Summary
The continuously growing rodent incisor is an emerging model for the study of renewal of mineralized tissues by adult stem cells. Although the Fgf, Shh, Wnt, and Bmp pathways have been studied in this organ previously, relatively little is known about the role of Notch signaling during incisor renewal. Notch signaling components are expressed in enamel-forming ameloblasts and the underlying stratum intermedium (SI), which suggested distinct roles in incisor renewal and enamel mineralization. Here, we injected adult mice with inhibitory antibodies against several components of the Notch pathway. This blockade led to defects in the interaction between ameloblasts and the SI cells, which ultimately affected enamel formation. Furthermore, Notch signaling inhibition led to the down-regulation of desmosome-specific proteins such as PERP and desmoplakin, consistent with the importance of desmosomes in the integrity of ameloblast-SI attachment and enamel formation. Together, our data demonstrate that Notch signaling is critical for proper enamel formation during incisor renewal, in part by regulating desmosome-specific components, and that the mouse incisor provides a model system to dissect Jag-Notch signaling mechanisms in the context of mineralized tissue renewal.
Keywords: Notch, Jag, tooth development, ameloblast, stratum intermedium, amelogenesis
Introduction
The mouse incisor provides a valuable model for the study of tooth development and renewal. This remarkable organ grows continuously throughout the animal’s life, and the highly calcified enamel is deposited exclusively on the labial (i.e. toward the lip) surface (Fig. 1A). Continuous growth is fueled by adult stem cells in the cervical loop (CL) region, and the well-characterized labial CL is comprised of the outer enamel epithelium, inner enamel epithelium, transit-amplifying region, and the stellate reticulum.(1–4) The cells from the labial CL differentiate into presecretory and secretory ameloblasts that ultimately form enamel (Fig. 1A”).
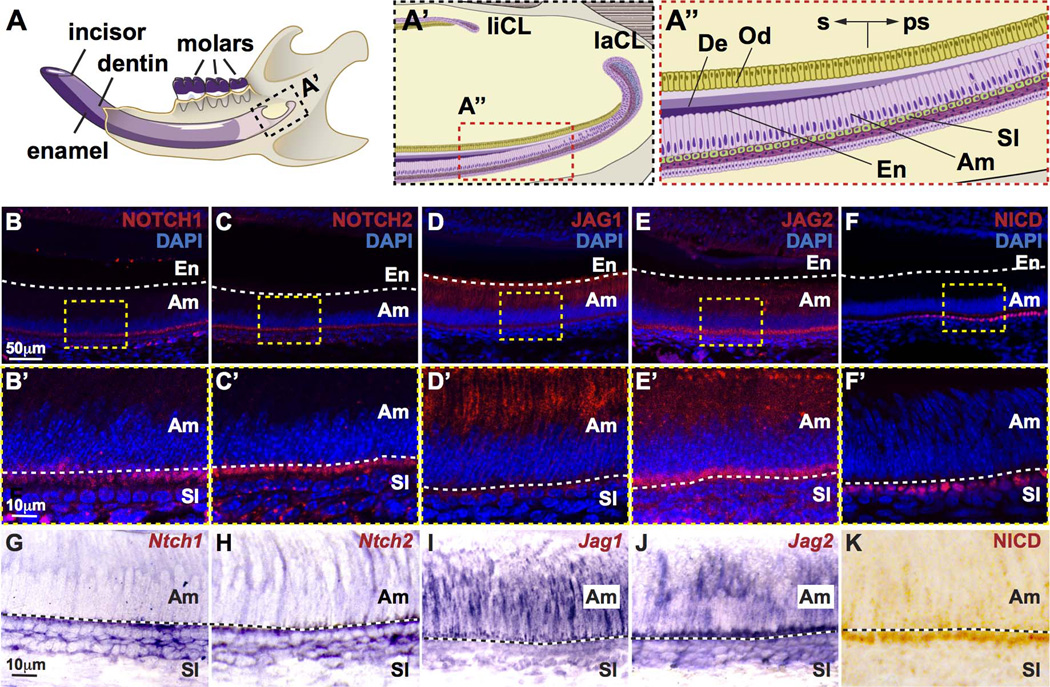
Figure 1. Expression of Notch signaling pathway components during incisor renewal.
(A) Illustration of the mouse hemi-mandible showing the incisor and molars, as well as the mineralized dentin and enamel comprising the incisor. (A’) Proximal region of the incisor showing the labial and lingual cervical loop (laCL and liCL, respectively). (A”) Magnified view of the ameloblast layer (Am), stratum intermedium (SI), enamel (En), and dentin (De). The location of prescretory (ps) and secretory (s) ameloblasts are shown. (B–F) Immunofluorescence staining for Notch receptors NOTCH1 and NOTCH2, Notch ligands JAG1 and JAG2, and the active Notch intracellular domain, NICD, are shown in ps and s ameloblasts. (B’–F’) Magnified views of B–F. (G–J) In situ hybridization was performed to detect RNA expression of Notch1, Notch2, Jag1, and Jag2. (K) Colorimetric visualization of NICD immunostaining.
The role of signaling pathways during tooth development has been well characterized, and many of the signals that regulate development, including the Fgf, Shh, Bmp and Wnt pathways, are also active during renewal.(5) However, the role of Notch signaling during tooth development and renewal has been relatively understudied compared to the other major pathways. Components of the Notch signaling pathway, which in mammals is comprised of four transmembrane Notch receptors (Notch1–4) and 5 canonical ligands (Jag1, Jag2, Dll1, Dll3, and Dll4), are expressed in teeth, and several studies have pointed to the importance of Notch signaling in tooth development and renewal.(6–8) First, the addition of JAG1 in culture to HAT-7 dental epithelial-like cells caused differentiation into cells resembling the SI, a layer of cells subjacent to ameloblasts, and this effect was neutralized with an anti-JAG1 antibody.(7) Second, Jag2-null mice at embryonic stages showed abnormal molar shapes and additional cusps, as well as inhibition of ameloblast differentiation and enamel matrix deposition.(8) Lastly, the inhibition of Notch signaling in vitro utilizing the broad gamma-secretase inhibitor, DAPT, resulted in apoptosis of dental epithelial stem cells in mouse incisors.(6) However, lethality in mice harboring mutations in Notch pathway components or lethality due to the use of broad gamma-secretase inhibitors have hampered studies into the role of Notch signaling during enamel formation.
The ameloblast-SI interface is integral to the formation of enamel, as evidenced by the inactivation of genes important in ameloblast-SI adhesion such as Pvrl1, Perp, and Cdh1.(9–11) The inactivation of Pvrl1 (also called nectin-1) led to hypomineralized incisor enamel, in part, because of increased separation between the ameloblasts and SI due to indirect effects on desmosome structure.(10) Furthermore, a compromise in desmosome structure was caused by inactivation of Perp, a gene encoding a desmosome-associated protein, and this resulted in ameloblast detachment from the SI, leading to developmental enamel defects.(11) These studies highlight the importance of the ameloblast-SI interface in enamel formation, but the signaling mechanisms involved are not currently known.
