Abstract
An endochitinase gene ‘ech42’ from the biocontrol fungus ‘Trichoderma virens’ was introduced to Brassica juncea (L). Czern and Coss via Agrobaterium tumefaciens mediated genetic transformation method. Integration and expression of the ‘ech42’ gene in transgenic lines were confirmed by PCR, RT-PCR and Southern hybridization. Transgenic lines (T1) showed expected 3:1 Mendelian segregation ratio when segregation analysis for inheritance of transgene ‘hpt’ was carried out. Fluorimetric analysis of transgenic lines (T0 and T1) showed 7 fold higher endochitinase activity than the non-transformed plant. Fluorimetric zymogram showed presence of endochitinase (42 kDa) in crude protein extract of transgenic lines. In detached leaf bioassay with fungi Alternaria brassicae and Alternaria brassicicola, transgenic lines (T0 and T1) showed delayed onset of lesions as well as 30–73 % reduction in infected leaf area compared to non-transformed plant.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-016-0340-8) contains supplementary material, which is available to authorized users.
Keywords: Trichoderma virens, ‘ech42’ gene, Brassica juncea, Agrobacterium -mediated transformation, Endochitinase activity, Fungal resistance
Introduction
Brassica juncea L. Czern and Coss (Indian mustard) is one of the major oil seed crops cultivated in many countries, including India, where mustard is cultivated in around 6 million hectares of land (Yadava and Singh 1999). This crop is highly susceptible to various fungal pathogens and insects, whereas bacterial and viral diseases have little effect on its yield (Abdel-Farida et al. 2009). Alternaria blight caused by A. brassicae (Berk.) Sacc. (Black leaf spot disease) and A. brassicicola (Grey leaf spot disease) is one of the major diseases of this crop causing up to 47 % yield losses (Singh et al. 1999; Meena et al. 2002). With the increasing demand for brassica oil especially in developing countries, it is necessary to minimize the yield losses caused by biotic stresses. Genetic engineering is an alternative for developing disease resistant plants where the resistance source is not available or there are sexual compatibilities between the source and the cultivar. (Grover and Gowthaman 2003; Chhikara et al. 2012).
Genes from the mycoparasitic Trichoderma spp. have been used to impart tolerance to biotic and abiotic stresses in many crops (Nicolas et al. 2014). Among the Trichoderma genes transferred to plants, an endochitinase of 42 kDa has been very popular due to its broad spectrum of activity against many fungal pathogens. Endochitinases are known to degrade fungal chitin by hydrolyzing the glycosidic bond between chitin monomers. In addition, the endochitinases also release chitin oligomers which act as elicitor molecules triggering broad spectrum plant defense (Emani et al. 2003). There are very few reports on development of transgenic Brassica species for resistance to fungal pathogens. For example, B. napus transformed with two genes sporamin and chitinase PjChi-1 (derived from sweet potato and Paecliomyces javanicus, respectively) showed enhanced resistance against both Plutella xylostella and Sclerotinia sclerotiorum (Liu et al. 2011). Wu et al. (2009) introduced a plant defensin gene Ovd, cloned from Orychophragmus violaceus (L.) into B. napus, and the transgenic plants showed resistance to S. sclerotiorum. Transgenic B. napus expressing B. napus mitogen-activated protein kinase (BnMPK4) showed resistance to S. sclerotiorum (Wang et al. 2009). Chitinase and glucanase gene from tomato have been introduced into B. juncea and the transgenic plants showed resistance to A. brassicae (Mondal et al. 2003, 2007). We have earlier cloned and transferred an endochitinase gene from T. virens to tobacco and tomato and the transgenic lines exhibited enhanced tolerance to some fungal pathogens (Shah et al. 2010).
In the present study, our objectives were to express an endochitinase ‘ech42’ gene from Trichoderma virens in transgenic B. juncea lines and to assess tolerance of transgenic lines against two fungal pathogens, A. brassicicola and A. brassicae, causing Alternaria blight.
Materials and methods
Plant material
Seeds of Indian mustard (Brassica juncea L. Czern and Coss) cv. Pusa Jaikisan from Indian Agriculture Research Institute (IARI), New Delhi were used for the transformation experiments.
