Abstract
As an extended gamut of integral membrane (extrinsic) proteins, and based on their transporting specificities, P-type ATPases include five subfamilies in Arabidopsis, inter alia, P4ATPases (phospholipid-transporting ATPase), P3AATPases (plasma membrane H+ pumps), P2A and P2BATPases (Ca2+ pumps) and P1B ATPases (heavy metal pumps). Although, many different computational methods have been developed to predict substrate specificity of unknown proteins, further investigation needs to improve the efficiency and performance of the predicators. In this study, various attribute weighting and supervised clustering algorithms were employed to identify the main amino acid composition attributes, which can influence the substrate specificity of ATPase pumps, classify protein pumps and predict the substrate specificity of uncharacterized ATPase pumps. The results of this study indicate that both non-reduced coefficients pertaining to absorption and Cys extinction within 280 nm, the frequencies of hydrogen, Ala, Val, carbon, hydrophilic residues, the counts of Val, Asn, Ser, Arg, Phe, Tyr, hydrophilic residues, Phe-Phe, Ala-Ile, Phe-Leu, Val-Ala and length are specified as the most important amino acid attributes through applying the whole attribute weighting models. Here, learning algorithms engineered in a predictive machine (Naive Bays) is proposed to foresee the Q9LVV1 and O22180 substrate specificities (P-type ATPase like proteins) with 100 % prediction confidence. For the first time, our analysis demonstrated promising application of bioinformatics algorithms in classifying ATPases pumps. Moreover, we suggest the predictive systems that can assist towards the prediction of the substrate specificity of any new ATPase pumps with the maximum possible prediction confidence.
Keywords: P-type ATPase, Arabidopsis, Attribute weighting models, Supervised clustering algorithms
Introduction
The P-type ATPases are an extended gamut of membrane proteins. In order to phosphorylation, they use a phosphate group of ATP at a key conserved aspartate residue during translocation of ions (Dipolo and Beaugé 2005). The P-type ATPases play essential roles in a variety of cellular processes, especially maintaining the electrochemical gradient of several ions (Na+, K+, H+ and Ca2+) across the cell membrane as a driving force for the secondary transporters and extrusion of them if they accumulate reaching high concentration. P-type ATPases mediate cellular signaling and, as quoted by both Tang et al. (1996) and Gomes et al. (2000), they might be engaged in bringing about lipid asymmetry in a membrane generation.
Such proteins have different transporting specificities and can translocate a variety of small cations, covering proton (H+), abundant metal ions (Ca2+, Na+, K+), less abundant heavy metals (Cu+, Zn2+), and perhaps also phospholipids against their electrochemical gradient (Møller et al. 1996; Axelsen and Palmgren 1998; Palmgren and Harper 1999; Mattle et al. 2013).
The superfamily of P-type ATPases has been classified into five major phylogenetic subfamilies according to the substrate being transported (Axelsen and Palmgren 1998) which are include heavy metal ATPases (P1B), Ca2+-ATPases (belong to two phylogenetic subfamilies, P2A-P2B ATPases), (P3A) H+-ATPases, the supposed aminophospholipid ATPases (P4), as well as aless reputed subfamily (P5) with a single member in Arabidopsis. Cronin and coworkers (Cronin et al. 2000) demonstrated that as a Ca2+ ATPase and within the scope of a membrane system, SPF1 (P5 ATPase) could play a significant role in taking hold of Ca2+ homeostasis utilizing a secretory route (Cronin et al. 2000).
Generally, regarding the overall genome sequence of Arabidopsis as a model plant, forty-five P-type ATPases have been unveiled which are the largest known ATPases in a single organism. Furthermore, three P-type ATPases like proteins with unknown substrate specificity have been reported in Arabidopsis (Axelsen and Palmgren 2001). At present, the wealth of amino acid sequences, as members of the P-type ATPase family, can provide a comprehensive overview of characteristics, structure, and substrate specificity of protein as well as the structure-function relationship.
The exponential growth of both protein sequences and structures via genome sequencing and high-throughput structure determination methods heightened need for reliable computational procedures to assign a reliable function to proteins of unknown function (Caitlyn et al. 2015). Williams and Mills conducted a survey and showed that out of a wide range of organisms, phylogenetic and structural analysis of P1B-ATPase protein sequences can be carried out to forecast the specificities associated with the metal-substrate in unlabeled plant pumps (Williams and Mills 2005).
To date, many computational methods have been developed to predict the active sites and biochemical functions of unknown proteins (Watson et al. 2005; Lee et al. 2007; Loewenstein et al. 2009; Gherardini and Helmer-Citterich 2008; Skolnick and Brylinski 2009; Sleator and Walsh 2010; Chi and Hou 2011; Wilkins et al. 2012), although, more studies are needed to improve the efficiency and performance of function predictors.
Having derived and scrutinized attributes relating physicochemical and structural properties of protein sequences, one can apply the attribute weighting and supervised algorithms to find important amino acid attributes, modeling and predicting protein function. Thus, the aforementioned types of algorithms could shed light on how to delve into the functionality of protein molecular mechanism. In this paper, a combination of attribute weighting and supervised clustering-algorithms is used to uncover the protein properties as well as predicting the substrate specificity of P-type ATPase among uncharacterized substrate specificity. These protein properties are the most important attributes in classification of P-type ATPases (heavy metal pumps, Ca2+pumps, plasma membrane H+ pump and phospholipid-transporting ATPase).
Materials and methods
The P-type ATPase sequences in Arabidopsis (forty five) were extracted from ExPASY (http://www.expasy.org) database. The substrate specificity of Q9LT02 (just member of P5) is unknown. So all sequences except Q9LT02 sequence are categorized into four groups: 7 (heavy metal pump), 14 (Ca+2 pump), 11 (plasma membrane H+ pump) and 12 (phospholipid-transporting ATPase). Eight-hundred and ninety-six amino acid attributes derived from protein sequences were extracted using CLC bio software (CLC bio, Aarhus, Denmark) which include length, weight, isoelectric point, count and frequency of each element (carbon, nitrogen, sulphur, oxygen, and hydrogen), count and frequency of each amino acid, count and frequency of negatively charged, positively charged, hydrophilic and hydrophobic residues, count and frequency of dipeptides, number of α-helix and β-strand, and other secondary protein features. All amino acid attributes were classified as continuous variables, except the substrate specificity of ATPase pumps and N-terminal amino acids, which were classified as categorical. A dataset of these protein attributes was imported into Rapid Miner (RapidMiner 5.0.001, Rapid-I GmbH, Stochumer Str. 475, 44,227 Dortmund, Germany), and the substrate specificity of ATPase pumps was set as the target or label attribute. Then, the steps detailed below were applied to the dataset.
