Abstract
Transformation of commercially important indica cultivars remains challenging for the scientific community even though Agrobacterium-mediated transformation protocols for a few indica rice lines have been well established. We report successful transformation of a commercially important restorer line JK1044R of indica rice hybrid JKRH 401. While following existing protocol, we optimized several parameters for callusing, regeneration and genetic transformation of JK1044R. Calli generated from the rice scutellum tissue were used for transformation by Agrobacterium harboring pCAMBIA2201. A novel two tire selection scheme comprising of Geneticin (G418) and Paramomycin were deployed for selection of transgenic calli as well as regenerated plantlets that expressed neomycin phosphotransferase-II gene encoded by the vector. One specific combination of G418 (30 mg l−1) and Paramomycin (70 mg l−1) was very effective for calli selection. Transformed and selected calli were detected by monitoring the expression of the reporter gene uidA (GUS). Regenerated plantlets were confirmed through PCR analysis of nptII and gus genes specific primers as well as dot blot using gus gene specific as probe.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-015-0334-y) contains supplementary material, which is available to authorized users.
Keywords: Rice (Oryza sativa), Agrobacterium tumefaciens, Genetic transformation; embryonic callus, NptII selection, Geneticin (G418), Paramomycin, UidA (GUS), Restorer line
Introduction
Rice is a staple food for nearly half of the world’s population mainly living across Asia, thus more than 90 % of this rice is being produced and consumed in this continent (Papademetriou 2000). It is widely grown in different environments depending upon water availability. Rice production and productivity is constrained by several biotic and abiotic stresses. Insect pests and diseases cause major damages to rice production. Abiotic stresses such as drought, salinity, water logging and temperature aberrations also cause serious economic losses in rice production and productivity.
India is the second country to commercialize hybrid rice (Praveen and Dinesh 2012). At present, in India hybrid rice cultivation is spread over about 2 million hectares out of 44 million hectares under rice cultivation (Anon 2014). Use of transgenic technologies for developing resistance to various biotic and abiotic stresses in the background of hybrid parental lines is important. Although, Agrobacterium-mediated transformation system for Japonica subspecies (Hiei et al. 1994) and a few Indica rice is well established efficiency of transformation protocol remains genotype dependent (Datta et al. 1999, 2000, 2001; Khanna and Raina 1999; Tu et al. 1998; Zhang et al. 1998; Lin and Zhang 2005; Park et al. 1996). Generally gene transformation activities taken up in tissue culture responsive inbreed lines, direct gene transfer to elite hybrid parental line will retains the agronomic value of the final commercial product, saving time in backcross breeding. Recently there are reports of Agrobacterium transformation using maintainer line (Baisakh et al. 2000), restorer rice line (Tu et al. 2003) and male-sterile line (Yang et al., 2013) to incorporate agronomical important traits in hybrids. Thus, it is important to develop transformation protocol for parental lines of commercially important hybrids.
In rice, antibiotic and herbicide resistance genes are the most efficient and widely used selectable markers (Miki and McHugh 2004). Hygromycin phosphotransferase-II (hptII) gene is the most commonly used antibiotic selectable marker in rice genetic transformation. However, the antibiotic Hygromycin that selects transgenic plants expressing the hptII gene is expensive in the context of developing countries such as India. The other most abundantly used gene for selection of transgenic plants is neomycin phosphotransferase–II (nptII). Neomycin phosphotransferase–II gene has been used as a selection marker in many commercialized transgenic plants and may face less regulatory hurdle for future commercialization of transgenic crops. Neomycin phosphotransferase–II has been approved by the US Food and Drug Administration (FDA) as a food additive for tomato, cotton and oilseed rape (US Food and Drug Administration, FDA 1994). The nptII gene codes for the aminoglycoside 3′-phosphotransferas enzyme and it is inactivated by phosphorylation by a range of aminoglycoside antibiotics such as Neomycin, Kanamycin, Paramomycin, Ribostamycin, Butirosin and Geneticin (G418). However, native rice callus shows various degrees of in-built natural resistance to Kanamycin (Caplan et al. 1992). Raineri et al. (1990) used 200 mg l−1 Kanamycin to select rice callus derived from mature embryos, but no transgenic plants were recovered. Selection on G418 is more effective than Kanamycin for transgenic plants expressing nptII gene, perhaps because G418 is more toxic than Kanamycin to rice cells (Twyman et al. 2002). Chan et al. (1992, 1993) used G418 for Agrobacterium-mediated transformation of rice however, a total of just four transgenic plants were produced following selection on 40 mg l−1 G418. In the present investigation, we tested various combinations of G418 and Paramomycin to develop an efficient Agrobacterium mediated transformation, selection and regeneration system for the rice restorer line JK1044R.
Materials and methods
Rice cultivar
Dried mature seed of indica rice cultivar JK1044R which is a proprietary restorer line of registered and notified hybrid JKRH 401 (IET 18181) by Protection of Plant Varieties & Farmers’ Rights Authority (PPVFR) was used in the present study.
Ex-plant preparation
De-husked seeds were initially washed in sterile double distilled water added with 5 % Tween-20 for 5 min followed by 0.2 % Bavistin for 10 min prior to 70 % ethanol for 2 min. Lastly, these seeds were surface sterilized with 0.1 % mercuric chloride for 2 min. After each treatment seeds were thoroughly rinsed for 2–3 times with sterilized double distilled water.
Nutrient medium for callus development and embryogenesis
Sterilized de-husked seeds were cultured on six different rice callusing media (RCM) (Table 1). Four combinations of rice regeneration media (RR) were used (Table 1). In all the combinations of RCM and RR media, N6 (Chu et al. 1975) or MS (Murashige and Skoog 1962) were used as the basal media. All the RCM combinations were additionally supplemented with 0.05 % casein hydrolase, 0.01 % Myo-inositol. Regenerating embryos from the appropriate RR media were transferred to shoot multiplication medium (RSM) (Table 1). The regenerated shoots were placed on ½ MS medium without any growth hormone for rooting. Gelrite was used as the solidifying agent at 0.5 %. The media pH was adjusted to 5.8 prior to autoclaving for 15 min at 121 °C.
Table 1.