We set out to determine the in vivo role of Notch signaling during incisor renewal utilizing highly specific monoclonal antibodies generated against JAG1, JAG2, NOTCH1, and NOTCH2.(12) The use of these blocking antibodies allowed us to target distinct components of the Notch signaling pathway in adult mice. We found that inhibition of JAG1, JAG2, NOTCH1, and NOTCH2 alone and in combination led to defects in the ameloblast-SI interface and, ultimately, enamel formation. Moreover, the down-regulation of Perp and desmoplakin with Notch signaling inhibition demonstrated a role for Notch signaling in desmosome integrity. Thus, we have identified a link between Notch signaling and the regulation of desmosome-specific components that is essential for formation of proper enamel during incisor renewal. Furthermore, we demonstrate that the mouse incisor provides a model for analysis of Jag-Notch signaling mechanisms during mineralization.
Materials and Methods
Animals
All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) at UCSF and the mice were handled in accordance with the principles and procedure of the Guide for the Care and Use of Laboratory Animals under the approved protocol AN084146-02F. Wild-type CD-1 or B6 mice (Jackson laboratories) at 3 months of age were injected intra-peritoneally with 2 mg/kg antibodies against NOTCH1 (i.e., anti-N1),(12,13) NOTCH2 (i.e., anti-N2),(12,13) JAG1 (i.e., anti-J1)(13,14), and JAG2 (i.e., anti-J2)(14), alone and in combination (i.e., anti-N1N2, anti-J1J2), for 6 days every other day (all antibodies were provided by Genentech). The specificities of all inhibitory antibodies utilized have been tested and confirmed.(12–14) Lethality was observed at day 7 in anti-N1N2 or anti-J1J2 treated animals. Anti-gD isotype (i.e., the Fc region) or PBS was injected in control mice. We did not observe any differences between PBS- and anti-gD-injected mice therefore the phenotypes described in our manuscript are likely not due to ill-defined activities of the antibody backbone (i.e., the Fc region). Furthermore, distinct phenotypic differences were observed with the different antibodies, all of which possess the same Fc, demonstrating that the Fc region is not sufficient to account for the phenotypes. All control images presented in this manuscript are from PBS-injected specimens.
Histology, immunohistochemistry, and in situ hybridization
Mice were euthanized following standard IACUC protocol, the mandibles isolated, fixed overnight in 4% paraformaldehyde at 4°C, demineralized in 0.5 M EDTA for 2 weeks, dehydrated, embedded in paraffin wax, and serially sectioned at 7 µm. Histological sections were stained with haematoxylin and eosin (H&E). Immunohistochemistry was performed according to standard protocols. Antigen retrieval was performed by boiling the slides in Trilogy (Cell Marque) for 15 min and cooled at room temperature for 20 min after deparaffinization and rehydration. Primary antibodies used were as follows: anti-NOTCH1 (D1E11; 1:200; Cell Signaling), anti-NOTCH2 (1:200; Santa Cruz), anti-JAG1 (1:200; Abcam), anti-JAG2 (1:200; Santa Cruz), anti-NICD (Val1744; 1:200; Cell Signaling), anti-PERP (1:100; Abcam), anti-desmoplakin (DSP; 1:50; AbD Serotec), anti-amelogenin (AMEL; 1:200; Abcam), and anti-ameloblastin (AMBN; 1:200; Abcam). Goat anti-rabbit, goat anti-mouse, or donkey anti-goat AlexaFluor 488 or 555 secondary antibodies were used (1:250, Invitrogen). For colorimetric immunostaining (i.e., NICD), goat anti-mouse HRP conjugated secondary antibody (1:250; Abcam) was used in combination with VECTASTAIN Elite ABC Kit (Vector Labs). For in situ hybridization analyses, sections were hybridized to DIG-labeled RNA probes for in situ detection of RNA transcripts. Sections were treated with 10 µg/mL of proteinase K and acetylated prior to hybridization with probe. DIG-labeled RNA probes were synthesized from plasmids containing cDNA fragments of Notch1,(15) Notch2,(15) Jag1,(16) and Jag2.(16,17) Please refer to Supplemental Table 1 for details of RNA probes.
RNA isolation and qPCR
Total RNA was isolated using the RNeasy kit (Qiagen). DNA was removed in-column with RNase-free DNAse (Qiagen). All qPCR reactions were performed using the GoTaq qPCR Master Mix (Promega) in a Mastercycler Realplex (Eppendorf). PrimeTime Primers (Integrated DNA Technologies, Inc.) for the following genes were utilized for SYBR Green real-time qPCR: Perp (Acc. #:NM_022032; Cat. #:Mm.PT.58.45805759), Dsp (NM_023842; Mm.PT.58.33654353), Trp63 (NM_001127262; Mm.PT.58.13970687), Irf6 (NM_016851; Mm.PT.58.12061624), Amelx (NM_001081978; Mm.PT.58.5718729), Ambn (NM_009664; Mm.PT.58.8649071), Hes1 (NM_008235; Mm.PT.58.41697865), and Hey1 (NM_010423; Mm.PT.58.30455891). All qPCR primers were pre-tested and validated by the company and detected all variants of the genes of interest. qPCR conditions were as follows: 95°C, 2 minutes; 40 cycles at 95°C,15 seconds; 58°C,15 seconds; 68°C, 20 seconds; followed by a melting curve gradient. Expression levels of the genes of interest were normalized to levels of Rpl19 (IDT, Inc.; NM_001159483; Mm.PT.58.12385796).
Microscopy
Fluorescent and bright field images were taken using a Leica DM5000B with a Leica DFC500 camera. For confocal images, a Leica SP5 Upright Confocal was used.
Micro-computed tomography (µCT)
Mice were treated for 21 days with single antibodies (i.e., anti-N1, anti-N2, anti-J1, anti-J2) or 6 days with double antibodies (i.e., anti-N1N2, anti-J1J2). PBS-treated mice for 21 days were used as controls, as we observed no differences between 6- or 21-day PBS treatments. The left hemi-mandible was isolated, fixed in 4% formalin for 48h, and stored and imaged in 70% ethanol. µCT analysis was performed on a MicroXCT-200 (Xradia, Pleasanton, CA) through the Micro-CT Imaging Facility at UCSF. Each specimen was scanned at 75 KVp and 6W at 4× magnification, then reconstructed in 3-dimensions. Cross-section images of mouse hemi-mandibles at the level of the mandibular first molar distobuccal cusp (e.g., Fig. 3A’–G’) were analyzed using ImageJ software to quantify the intensity of enamel in the incisor and molar, because intensity is indicative of mineralization density.(18) Briefly, a line was drawn from the dentino-enamel junction to the outer margin of enamel in incisors and molars, and quantified using the Plot Profile option in the Analyze menu. Since the mouse molar, unlike the incisor, is not renewed in adult mice, we reasoned that adult molar enamel would not be affected by inhibitory antibodies, and therefore, molar enamel intensities could serve to normalize incisor enamel intensities to correct for any inter-specimen µCT and processing variation.