Bacterial strain and plasmid
Agrobacterium tumefaciens strain LBA4404 harboring binary vector pCAMBIA1301:ech42 containing the chitinase coding region (GenBank Acc.No. EU035808) (Shah et al. 2010) was used for transformation of B. juncea.
Transformation of B. juncea
Seeds of B. juncea were germinated aseptically in vitro on MS (Murashige and Skoog 1962) medium. The cotyledonary petioles from 5 day old seedlings were used as explants and pre-cultured for 2 day on SRM i.e. shoot regeneration medium [MS medium supplemented with 1 mg l−1 BA (6 -Benzyl amino purine) and 1 mg l−1 Kn (Kinetin)]. Transformation of B. juncea with Agrobacterium tumefaciens strain LBA4404 harboring binary vector pCAMBIA1301:ech42 was carried out as described earlier by Kamble et al. (2013). Putative transformed plants (T0) were self-pollinated and their seeds were germinated on MS medium containing 2.5 mg l−1 hygromycin. The surviving seedlings (T1) resistant to hygromycin was counted and data analyzed using the Chi-square test to determine the number of functional ‘hpt’ gene loci in the B. juncea genome.
Molecular analysis of putative transgenic plants
Polymerase chain reaction (PCR)
Genomic DNA was isolated from 25 plants each from 25 randomly selected T0 lines obtained from independent transformation events and one non-transformed plant (control) according to Dellaporta et al. (1983). PCR was carried out according to Shah et al. (2010). PCR analysis was also carried out with 5 T1 plants each from 5 different T1 lines to confirm the stable integration of transgene.
Reverse transcription PCR (RT-PCR)
Transcription of ‘ech42’ gene in PCR positive T0 lines was confirmed by RT- PCR. Total RNA from 10 transformed lines as well as one non-transformed (control) plant was isolated using Tri-reagent. One μg of DNase-treated and purified RNA was taken for cDNA synthesis using affinity script multi temperature cDNA synthesis kit (Stratagene, USA). Two μl of this reaction mix was used for PCR amplification of ‘ech42’and a housekeeping gene ‘actin’ (Shah et al. 2010).
Southern blot hybridization
Southern blot hybridization was carried out to confirm the integration and copy number of ‘ech42’ gene in transgenic B. juncea lines. Genomic DNA (50 μg) from 5 randomly selected PCR positive T0 lines were digested with HindIII (NEB) and size separated on 1 % agarose gel at 25 V for 16 h. PCR amplified product of ‘ech42’ gene (1.2 kb) from plasmid pCAMBIA1302:ech42 was used for probe preparation using DIG-DNA labeling kit (Roche Biochemicals, Germany). Pre-hybridization, hybridization, washing and detection were carried out using chemilluminescent detection system (Roche Biochemicals, Germany).
Endochitinase activity and fluorimetric zymogram
Endochitinase activity in transgenic plants (each from different T0 and T1 lines) was measured by fluorimetric assay and fluorimetric zymogram was performed for visualization of chitinase activity as described earlier by Shah et al. (2010). All the reagents used were from Sigma and assay was repeated at least twice with three replicates.
Detached leaf bioassay
Cultures of Alternaria brassicae (ITCC No. 5097) and A. brassicicola (ITCC No. 1707) were obtained from the Indian Type Culture Collection, Division of Plant Pathology, IARI, New Delhi. Bioassay for sensitivity of transgenic lines against the fungal pathogens were done as described earlier (Shah et al. 2010). Three independent experiments with three replicates were carried out and the data was analyzed using Analysis of Variance (ANOVA) and Pearson’s correlation using Microcal™ Origin pro 6.1.
Results
Transformation of B. juncea with ‘ech42’ gene
Prior to infection with Agrobacterium, cotyledonary petioles were pre-cultured on SRM for 2 days (Fig. 1a). Followed by infection with Agrobacterium, the explants were transferred to selection medium. Cotyledonary petioles showed initiation of shoot at the proximal end after 15–20 days of infection (Fig. 1b). The regenerated shoots were subcultured 3-4 times on selection medium i.e. SRM supplemented with 2.5 mg l−1 hygromycin (Fig. 1c). Several shoots were bleached at this stage. Hygromycin resistant shoots were transferred to rooting medium (Fig. 1d, e) and well rooted plants were hardened in paper cups (Fig. 1f). Total 8 independent transformation events were carried-out and 25 hygromycin resistant lines were obtained. Some plants showed reduction in leaf size and stunted growth compared to other transgenic plants, even though they produced seeds. Five plants, each from 5 different T0 lines showing high endochitinase activity were allowed to grow in growth chamber, selfed and seeds collected for raising T1 generation (Fig. 1g). The average transformation efficiency obtained ranged from 2.1–4.2 % (Table 1).