Media matrix used for the present study
| Media | Basal Medium | Carbon Source (%) | Auxin (mg L−1) | Cytokinin | Proline | Yeast Extract | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Maltose | Mannitol | Sucrose | Sorbitol | 2,4-D | NAA | BAP | KIN | (mg L−1) | (mg L−1) | ||
| RCM1 | MS | 5.00 | - | - | - | 3.00 | - | - | - | - | - |
| RCM2 | MS | 4.00 | - | - | - | 2.50 | - | 0.25 | 0.25 | 2.87 | - |
| RCM3 | MS | 2.00 | 1.00 | - | - | 2.00 | 0.50 | 0.50 | - | 2.87 | 500.00 |
| RCM4 | N6 | 5.00 | - | - | - | 3.00 | - | - | - | - | - |
| RCM5 | N6 | 4.00 | - | - | - | 2.50 | 0.25 | 0.25 | 2.87 | - | |
| RCM6 | N6 | 2.00 | 1.00 | - | - | 2.00 | 0.50 | 0.50 | 2.87 | 500.00 | |
| RR1 | MS | 3.00 | - | - | - | 1.00 | 2.50 | - | - | ||
| RR2 | MS | - | - | 2.00 | 1.00 | - | - | 0.10 | 2.50 | - | - |
| RR3 | N6 | 3.00 | - | - | - | - | - | 1.00 | 2.50 | - | - |
| RR4 | N6 | - | - | 2.00 | 1.00 | - | - | 0.10 | 2.50 | - | - |
| RSM | MS | - | - | 3.00 | - | - | - | 4.00 | 2.00 | - | - |
All RCM medium were supplemented with 0.05 % Casein hydrolase, 0.01 % Myo-inositol and 0.5 % Gelrite. Whereas RR medium contain only 0.01 % Myo-inositol and 0.5 % Gelrite
All the tissue culture steps for seed germination/callusing/embryo development were carried out in dark at 27 ± 1 °C, whereas plant regeneration, rooting and hardening steps were carried out under light (16/8 h of light/dark cycle). Sub-culturing frequency was every 15 days. Embryogenic callus formation efficiency was recorded after six weeks of incubation.
Callus age on regeneration efficiency
Calli were grown on N6 based medium RCM6 medium for 4, 5, 6, 7 and 8 weeks. These calli were then transferred to MS based RR1 medium and the shoot induction frequency was estimated. For each experiment 50 calli were used per age group and the data shown represent the average of three independent replication.
Sensitivity of rice callus unit to selection agent
To determine the lethal dose of selection agents 25 non-transformed calli in three replication on RCM6 medium were sub-cultured onto RCM6 media supplemented with different concentrations of the filter sterilized Geneticin (G418) and Paramomycin, either singly or in combinations. The number of dead (black necrotic) and healthy calli were identified after 4 weeks to ascertain dose sensitivity of antibiotics.
Vector and strains
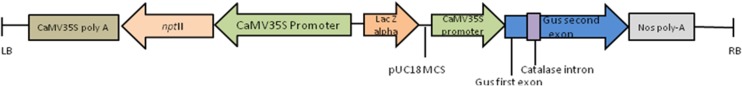
The binary vector pCAMBIA2201 (Cambia, Canberra, Australia) (Hajdukiewicz et al. 1994) (Fig. 1) was used for standardizing the transformation protocol using the novel selection scheme agents such as G418 and Paramomycin. This vector harbours nptII gene as a plant selection marker and an intron β-glucuronidase (uidA gene) as a reporter gene (Jefferson et al. 1987) both driven by CaMV35S promoter. The vector DNA was electroporated into EHA105 (Hood et al. 1986) cells using Multiporator (Effendorf, Germany) following manufacturer’s protocol. The electroporated EHA105 cells were grown on AB agar minimal medium (Chilton et al. 1974) supplemented with 20 mg l−1 Rifampicin, 50 mg l−1 Kanamycin for 3 days at 28 °C.
Fig. 1.
Schematic representation of the T-DNA region in the pCAMBIA 2201 vector used for genetic transformation. Binary construct with nptII selection gene and gus (with intron) reporter gene both controlled by CaM35S promoter
Agrobacterium culture
Single colony of EHA105 carrying binary vector pCAMBIA2201 was cultured in dark in a 30 ml glass tube containing 5 ml AB liquid medium (pH 7.2) supplemented with 20 mg l−1 Rifampicin, 50 mg l−1 Kanamycin overnight on a rotary shaker (120 rpm) at 28 °C. An inoculum of 250 μl of this overnight culture was further injected into a 100 ml conical flash containing 20 ml of AB broth under similar antibiotics and culture condition. The culture was grown till an OD600 of 0.6 to 0.8 followed by centrifuged at 5000 rpm. The pellet was re-suspended into liquid infection media (half strength MS medium supplemented with 100 μM Acetosyringone and tobacco leaf bits taken from plants grown under sterile conditions). The OD600 of this suspension was adjusted to 1.0 (corresponds to a density of 109 cells/ml) and was used for calli infection.
Infection and co-cultivation
One day prior to infection, actively growing calli on RCM6 (2–3 cm in diameter) were transferred to co-cultivation medium adjusted to pH 5.2 (half strength MS basal medium supplemented with 1 % Glucose, 2 % Sucrose, 1 mg l−1 NAA, 200 μm Acetosyringone, 40 mg l−1 Cysteine, 1.06 g l−1 MES monohydrate, 0.01 % Myo-inositol and 1 % Agar). The calli were desiccated just before infection by placing it on sterile blotting paper under sterile air flow of laminar cabinet for 10 min. Callus units were immersed in bacterial suspension, for 15 min and blot-dried prior to transferring those to a fresh co-cultivation medium. The infected calli were co-cultivated for 2 days at 25 °C in dark. Post co-cultivation, callus units were rinsed thoroughly in liquid RCM6 medium fortified with 0.02 % Triton-X, 250 mg l−1 Carbenicillin (Duchefa Biochemic, The Netherlands) and 250 mg l−1 Cefotaxime for 10 min to eliminate Agrobacterium.
Selection regeneration and root induction
The calli were transferred from co-cultivation medium to the resting medium (RCM6 + 250 mg l−1 Carbenicillin +250 mg l−1 Cefotaxime) for 4–5 days to facilitate recovery and growth of the transformed cells. Subsequent to resting calli were transferred onto RCM6 agar selection medium supplemented with 70 mg l−1 Paramomycin (Duchefa Biochemic, The Netherlands), 30 mg l−1 G418 (Duchefa Biochemic, The Netherlands), 100 mg l−1 Carbenicillin and 100 mg l−1 Cefotaxime. Calli were sub-cultured every fort-night (thrice) into the same selection media till the emergence of embroygenic calli.
Selected embryogenic calli were transferred onto the rice regeneration medium (RR1) supplemented with 100 mg l−1 Carbenicillin and 50 mg l−1 G418 at 26 ± 1 °C with 16/8 h light/dark cycles and sub-cultured onto the same media at an interval of 15 days. After four weeks tiny little shoots emerged from embryos which were sub-cultured onto rice shoot multiplication medium (RSM) supplemented with 100 mg l−1 Carbenicillin and 50 mg l−1 G418 for selection of putative transgenic rice shoots. Putative transgenic rice shoots retaining dark green colour were excised and transferred to half strength MS basal medium without any growth regulators and selection agent for rooting.