Data cleansing
Useless amino acid attributes were removed from the dataset to improve processing performance. Regarded as inefficient or redundant, attributes displaying less than or equal to a given standard deviation (SD) threshold (0.1) along with the correlated features were thoroughly excluded and written off from the database as well. The finalized list containing the efficient attributes was labeled as FCdb (final cleaned database).
Attribute weight scrutiny
To figure out the most effective amino acid attributes contributing to different ATPase pumps, the following tenvarious algorithms used in attribute weight scrutiny were employed in the finalized dataset (FCdb):
Weight based on Information gain. Figuring the information gain in class distribution, the algorithm calculates the correlation of a feature.
Weight by Information Gain ratio. Figuring the information gain ratio in class distribution, the algorithm calculates the correlation of a feature.
Rule weight scrutiny. Estimating the error rate of an OneR Model on a sample set while having a feature excluded, the algorithm calculated the correlation of the supposed feature.
Weight Deviation scrutiny. Scrutinizing standard deviations of all amino acid attributes, the algorithm defined normalized assessments by attributing the average, the minimum, or the maximum to the features.
Chi Squared statistic weight scrutiny. Regarding the class attribute, the algorithm calculated the correlation of a feature by figuring the chi-squared statistic value for each attribute used in the input sample set.
Gini index weight scrutiny. Estimating the Gini index of the class distribution, the algorithm calculated the correlation of an attribute if the given sample would have been segregated in regards with the feature.
Uncertainty Weight scrutiny. Evaluating the proportional uncertainty regarding the class, the algorithm computed an attribute correlativity.
Weight scrutiny by Relief. Setting samples and drawing analogy between the values of the present feature and the closest example given both similarity and distinction in class, the algorithm estimated the features correlativity.
Weight by SVM (Support Vector Machine). The algorithm used the typical vector coefficients of a lineal SVM for the feature assessment.
PCA weightEvaluation (Principle Component Analysis). The algorithm used the principal component factors for the feature assessment.
Attribute evaluation
Attribute assessment models are able to compute each attribute score, namely, a measure of its importance in the P-type ATPase substrate specificity.
Selection of the entire variables weighting more than 0.50 resulted in creating ten new datasets. They were named based on the models used for attribute weighting (Relief, Information gain, Uncertainty, Information gain ratio, Chi Squared, Rule, Deviation, SVM and PCA, Gini index) and thus subjected to the proceeding model of supervision. Each supervisory process using models in clustering was fulfilled for about 11 instances; in the first time, the aforementioned model was managed and performed, utilizing the principal dataset (FCdb) and on the ten newly created datasets afterwards (resulting from different algorithms used in attribute weighting).
Supervised categorization
The process of supervised clustering achievement (Baysian models and Decision Trees) is indicated as follows:
Decision Trees. Given the procedure previously described (Ebrahimi et al. 2010, 2011, 2014; Ashrafi et al. 2011), sixteen models for tree induction were employed ranging from DT Random Forest Accuracy, DT Parallel Info Gain, DT Parallel Gain Ratio, DT Gini Index,DT Random Forest Gain Ratio, DT Random Forest Gini Index, DT Parallel Gini Index, Decision Tree (DT) Accuracy, DT Gain Ratio, DT Stump Info Gain, DT Info Gain, DT Parallel Accuracy, DT Stump Gain Ratio, DT Stump Accuracy, DT Stump Gini Index, to DT Random Forest Info Gain. The entire models were applied to eleven datasets containing both the FCdb dataset and ten datasets filtered through Rule, PCA, Info Gain Ratio, Info Gain, Uncertainty, Chi Squared, Relief, Gini Index, Deviation, and SVM algorithms utilized in attributes scrutiny. Examined for comparing different models, performances the models displayed in ATPase substrate specificity prediction were computed based on structural attributes of ATPase pump. In the present survey, the definition of performance relied upon the extent to which the model demonstrated accuracy. The calculation of accuracy was not accomplished unless the percentage of correct predictions was taken in respect with the total number of instances. In other words, should a predicted attribute shows accuracy in value, which is equivalent to that of the labeled, it would be inferred as the substantiation of the correct prediction.
Naive Bayes classifier
Naive Bayes classifier is a conditional probability model based on Bayes’ rule with independence assumptions between the features. Naive Bayes is well documented and consequently, often works reasonably well and outperforms more sophisticated learning algorithms in classification, even if the features are not statistically independent (Domingos and Pazzani 1997). In this survey, two models, including Naive Bayes and kernel based Naive Bayes, were applied to predict the substrate specificity of ATPase pumps.
Outcomes
Data cleansing
The primary dataset included 44 samples (protein sequences) with 852 amino acid features. Among these examined samples, 7, 12, 11 and 14 cases were categorized as heavy metal pump, Ca+2 pump, plasma membrane H+ pump, and phospholipid-transporting ATPases, respectively. Having gone through a purgatory process by removing duplicates and inefficient/correlated features (data cleansing), the survey was conducted onward with 44 samples and 506 features left.
Feature weight scrutiny
After achieving the Data normalization, models were managed to run and meet the weight range requirements (between 0 and 1).
Weight scrutiny through PCA
Employing the PCA model, the researchers found out that the sole feature showing the weight of 1.00 (according to Table 1) was non-reducedcoefficient of Cys extinction within 280Â nm.
Table 1.