Fig. 1.
Development of transgenic B. juncea with ‘ech42’ gene (a) Cotyledon with petiole cultured on shoot regeneration medium (b) Explants showing regeneration on selection medium after infection with Agrobacteria. (c) Multiple shoots growing on selection medium (d) Elongated shoot transferred to rooting medium (e) Shoot with well developed roots (f) Transgenic plants transferred to soil for hardening (g) Hardened plants in pots showing flowering and pod setting
Table 1.
Transformation efficiency (%) of B. juncea cv. Pusa jaikisan following co-cultivation with A. tumefaciens LBA4404 with ‘ech42’ gene
| Batch No. | No. of explants co-cultivated | No. of PCR positive plants | Transformation (%) |
|---|---|---|---|
| 1 | 96 | 2 | 2.1 |
| 2 | 96 | 4 | 4.2 |
| 3 | 96 | 4 | 4.2 |
| 4 | 100 | 3 | 3.0 |
| 5 | 86 | 2 | 2.3 |
| 6 | 96 | 3 | 3.1 |
| 7 | 96 | 2 | 2.1 |
| 8 | 150 | 3 | 3.0 |
Molecular analysis of transgenic plants
PCR and RT-PCR
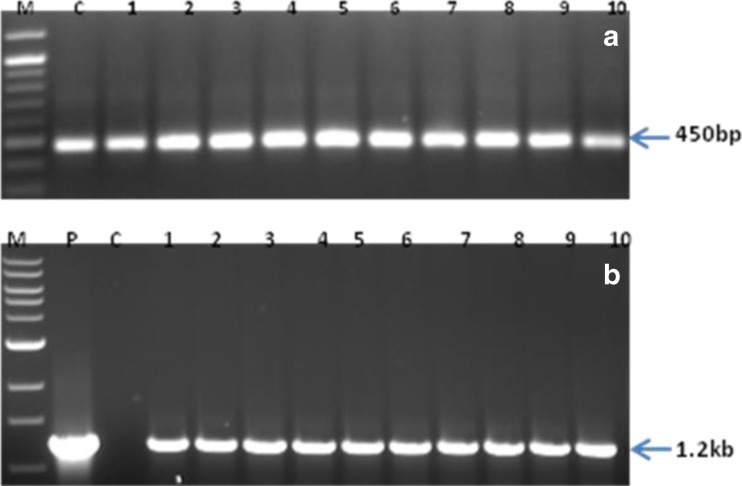
Out of 25 putative transgenic T0 lines, 20 lines showed presence of expected size PCR products confirming integrations of ‘ech42’ (1.2 kb) (Fig. S1). These lines were further tested for endochitinase activity. PCR analysis of five T1 lines also showed presence of ‘ech42’gene (Fig. S2). RT-PCR results confirmed expression of ‘actin’ as well as ‘ech42’ gene in 10 randomly selected PCR positive T0 lines (Fig. 2a, b).
Fig. 2.
