Abstract
Genomic instability is a well-known hallmark of cancer. Recent genome sequencing studies have led to the identification of novel phenomena called chromothripsis and chromoanasynthesis in which complex genomic rearrangements are thought to be derived from a single catastrophic event rather than by several incremental steps. A new term chromoanagenesis or chromosomal rebirth was coined recently to group these two one-step catastrophic events together. These phenomena suggest an evolutionary modality for cancer cells to circumvent individual mutational events with one simultaneous shattering of chromosomes resulting in the random reassembling of segmented genetic material to form complex derivative chromosomes. We report a case of possible chromoanagenesis in a patient with diffuse large B-cell lymphoma. Chromosome analysis from the biopsy showed a complex karyotype with multiple numerical and structural rearrangements including a translocation of chromosomes 3 and 7 involving the BCL6 gene region, with the derivative chromosome further rearranging with chromosomes 14, 7, and 22 with involvement of the IGH gene region. Fluorescence in situ hybridization studies confirmed these findings. Chromosomal microarray studies showed multiple complex copy number variations including a chromosome 12 abnormality, the complexity of which appears to suggest the phenomenon of chromoanagenesis. Our case further illustrates that lymphomagenesis can be complex and may arise from a catastrophic event resulting in multiple complex chromosome rearrangements.
Introduction
Genomic instability is a well-described distinctive feature of cancer. Thus, uncovering pathways describing acceleration of such instability is not surprising. Previously, genomic instability was thought to arise through a gradual multistep process resulting in sequential accumulation of many independent genomic lesions [1], [2], [3]. Such lesions may include somatic point mutations, copy number alterations such as chromosomal gains and losses, and balanced structural rearrangements such as translocations and inversions [4], [5], [6], [7]. Although there is well-established evidence for this gradualism in cancer development, there were reasons to hypothesize that cancer cells might acquire all genomic lesions at once to circumvent the defensive responses from the genome. Recent genome sequencing studies have led to identification of three novel phenomena called chromothripsis, chromoanasynthesis, and chromoplexy [8], [9], [10], [11]. Although the underlying mechanisms of these novel phenomena are not known and were hypothesized to be different, the resulting complex genomic rearrangements are thought to be derived from a single catastrophic event rather than by several incremental steps [12], [13], [14], [15], [16]. Holland and Cleveland [17] suggested the term chromoanagenesis or chromosomal rebirth to group these one-step catastrophic events together. We report a case of possible chromoanagenesis in a patient with diffuse large B-cell lymphoma (DLBCL) arising from follicular lymphoma (FL).
Material and Methods
Case Report
A 59-year-old Caucasian woman with history of hypothyroidism presented to the clinic with rapidly enlarging goiter causing significant dyspnea. She noticed swelling in the neck 2 months before presenting to the clinic. A computed tomographic scan showed extensive infiltration and enlargement of the thyroid gland with significant effect on the trachea, limited to 4.3 mm in width at the thoracic inlet. Thyroid biopsy showed sheets of large dysplastic B-cells, diagnostic of DLBCL. The neoplastic B-cells expressed CD10, BCL6, MUM1, and BCL2 and lacked expression of CD30, Cyclin D1, and EBER. Flow cytometric analysis also showed the clonal B-cells (45% of total cells) expressed CD19, CD20, and surface lambda light chain and lacked CD45. Background nodular follicular dendritic meshwork (CD21 +) suggested that the DLBCL may have arisen from FL. Bone marrow biopsy was normal and unremarkable.
Chromosome Analysis
Cytogenetic analysis was carried out on biopsy tissue. Culture initiation, maintenance, and harvest were done using standard methods [18]. Chromosomes were G-banded [19] and then analyzed using a Cytovision image analysis system (Applied Imaging, Santa Clara, CA).