This table presents the most important attributes confirmed, based on different weighting algorithms, to be involved in the substrate specificity of ATPase pumps (Values closer to 1 shows higher effectiveness of features). Weighting algorithms were PCA, relief, uncertainty, gini index, chi squared, deviation, rule, correlation, gain ratio, and information gain
| Weighting method | Attribute | Weight |
|---|---|---|
| Weighting by PCA | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 1.00 |
| Weighting by SVM | Val frequency | 0.9 |
| Phe-Phe count | 0.7 | |
| Val count | 0.7 | |
| Non-reduced absorption at 280 nm | 0.7 | |
| Val-Ala count | 0.9 | |
| Gly-Ser count | 0.8 | |
| Gly-Leu count | 1.0 | |
| Met-Ala count | 0.7 | |
| Ile-Asn count | 0.8 | |
| Lys-Ala count | 0.9 | |
| Ala-Val count | 0.8 | |
| Lys-Ala frequency | 0.7 | |
| Val-Gly count | 0.7 | |
| Ile-Gly count | 0.8 | |
| Ser-Glu count | 0.8 | |
| Cys-Pro count | 0.7 | |
| Gly-Thr count | 0.7 | |
| Met-Gly count | 0.7 | |
| Glu-Gly count | 0.7 | |
| Gly-Ala count | 0.8 | |
| Weighting by relief | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 0.8 |
| Val frequency | 0.7 | |
| Val count | 0.7 | |
| Hydrophilic residues frequency | 0.7 | |
| Ser count | 0.7 | |
| Phe count | 0.7 | |
| Tyr count | 0.9 | |
| Hydrophilic residues count | 1.0 | |
| Length | 0.7 | |
| Pro-Gly count | 0.7 | |
| Glu-Tyr count | 0.7 | |
| Phe-Arg count | 0.7 | |
| Tyr-Leu count | 0.7 | |
| Met-Tyr count | 0.8 | |
| Cys-Tyr count | 1.0 | |
| Leu-Tyr count | 0.7 | |
| Trp-Cys count | 0.8 | |
| Weighting by uncertainty | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 0.9 |
| Hydrogen frequency | 0.7 | |
| Val frequency | 0.7 | |
| Phe-Phe count | 0.7 | |
| count of Val | 0.9 | |
| Carbon frequency | 1.0 | |
| Hydrophilic residues frequency | 0.8 | |
| Non-reduced Absorption at 280 nm | 0.7 | |
| Val-Ala count | 0.7 | |
| Ser count | 0.7 | |
| Phe count | 0.8 | |
| Tyr count | 0.7 | |
| Hydrophilic residues count | 0.9 | |
| Length | 1.0 | |
| Trp frequency | 0.7 | |
| Ser frequency | 0.8 | |
| Gly frequency | 0.7 | |
| Hydrophobic residues frequency | 0.7 | |
| Cys count | 0.7 | |
| Phe-Asn count | 0.7 | |
| Glu-Tyr count | 0.7 | |
| Phe-Arg count | 0.7 | |
| Tyr-Glu count | 0.7 | |
| Arg-Thr count | 0.7 | |
| Sulphur count | 0.7 | |
| Leu count | 0.7 | |
| Hydrophilic other count | 0.7 | |
| Hydrophobic residues count | 0.8 | |
| Gln-Ser count | 0.7 | |
| Val-Gln count | 0.7 | |
| Met-Tyr count | 0.7 | |
| Cys-Tyr count | 0.7 | |
| Leu-Leu count | 0.7 | |
| Cys-Glu count | 0.7 | |
| Ile-Phe count | 0.7 | |
| Weighting by gini index | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 1.0 |
| Hydrogen frequency | 0.9 | |
| Val frequency | 1.0 | |
| Val count | 0.9 | |
| Carbon frequency | 1.0 | |
| Hydrophilic residues frequency | 0.8 | |
| Non-reduced Absorption at 280 nm | 0.9 | |
| Phe-Leu count | 0.9 | |
| Val-Ala count | 0.8 | |
| Ser count | 0.8 | |
| Arg count | 0.9 | |
| Phe count | 0.9 | |
| Tyr count | 1.0 | |
| Hydrophilic residues count | 0.9 | |
| Length | 0.9 | |
| Gly frequency | 0.8 | |
| Cys count | 0.8 | |
| Hydrophilic other count | 0.8 | |
| Hydrophobic residues count | 0.9 | |
| Phe-Met count | 0.8 | |
| Trp-Tyr count | 0.9 | |
| Cys-Tyr count | 0.9 | |
| Leu-Tyr count | 0.9 | |
| Ser-Ser count | 0.8 | |
| Ala frequency | 0.7 | |
| Phe-Phe count | 0.7 | |
| Ala-Ile count | 0.7 | |
| Ile frequency | 0.7 | |
| Ser frequency | 0.7 | |
| Phe-Asn count | 0.7 | |
| Pro-Gly count | 0.7 | |
| Gly-Ser count | 0.7 | |
| Gly-Leu count | 0.7 | |
| Tyr-Leu count | 0.7 | |
| Thr count | 0.7 | |
| Leu count | 0.7 | |
| Arg frequency | 0.7 | |
| Asp-Ala count | 0.7 | |
| Val-Gln count | 0.7 | |
| Ile-Ala count | 0.7 | |
| Tyr-Arg count | 0.7 | |
| Leu-Phe count | 0.7 | |
| Aliphatic index | 0.7 | |
| Cys-Asn count | 0.7 | |
| Ile-Tyr count | 0.7 | |
| Phe-Tyr count | 0.7 | |
| Asn-Leu count | 0.7 | |
| Weight scrutiny through Chi squared | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 1.0 |
| Hydrogen frequency | 0.7 | |
| Val frequency | 0.7 | |
| Val count | 0.8 | |
| Carbon frequency | 1.0 | |
| Hydrophilic residues frequency | 0.8 | |
| Non-reduced absorption at 280 nm | 0.7 | |
| Asp count | 0.7 | |
| Ser count | 0.7 | |
| Phe count | 0.8 | |
| Hydrophilic residues count | 0.9 | |
| Length | 0.9 | |
| Trp frequency | 0.7 | |
| Ser frequency | 0.9 | |
| Glu count | 0.7 | |
| Hydrophilic other count | 0.7 | |
| Hydrophobic residues count | 0.7 | |
| Leu-Leu count | 0.7 | |
| Ile-Phe count | 0.7 | |
| Weight Scrutiny through deviation | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 1.00 |
| Weight scrutiny through rule | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 1.0 |
| Val count | 0.7 | |
| Carbon frequency | 0.8 | |
| Non-reduced absorption at 280 nm | 0.9 | |
| Asp count | 0.7 | |
| Ser count | 0.7 | |
| Arg count | 0.7 | |
| Phe count | 0.8 | |
| Hydrophilic residues count | 0.9 | |
| Length | 0.9 | |
| Ser frequency | 0.8 | |
| Hydrophobic residues frequency | 0.7 | |
| Sulphur count | 0.7 | |
| Leu count | 0.7 | |
| Negatively Charged (D & E) count | 0.7 | |
| Hydrophilic other count | 0.8 | |
| Hydrophobic residues count | 1.0 | |
| Index of Aliphatic | 1.0 | |
| Point of Isoelectric | 1.0 | |
| Weight Scrutiny through Gain Ratio | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 1.0 |
| Hydrogen frequency | 0.9 | |
| Ala frequency | 0.7 | |