Rooting and hardening
After sufficient root development, plants were transferred to a plastic cup (200 ml) containing sterile vermiculite and coco-peat mix (50:50) and kept covered with a light-transparent transparent polyethylene plastic for acclimatization under light at 26 ± 1 °C with 16/8 h light/dark cycles. The plants were watered with 1X Yoshida solution (Yoshida et al. 1976) when needed. After the removal of the plastic cover and the appearance of the first new leaf, the plants were finally transplanted into pots filled with equal volume of sterile clay and loam soil. Chemical fertilizers (N, P and K) were applied to pots and were maintained in a greenhouse at 28 ± 2 °C till harvest.
Histochemical GUS assay
β-glucuronidase (GUS) gene activity was detected using freshly prepared histochemical solution having substrate X-Gluc (5-bromo, 4-chloro, 3-indolyl-D-glucuronide) (Gold Biotechnology, Inc. Cat # G1281C1, St. Louis, USA) in callus unit at resting medium (10 days after infection), embryonic calli on selection medium added with Paramomycin and G418 (25 days after infection) and leaves of hardened plants in greenhouse as described by Chaudhury et al. (1995). Histochemical reaction was observed at 37 °C after 24 h of incubation and the tissue cleared in 100 % methanol was examined for presence of blue colour. Non-transformed control calli, leaves and Agrobacterium suspension were served as negative controls.
PCR analyses
Genomic DNA was extracted from the leaf tissue collected from putative transgenic (T0 generation) rice plants grown in greenhouse following the procedure of Dellaporta et al. (1983). The 575 bp region of nptII fragment was amplified with nptII-specific primer sequences: 5′-TCCGCTTGCTGAAAATGTCC-3′, and 5′-CTGTTGTGCCCAGTCATAGC-3′; and 478 bp region of gus with gus–specific primer sequences: 5′-GAAGTTCATCTGCACCACCG-3′; and 5′-GGTGCTCAGGTAGTGGTTGT-3′. Each PCR reaction was performed in 25 μL (total volume) of reaction mixture comprising 100 ng template DNA, 0.2 μl (1 Unit) of Taq DNA polymerase (MBI Fermentas, UK), 2.5 μl of 10X PCR buffer (MBI Fermentas, UK), 2.5 μl of 2 mM dNTPs mixture (MBI Fermentas, UK), forward primer and reverse primer 0.5 μl (10pM/μl) each and made up the volume using molecular grade double distilled water. Thermal cycling condition used included 3 min at 94 °C for initial de-naturation followed by 35 cycles of 94 °C for 30 s, 58–62 °C for 30s, 72 °C for 30 s and a final extension at 72 °C for 5 min, finally halting the reaction at 4 °C. PCR was performed by using Mastercycler (EP Gradient Eppendorf). DNA from non-transformed (wild type) plant was included in the experiments as a control. Amplified DNA fragments of nptII and gus were separated through electrophoreses on a 1.2 % agarose gel, detected by ethidium bromide staining, visualized and documented on Molecular Imager Gel Doc XR System (Bio-Rad Laboratories, CA, USA).
DOT blot analysis
Rice genomic DNA was extracted from leaf tissues as described earlier. The total genomic DNA was denatured by treating with 4 M NaOH and was blotted onto a nylon membrane (Hybond N+). Pre-hybridization and hybridization procedures were followed according to Sambrook et al. Probe preparation (478 bp PCR amplified product from gus region), labeling and detection was done using DIG High Prime DNA labeling and detection starter kit II (Roche Applied Science, USA) as per manufacturer’s instructions. The final detection was done by chemiluminescence (1:100) dilution of CDP-Star Reagent (Roche Diagnostics) followed by exposure to X-ray films.
Data analysis
Arcsine transformation was done for the percentage data. The transformed data were analyzed using standard ANOVA-single factor (Excel; Microsoft), procedures and the difference between the treatments means were compared using the Fisher’s Least Significant Difference test (LSD). All differences were judged to be significant at p < 0.05. From total number of calli kept for regeneration, the number of calli that eventually regenerate into plantlets were counted and expressed as callus regeneration ability (%).
Results and discussions
Callusing and plant regeneration
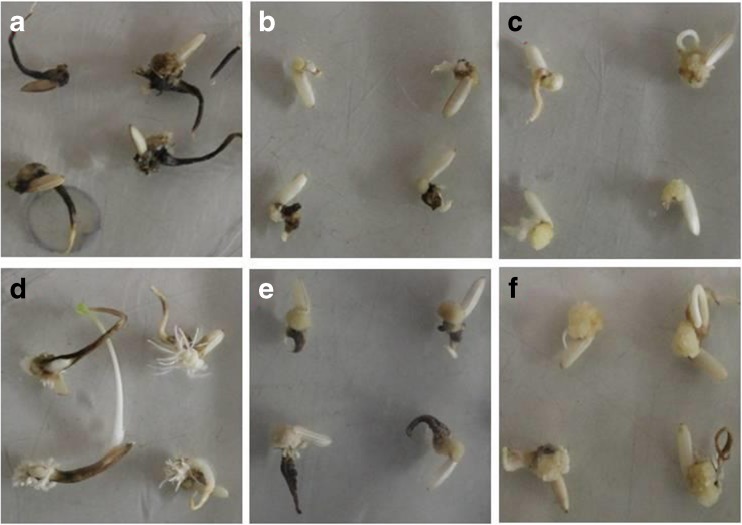
High quality callus formation from suitable explants is the major pre-requisite for developing an efficient somatic embryo mediated plant transformation protocol. In order to identify the suitable medium for getting high quality callus, a series of rice callusing medium were tested. Six different media (Table 1) with various concentrations of growth regulators, sugars and amino acid sources were investigated. Surface sterilized JK1044R seeds were placed on each of these media and callus initiation, and quality were observed. On most of the RCM media, swollen scutellum of rice seeds developed irregular callus. Callus initiation and embryogenic callus formation frequency varied among the six combination tested. However, each medium produced different kind of callus (Fig. 2). As evident from Fig. 2, RCM1 produced necrotic and rhizogenic calli; RCM2 necrotic and slow growing callus; RCM3: low rate of scutellar calli; RCM4: non-scutellar and rhizogenic callus; RCM5: slow growing callus with root emergence; RCM6: compact rough surface callus with pale yellow colour and high rate of scutellarcalli.RCM4, RCM5 and RCM6, all of which has a N6 base, provided higher efficiency of callus induction and stimulation of embryogenic callus development compared to RCM1, RCM2and RCM3. After 4 weeks of subculture under dark condition, pockets of friable calli (small clumps <5 mm diameter) of creamy white colour were observed only in RCM6 medium. These embryogenic calli from RCM6 were found highly regenerable and hence chosen for transformation activities.
