood. 2009;113:4074–4077. doi: 10.1182/blood-2007-11-125476. [DOI] [PubMed] [Google Scholar]
- 7.Schnittger S, Bacher U, Haferlach C, Alpermann T, Kern W, Haferlach T. Diversity of the juxtamembrane and TKD1 mutations (exons 13-15) in the FLT3 gene with regards to mutant load, sequence, length, localization, and correlation with biological data. Genes Chromosomes Cancer. 2012;51:910–924. doi: 10.1002/gcc.21975. [DOI] [PubMed] [Google Scholar]
- 8.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 9.Miething C, Feihl S, Mugler C, Grundler R, von Bubnoff N, Lordick F, et al. The Bcr-Abl mutations T315I and Y253H do not confer a growth advantage in the absence of imatinib. Leukemia. 2006;20:650–657. doi: 10.1038/sj.leu.2404151. [DOI] [PubMed] [Google Scholar]
- 10.Fischer T, Stone RM, Deangelo DJ, Galinsky I, Estey E, Lanza C, et al. Phase IIB trial of oral Midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J Clin Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blau O, Berenstein R, Sindram A, Blau IW. Molecular analysis of different FLT3-ITD mutations in acute myeloid leukemia. Leuk lymphoma. 2013;54:145–152. doi: 10.3109/10428194.2012.704999. [DOI] [PubMed] [Google Scholar]
- 12.Chillon MC, Santamaria C, Garcia-Sanz R, Balanzategui A, Sarasquete ME, Alcoceba M, et al. Long FLT3 internal tandem duplications and reduced PMLRARalpha expression at diagnosis characterize a high-risk subgroup of acute promyelocytic leukemia patients. Haematologica. 2010;95:745–751. doi: 10.3324/haematol.2009.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stirewalt DL, Kopecky KJ, Meshinchi S, Engel JH, Pogosova-Agadjanyan EL, Linsley J, et al. Size of FLT3 internal tandem duplication has prognostic significance in patients with acute myeloid leukemia. Blood. 2006;107:3724–3726. doi: 10.1182/blood-2005-08-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, et al. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullinger L, Dohner K, Kranz R, Stirner C, Frohling S, Scholl C, et al. An FLT3 gene-expression signature predicts clinical outcome in normal karyotype AML. Blood. 2008;111:4490–4495. doi: 10.1182/blood-2007-09-115055. [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]