| Val frequency | 1.0 | |
| Val count | 0.9 | |
| Carbon frequency | 1.0 | |
| Hydrophilic residues frequency | 0.9 | |
| Non-reduced absorption at 280 nm | 0.9 | |
| Ala-Ile count | 0.7 | |
| Phe-Leu count | 0.9 | |
| Val-Ala count | 0.8 | |
| Asp count | 0.8 | |
| Ser count | 0.9 | |
| Arg count | 0.9 | |
| Phe count | 0.9 | |
| Tyr count | 1.0 | |
| Hydrophilic residues count | 0.9 | |
| Length | 0.9 | |
| Ile frequency | 0.8 | |
| Ser frequency | 0.7 | |
| Gly frequency | 0.8 | |
| Hydrophobic residues frequency | 0.7 | |
| Cys count | 0.9 | |
| Pro-Gly count | 0.7 | |
| Glu-Thr count | 0.7 | |
| Tyr-Leu count | 0.7 | |
| Tyr-Glu count | 0.7 | |
| Arg-Thr count | 0.7 | |
| Thr count | 0.7 | |
| Glu count | 0.8 | |
| Leu count | 0.7 | |
| Hydrophilic other count | 0.7 | |
| Hydrophobic residues count | 0.9 | |
| Ser-Trp count | 0.8 | |
| Phe-Met count | 0.9 | |
| Met-Ala count | 0.8 | |
| Val-Gln count | 0.8 | |
| Trp-Met count | 0.7 | |
| Ile-Ala count | 0.8 | |
| Trp-Tyr count | 0.9 | |
| Cys-Tyr count | 0.9 | |
| Leu-Phe count | 0.7 | |
| Leu-Tyr count | 0.9 | |
| Aliphatic index | 0.7 | |
| Trp count | 0.7 | |
| Ser-Ser count | 0.9 | |
| Lys-Ala count | 0.8 | |
| Ser-Val frequency | 0.7 | |
| Lys-Ala frequency | 0.7 | |
| Met count | 0.7 | |
| Glu-Met count | 0.7 | |
| Weighting by info gain | Non-reduced coefficient pertaining to Cys extinction within 280 nm | 0.8 |
| Hydrogen frequency | 0.7 | |
| Ala frequency | 0.7 | |
| Val frequency | 1.0 | |
| Val count | 0.7 | |
| Carbon frequency | 0.8 | |
| Hydrophilic residues frequency | 0.7 | |
| Non-reduced absorption at 280 nm | 0.9 | |
| Phe-Leu count | 0.7 | |
| Val-Ala count | 0.8 | |
| Asp count | 0.8 | |
| Ser count | 0.7 | |
| Arg count | 0.8 | |
| Phe count | 0.8 | |
| Tyr count | 0.8 | |
| Hydrophilic residues count | 0.8 | |
| Length | 0.8 | |
| Gly frequency | 0.7 | |
| Hydrophobic residues frequency | 0.7 | |
| Cys count | 0.7 | |
| Pro-Gly count | 0.7 | |
| Glu-Thr count | 0.7 | |
| Thr count | 0.7 | |
| Leu count | 0.7 | |
| Hydrophobic residues count | 0.7 | |
| Ser-Trp count | 0.7 | |
| Phe-Met count | 0.7 | |
| Met-Ala count | 0.7 | |
| Trp-Tyr count | 0.7 | |
| Cys-Tyr count | 0.7 | |
| Leu-Phe count | 0.7 | |
| Leu-Tyr count | 0.7 | |
| Ser-Ser count | 0.7 |
Weight scrutiny through SVM
Twenty features were shown to outweigh 0.70 by SVM algorithm: As indicated in Table 1, the features were non-reduced absorption within 280 nm, the Val, Lys-Ala frequencies, the Phe-Phe, Val, Val-Ala, Gly-Ser, Gly-Leu, Met-Ala, Ile-Asn, Lys-Ala, Ala-Val, Val-Gly, Ile-Gly, Ser-Glu, Cys-Pro, Gly-Thr, Met-Gly, Glu-Gly, Gly-Ala accounts.
Weight scrutiny through Relief
The application of this model to the dataset, as shown in Table 1, unveiled that seventeen features outweighed 0.70: non-reduced coefficient pertaining to Cys extinction within 280 nm, length, the Val frequencies, hydrophilic residues and the counts of Val, Ser, Phe, Tyr, hydrophilic residues, Pro-Gly, Glu-Tyr, Phe-Arg, Tyr-Leu, Met-Tyr, Cys-Tyr, Leu-Tyr, Trp-Cys.
Weight scrutiny through uncertainty
When this model was applied, thirty five attributes outweighed 0.70 using the algorithm of uncertainty: non-reduced coefficient pertaining to Cys extinction within 280Â nm, non-reduced absorption at 280 nm, length, the frequencies of hydrogen, Val, carbon, hydrophilic residues, Trp, Ser, Gly, hydrophobic residues and the counts of Phe-Phe, Val, Val-Ala, Ser, Phe, Tyr, hydrophilic residues, Cys, Phe-Asn, Glu-Tyr, Phe-Arg, Tyr-Glu, Arg-Thr, sulphur, Leu, hydrophilic Other, hydrophobic residues, Gln-Ser, Val-Gln, Met-Tyr, Cys-Tyr, Leu-Leu, Cys-Glu, Ile-Phe (Table 1).
Weight scrutiny through Gini index
Forty seven features outweighed 0.80 using the algorithm of uncertainty: non-reduced coefficient pertaining to Cys extinction within 280 nm, non-reduced absorption within 280 nm, length, aliphatic index, the frequency of hydrogen, Val, carbon, hydrophilic residues, Gly, Ala, Ile, Ser, Arg and the counts of Val, Phe-Leu, Val-Ala, Ser, Arg, Phe, Tyr, hydrophilic residues, Cys, hydrophilic Other, hydrophobic residues, Phe-Met, Trp-Tyr, Cys-Tyr, Leu-Tyr, Ser-Ser, Phe-Phe, Ala-Ile, Phe-Asn, Pro-Gly, Gly-Ser, Gly-Leu, Tyr-Leu, Thr, Leu, Asp-Ala, Val-Gln, Ile-Ala, Tyr-Arg, Leu-Phe, Cys-Asn, Ile-Tyr, Phe-Tyr, Asn-Leu (Table 1).
Weight scrutiny through Chi squared
Nineteen features outweighed 0.70 using the algorithm of Chi Squared: non-reduced coefficient pertaining to Cys extinction within 280 nm, non-reduced absorption within 280 nm, length, The frequency of hydrogen, Val, carbon, hydrophilic residues, Trp, Ser, The counts of Val, Asp, Ser, Phe, hydrophilic residues, Glu, hydrophilic Other, hydrophobic residues, Leu-Leu, Ile-Phe (Table 1).
Weight scrutiny through deviation
The algorithm of deviation brought into light the sole feature which outweighed 0.70 where non-reduced coefficient pertaining to Cys extinction within 280Â nm showed the weight equivalence of 1.00 as is indicated in Table 1.
Weight scrutiny through rule
Seven attributes outweighed 0.70 using the algorithm of Rule: non-reduced coefficient pertaining to Cys within 280 nm, non-reduced absorption within 280 nm, length, aliphatic index and isoelectric point, the frequency of carbon, Ser, hydrophobic residues, the counts of Val, Asp, Ser, Arg, Phe, hydrophilic residues, sulphur, Leu, negatively charged (D & E), hydrophilic other, hydrophobic residues (Table 1).