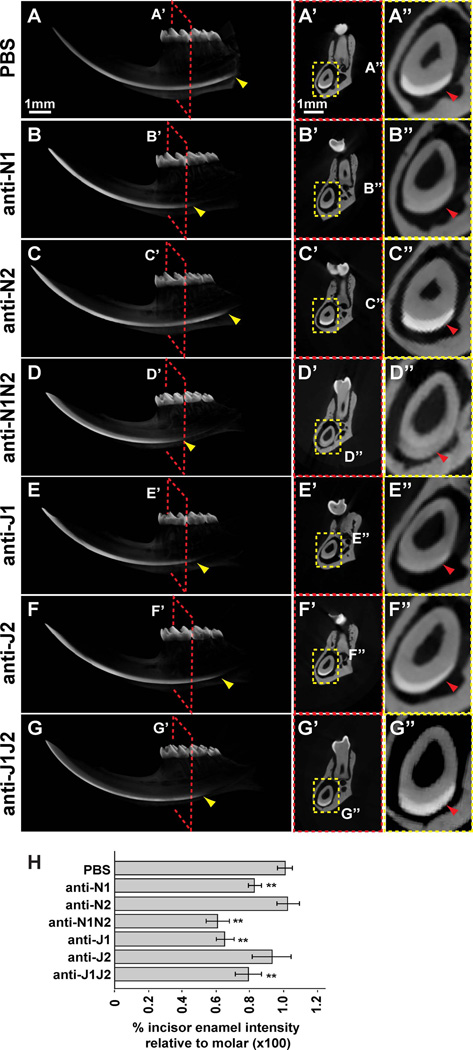
Figure 3. µCT analyses shows varying sites of initial incisor enamel mineralization and intensities.
(A–G) The left hemi-mandibles of PBS- and antibody-treated mice were analyzed by µCT. The initial mineralization sites of incisor enamel were altered with antibody treatments (yellow arrowheads), with the exception of mice treated with anti-N2 (C) or anti-J2 (F). (A’–G’) Cross-section of the hemi-mandibles underneath the distobuccal cusp of the mandibular first molar. (A”–G”) Magnified views of the incisor from A’–G’ showing mineralized enamel (red arrowheads). (H) The intensity of incisor and molar enamel was determined from A’–G’. Incisor enamel intensity was normalized to molar enamel intensity to correct for any inter-specimen processing variations. Incisors of mice treated with anti-N2 or anti-J2 did not show significant differences in normalized intensities compared to PBS-treated specimens. The remaining treatments showed a decrease in incisor enamel intensities. N1, NOTCH1; N2, NOTCH2; J1, JAG1; J2, JAG2. **, p-value<0.01.
Scanning Electron Microscopy (SEM)
Mouse hemi-mandibles were dissected free of soft and connective tissue, fixed in 4% PFA in PBS overnight, then dehydrated in a graded ethanol series and dried in a vacuum desiccator. Hemi-mandibles were then embedded in epoxy resin (resin 105 and hardener 205 at a ratio of 5:1 w/w, WestSystem, Bay City, MI, USA), ground to the desired thickness on a plate grinder (EXAKT 400CS, Norderstedt, Germany) using 800 grit silicon carbide paper and polished with 2000 and 4000 grit silicon carbide paper (Hermes Abrasives, Mississauga, ON, Canada). The exposed tissue was etched with 10% phosphoric acid for 30 seconds, rinsed with water and dried in a vacuum desiccator. Samples were mounted on SEM stubs with carbon tape, surfaces coated with 7nm gold using a sputter coating machine (Desk II, Denton Vacuum, Moorestown, NJ, USA), and imaged in a Philips SEM instrument (XL30 ESEM, Philips, Andover, MA, USA) operating at a beam energy of 20 keV in secondary electron or backscatter mode. Images were processed using Adobe Photoshop CS5.1 to adjust upper and lower limits of input levels in grayscale mode, and to apply auto balance and auto contrast settings.
Transmission Electron Microscopy (TEM)
Hemi-mandibles were dissected and immediately fixed for 1h at room temperature and overnight at 4°C in Karnovsky fixative (2% glutaraldehyde and 3% paraformaldehyde in 0.1M cacodylate buffer at pH 7.4). Samples were washed in cacodylate buffer and then demineralized for 4 days in PBS containing 12.5% EDTA and 0.8% glutaraldehyde at 4°C with rocking and daily solution change. Hemi-mandibles were post-fixed for 2 h in PBS containing 1% osmium tetraoxide, 0.5% potassium dichromate, and 0.5% potassium ferrocyanide. Samples were washed in PBS and stained in 2% uranyl acetate in water for 2 h in the dark on a rocking table. Following staining, samples were washed with water, dehydrated in an ethanol gradient followed by propylene oxide and embedded in EMbed 812 resin (Electron Microscopy Sciences). Prior to embedding, each of the mandibular incisors was cut perpendicular to the midline at approximately the level of the first molar. Sections (~80 nm thick) were cut using a Leica Ultracut ultramicrotome, transferred to formvar-coated Cu grids, and post-stained with Reynold's lead citrate and 2% uranyl acetate in 50% ethanol for 5 and 15min, respectively. Grids were examined on an FEI Tecnai 20 TEM operating at 100 kV and imaged on an AMT 16000-S CCD camera. Images are presented either in the native state or after contrast enhancement using Photoshop (Adobe).
Statistical analysis
All experiments were performed independently at least three times (i.e., N=3) in triplicates where possible, and when applicable, presented as an average ± standard deviation or standard error of the mean. Student t-test was used to determine p-values and P<0.05 was deemed to be significant.
Results
The Notch signaling pathway is active in ameloblasts and the SI region
We first confirmed the expression of principal members of the Notch signaling pathway, including Notch1, Notch2, Jag1, Jag2, and NICD (Notch intracellular domain), in adult mouse incisor ameloblasts and the SI region using immunofluorescence staining (Fig. 1B–F’) and in situ hybridization (Fig. 1G–J). As previously shown, NOTCH1 appeared to be localized largely in the SI with some staining on the basal surface of ameloblasts (Fig. 1B,B’,G), whereas JAG1 was localized to ameloblasts (Fig. 1D,D’).(7) NOTCH2 expression was very similar to NOTCH1 and was primarily present in the SI cells (Fig. 1C,C’,H). JAG2, like JAG1, was expressed in ameloblasts, although JAG2 appeared to also appeared to be expressed in SI cells (Fig. 1E,E’,J). NICD, the activated form of NOTCH1, was mainly localized to the ameloblast-SI interface (Fig. 1F,F’,K).