Fig. 2.
Callus initiation from scutellum region of rice seeds on rice callusing medium (RCM) a RCM1- Necrotic and rhizogenic callus; b RCM2- Necrotic and slow growing callus; c RCM3- Low rate of scutellar calli; d RCM4- Non-scutellar and rhizogenic callus; e RCM5- Slow growing callus with root emergence; f RCM6- Compact rough surface callus with pale yellow colour, high rate of scutellar callus
For rice tissue culture and embryogenic callus induction, MS media with 2,4-D was used widely (Visarda and Sarma 2004; Saharan et al. 2004; Lin and Zhang 2005; Tariq et al. 2008; Syaiful et al. 2009; Wani and Gosal 2010). However, N6 as basal media for callus initiation and embryo formation showed enhanced efficiency and responsiveness to embryogenic callus formation than MS containing media. In the current study, RCM6, with a N6 base, and with a combination of auxin and cytokinin is found to induce better callusing and regeneration. As reported by many authors, our study also demonstrated the improvement of embryogenesis in presence of amino acid source like L-Proline, Yeast extract and Casein hydrolase (KaviKishor et al. 1999; Khaleda and Al-Forkan 2006; Saharan et al. 2004).
Four different media, RR1, RR2, RR3 and RR4 (Table 1) were investigated for plant regeneration capability from the embryogenic callus developed in RCM6 media. RR1 and RR2 were based on MS basal media whereas RR3 and RR4 were based on N6. We formed these media by supplementing the corresponding basal media with various concentrations of 6-BAP, Kinetin and NAA. The regeneration frequencies were found to be ranging from 32.3 % to 70.2 % (Table 2). Interestingly, RR1 was found to be most effective and showed highest regeneration frequency (70.2 %). In general, among all regeneration enabled with MS basal media was found to be more effective for the regeneration of shoots than that of N6.
Table 2.
Embryogenesis and Regeneration frequency in JK1044R
| Media | Total number of embryonic callus unit | Regeneration Percentage | |
|---|---|---|---|
| Cultured callus unit | Responsive callus unit | ||
| RR1 | 127 | 89 | 70.2 ± 1.7c |
| RR2 | 124 | 40 | 32.3 ± 2.8a |
| RR3 | 123 | 61 | 49.9 ± 5.7abc |
| RR4 | 128 | 51 | 39.5 ± 3.7ab |
Means followed by the same letter are not significantly different from each other (LSD > 0.05)
Callus age on regeneration efficiency
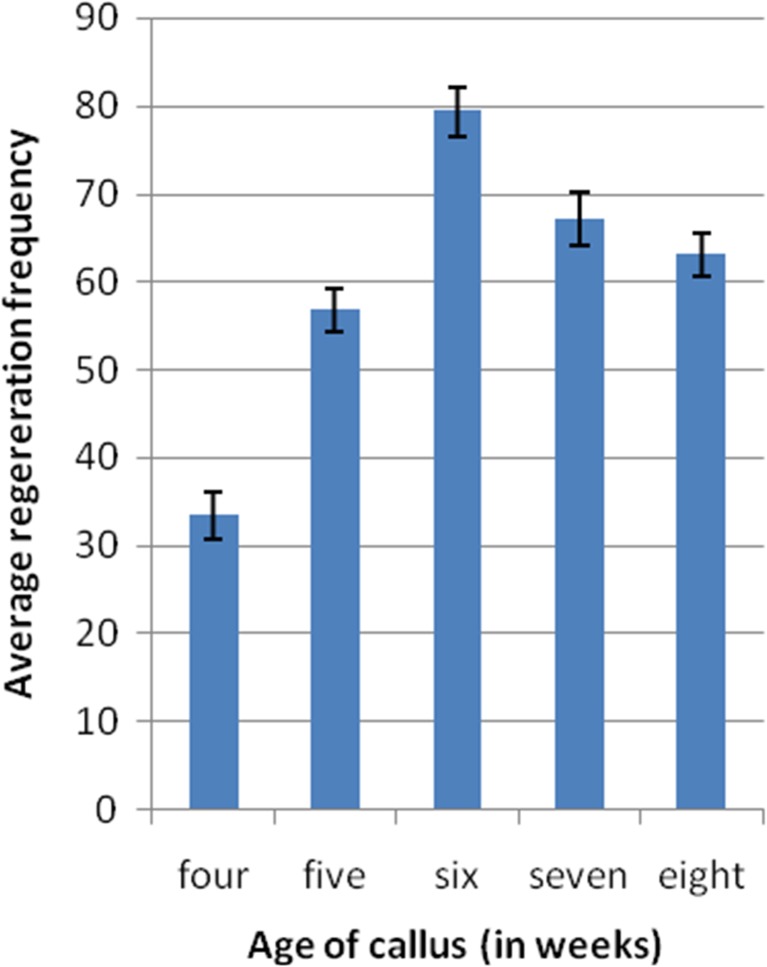
Age of callus often determine the regenerability, regeneration efficiency was calculated as a percentage of the number of plant-let regenerated/total number of callus explants. We multiplied embryogenic callus on RCM6 medium for various time durations and then transferred them to RR1 regeneration medium for shoot development. It was observed (Fig. 3) that 6 week old embryonic callus provided highest regeneration frequency of 77.3 % when compared to other age callus. Based on high competency to regenerate via somatic embryogenesis only 6 weeks old calli were only used for further transformation studies. In contrast, 61.3 % regeneration was reported by 6 days old callus with indica rice cultivar BPT5204 (Manimaran et al. 2013). The slight difference observed in callus regeneration frequency with respect to both cultivars might be due to the genotypic variation or culture media and/or growth conditions. However, in both cases, regeneration frequencies were gradually declined as the age of the callus increased (Fig. 4).
Fig. 3.