Fluorescence In Situ Hybridization (FISH)
FISH was performed on the cultured biopsy specimen using directly labeled break-apart probe BCL6 (5’ labeled in spectrum orange and 3’ in spectrum green); dual-color, dual-fusion translocation probe IGH/BCL2 (IGH labeled in spectrum green and BCL2 in spectrum orange); and whole chromosome paint probes for chromosomes 7 (labeled in spectrum green), 14 (spectrum orange), and 12 (spectrum green) (Cytocell, Windsor, CT). The probes were hybridized to interphase nuclei and metaphase chromosomes using standard procedures, followed by counterstaining with 4,6-diamidino-2-phenylindole, and then analyzed using a Cytovision image analysis system (Applied Imaging, Santa Clara, CA). For interphase analysis, a minimum of 100 nuclei were scored, and for metaphase analysis, a minimum of 10 metaphases were scored.
Single Nucleotide Polymorphism (SNP) Oligonucleotide Microarray
Given the complex nature of the abnormalities observed, chromosome microarray studies were carried out using Affymetrix CytoScan HD microarray. The Affymetrix CytoScan HD Assay uses a high density combined CGH and SNP array platform, which assesses approximately 2,696,550 markers, including approximately 750,000 SNP markers. Each oligonucleotide is approximately 25 bp long. Intragenic probe spacing is approximately 1 probe every 880 bp, and intergenic probe spacing is approximately 1 probe every 1700 bp. To perform the assay, gDNA is digested with the Nsp1 restriction enzyme and digested DNA is then ligated to Nsp1 adapters. The ligation product is then amplified via polymerase chain reaction to produce amplicons in the 200- to 1100-bp range. The amplicons are then purified and digested with DNAse I to produce 25- to 125-bp fragments. The fragments are end-labeled with a modified biotinylated base, and the sample is then hybridized to the array. The array is washed and then stained with a streptavidin-coupled dye and a biotinylated antistreptavidin antibody. The array is then scanned with the GeneChip Scanner, and the signal intensity for each marker is assessed. Using the Chromosome Analysis Suite (ChAs 3.0) software, the signal for the sample is then compared with a reference set, which is based on the average of more than 400 samples. Differences in signal between the sample and reference are expressed as a log2 ratio and represent relative intensity for each marker. A discrete copy number value is determined from the relative intensity data and is displayed. Genotype information for the SNP markers is visualized with the Allele Track [20].
Results
Cytogenetic Studies
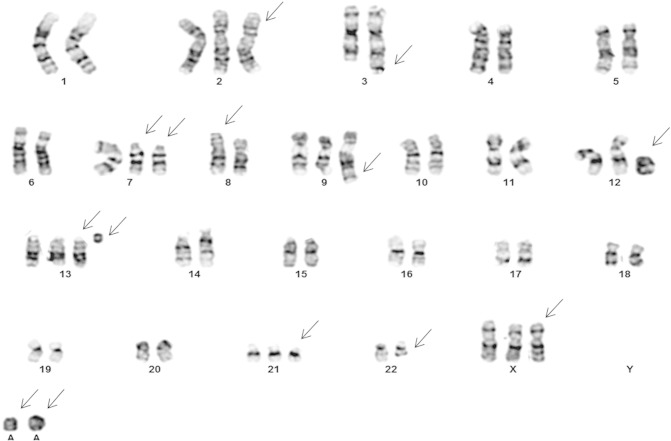
Chromosome analysis from the biopsy tissue showed a complex hyperdiploid karyotype with multiple structural and numerical abnormalities including rearrangements involving the 3q and 14q regions. Based on G-banded analysis of 20 cells, the karyotype was interpreted as 51~56,XX,+X,+X,+2,t(3;7)(q29;p11.2),der(7)t(3;7)t(14;7;22)(q32;p11.2;q12),+der(7)t(14;7;22),der(8)t(8;18)(p12;q21),+der(9)t(5;9)(q13;q22),+13,der(14)t(14;7;22),+21,+1~4r[cp20] (Figure 1). Chromosome analysis of the bone marrow sample showed a normal female karyotype.