Inhibition of Notch signaling leads to defects in the ameloblast-SI interface
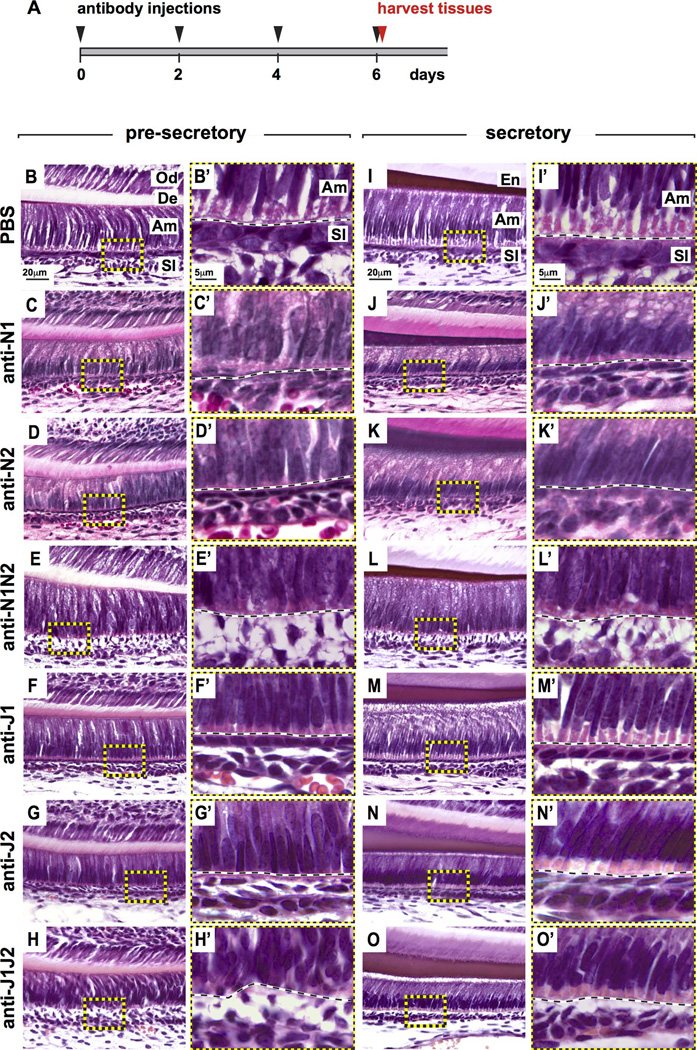
Next, we set out to test the role of Notch signaling during incisor renewal. For these studies, we injected mice with blocking monoclonal antibodies against NOTCH1, NOTCH2, JAG1, and JAG2,(12) either alone or in combination for 6–15 days (Fig. 2A). Inhibition of Notch signaling led to varying degrees of defects in the ameloblast-SI interface at the pre-secretory and secretory stages of amelogenesis (Fig. 2B–O’). Single antibody treatments resulted in flattening of the SI layer in the apical-basal direction (Fig. 2C–D’, F–G’, J–K’, M–N’). Combination treatment of NOTCH1 and NOTCH2 (i.e., anti-N1N2) or JAG1 and 2 (i.e., anti-J1J2) led to more severe defects in the ameloblast-SI interface, with an increase in separation between the ameloblasts and SI leading to partial or full detachment (Fig. 2E,E’,H,H’,L,L’,O,O’).
Figure 2. Defects to the ameloblast-SI interface with inhibition of different components of the Notch signaling pathway.
(A) Experimental design showing the injection of antibodies and harvesting of tissues. All tissues henceforth were collected on day 6 of treatment 3 hours after the final antibody injection. (B–O’) Hematoxylin and eosin staining of sagittal sections of the presecretory and secretory stages of the continuously growing mouse incisor with Notch inhibition. Differences in the ameloblast (Am)-stratum intermedium (SI) interface at the presecretory and secretory stages were observed with inhibition of Notch signaling compared to PBS-injected controls. (A’–N’) Higher magnification views of the boxed regions (A–N). Varying degrees of Am-SI detachment were observed with the different treatments. Od, odontoblasts; De, dentin; Am, ameloblasts; SI, stratum intermedium; SR, stellate reticulum; N1, NOTCH1; N2, NOTCH2; J1, JAG1; J2, JAG2.
We performed µCT analyses on hemi-mandibles collected from mice treated with single antibodies (i.e., anti-N1, anti-N2, anti-J1, anti-J2) for 21 days and combined antibodies (i.e., anti-N1N2, anti-J1J2) for 6 days (Fig. 3). We observed that the sites of initial incisor enamel mineralization differed with various antibody treatments, such that all antibody treatments with the exception of anti-N2- and anti-J2-treatment caused a delay in enamel mineralization (Fig. 3A–G). Second, we analyzed the intensities of incisor enamel directly underneath the distobuccal cusp of the mandibular first molar (Fig. 3). Since ameloblasts migrate at ~400 microns/day,(9,19,20) 80–100% of the length of the mouse incisor (~10 mm in length) would be renewed in a 21-day span (a conservative estimate of 400 microns × 21 = 8.4 mm), whereas a 6-day span would lead to the renewal of 2.4 mm of the incisor. We found clear differences in incisor enamel intensity between treatments with decreased incisor enamel intensities in mice treated with anti-N1N2 or anti-J1J2 (Fig. 3). Together with our histological analysis (Fig. 2), these data led us to focus on combined antibody treatments (i.e., anti-N1N2 and anti-J1J2) in order to analyze the most severe and reproducible defects.