RT-PCR analysis of transgenic B. juncea lines (T0) (a) actin as ‘house- keeping’ gene (b) an ‘ech42’ gene. Lane M- 100 bp ladder (a), 1 kb ladder (b), C-control (non-transgenic) plant, P- positive control- pCAMBIA1301-ech42 plasmid, Lane 1–10 -Transgenic lines
Southern blot hybridization
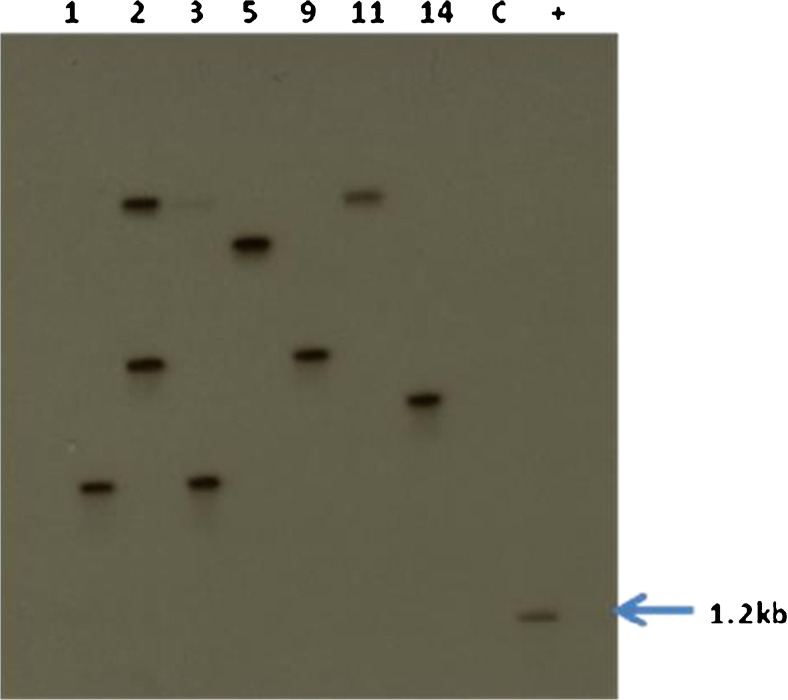
Seven randomly selected T0 lines of B. juncea were subjected to Southern hybridization. Each transgenic line had a single band except line 2, where two copies of the gene were seen. Non-transformed (control) plants did not show any hybridization signal to the probe. All the transgenic lines showed a band of more than 5.3 kb as expected (Fig. 3).
Fig. 3.
Southern blot hybridization of transgenic B. juncea lines (T0) - 1, 2, 3, 5, 9, 11 and 1, C- control (non-transformed) plant, (+) - positive control - HindIII digested PCR product of ‘ech42 gene
Segregation analysis of T0 progeny
The healthy seedlings (T1) surviving on selection medium containing 2.5 mg l−1 of hygromycin obtained by germinating seeds of T0 lines were classified as hygromycin resistant and pale seedlings were classified as hygromycin sensitive. Segregation ratio of 3:1 in T1 generation was seen at P ≤ 0.05, n = 1 (Table 2).
Table 2.
Inheritance of hygromycin resistance (hpt) in T1 generation of transgenic B. juncea lines
| Line | HygR seedlings | HygS seedlings | χ2 test (3:1) |
|---|---|---|---|
| Non-transformed | 0 | 20 | - |
| 1 | 15 | 6 | 0.142 |
| 3 | 17 | 5 | 0.06 |
| 5 | 14 | 6 | 0.26 |
| 11 | 32 | 13 | 0.363 |
| 14 | 37 | 13 | 0.027 |
At p ≤ 0.05 and n = 1, χ2 value is 3.84, hence χ2value from all lines were found to be not significant (HygR: Hygromycin resistant; HygS: Hygromycin sensitive)
Endochitinase activity in T0 and T1 plants and fluorimetric zymogram
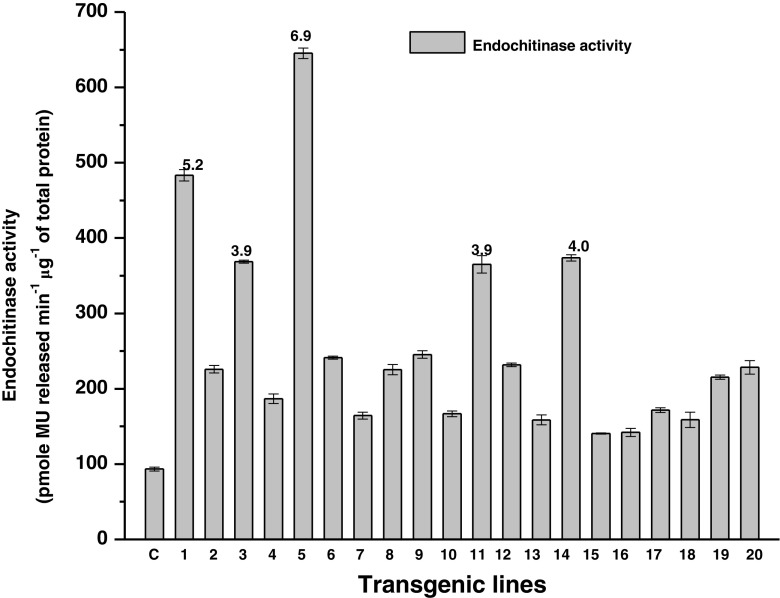
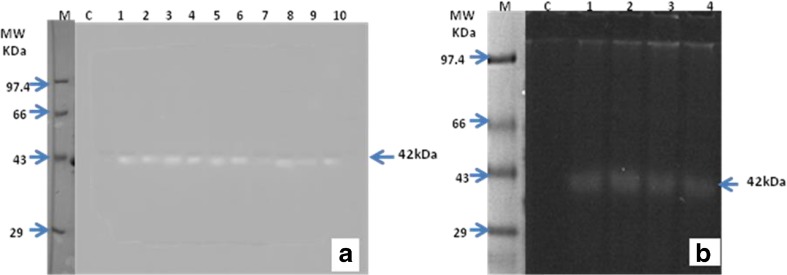
Transgenic lines (T0 and T1) showed enhanced endochitinase level compared to non-transformed B. juncea plants. T0 lines showed 1.5–6.9 fold higher endochitinase activity (Fig. 4) while T1 lines showed 4.2–6.7 fold higher endochitinase activity compared with non-transformed plant (Table 3). Total protein of 10 T0 and 4 T1B. juncea lines were separated on SDS-PAGE gel. After renaturation, the enzyme was probed with the fluorogenic substrate, [4MU-β-(glcNAc)3]. Fluorescence at position of 42 kDa was detected under UV transilluminator (Fig. 5 a, b).