Effect of embryonic callus age on regeneration frequency
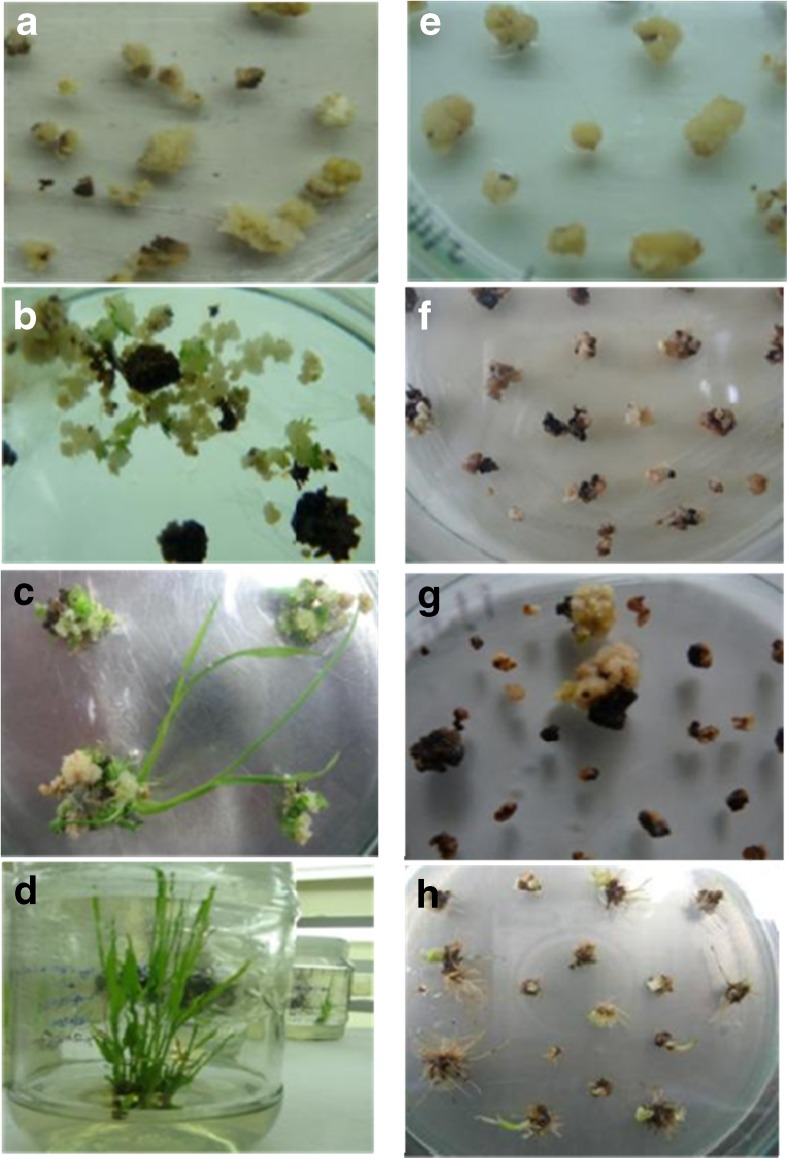
Fig. 4.
Different stages of embryogenic callus (a, b, c, d) compare to non-embryogenic callus (e, f, g, h) a Friable embryogenic callus units with dry appearance and white patches; b at selection stage; c: regeneration stage; d multiple shooting stage e: Non-embryogenic callus; f at selection stage most of them turn back; g no complete regeneration observed; h Non-embryogenic callus lead rhizogenic callus and root emergence
Plant selection marker
As described earlier, we wanted to use of nptII for plant selection. This is to avoid the higher cost of Hygromycin in our context. But for monocot species use of nptII gene as a plant selectable marker using Kanamycin as antibiotic have been reported to be ineffective, including rice. This is because, few genotypes of rice have even shown to display natural resistance to the Kanamycin antibiotic. So, we wanted to test other antibiotics such as Paramomycin and Geneticin (G418) either alone or in combination for effective transgenic calli/tissue selection.
The control callus units of JK1044R were found to be highly sensitive to G418 in combination with Paramomycin. Increasing the concentration of G418 resulted in gradual decrease in percentage of callus survival. Based on antibiotic sensitivity assay minimal inhibitory concentration of antibiotics was observed on combination of G418 30 mg l−1 with Paramomycin 70 mg l−1 when applied at proliferating stage of transgenic callus during selection. RCM6 was supplemented with various concentrations of G418 and/or Paramomycin and well established calli from RCM6 medium were transferred onto it and grown for four weeks with a sub-culturing in between. As provided in the Table 3, out of ten different treatments, only two treatments, treatment 7 (Paramomycin alone at 100 mg l−1) and 9 (70 mg l−1 Paramomycin with 30 mg l−1 G418) killed all the non-transgenic calli.
Table 3.
Sensitivity of rice calli to selection agent G418, Paramomycin and in combination on RCM6 Medium
| Antibiotic Concentration (mg l−1) | Average percentage of survived embryonic callus | Arcsine transformed percentage ± SE | ||
|---|---|---|---|---|
| Geneticin (G418) | Paramomycin | |||
| Treatment 1 | 0.0 | 0.0 | 98.6 | 86.2 ± 3.8e |
| Treatment 2 | 50.0 | 0.0 | 20.0 | 26.5 ± 1.6d |
| Treatment 3 | 75.0 | 0.0 | 9.3 | 17.7 ± 1.2bc |
| Treatment 4 | 100.0 | 0.0 | 1.3 | 3.8 ± 3.8a |
| Treatment 5 | 0.0 | 80.0 | 14.6 | 22.4 ± 2.1cd |
| Treatment 6 | 0.0 | 90.0 | 5.3 | 13.2 ± 1.6b |
| Treatment 7 | 0.0 | 100.0 | 0.0 | 0.0 ± 0.0a |
| Treatment 8 | 20.0 | 80.0 | 6.6 | 14.8 ± 1.6b |
| Treatment 9 | 30.0 | 70.0 | 0.0 | 0.0 ± 0.0a |
| Treatment 10 | 40.0 | 60.0 | 1.3 | 3.8 ± 3.8a |
All these data were resulted from three replicates with 25 calli per replication; data are presented after arcsine transformations as avg. means ± standard error followed by the same letter are not significantly different from each other (LSD > 0.05)
Calli kept on RCM6 with treatment 9 showed normal growth of callus for an initial period of 15 days and subsequently turned brown. However calli on RCM6 medium with treatment 7 turned brown immediately after 15 days. In addition it was also observed that treatment 2 (50 mg l−1 G418) restricted growth of calli and killed approx. 80 % of the calli. Therefore in our transformation experiment the infected explants were selected with treatment 9 on rice callusing medium for 8 weeks. Subsequently a mild dose of G418 (treatment 2) was used on the selected calli for plant regeneration in RR1 media and shoot multiplication in RSM media.