Weight scrutiny through gain ratio
As the algorithm implemented in the dataset, 51 attributes outweighed or were on a par with 0.70: non-reduced coefficient pertaining to Cys extinction within 280 nm, non-reduced absorption within 280 nm, length, aliphatic index, the frequency of hydrogen, Ala, Val, carbon, hydrophilic residues, Ile, Ser, Gly, hydrophobic residues, Ser-Val, Lys-Ala, the counts of Val, Ala-Ile, Phe-Leu, Val-Ala, Asp, Ser, Arg, Phe, Tyr, hydrophilic residues, Cys, Pro-Gly, Glu-Thr, Tyr-Leu, Tyr-Glu, Arg-Thr, Thr, Glu, Leu, hydrophilic other, hydrophobic residues, Ser-Trp, Phe-Met, Met-Ala, Val-Gln, Trp-Met, Ile-Ala, Trp-Tyr, Cys-Tyr, Leu-Phe, Leu-Tyr, Trp, Ser-Ser, Lys-Ala, Met, Glu-Met (Table 1).
Weight scrutiny through Info gain
Thirty three features outweighed 0.70 using Info Gain: non-reduced coefficient pertaining to Cys extinction within 280 nm, non-reduced absorption within 280 nm, length, the hydrogen frequency, Val, Ala, carbon, hydrophilic residues, Gly, hydrophobic residues, the counts of Val, Phe-Leu, Val-Ala, Asp, Ser, Arg, Phe, Tyr, hydrophilic residues, Cys, Pro-Gly, Glu-Thr, Thr, Leu, hydrophobic residues, Ser-Trp, Phe-Met, Met-Ala, Trp-Tyr, Cys-Tyr, Leu-Phe, Leu-Tyr, Ser-Ser (Table 1).
Key structural protein attributes distinguishing the substrate specificity of ATPase pumps regarding the outcome of the algorithms used in attribute weighting
With regards to the thorough outcome of applying models for attribute weighting (Table 1), nineteen protein attributes were proclaimed as the key distinctive features in ATPase pumps structures including non-reduced coefficient pertaining to Cys extinction within 280 nm, non-reduced absorption within 280 nm, the frequencies of hydrogen, Ala, Val, carbon, hydrophilic residues, the counts of Val, Asn, Ser, Arg, Phe, Tyr, hydrophilic residues, Phe-Phe, Ala-Ile, Phe-Leu, Val-Ala and length.
Supervised clustering
Tree of decision
Sixteen various models of Decision Tree were employed to cover the 11 datasets. The entire model trees were characterized with leaves and roots; minimum and maximum performances were considered 48.5 % &93.5 %, respectively as shown in Table 2. Three Decision Tree models were the epitome ones which included Random Forest model with Gain ratio criterion run on Chi Squared dataset, Random Forest models with Gini index criterion run on Gini index dataset, Decision Tree model with Info Gain criterion run on Relief dataset, respectively.
Table 2.
Percentage of performance which indicate the suitability of sixteen Tree Induction models such as DT Accuracy, DT Gain Ratio, DT gini index, DT info gain, DT parallel Accuracy, DT parallel Gain Ratio, DT parallel Gini Index, DT parallel Info Gain, DT stump Accuracy, DT stump Gain Ratio, DT stump Gini Index, DT stump Info Gain, DT Random Forest Accuracy, DT Random Forest Gain Ratio, DT Random Forest Gini Index, DT Random Forest Info Gain and Naı¨ve bayes such as Naive bayes and Naive Bayes Kernel models which were run on eleven datasets including Chi Squared, Info Gain, Deviation, Gini Index, Info Gain Ratio, PCA, Relief, Rule, Uncertainty, FCdb, and SVM
| Tree induction models | Accuracy regarding tree of decision | Ratio gain regarding tree of decision | Gini index regarding tree of decision | Info gain regarding tree of decision | Tree of decision regarding parallel Accuracy | Tree of decision regarding parallel gain Ratio | Tree of decision regarding parallel gini Index | Tree of decision regarding parallel Info gain | Tree of decision regarding stump accuracy | Tree of decision regarding stump gain Ratio | Tree of decision regarding stump gini Index | Tree of decision regarding stump info gain | Tree of decision regarding the Accuracy of random forest | Tree of decision regarding the gain Ratio in random Forest | Tree of decision regarding random Forest gini Index | Tree of decision regarding the Info Gain in random Forest | Bayse Kernel | Naive Bayse |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Database | ||||||||||||||||||