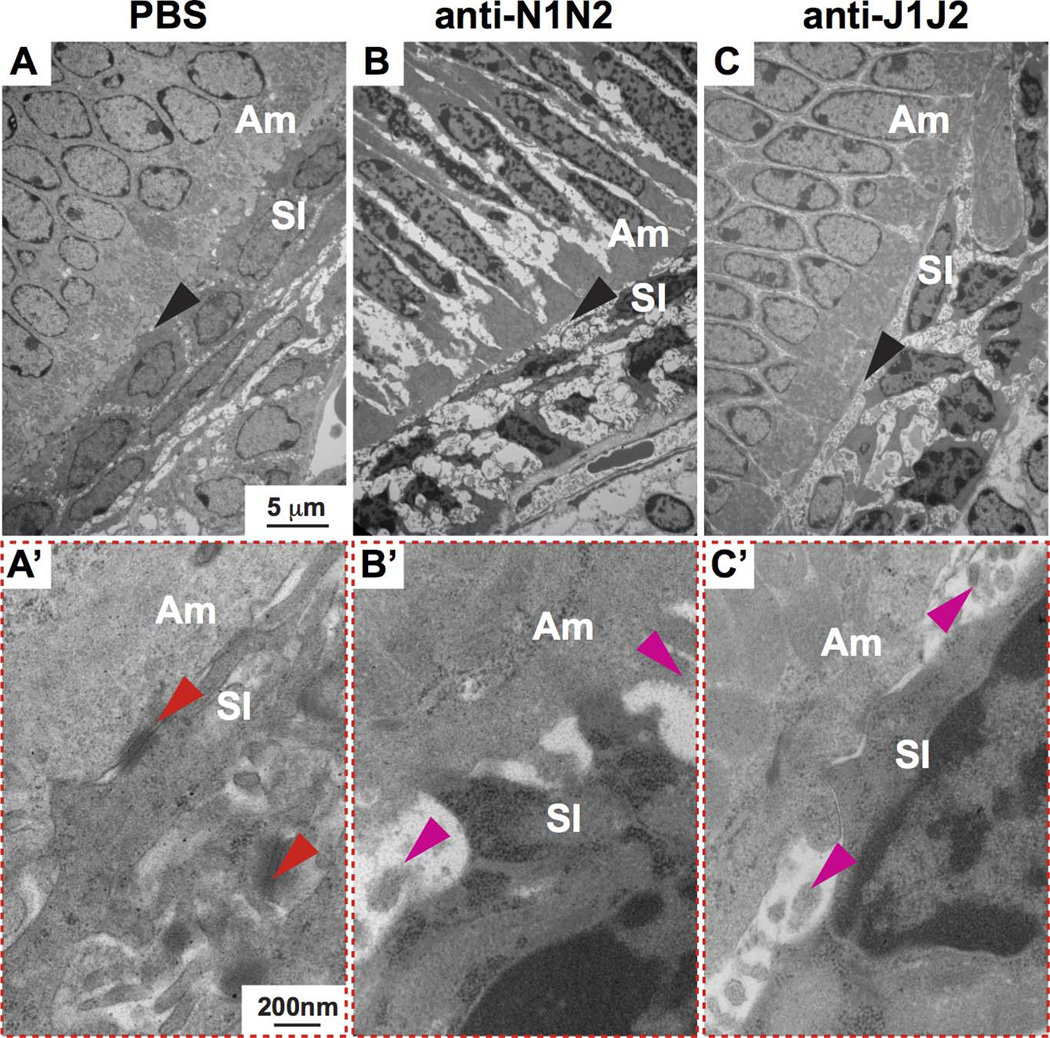
To further assess the morphology of the SI cells, we performed transmission electron microscopy (TEM; Fig. 4). Anti-N1N2 and anti-J1J2 treatment led to shrinkage and flattening of the SI cells directly subjacent to ameloblasts and to increased spacing at the ameloblast-SI interface as well as between SI cells. Notch signaling inhibition also affected ameloblast-ameloblast attachment, as evidenced by increased spacing between ameloblasts, and often the spaces were filled with what appeared to be either cellular debris or cell processes (Fig. 4B’,C’). Desmosomes were scattered throughout ameloblast-SI and SI-SI interfaces in control mice (Fig. 4A), but no desmosomes could be identified after anti-N1N2 and anti-J1J2 treatment (Fig. 4B,C).
Figure 4. Transmission electron microscopy (TEM) reveals defects in the ameloblast (Am)-SI interface and SI-SI attachment.
(A–C) TEM analysis of sagittal sections through the mouse incisor with Notch signaling inhibition. Black arrowheads indicate Am-SI interfaces. (A’–C’) Magnified views of the Am-SI interfaces. In PBS controls (A,A’), the characteristic zipper-like structures of normal desmosomes are evident (red arrowheads indicate normal desmosomes in the Am-SI interface and between SI cells. (B’,C’) Unlike controls, anti-N1N2 or anti-J1J2 treatment led to the absence of any identifiable desmosomes in the Am-SI interface or between SI cells. Furthermore, there was increased separation between the ameloblasts and SI, with the space often being filled with yet unknown cellular debris (pink arrowheads). SI, stratum intermedium; N1, NOTCH1; N2, NOTCH2; J1, JAG1; J2, JAG2.
Notch signaling inhibition leads to downregulation of desmosome-specific proteins in the ameloblast-SI interface
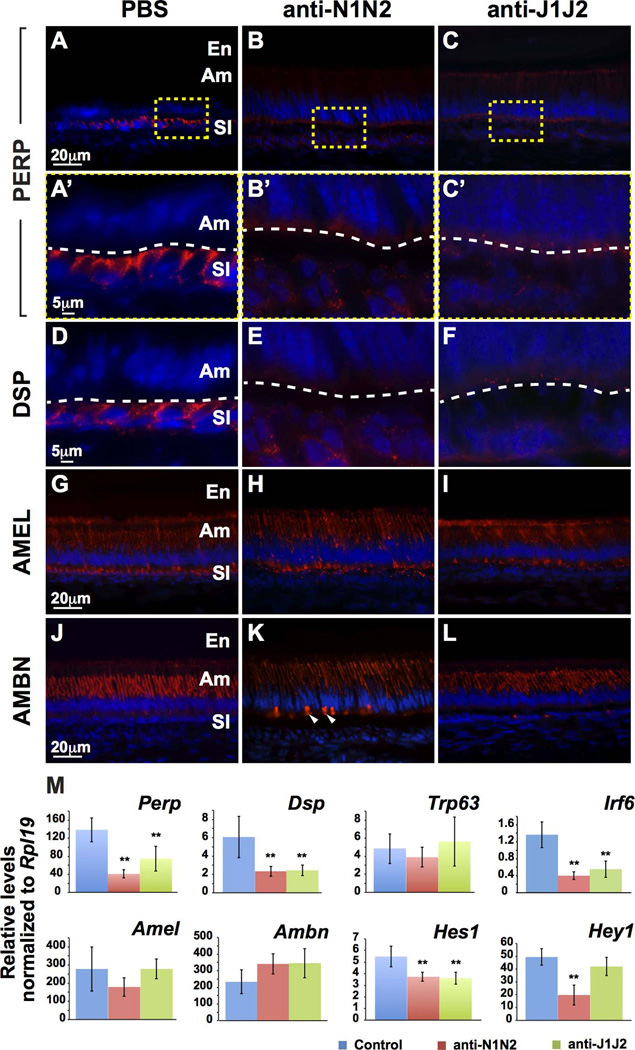
The involvement of desmosome-specific components in Notch signaling was further analyzed by immunofluorescence staining (Fig. 5). PERP, a desmosome-associated protein, and desmoplakin (DSP), a desmosome-specific protein, were downregulated in mice treated with anti-N1N2 or anti-J1J2 (Fig. 5A–F). The expression of amelogenin (X- and Y-linked; AMEL) and ameloblastin (AMBN), two enamel matrix-specific proteins, appeared similar to controls (Fig. 5G–L), although some intense AMBN staining on the basal end of ameloblasts was observed with anti-N1N2 treatment (Fig. 5K).