Even though many amino-glycoside antibiotics work against the nptII gene, degree of susceptibility of explants during selection differs. Dekeyser et al. 1989 found the performance of G418 as a selectable agent for rice was better than other antibiotics, but with poor transformation efficiency. Similarly, Paramomycin has been used to select transgenic callus carrying nptII and found to be more effective than other aminoglycosides selection in rapeseed (Guerche et al. 1987), tobacco (Bellini et al. 1989), citrus (Vardi et al. 1990) and sunflower (Escandon and Hahne 1991). Use of both G418 and Paramomycin as transgenic shorting agent has been utilized in sweetpotato. Where in in-vitro screening of transgenic calli was carried out under medium supplemented with G418 and in vivo screening of transgenic plants were accomplished using Paramomycin (Shin et al. 2007). We did not find any report of using Paramomycin as a selection agent for rice. However, instead of using Paramomycin alone, we preferred a combination of Paramomycin and G418 since it may provide better selection and regeneration efficiency (data not shown). The mode of action of G418 can be simplified as it irreversibly binds to 70S and 80S ribosomal subunit thereby blocks polypeptide synthesis in turn inhibit protein synthesis disrupting proofreading. Whereas, Paramomycin not only inhibits an early stage translation but also plays a major role in subsidizing translocation of amino-actyl-tRNA (Eustice and Wilhelm 1984). When the treatment 9 was used in the selection media against transformed rice calli for 8 weeks, it resulted in survival of only the transgenic calli.
Genetic transformation
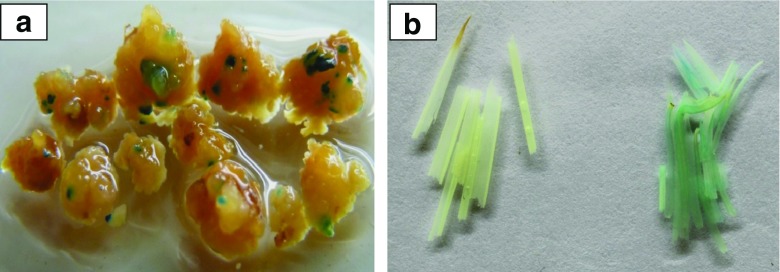
JK1044R callus units proliferated on RCM6 were infected with EHA105 cells carrying with plant transformation plasmid pCAMBIA2201as per the protocol provided in the Materials and Methods. A total of 80 callus units were infected in two different experiments, calli were analyzed for destructive GUS staining protocol six days after infection. Based on the GUS analysis transformation percentage was estimated to be 30 %. One of the highlight of this protocol is desiccation of calli for 10 min prior to infection. Adopting this additional step helped in achieving higher transformation efficiency (data not provided). This is agreement with Urushibara et al. (2001), they reported that desiccation treatment increased transformation efficiency by more than 10 folds in calli derived from rice suspension cultures. The expression of GUS gave prominent dark-blue foci (Fig. 5a) which was easily detected in infected embryonic callus units. Small cell aggregates stained dark blue, confirming T-DNA transfer. Most likely this light blue colour of GUS expression occurred in transformed cells and not in EHA105 Agrobacterium cells since the GUS gene was interrupted with a plant intron and can only be processed within a plant cell (Ohta et al. 1990).
Fig. 5.
GUS expression at two stage 5a GUS foci in callus on resting medium (after 6-days co-cultivation); 5b GUS expression in leaf tissues of green shoots developed on RSM medium compared with non-transformed control leaf tissue from seedling
The leaf tissues of green shoots developed on RSM medium developed blue colour when subjected to GUS staining and control showed no detectable blue staining under the same staining conditions. A total of 20 independent putative transgenic plants were regenerated. In our study 15 out of all the regenerated rice shoots expressed uniform faint GUS activity which was spread throughout the leaf lamina of different transgenic events. Although the GUS expression in leave tissues were dull in nature but it was uniform throughout the surface. Similarly, Zhao et al., 2001 reported faint blue staining during early sheath development in transgenic rice expressing gus gene. Degree of GUS staining observed in plant tissue are not only dependent on activity of promoter but also contributed by cell size, metabolic activity and substrate accessibility to various cell types, and thus any of these factors can be held responsible for reporting faint blue colour of transgenic rice leaves (Battraw and Hall 1990).
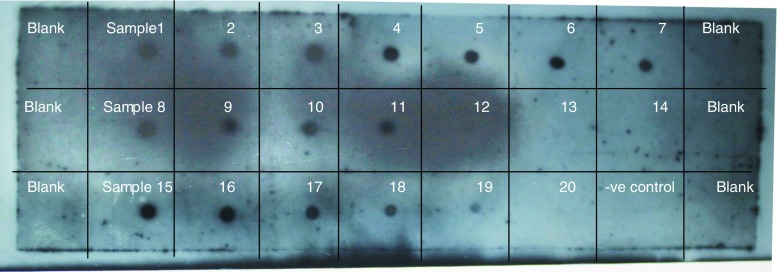
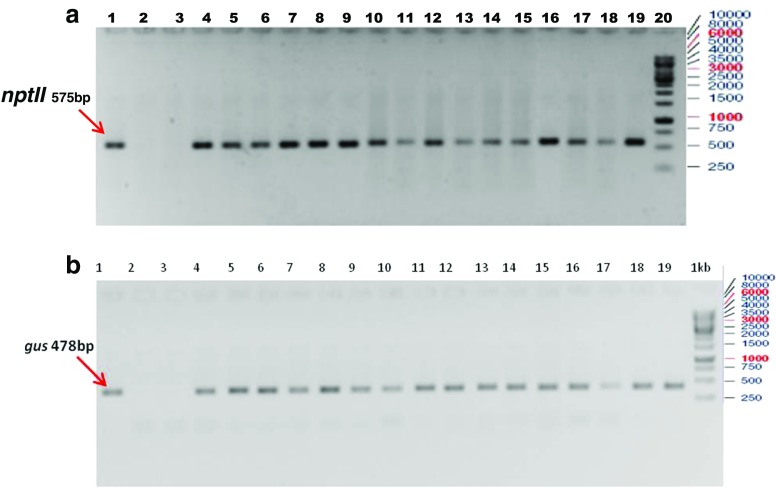
DNA dot blot using a gus specific probe (Fig. 6) indicated 16 out of 20 putative transgenic plantlets had the T-DNA in their genome. To further confirm, PCR experiments with gene specific primers of gus and nptII were conducted. As in Fig. 7a and b, gus and nptII gene specific primers amplified 575 bp and 478 bp fragments respectively proving the presence of transgene in the sampled genomes. Taken together, PCR experiments and dot blot analyses of genomic DNA of putative transgenics confirmed the integration of transgene into the genome. We generated 16 transgenic plants out of 435 callus units infected in four different experiments. In the present study transformation efficiency was calculated as 3.6 % based on the number of successfully established plants in greenhouse which showed positive result to GUS, PCR and dot blot analysis and not on the basis of only GUS expression in callus unit. All the regenerated plantlets exhibited normal root growth and were morphologically similar to seed-grown plants. Total timeline for generating transformants was found to be about 28–30 weeks, from seeds to seeds (Supplementary Fig. 1). Upon maturity, seed set was observed in all the regenerated transgenic lines indicating tissue culture and selection did not affect their reproductive potential.
Fig. 6.