Fig. 4.
Fluorimetric assay for transgenic B. juncea lines (T0). Error bars indicates ± standard error of means. The number above the error bar represents fold increase in the endochitinase activity over the control plant. The values were statistically significant at P ≤ 0.05 over the control (non-transgenic) plant
Table 3.
Fluorimetric assay for transgenic B. juncea T1 lines for measuring the level of endochitinase activity
| lines | Endochitinase activity (pmol MU released min−1 μg−1 of total protein ± SE) | Fold increase over non-transformed T1 plants |
|---|---|---|
| Control | 85.5 ± 1.2 | 1 |
| 1 | 470.3 ± 1.4 | 5.5 |
| 3 | 369.3 ± 1.6 | 4.3 |
| 5 | 573.3 ± 2.6 | 6.7 |
| 11 | 374.0 ± 2.5 | 4.3 |
| 14 | 363.3 ± 3.9 | 4.2 |
Fig. 5.
Fluorimetric zymogram for detection of 42 kDa endochitinase in transgenic B. juncea lines. M- protein marker; C- control plant; Lanes 1–10- T0 lines (a), Lanes 1– 4 T1 lines (b). Marker lane was cut and overlapped with the zymogram
Detached leaf bioassay
Five T0 and T1 transgenic lines (line 1, 3, 5, 11 and 14) showing high endochitinase activity were challenged with two fungi, A. brassicola and A. brassicae. The transgenic lines showed delay in lesion formation compared to control plant (Fig. 6). In control plants, lesion was formed within 2 days after incubation with the fungus while transgenic plants showed lesion formation after 4–5 days. When challenged with A. brassicicola, T0 lines showed 34–73.7 % reduction in lesion area and there was a strong negative correlation between endochitinase activity and % lesion area in T0 lines (R = − 0.90) (Fig. 7a). T1 lines also showed 31–65 % reduction in lesion area as compared to control plants, and a negative correlation between endochitinase activity and % lesion area (R = − 0.91) (Fig. 7c). When challenged with A. brassicae, T0 lines showed 30.9–62 % reduction in lesion area with negative correlation between endochitinase activity and lesion size (R = − 0.96) (Fig. 7b) while T1 lines showed 30.5–73 % reduction in lesion area with a negative correlation (R = − 0.91) between endochitinase activity and lesion size (Fig. 7d).
Fig. 6.