Figure 5. Expression of PERP and desmoplakin at the secretory stage is altered with Notch signaling inhibition.
(A–L) Immunofluorescence staining of PERP, DSP, AMEL, and AMBN. Expression of PERP (A–C’) and DSP (D–F) is downregulated with Notch signaling inhibition. Expression of AMEL (G–I) is unaffected with Notch signaling inhibition, whereas AMBN (J–L) is mislocalized in the basal ameloblast with anti-N1N2 but not anti-J1J2. (M) qPCR analysis confirmed the downregulation of Perp, Dsp, and Irf6. Expression of Amel and Ambn was not significantly altered with Notch signaling inhibition. Decreased expression of Hes1 and Hey1 demonstrated Notch signaling inhibition. Am, ameloblasts; SI, stratum intermedium; SR, stellate reticulum; DSP, desmoplakin; AMEL, amelogenin; AMBN, ameloblastin; N1, NOTCH1; N2, NOTCH2; J1, JAG1; J2, JAG2. **, p-value<0.01.
Differences in expression levels of several genes in the ameloblast-SI region were identified by qPCR (Fig. 5M). Perp and Dsp were downregulated with anti-N1N2 or anti-J1J2 treatments. Interestingly, Trp63, which was previously shown to transactivate Perp directly in keratinocytes and LS-8 oral epithelial-like cells,(11,21) was not affected by Notch signaling inhibition. Irf6, which is a primary Notch signaling target in keratinocytes,(22) was downregulated with Notch signaling inhibition. Expression of Amel and Ambn was not affected, confirming the immunofluorescence data (Fig. 5C–L). Finally, confirmation of Notch signaling inhibition was validated by decreased expression of Hes1 and Hey1 (Fig. 5M). Surprisingly, although Hey1 was downregulated with anti-J1J2 treatment it was not significantly downregulated, perhaps highlighting the distinct downstream effects of Notch and Jag blockade.
Notch signaling inhibition leads to enamel defects
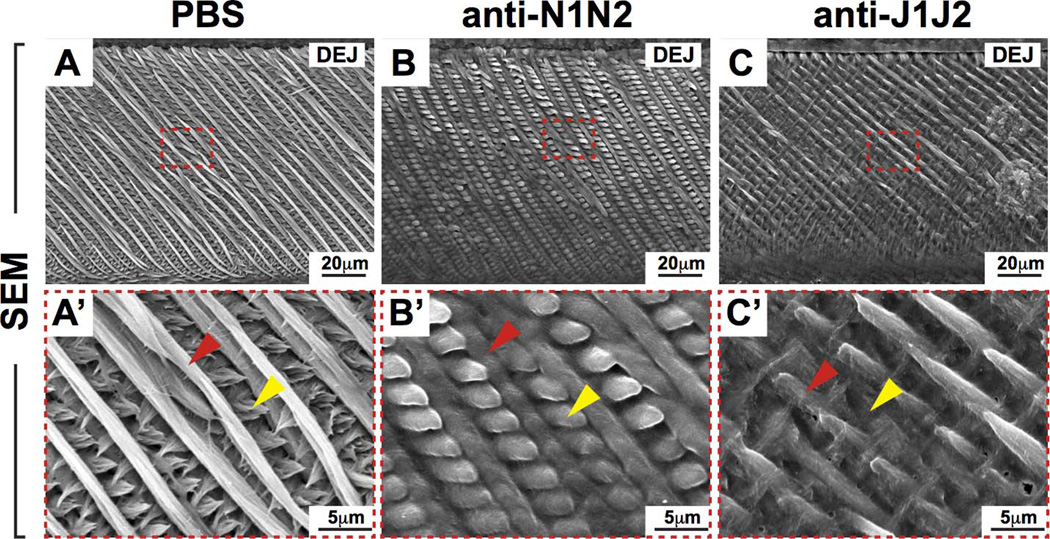
To determine the effects on enamel of the defects in the ameloblast-SI interface that occurred after Notch signaling inhibition, we analyzed the mineralized incisor enamel by scanning electron microscopy (SEM; Fig. 6). Notch signaling inhibition had major effects on the microarchitecture of the enamel rods. Normally, enamel is comprised of mineralized rods that span the dentin-enamel junction (DEJ) to the enamel surface. As enamel matures, the rods increase in diameter and are interconnected by smaller inter-rod enamel. In controls, the enamel rods were highly organized, running parallel in the same plane from the DEJ to the enamel surface (Fig. 6A,A’) similar to that observed in human teeth (data not shown). With anti-N1N2 treatment, the primary enamel rods remained relatively unchanged and normal, whereas the inter-rods appeared to be rounded and enlarged (Fig. 6B,B’) compared to controls. Anti-J1J2 treatment resulted in subtler differences compared to controls (Fig. 6C,C’). The primary rods appeared interrupted and shortened, and may indicate a change in the angle of matrix deposition and mineralization as this was observed in all three anti-J1J2 specimens analyzed (Fig. 6C.C’). However, our observation may also be partly due to a change in the orientation of the specimen during preparation for SEM. The inter-rods appeared to be decreased in size compared in controls and clearly smaller and less rounded than in anti-N1N2 specimens (Fig. 6). The distinct differences in the micro-architecture of mineralized enamel between Notch (i.e., anti-N1N2) or Jag (i.e., anti-J1J2) blockade underscore the complexity of the roles of Notch signaling pathways in enamel formation.
Figure 6. Scanning electron microscopy (SEM) of adult mouse incisors.
(A–C) SEM analysis of incisor enamel in mice treated with PBS or Notch antibodies in sagittal views. (A’–C’) Magnified views of red-boxed regions (A–C). Red arrowheads point to primary enamel rods and yellow arrowheads point to inter-rod enamel. Note the enlarged inter-rod enamel in anti-N1N2 incisors (B,B') compared to controls. In anti-J1J2 incisors, note the interrupted primary rod enamel and smaller size of inter-rod enamel (C,C') compared to controls. These observations highlight distinct roles of N1N2 and J1J2 in incisor enamel formation. DEJ, dentin-enamel junction; N1, NOTCH1; N2, NOTCH2; J1, JAG1; J2, JAG2.