Dot blot hybridization of transgenic rice DNA along with negative non-transformed plant DNA using gus gene specific probe).Sample No. 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 15, 16, 17, 18 and 19 produced signals
Fig. 7.
a PCR analysis of transgenic rice events showing nptII gene fragment (575 bp); Lanes 1 pCAMBIA2201 plasmid DNA, lanes 2: untransformed negative control, lane 3: Blank (water control), lane: 4 to 19 different transgenic events, and lane 20; 1 kb DNA ladder.b PCR analysis of transgenic rice events showing gus gene fragment (478 bp); Lanes 1: pCAMBIA2201 plasmid DNA, lane 2: untransformed negative control, lane 3: Blank (water control), lane 4 to 19 different transgenic events, and lane 20: 1 kb DNA ladder.
This is the first report to demonstrate the use of widely used nptII gene in rice transformation protocol. A two tier unique non-Kanamycin based selection system is developed which can be used for the development of transgenic events in other monocot plant species with inherent resistance to Kanamycin. The developed protocol was used to produce independent transgenic lines (result not provided) for one more indica rice genotypes, IR 64 and the results were mostly consistent. The current study established a comprehensive protocol to achieve reproducible and efficient genetic transformation, selection and regeneration protocol for elite indica rice genotypes. This protocol is used further for transformation of restorer line with agronomical important traits.
Electronic supplementary material
(JPEG 165 kb)
Acknowledgments
We are thankful to Mr. Sanjay Gupta, Director JK Agri. Genetics Ltd., Hyderabad, India and all research scientists and staffs of organization. Authors, also acknowledge the partial funding support of Biotechnology Industry Research Assistance Council (BIRAC), New Delhi, India and Department of Biotechnology, Government of India (BIPP grant no. BT/BIPP/0320/07/10)
References
- Anon, (2014). [Online] Available at: http://oryza.com/news/rice-news/usda-post-forecasts-india-my-2014-15-rice-production-104-million-tons [Accessed 1 Jul. 2014].
- Baisakh N, Datta K, Rashid H, Oliva N, Datta SK. Agrobacterium tumefaciens-mediated transformation of an elite indica rice maintainer line IR68899B with a reconstructed T-DNA carrying multiple genes. Rice Genet Newsl. 2000;17:122–125. [Google Scholar]
- Battraw MJ, Hall TC. Histochemical analysis of CaMV 35S promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Mol Biol. 1990;15:527–538. doi: 10.1007/BF00017828. [DOI] [PubMed] [Google Scholar]
- Bellini CP, Guerche A, Spielmann J, Goujaud C, Lesaint CM. Genetic analysis of transgenic tobacco plants obtained by liposome-mediated transformation: absence of evidence for the mutagenic effect of inserted sequences in sixty characterized transformants. J Hered. 1989;80:361–367. [Google Scholar]
- Caplan A, Dekeyser R, van Montagu M. Selectable markers for rice transformation. Methods Enzymol. 1992;216:426–441. doi: 10.1016/0076-6879(92)16039-M. [DOI] [PubMed] [Google Scholar]
- Chan MT, Lee TM, Chang HH. Transformation of indica rice (Oryza sativa L.) mediated by Agrobacterium tumefaciens. Plant Cell Physiol. 1992;33:577–583. [Google Scholar]
- Chan MT, Chang HH, Ho SL, Tong WF, Yu SM. Agrobacterium-mediated production of transgenic rice plants ex-pressing a chimeric α-amylase promoter/β-glucuronidase gene. Plant Mol Biol. 1993;22:491–506. doi: 10.1007/BF00015978. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Maheshwari SC, Tyagi AK. Transient expression of gus gene in intact seed embryos of indica rice after electroporation-mediated gene delivery. Plant Cell Rep. 1995;14:215–220. doi: 10.1007/BF00233636. [DOI] [PubMed] [Google Scholar]
- Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Wang CC, Sun CS, Msu C, Yin KC, Chu CY, Bi FY. Establishment of an efficient medium for anther cultures of rice through comparative experiments on nitrogen sources. Sci Sinica. 1975;18:659–668. [Google Scholar]
- Datta K, Velazhahan R, Oliva N, Ona I, Mew T, Khush GS, Muthukrishnan S, Datta SK. Over-expression of cloned rice thaumatin like protein (PR-5) in transgenic rice plants enhances environmental-friendly resistance to Rhizoctonia solani causing sheath blight disease. Theor Appl Genet. 1999;98:1138–1145. doi: 10.1007/s001220051178. [DOI] [Google Scholar]
- Datta K, Koukolíková-Nicola Z, Baisakh N, Oliva N, Datta SK. Agrobacterium-mediated engineering for sheath blight resistance of indica rice cultivars from different ecosystems. Theor Appl Genet. 2000;100:832–839. doi: 10.1007/s001220051359. [DOI] [Google Scholar]
- Datta K, Tu JM, Oliva N, Ona I, Velazhahan R, Mew TW, Muthukrishnan S, Datta SK. Enhanced resistance to sheath blight by constitutive expression of infection related rice chitinase in transgenic elite indica rice cultivar. Plant Sci. 2001;160:405–414. doi: 10.1016/S0168-9452(00)00413-1. [DOI] [PubMed] [Google Scholar]
- Dekeyser R, Claes B, Marichal M, Montagu MV, Caplan A. Evaluation of selectable markers for rice transformation. Plant Physiol. 1989;90:217–223. doi: 10.1104/pp.90.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA mini preparation: version II. Plant Mol Biol Report. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Escandon A, Hahne G. Genotype and composition of culture medium are factors important in the selection for transformed sunflower (Helianthus annus) callus. Physiol Plant. 1991;81:367–376. doi: 10.1111/j.1399-3054.1991.tb08745.x. [DOI] [Google Scholar]
- Eustice DC, Wilhelm JM. Mechanisms of action of aminoglycoside antibiotics in eucaryotic protein synthesis. Antimicrob Agents Chemother. 1984;26(1):53–60. doi: 10.1128/AAC.26.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerche P, Charbonnier M, Jouanin L, Tourneur C, Paszkowski J, Pelletier G. Direct gene transfer by electroporation in Brassica napus. Plant Sci. 1987;52:111–116. doi: 10.1016/0168-9452(87)90112-9. [DOI] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hood EE, Helmer GL, Fraley RT, Chilton MD. The hyper virulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bac. 1986;168:1291–1301. doi: 10.1128/jb.168.3.1291-1301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo j. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KaviKishor PB, Sangam S, Naidu KP (1999) Sodium, potassium, sugar alcohol and proline mediated somatic embryogenesis and plant regeneration in recalcitrant rice callus. Plant Tissue Culture and Biotechnology: Emerging Trends, Proceedings of a symposium held in Hyderabad, India, pp 78–85