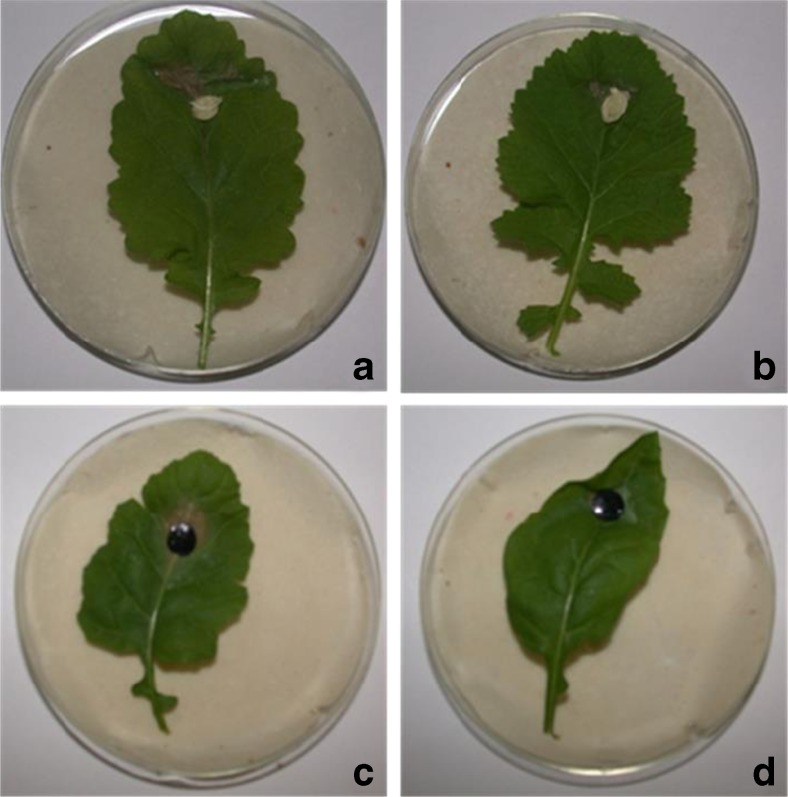
A representative photograph of transgenic B. juncea lines showing resistance to two fungi, Alternaria brassicicola (a- nontransgenic plant; b-transgenic plant) and Alternaria brassicae (c- nontransgenic plant; d- transgenic plant) in detached leaf bioassay
Fig. 7.
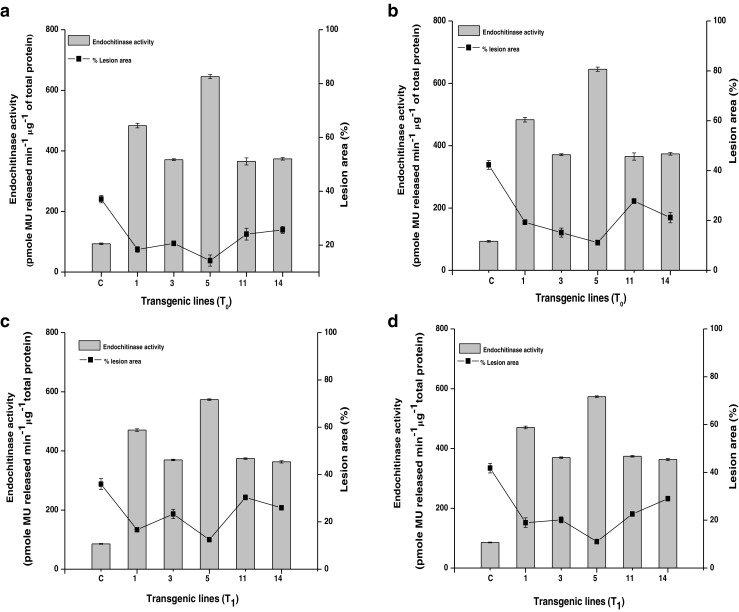
Endochitinase activity and average lesion area in control and transgenic B. juncea lines. (a and b: T0 lines challenged with A. brassicicola and A. brassicae respectively); (c and d: T1 lines challenged with A. brassicicola and A. brassicae respectively). Error bars indicates ± standard error of means. The values for the all the transgenic lines are significantly different from the control plant at p < 0.01
Discussion
In the present study, we have developed transgenic B. juncea lines expressing an endochitinase ‘ech42’ gene from T. virens. In a detached leaf bioassay, these lines showed enhanced tolerance against two fungi, A. brassicicola and A. brassicae, major pathogens of Brassica species. Integration, expression and copy number of transgenes in the transgenic lines (T0 and T1) were confirmed by PCR, RT-PCR and Southern blot analysis respectively. In Southern blot, all but one T0 lines showed the presence of a single copy of the transgene. This confirms stable integration of ‘ech42’gene in the plant genome. Though line 2 showed 2 copies of ‘ech42’ gene, the plant did not show any significant difference in endochitinase activity as well as fungal resistance compared to other transgenic lines. An enhancement in endochitinase activity to the tune showed 1.5 to 6.9 fold in T0 lines and 4.2 to 6.7 fold in T1 lines. The variation in the endochitinase activity in different transgenic lines could be due to the varied level of activity of 35S promoter which is known to be influenced by several regulatory controls (Benefy and Chua 1990; Emani et al. 2003). In the present study, some transgenic lines of B. juncea showed different leaf morphology (reduction in leaf size) and stunted growth which is known in apple expressing ‘ech42’ gene from Trichoderma atroviride (Bolar et al. 2000).