Discussion
The lethality associated with embryonic inactivation of Notch signaling has hampered efforts to determine the role of Notch signaling during adult tooth renewal.(23–28) In this study, we inhibited Notch signaling by injecting adult mice with highly specific blocking antibodies raised against NOTCH1, NOTCH2, JAG1, and JAG2.(12) This approach allowed us to study the effects of Notch signaling inhibition on tooth renewal in adult animals. We found that Notch signaling is critical for ameloblast-SI and SI-SI adhesion, as well as enamel mineralization, in part, because of its effects on specific components of desmosomes.
Desmosomes are transmembrane, macromolecular complexes that provide strong cell-cell adhesion and are anchored to intermediate filaments.(29–31) Desmosomes consist of members of at least three distinct protein families: the cadherins, such as desmogleins and desmocollins, the armadillo proteins, including plakoglobin and the plakophilins, and the plakins. In vivo evidence for the importance of the ameloblast-SI interface and desmosomes in enamel formation has been limited to analyses of Perp- and Pvrl1- (i.e., nectin-1) null mice.(10,11) Perp encodes a transmembrane protein that is specifically associated with desmosomes, and its inactivation leads to desmosome defects.(11,21) On the other hand, Pvrl1 is known to be important in both adherens and tight junctions,(32,33) and its inactivation led to indirect changes in desmosome density and size. While some differences exist between the enamel phenotypes in Perp- and Pvrl1-null mice, both published studies show that the ameloblast-SI interface and desmosomes are integral for proper formation of enamel during development.
Notch signaling inhibition led to defects in the ameloblast-SI interface and SI cells, as well as the absence of desmosomes. With single antibody injections, the SI layer appeared flattened in the apical-basal direction (Fig. 2). With anti-N1N2 or anti-J1J2 treatment, the flattening of the SI layer was more pronounced, and some of the SI cells appeared to detach completely from the ameloblast layer (Fig. 2D,G,K,N). Incisor enamel mineralization was also altered with single and combined antibody treatments (Fig. 3). TEM analysis showed that adhesion between ameloblasts and SI cells, as well as between SI cells, was defective with Notch signaling inhibition (Fig. 4). However, unlike the decreases in desmosome size and number observed in Perp-null mice,(11) there was an absence of desmosomes at the ameloblast-SI interface and between SI cells with anti-N1N2 or anti-J1J2 treatment (Fig. 4). This observation reveals the importance of Notch signaling in desmosome formation and/or maintenance.
Although it has been shown previously that Notch signaling regulates the major desmosome cadherin expressed in the hair shaft cortex, desmoglein 4 (Dsg4),(34) we provide the first evidence that Notch signaling regulates desmosome-specific factors such as Perp and Dsp in tooth renewal (Fig. 5M). Besides the defects in the ameloblast-SI interface, the SI cells appeared abnormal, and in many cases, flattened compared to controls. However, there was no evidence of apoptosis (data not shown), which suggests additional roles for Notch signaling besides effects on desmosomes and adhesion in the ameloblast-SI interface. Furthermore, the differential expression of NOTCH1/2 (i.e., primarily in ameloblasts) and JAG1/2 (i.e., primarily in SI cells) and the subtle phenotypic differences with single and combinatorial NOTCH1/2 and JAG1/2 blockade (Figs. 2–6) demonstrate that the adult mouse incisor provides a powerful model system to finely dissect molecular mechanisms of Notch signaling during mineralization.
Expression levels of Amel and Ambn did not change with Notch signaling inhibition (Fig. 5M). Interestingly, AMBN immunofluorescence showed distinct, robust staining on the basal region of ameloblasts with NOTCH1/2 blockade (Fig. 5H), however it is unclear what this staining may represent. These effects on ameloblasts may be related to the changes to ameloblast-ameloblast adhesion observed on TEM analysis, as well as to the abnormal ameloblast morphology with Notch signaling inhibition. Moreover, there is a precedent for a link between enamel matrix proteins and Notch signaling as mice that overexpressed the P70T amelogenin transgene showed increased levels of Notch1 in developing molars.(35) Together, these findings underscore the complexity of Notch signaling during tooth renewal.
The relationship between Notch signaling and Trp63 is complex, but Notch signaling and Trp63 have been shown previously to antagonize each other.(36,37) Our experiments showed that Trp63 expression was not affected with inhibition of Notch signaling (Fig. 5M). We and others have previously shown that Perp is a direct target of the p53-paralog p63 (or Trp63) in various cells types.(11,21,38) Taken together with the observation that Notch signaling inhibition leads to a decrease in Perp expression (Fig. 5M), our data point to a mechanism in which Notch signaling regulates Perp expression directly through Notch regulatory elements within the Perp promoter and not through TRP63 during enamel formation. Additional support for this hypothesis is provided by the presence of 2 putative CSL binding elements at positions −6854 (i.e., TTCCCACG) and −6059 (i.e., GTGGGAA) upstream of the Perp transcription start site. CSL (also known as CBF1/RBP-J in mammals, Suppressor of Hairless [Su(H)] in Drosophila and Xenopus and Lag-1 in Caenorhabditis elegans) is a DNA binding factor that represses and activates transcription in the absence and presence of Notch signaling, respectively.(39,40) CSL is also considered to be the primary target of Notch signaling in mammalian cells and the NICD-CSL complex activates transcription through recruitment of the histone acetyltransferase PCAF.(39,40) It will be of interest to test whether the 2 putative CSL binding elements can transactivate Perp, which is required for proper desmosome formation and/or maintenance. Thus, Notch signaling appears to regulate Perp through at least two distinct mechanisms: first, directly through Notch responsive elements in the Perp promoter, and second, through transactivation by TRP63.
Together, our data point to a model in which Notch signaling is upstream of Perp and Dsp. Notch signaling also appears to regulate Perp through at least two distinct pathways: 1) directly through Notch responsive elements present in the Perp promoter, and 2) via TRP63 transactivation. Disruption in Notch signaling leads to defective enamel formation, in part, due to compromised desmosome formation and/or maintenance at the ameloblast-SI interface. This study highlights the importance of the ameloblast-SI interface in enamel formation and demonstrates the requirement of Notch signaling in enamel formation during tooth renewal.
Supplementary Material
Acknowledgments
We thank the members of the Klein laboratory and Lisa Choy-Tomlinson at Genentech for their technical assistance and discussions. Micro-CT imaging work was performed by Sabra Djomehri at the Division of Biomaterials and Bioengineering Micro-CT Imaging Facility, UCSF, which is supported by the Department of Health and Human Services/NIH S10 Shared Instrumentation Grant (S10RR026645), and the Departments of Preventive and Restorative Dental Sciences and Orofacial Sciences, School of Dentistry, UCSF.
The authors are funded by the National Institutes of Health (R01-DE021420 to O.D.K. and R00-DE022059 to A.H.J.).
Disclosure
C.W.S. is an employee of Genentech, Inc. and owns shares of Roche.