- Khaleda L, Al-Forkan M. Stimulatory effects of casein hydrolysate and proline in in vitro callus induction and plant regeneration from five Deepwater rice (Oryza sativa L.) Biotech. 2006;5(3):379–384. doi: 10.3923/biotech.2006.379.384. [DOI] [Google Scholar]
- Khanna HK, Raina SK. Agrobacterium-mediated transformation of indica rice cultivars using binary and super binary vectors. Aust J Plant Physiol. 1999;26:311–324. doi: 10.1071/PP98160. [DOI] [Google Scholar]
- Lin YJ, Zhang Q. Optimizing the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- Manimaran P, Kumar GR, Reddy MR, Jain S, Rao TB, Mangrauthia SK, Sundaram RM, Ravichandran S, Balachandran SM. Infection of early and young callus tissues of indica rice BPT 5204 enhances regeneration and transformation efficiency. Rice Sci. 2013;20:415–426. doi: 10.1016/S1672-6308(13)60153-5. [DOI] [Google Scholar]
- Miki B, McHugh S. Selection marker genes in transgenic plants: applications, alternatives and biosafety. J Biotechnol. 2004;170:193–232. doi: 10.1016/j.jbiotec.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Ohta S, Mita S, Hattori T, Nakamura K. Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 1990;31:805–813. [Google Scholar]
- Papademetriou MK (2000) Rice production in the Asia-Pacific region: issues and perspectives. Bridging Rice Yield Gap Asia-Pacific Region 220
- Park SH, Pinson SRM, Smith RH. T-DNA integration into genomic DNA of rice following Agrobacterium inoculation of isolated shoot apices. Plant Mol Biol. 1996;32:1135–1148. doi: 10.1007/BF00041397. [DOI] [PubMed] [Google Scholar]
- Praveen P, Dinesh TK. Modern techniques and agronomic packages for hybrid rice cultivation in India. Adv Agri Bota. 2012;4(1):17–21. [Google Scholar]
- Raineri DM, Bottino P, Gordon MP, Nester EW. Agrobacterium mediated transformation of rice Oryza sativa L. Nat Biotechnol. 1990;8:33–38. doi: 10.1038/nbt0190-33. [DOI] [Google Scholar]
- Saharan V, Yadav RC, Yadav NR, Ram K. Studies on improved agrobacterium mediated transformation in two indica rice (Oryza sativa L) varieties. Afr J Biotechnol. 2004;3:572–575. [Google Scholar]
- Shin Y-M, Choe G, Shin B, Yi G, Yun PY, Yang K, Lee JS, Kwak SS, Kim KM. Selection of nptII transgenic sweetpotato plants using G418 and paromomycin. J Plant Biol. 2007;50(2):206–212. doi: 10.1007/BF03030631. [DOI] [Google Scholar]
- Syaiful BP, Siti NAA, Maheran AA, Sariah M, Othman O. Somatic embryogenesis from scutellar embryo of Oryza sativa L. var. MR 219. Pertanika J Tropl Agri Sci. 2009;32:185–194. [Google Scholar]
- Tariq M, Ali G, Hadi F, Ahmad S, Ali N, Shah AA. Callus induction and in vitro plant regeneration of rice (Oryza sativa L.) under various conditions. Pak J Biol Sci. 2008;11:255–259. doi: 10.3923/pjbs.2008.255.259. [DOI] [PubMed] [Google Scholar]
- Tu J, Ona I, Zhang Q, Mew TW, Khush GS, Datta SK. Transgenic rice variety ‘IR72’ with Xa21is resistant to bacterial blight. Theor Appl Genet. 1998;97:31–36. doi: 10.1007/s001220050863. [DOI] [Google Scholar]
- Tu J, Datta K, Oliva N, Zhang G, Xu C, Khush GS, Zhang Q, Datta Sk. Site-independently integrated transgenes in the elite restorer rice line minghui 63 allow removal of a selectable marker from the gene of interest by self-segregation. Plant Biotechnol J. 2003;1:155–165. doi: 10.1046/j.1467-7652.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- Twyman RM, Stoger E, Kohli A, Capell T, Christou P. Selectable and screenable markers for rice transformation. In: Jackson JF, Linskens HF, editors. Molecular methods of plant analysis, volume 22 (testing for genetic manipulation in plants) NY: Springer-Verlag; 2002. pp. 1–18. [Google Scholar]
- Urushibara S, Tozawa Y, Kawagishi-Kobayashi M, Wakasa K. Efficient transformation of suspension-cultured rice cells mediated by Agrobacterium tumefaciens. Breed Sci. 2001;51:33–38. doi: 10.1270/jsbbs.51.33. [DOI] [Google Scholar]
- US Food and Drug Administration (FDA) Secondary food additives permitted in food for human consumption: food additives permitted in feed and drinking water of animals; aminoglycoside 3-phosphotransferase II; final rule. Fed Regist. 1994;59:26700–26711. [Google Scholar]
- Vardi A, Bleichman S, Aviv D. Genetic transformation of citrus protoplasts and regeneration of transgenic plants. Plant Sci. 1990;69:199–206. doi: 10.1016/0168-9452(90)90118-8. [DOI] [Google Scholar]
- Visarda KBRS, Sarma NP. Transformation of indica rice through particle bombardment: factors influencing transient expression and selection. Biol Plant. 2004;48(1):25–31. doi: 10.1023/B:BIOP.0000024271.38723.a6. [DOI] [Google Scholar]
- Wani SH, Gosal SS. An efficient and reproducible method for regeneration of whole plants from mature seeds of a high yielding indica rice (Oryza sativa L.) variety PAU 201. New Biotechnol. 2010;28(4):418–422. doi: 10.1016/j.nbt.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Yang R, Zhou Y, Cao Y, Yin Z, Yang L, Li J. The transformation of the photo-thermo sensitive genic male-sterile line 261S of rice via an expression vector containing the anti-waxy gene. Breed Sci. 2013;63(2):147–153. doi: 10.1270/jsbbs.63.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Routine procedures for growing rice plants in culture solution. In: Cock J H, Gomez K A. Laboratory Manual for Physiological Studies of Rice. Los Banos, Philippines: IRRI: 61–66.
- Zhang SP, Song WY, Chen LL, Ruan DL, Taylor N, Ronail P, Beachy R, Na dFauquet C (1998) Transgenic elite indica rice varieties, resistant to Xanthomonas oryzae pv. Orzae. Mol Breed 4: 551–558.
- Zhao ZY, Gu W, Tishu C, Tagliani L, Hondred D, Bond D, Schroeder S, Rudert M, Pierce D. High throughput genetic transformation mediated by Agrobacterium tumefaciens in maize. Mol Breed. 2001;8:323–233. doi: 10.1023/A:1015243600325. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPEG 165 kb)