In detached leaf bioassay, transgenic B. juncea lines (T0 and T1) showed moderate reduction in disease severity, even though the endochitinase activity level in the plants was higher compared to non-transgenic plants. Similar results have been reported by Mora and Earle (2001) in transgenic broccoli plants with ‘ech42’ gene from T. harzianum when challenged with A. brassicicola. Mondal et al. (2003) reported over-expression of ‘glucanase’ gene in transgenic B. juncea lines showing a 10–15 days delay in onset of fungal infection. Overexpression of ‘ech42’ gene from T. virens in transgenic tobacco and tomato plants also showed strong negative correlation between endochitinase activity and percentage leaf area infected with two fungi, B. cinerea and A. alternata respectively (Shah et al. 2010). In the present study, leaves of transgenic B. juncea lines showed 4–5 day delay in the onset of fungal infection while leaves of control plant showed lesions within 2 days.
Whether or not plants transformed with chitinase genes are protected against fungal pathogens appears to depend on multiple factors, including the source of the transgene, the crop transformed, the pathogen tested, and the rigor of the statistical analysis applied (Mora and Earle 2001). In the present study, the range of endochitinase activity in T1 generation provided similar levels of protection as in T0B. juncea lines against A. brassicae and A. brassicicola confirming stable expression of ‘ech42’ gene in subsequent generations and the transgenic B. juncea lines were found to be equally tolerant to both A. brassicae and A. brassicicola. Considering the importance of B. juncea as an important oil seed crop in the Indian subcontinent and its yield losses due to fungal diseases, an endochitinase gene ‘ech42’ from T. virens could be a potential candidate gene for development of disease resistant B. juncea plants in combination with other disease resistant genes.
Electronic Supplementary Material
(DOC 313 kb)
Acknowledgments
The authors thank Dr. Sudhir Singh and Dr. Archana Joshi - Saha for critically reading this manuscript and valuable suggestions.
Contributor Information
Suchita Kamble, Phone: +91-22-25592638, Email: snkamble@barc.gov.in.
Prasun K. Mukherjee, Phone: +91-22-25590367, Email: prasunm@barc.gov.in
Susan Eapen, Phone: +91-22-25592638, Email: eapenhome@yahoo.com.