| Chi squared | 75.50 % | 77.50 % | 75.50 % | 87.50 % | 73.50 % | 75.00 % | 73.00 % | 85.00 % | 54.50 % | 50.00 % | 52.50 % | 50.50 % | 91.00 % | 93.50 % | 89.00 % | 91.00 % | 90.50 % | 93.00 % |
| Info gain | 75.50 % | 77.50 % | 75.50 % | 85.50 % | 75.50 % | 77.50 % | 73.50 % | 85.50 % | 54.50 % | 50.00 % | 52.50 % | 50.50 % | 87.00 % | 88.50 % | 88.50 % | 89.00 % | 50.50 % | 88.00 % |
| Deviation | 81.00 % | 83.50 % | 79.00 % | 79.00 % | 81.00 % | 83.50 % | 79.00 % | 79.00 % | 59.50 % | 59.50 % | 59.50 % | 59.50 % | 84.50 % | 83.50 % | 86.00 % | 81.50 % | 81.00 % | 83.50 % |
| Gini index | 75.50 % | 77.50 % | 75.50 % | 85.50 % | 77.50 % | 77.50 % | 78.00 % | 83.50 % | 54.50 % | 50.00 % | 52.50 % | 50.50 % | 91.00 % | 87.00 % | 92.00 % | 90.50 % | 28.57 % | 81.50 % |
| Info gain ratio | 75.50 % | 77.50 % | 75.50 % | 85.50 % | 75.50 % | 77.50 % | 75.50 % | 85.50 % | 54.50 % | 50.00 % | 52.50 % | 50.50 % | 89.00 % | 90.50 % | 88.50 % | 86.00 % | 28.57 % | 83.50 % |
| PCA | 81.00 % | 83.50 % | 79.00 % | 79.00 % | 81.00 % | 83.50 % | 79.00 % | 79.00 % | 59.50 % | 59.50 % | 59.50 % | 59.50 % | 84.50 % | 83.50 % | 86.00 % | 81.50 % | 81.00 % | 83.50 % |
| Relief | 80.00 % | 80.00 % | 85.00 % | 92.00 % | 82.00 % | 84.50 % | 78.00 % | 92.00 % | 54.50 % | 52.00 % | 52.50 % | 52.50 % | 89.00 % | 88.50 % | 84.00 % | 80.50 % | 25.81 % | 79.00 % |
| Rule | 75.50 % | 75.00 % | 70.50 % | 85.00 % | 75.50 % | 75.00 % | 72.50 % | 89.00 % | 54.50 % | 50.00 % | 54.50 % | 48.50 % | 87.00 % | 87.00 % | 81.50 % | 84.50 % | 93.00 % | 95.50 % |
| Uncertainty | 75.50 % | 77.50 % | 75.50 % | 87.50 % | 75.50 % | 77.50 % | 78.00 % | 87.50 % | 54.50 % | 50.00 % | 52.50 % | 50.50 % | 88.50 % | 86.50 % | 85.00 % | 91.00 % | 28.57 % | 86.00 % |
| FCdb | 75.50 % | 77.50 % | 75.50 % | 85.50 % | 73.00 % | 75.50 % | 78.00 % | 83.50 % | 54.50 % | 50.00 % | 52.50 % | 50.50 % | 69.00 % | 89.00 % | 89.00 % | 86.50 % | 23.53 % | 57.50 % |
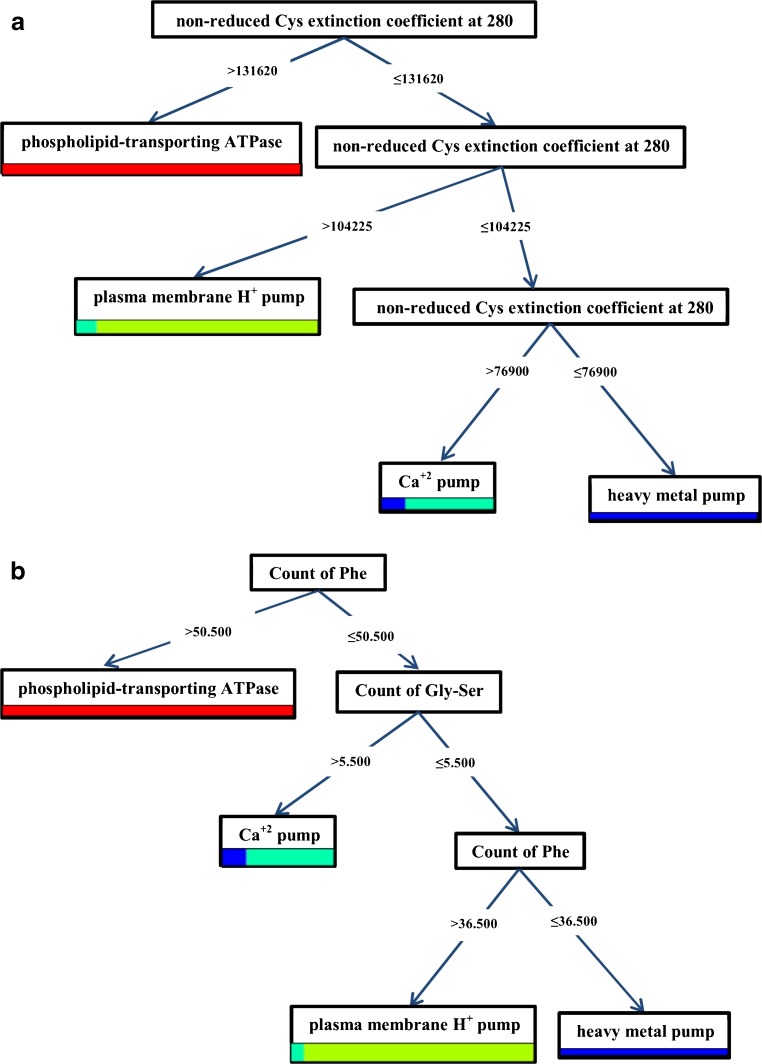
The simplest trees were generated by the Random Forest model with Gain Ratio criterion run on Chi Squared. Interestingly, non-reduced coefficient pertaining to Cys extinction within 280 nm was the only protein feature applied to generate the first tree. The proteins, where the feature value outweighed 131,620, were observed as the phospholipid-transporting ATPases; but when the value outweighed 104,225 and was lower than or on a par with 131,620, such proteins were categorized as plasma membrane H+ pumps. In the case this feature value outweighed 76,900 and was lower than or on a par with 104,225, the protein was assigned as the Ca+2 pumps; otherwise, it belonged to the heavy metal pump class. Performance was 93.5 % (Fig. 1a).
Fig. 1.
The simplest trees generated by the Random Forest model with Gain Ratio criterion run on Chi Squared dataset
Counts of Phe and Gly-Ser were the protein attributes which applied in the construction of the second tree. When the Phe count was higher than 50.5, the proteins were categorized as phospholipid transporting ATPases; but when the value was lower than or on a par with 50.5 along with the Gly-Ser count outweighing 5.5, such proteins were recognized as Ca+2 pumps. If the Gly-Ser count was lower than or on a par with 5.5, and the Phecount outweighed 36.5, these proteins were assigned to the plasma membrane H+ pump class; otherwise, they belonged to the heavy metal pump group. The performance was 92.0 % (Fig. 1b).
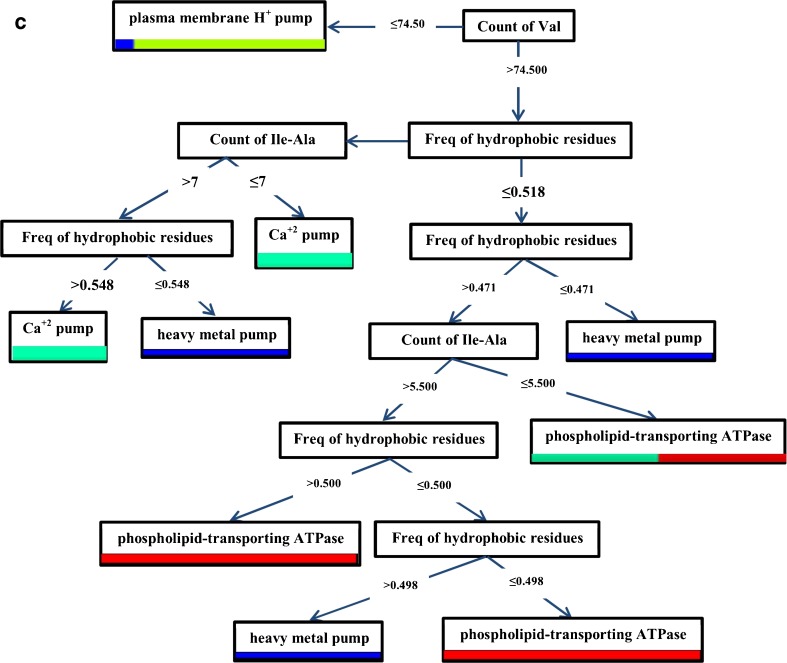
In the third tree, count of Val, the frequency of hydrophobic residues and Ile-Ala count were the protein attributes applied to build this tree. Performance was 92.0 % (Fig.1c).
The best Decision Tree model in distinguishing P-type ATPase substrate specificity was Random Forest model with Gain Ratio criterion based on the Chi Squared dataset. It predicted that Q9LVV1 and O22180 are Ca+2 pump and heavy metal pump with 70 % and 60 % prediction confidence, respectively (Table 3).
Table 3.