Footnotes
Authors’ roles: Study design – AHJ, CWS, and ODK; Animal treatment - AHJ, MP, TW, AN; Histology, in situ hybridization, and immunostaining - AHJ, MP, TW, BM, YJL, AN, RE; TEM, AHJ, and EDS; SEM, AHJ and BG; qPCR data collection and analyses - AHJ and MP; Data interpretation - AHJ, CWS, and ODK; Manuscript preparation - AHJ, CWS, and ODK; Revisions - AHJ, CWS, and ODK; Approval of final versions of manuscript - All; AHJ and ODK are responsible for the integrity of data analysis.
References
- 1.Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147(1):105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsa S, Kuremoto K, Seidel K, et al. Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development. 2010;137(22):3743–3752. doi: 10.1242/dev.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seidel K, Ahn CP, Lyons D, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137(22):3753–3761. doi: 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biehs B, Hu J, Strauli NB, et al. Bmi1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol. 2013;5:846–852. doi: 10.1038/ncb2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jheon AH, Seidel K, Biehs B, Klein OD. From molecules to mastication: the development and evolution of teeth. WIREs Dev Biol. 2013 doi: 10.1002/wdev.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010;80(4–5):241–248. doi: 10.1016/j.diff.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Harada H, Ichimori Y, Yokohama-Tamaki T, et al. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 2006;340(2):611–616. doi: 10.1016/j.bbrc.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 8.Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137(18):3025–3035. doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li CY, Cha W, Luder HU, et al. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Developmental biology. 2012;366(2):357–366. doi: 10.1016/j.ydbio.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barron MJ, Brookes SJ, Draper CE, et al. The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Hum Mol Genet. 2008;17(22):3509–3520. doi: 10.1093/hmg/ddn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jheon AH, Mostowfi P, Snead ML, et al. PERP regulates enamel formation via effects on cell-cell adhesion and gene expression. J Cell Sci. 2011 doi: 10.1242/jcs.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 13.Huntzicker EG, Hotzel K, Choy L, et al. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. 2015;61(3):942–952. doi: 10.1002/hep.27566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafkas D, Shelton A, Chiu C, et al. Notch-controlled trans-differentiation in the adult lung. Nature. in revision. [Google Scholar]
- 15.Mitsiadis TA, Lardelli M, Lendahl U, Thesleff I. Expression of Notch 1, 2 and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J Cell Biol. 1995;130(2):407–418. doi: 10.1083/jcb.130.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsiadis TA, Henrique D, Thesleff I, Lendahl U. Mouse Serrate-1 (Jagged-1): expression in the developing tooth is regulated by epithelial-mesenchymal interactions and fibroblast growth factor-4. Development. 1997;124(8):1473–1483. doi: 10.1242/dev.124.8.1473. [DOI] [PubMed] [Google Scholar]
- 17.Mitsiadis TA, Hirsinger E, Lendahl U, Goridis C. Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol. 1998;204(2):420–431. doi: 10.1006/dbio.1998.9092. [DOI] [PubMed] [Google Scholar]
- 18.Djomehri SI, Candell S, Case T, et al. Mineral density volume gradients in normal and diseased human tissues. PLoS One. 2015;10(4):e0121611. doi: 10.1371/journal.pone.0121611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang WS, Tonna EA. Autoradiographic Analysis of Labeling Indices and Migration Rates of Cellular Component of Mouse Incisors Using Tritiated Thymidine (H3tdr) J Dent Res. 1965;44:42–53. doi: 10.1177/00220345650440012901. [DOI] [PubMed] [Google Scholar]
- 20.Smith CE, Warshawsky H. Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat Rec. 1975;183(4):523–561. doi: 10.1002/ar.1091830405. [DOI] [PubMed] [Google Scholar]
- 21.Ihrie RA, Marques MR, Nguyen BT, et al. Perp is a p63-regulated gene essential for epithelial integrity. Cell. 2005;120(6):843–856. doi: 10.1016/j.cell.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Restivo G, Nguyen BC, Dziunycz P, et al. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 2011;30(22):4571–4585. doi: 10.1038/emboj.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121(5):1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 24.Demehri S, Liu Z, Lee J, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6(5):e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphreys R, Zheng W, Prince LS, et al. Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients. Hum Mol Genet. 2012;21(6):1374–1383. doi: 10.1093/hmg/ddr575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang R, Lan Y, Chapman HD, et al. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12(7):1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCright B, Gao X, Shen L, et al. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128(4):491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- 28.Xue Y, Gao X, Lindsell CE, et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8(5):723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 29.Franke WW. Discovering the molecular components of intercellular junctions--a historical view. Cold Spring Harb Perspect Biol. 2009;1(3):a003061. doi: 10.1101/cshperspect.a003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green KJ, Simpson CL. Desmosomes: new perspectives on a classic. J Invest Dermatol. 2007;127(11):2499–2515. doi: 10.1038/sj.jid.5701015. [DOI] [PubMed] [Google Scholar]
- 31.Thomason HA, Scothern A, McHarg S, Garrod DR. Desmosomes: adhesive strength and signalling in health and disease. Biochem J. 2010;429(3):419–433. doi: 10.1042/BJ20100567. [DOI] [PubMed] [Google Scholar]
- 32.Fukuhara A, Irie K, Nakanishi H, et al. Involvement of nectin in the localization of junctional adhesion molecule at tight junctions. Oncogene. 2002;21(50):7642–7655. doi: 10.1038/sj.onc.1205875. [DOI] [PubMed] [Google Scholar]
- 33.Fukuhara A, Irie K, Yamada A, et al. Role of nectin in organization of tight junctions in epithelial cells. Genes Cells. 2002;7(10):1059–1072. doi: 10.1046/j.1365-2443.2002.00578.x. [DOI] [PubMed] [Google Scholar]
- 34.Bazzi H, Demehri S, Potter CS, et al. Desmoglein 4 is regulated by transcription factors implicated in hair shaft differentiation. Differentiation. 2009;78(5):292–300. doi: 10.1016/j.diff.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Li Y, Alawi F, Bouchard JR, Kulkarni AB, Gibson CW. An amelogenin mutation leads to disruption of the odontogenic apparatus and aberrant expression of Notch1. J Oral Pathol Med. 2011;40(3):235–242. doi: 10.1111/j.1600-0714.2010.00940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer. 2009;9(8):587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17(10):1600–1612. doi: 10.1038/cdd.2010.37. [DOI] [PubMed] [Google Scholar]
- 38.Ihrie RA, Bronson RT, Attardi LD. Adult mice lacking the p53/p63 target gene Perp are not predisposed to spontaneous tumorigenesis but display features of ectodermal dysplasia syndromes. Cell Death Differ. 2006;13(9):1614–1618. doi: 10.1038/sj.cdd.4401871. [DOI] [PubMed] [Google Scholar]
- 39.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;275(22):17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 40.Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26(23):5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.