References
- Abdel-Farida IB, Jahangira M, van den Hondelc CAMJJ, Kim HK, Choi YH, Verpoorte R. Fungal infection-induced metabolites in Brassica rapa. Plant Sci. 2009;176(5):608–615. doi: 10.1016/j.plantsci.2009.01.017. [DOI] [Google Scholar]
- Benefy PN, Chua NH. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Sci. 1990;250:959–966. doi: 10.1126/science.250.4983.959. [DOI] [PubMed] [Google Scholar]
- Bolar JP, Norelli JL, Wong KW, Hayes CK, Harman GE, Aldwinckle HS. Expression of endochitinase from Trichoderma harzianum in transgenic apple increases resistance to apple scab and reduces vigor. Pytopathol. 2000;90(1):72–77. doi: 10.1094/PHYTO.2000.90.1.72. [DOI] [PubMed] [Google Scholar]
- Chhikara S, Chaudhury D, Dhankher OP, Jaiwal PK. Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae. Plant Cell Tiss Organ Cult. 2012;108:83–89. doi: 10.1007/s11240-011-0015-7. [DOI] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Emani C, Garcia JM, Lopata-Finch E, Pozo MJ, Uribe P, Kim DJ, Sunilkumar S, Cook DR, Kenerley CM, Rathore KS. Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol J. 2003;1:321–336. doi: 10.1046/j.1467-7652.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- Grover A, Gowthaman R. Strategies for development of fungus-resistant transgenic plants. Curr Sci. 2003;84(3):330–340. [Google Scholar]
- Kamble S, Hadapad AB, Eapen S. Evaluation of transgenic lines of Indian mustard (Brassica juncea L. Czern and Coss) expressing synthetic cry1Ac gene for resistance to Plutella xylostella. Plant Cell Tiss Organ Cult. 2013;115:321–328. doi: 10.1007/s11240-013-0364-5. [DOI] [Google Scholar]
- Liu H, Guo X, Naeem MS, Liu D, Xu L, Zhang W, Tang G, Zhou W. Transgenic Brassica napus L. lines carrying a two gene construct demonstrate enhanced resistance against Plutella xylostella and Sclerotinia sclerotiorum. Plant Cell Tissue Organ Cult. 2011;106(1):143–151. doi: 10.1007/s11240-010-9902-6. [DOI] [Google Scholar]
- Meena PD, Chattopadhyay C, Singh F, Singh B, Gupta A. Yield loss in Indian mustard due to white rust and effect of some cultural practices on alternaria blight and white rust severity. Brassica. 2002;4:18–24. [Google Scholar]
- Mondal KK, Chatterjee SC, Viswakarma N, Bhattacharya RC, Grover A (2003) Chitinase mediated inhibitory activity of brassica transgenic on growth of Alternaria brassicae. Curr Microbiol 47(3):171–173 [DOI] [PubMed]
- Mondal KK, Bhattacharya RC, Koundal KR, Chatterjee SC. Transgenic Indian mustard (Brassica juncea) expressing tomato glucanase leads to arrested growth of Alternaria brassicae. Plant Cell Rep. 2007;26(2):247–252. doi: 10.1007/s00299-006-0241-3. [DOI] [PubMed] [Google Scholar]
- Mora AA, Earle ED. Resistance to Alternaria brassicicola in transgenic broccoli expressing a Trichoderma harzianum endochitinase gene. Mol Breed. 2001;8(1):1–9. doi: 10.1023/A:1011913100783. [DOI] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nicolas C, Hermosa R, Rubio B, Mukherjee P, Monte E. Trichoderma genes in plants for stress tolerance – status and prospects. Plant Sci. 2014;228:71–78. doi: 10.1016/j.plantsci.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Shah MR, Mukherjee PK, Eapen S. Expression of a fungal endochitinase gene in transgenic tomato and tobacco results in enhanced tolerance to fungal pathogens. Physiol Mol Biol Plants. 2010;16(1):39–51. doi: 10.1007/s12298-010-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh DN, Singh NK, Srivastava S. Biochemical and morphological characters in relation to alternaria blight resistance in rapeseed - mustard. Ann Agri Res. 1999;20:472–477. [Google Scholar]
- Wang Z, Mao H, Dong C, Ji R, Cai L, Fu H, Liu S. Overexpression of Brassica napus MPK4 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Mol Plant-Microbe Interact. 2009;22(3):235–244. doi: 10.1094/MPMI-22-3-0235. [DOI] [PubMed] [Google Scholar]
- Wu J, Wu L, Liu Z, Quin L, Wang M, Zhou L, Yang Y, Li X. A plant defensin gene from Orychophragmus violaceus can improve Brassica napus resistance to Sclerotinia sclerotiorum. Afr J of Biotechnol. 2009;8(22):6101–6109. [Google Scholar]
- Yadava JS, Singh NB (1999) Strategies to enhance yield potential of rapeseed mustard in India. 10th International Rapeseed Conference, (Eds.): Wratten N, Salisbury P, Canberra, Australia
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 313 kb)