Results of application of the predictive models to predict the substrate specificity of P-type ATPase like proteins (Q9LVV1 and O22180)
| Description | Confidence (heavy metal pump) | Confidence (Ca+2 pump) | Confidence (plasma membrane H+ pump) | Confidence (phospholipid-transporting ATPase) | Prediction (substrate specificity) | Predictive models |
|---|---|---|---|---|---|---|
| Q9LVV1 | 0.10 | 0.70 | 0.10 | 0.10 | Ca+2 pump | Tree |
| O22180 | 0.60 | 0.10 | 0.20 | 0.10 | heavy metal pump | Tree |
| Q9LVV1 | 0.00 | 1.00 | 0.00 | 0.00 | Ca+2 pump | Bayes SVM |
| O22180 | 1.00 | 0.00 | 0.00 | 0.00 | heavy metal pump | Bayes SVM |
| Q9LVV1 | 0.00 | 1.00 | 0.00 | 0.00 | Ca+2 pump | Bayes RULE |
| O22180 | 1.00 | 0.00 | 0.00 | 0.00 | heavy metal pump | Bayes RULE |
NaiveBayes
Regarding the usage of Rule and SVM dataset, the performances of Naive Bayes and kernel based Naive Bayes models were 95.5 % and 93 %, respectively. Naive Bayes based on the Rule and SVM datasets predicted that Q9LVV1 and O22180 are Ca+2 pump and heavy metal pump, respectively, with 100 % prediction confidence (Table 3).
Discussion
A putative or possible function is sometimes assigned when the function of a protein is unknown. These assignments are often incorrect due to the simple bioinformatics analyses including sequence and structural comparisons using programs such as BLAST (Altschul et al. 1997; Cameron et al. 2004) and Dali (Holm et al. 2006; Holm and Rosenstrom 2010), respectively. With the growing challenges of protein function prediction, the development of reliable computational methods is vital for assigning accurate function to proteins with confidence (Caitlyn et al. 2015).
Accordingly, a computational method to discriminate between P-type ATPase substrate specificity would be immensely helpful in engineering these pumps and predicting the substrate specificity of any new ATPases pumps. This study was undertaken for specifying the most significant amino acid features in order to classify and predict the substrate specificity of ATPase pumps. Different bioinformatics algorithms were applied to study 896 amino acid attributes of ATPase pumps.
Data cleansing algorithms were applied to the original dataset to remove correlated, useless or redundant attributes and consequently, access to a smaller and more manageable set of attributes as well as improve the efficiency of process (Ebrahimi and Ebrahimie 2010; Ebrahimi et al. 2009). After the cleansing algorithms were applied to the original dataset, approximately 43 % of the features were discarded, implying that nearly half of the variables are statistically useless and redundant.
Weighting algorithms provide useful information about those attributes which are most significant for prediction or classification. In this investigation, 70 % of the weighting algorithms selected features such as non-reduced coefficient pertaining to Cys extinction, non-reduced absorption within 280 nm, length, the hydrogen, Val, carbon, Ala, hydrophilic residues frequencies, and the counts of Phe-Phe, Val, Ala-Ile, Phe-Leu, Val-Ala, Asp, Ser, Arg, Phe, Tyr, hydrophilic residues as the most important protein attributes in classification of heavy metal pump, Ca2+ pump, plasma membrane H+ pump and phospholipid-transporting ATPase group of ATPase pumps.
The role of dipeptides, such as Phe-Phe, Phe-Leu and Val-Ala and Gly-Ser, in the substrate specificity of pumps has been shown in this survey. As far as the records are concerned, however, considerable discussions regarding the dipeptides importance in substrate specificity of ATPase pumps have been doomed to limited quantities. A number of researchers have reported the essential role of dipeptide composition in halostability (Ebrahimie et al. 2011), thermo stability (Ebrahimi et al. 2011) and α-linolenic acid content (Zinati et al. 2014).
Therefore, the substrate specificity of ATPase pumps can be distinguished from their amino acid and dipeptide compositions. It is therefore likely that variations in these key distinguishing features are associated with changes in substrate specificity of ATPase pump.
Decision Trees are powerful classification algorithms that have become a popular tool for many researchers. Here, for the first time, a variety of tree induction models were used for classification of heavy metal pumps, Ca2+pumps, plasma membrane H+ pumps as well as phospholipid-transporting ATPase; it was also predicted the substrate specificity of uncharacterized P-type ATPase.
The results indicated that the performances of these Decision Trees varied from 48.5 % to 93.5 %, implying that the capability of various Decision Tree in induction models are different to classify different pumps based on amino acid attributes.
The percentages of performance in Tree induction models were used for comparing different models and determining the most efficient and precise model. The results showed that the best performance obtained when Random Forest model with Gain ratio criterion run on Chi Squared dataset. Correspondingly, in this model, non-reduced coefficients pertaining to Cys extinction within 280 nm was considered the only protein feature which used in construction of the simplest tree. Cysteine plays a vital role in protein structural stability due to its side chain; it contains a reactive sulfhydryl group that can oxidize to make a covalent bond with another Cystein (formation of a disulfide bond) (Leichert and Jakob 2006). Moreover, non-reduced coefficients pertaining to Cys extinction within 280 nm was a significant attribute assigned by 10 weighting algorithms. Therefore, those attributes, obtained by feature selection, are the best choice to predict substrate specificity of P-type ATPase.
The most substantial result revealed from this study could be observed in the substrate specificity prediction of the two P-type ATPase like proteins based on its amino acid composition. Naive Bayes, as the best predictive machine learning algorithm in this survey, was able to predict two P-type ATPase like proteins of unknown substrate specificity. To our awareness, this is the first study for the application of the predictive models to predict the substrate specificity of P-type ATPase like proteins (Q9LVV1 and O22180) with a confidence rate up to 100 %. It proved the efficiency of the predictive models in predicting P-type ATPase substrate specificity. Furthermore, the features information of the tertiary and quaternary protein structure are not essential for prediction of substrate specificity. On the basis of predictive models results, Q9LVV1 and O22180 are thought to act as Ca+2 pump and heavy metal pump, respectively. Additional experimental methods are also needed to validate these putative substrate specificities.
These mentioned approaches have been taken to specify important structural attributes, prediction and classification of protein thermo-stability (Ebrahimi et al. 2009), P glycoprotein pump (Hammann et al. 2009) halo-stability (Ebrahimie et al. 2011), olive cultivars (Beiki et al. 2012), α-linolenic acid content (Zinati et al. 2014) as well as genotype discrimination (Nasiri et al. 2015).
As a conclusion, it should be noted that determination of pump substrate specificity through common laboratory procedures is highly demanding of time and labor as compared to this rapid bioinformatics approaches. Attribute weighting algorithms were able to apply cleansing process to a notable numbers of features, enhance the modeling accuracy rate and thus predict the substrate specificity of ATPase pumps. The findings of this study suggest that supervised algorithms (Decision Tree and Naive Bayes) can be used as efficient methods for classification and prediction of the substrate specificity of new ATPase pumps with the maximum possible prediction confidence.
Additionally, the findings substantiated that amino acid structural composition could be optimally exerted for the P-type ATPase substrate specificity precise determination. Furthermore, these models suggest which amino acids or dipeptides are of critical importance for translocation in different ATPases and gain insight to engineer according to the important attributes in this survey.
Acknowledgments
We would like to greatly thank Department of Agroecology of Agriculture and Natural Resources of Darab for supporting this research.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi E, Alemzadeh A, Ebrahimi M, Ebrahimie E, Dadkhodaei N, Ebrahimi M. Amino acid features of P1B-ATPase heavy metal transporters enabling small numbers of organisms to cope with heavy metal pollution. Bioinform Biol Insights. 2011;5:59–82. doi: 10.4137/BBI.S6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/PL00006286. [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiki AH, Saboor S, Ebrahimi M. A new avenue for classification and prediction of olive cultivars using supervised and unsupervised algorithms. PLoS One. 2012;7:44164. doi: 10.1371/journal.pone.0044164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caitlyn L, Mills PJ, Beuning MJ. Biochemical functional predictions for protein structures of unknown or uncertain function. Comput Struct Biotechnol J. 2015;13:182–191. doi: 10.1016/j.csbj.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M, Williams HE, Cannane A. Improved gapped alignment in BLAST. IEEE/ACM Trans Comput Biol Bioinform. 2004;1:116–129. doi: 10.1109/TCBB.2004.32. [DOI] [PubMed] [Google Scholar]
- Chi X, Hou J. An iterative approach of protein function prediction. BMC Bioinformatics. 2011;12:437. doi: 10.1186/1471-2105-12-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SR, Khoury A, Ferry DK, Hampton RY. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J Cell Biol. 2000;148:915–924. doi: 10.1083/jcb.148.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dipolo R, Beaug© L (2005) Sodium/calcium exchanger: influence of metabolic regulation on ion carrier interactions. Physiol Rev 86:155–203 [DOI] [PubMed]
- Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero-one loss. Mach Learn. 1997;29:103–130. doi: 10.1023/A:1007413511361. [DOI] [Google Scholar]
- Ebrahimi M, Ebrahimie E. Sequence-based prediction of enzyme thermo-stability through bioinformatics algorithms. Curr Bioinforma. 2010;5:195–203. doi: 10.2174/157489310792006693. [DOI] [Google Scholar]
- Ebrahimi M, Ebrahimie E, Ebrahimi M. Searching for patterns of thermo-stability in proteins and defining the main features contributing to enzyme thermo stability through screening, clustering, and decision tree algorithms. EXCLI J. 2009;8:218–233. [Google Scholar]
- Ebrahimi M, Ebrahimie E, Shamabadi N. Are there any differences between features of proteins expressed in malignant and benign breast cancers? J Res Med Sci. 2010;15:299–309. [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi M, Lakizadeh A, Agha-Golzadeh P, Ebrahimie E. Prediction of thermostability from amino acid attributes by combination of clustering with attribute weighting: a new vista in engineering enzymes. PLoS One. 2011;6:e23146. doi: 10.1371/journal.pone.0023146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi M, Agha-golzadeh P, Shamabadi N, Tahmasebi A, Alsharifi M, Adelson DL, Hemmatzadeh F, Ebrahimie E. Understanding the underlying mechanism of HA-subtyping in the level of physic-chemical characteristics of protein. PLoS One. 2014;9:e96984. doi: 10.1371/journal.pone.0096984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimie E, Ebrahimi M, Sarvestani NR. Protein attributes contribute to halo-stability, bioinformatics approach. Saline Systems. 2011;7:1. doi: 10.1186/1746-1448-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardini PF, Helmer-Citterich M. Structure-based function prediction: approaches and applications. Brief Funct Genomic Proteomic. 2008;7:291–302. doi: 10.1093/bfgp/eln030. [DOI] [PubMed] [Google Scholar]
- Gomes E, Jakobsen MK, Axelsen KB, Geisler M, Palmgren MG. Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell. 2000;12:2441–2454. doi: 10.1105/tpc.12.12.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammann F, Gutmann H, Jecklin U, Maunz A, Helma C, Drewe J. Development of decision tree models for substrates, inhibitors, and inducer of P-glycoprotein. Curr Drug Metab. 2009;10:339–346. doi: 10.2174/138920009788499021. [DOI] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Wilton C, Plewczynski D. Using Dali for structural comparison of proteins. Curr Protoc Bioinformatics. 2006;S14:5.5.1–5.5.24. doi: 10.1002/0471250953.bi0505s14. [DOI] [PubMed] [Google Scholar]
- Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8:995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- Leichert LI, Jakob U (2006) Global methods to monitor the thio--disul-fide state of proteins in vivo. Antioxid Redox Signal l8:763–772 [DOI] [PubMed]
- Loewenstein Y, et al. Protein function annotation by homology-based inference. Genome Biol. 2009;10:207. doi: 10.1186/gb-2009-10-2-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattle D, Sitse O, Autzen HE, Meloni G, Gourdon P, Nissen P. On allosteric modulation of p-type Cu +-ATPases. J Mol Biol. 2013;425:2299–2308. doi: 10.1016/j.jmb.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Møller JV, Juul B, Le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Nasiri J, Naghavi MR, Kayvanjoo AH, Nasiri M, Ebrahimi M. Precision assessment of some supervised and unsupervised algorithms for genotype discrimination in the genus Pisum using SSR molecular data. J Theor Biol. 2015;368:122–132. doi: 10.1016/j.jtbi.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Harper JF. Pumping with plant P-type ATPases. J Exp Bot. 1999;50:883–893. doi: 10.1093/jxb/50.Special_Issue.883. [DOI] [Google Scholar]
- Skolnick J, Brylinski M. FINDSITE: a combined evolution/structure-based approach to protein function prediction. Brief Bioinform. 2009;10:378–391. doi: 10.1093/bib/bbp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator RD, Walsh P. An overview of in silico protein function prediction. Arch Microbiol. 2010;192:151–155. doi: 10.1007/s00203-010-0549-9. [DOI] [PubMed] [Google Scholar]
- Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Wilkins AD, Bachman BJ, Erdin S, Lichtarge O. The use of evolutionary patterns in protein annotation. Curr Opin Struct Biol. 2012;22:316–325. doi: 10.1016/j.sbi.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Mills RF. P1B-ATPases – an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Zinati Z, Zamansani F, KayvanJoo A, Ebrahimi M, Ebrahimi M, Ebrahimie E, Mohammadi Dehcheshmeh M. New layers in understanding and predicting α-linolenic acid content in plants using amino acid characteristics of omega-3 fatty acid desaturase. Comput Biol Med. 2014;54:14–23. doi: 10.1016/j.compbiomed.2014.08.019. [DOI] [PubMed] [Google Scholar]