Figure 1.
Karyotype from thyroid biopsy showing complex chromosome abnormalities.
FISH
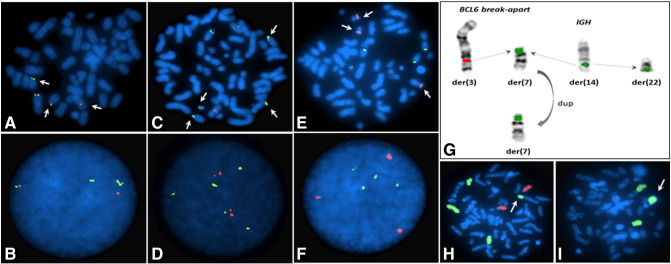
FISH studies with BCL6 break-apart probe showed BCL6 gene rearrangement in 36.5% of the nuclei (Figure 2, A and B), and with dual-color, dual-fusion IGH/BCL2 probes showed IGH gene rearrangement in 41.2% of the nuclei (Figure 2, C and D) and an additional copy of the BCL2 gene region in 7% of the nuclei (Figure 2, E and F). Metaphase FISH analysis showed that the 5’BCL6 remained on chromosome 3q whereas the 3’ BCL6 was translocated to a chromosome 7. Similarly, metaphase analysis showed complex structural rearrangement involving the IGH gene region with parts of IGH being translocated to chromosome 7 and also chromosome 22 (Figure 2G). These results confirm the complex nature of the derivative chromosome 7 observed on chromosome analysis. FISH analysis with whole chromosome paints confirmed the t(3;7) (Figure 2H) and that the ring chromosome was derived from a chromosome 12 (Figure 2I).
Figure 2.
FISH analysis showing multiple gene rearrangements. (A) BCL6 gene rearrangement on metaphase. (B) BCL6 gene rearrangement in interphase nuclei. (C) IGH gene rearrangement on metaphase. (D) IGH gene rearrangement in interphase nuclei. (E) BCL2 gene rearrangement in metaphase. (F) BCL2 gene rearrangement in interphase nuclei. (G) Schematic showing the complex IGH gene rearrangement. (H) Whole chromosome paint 7 and 14 showing the t(3;7). (I) Whole chromosome paint 12 confirming that the ring chromosome is derived from a chromosome 12.
Chromosomal Microarray Studies
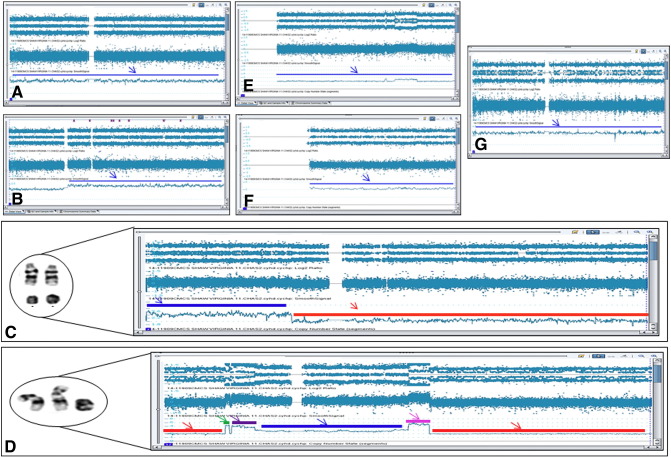
The results showed multiple abnormalities interpreted as arr(1-22,X)cx. The abnormalities observed included mosaic gain (14.7%) of chromosome 2, mosaic gain (40%) of chromosome 5q11.2q35.3, mosaic gain (45%) of chromosome 7p22p12.3, mosaic loss (7%) of chromosome 7p12.3q36.3, multiple aberrations indicating chromoanagenesis on chromosome 12, mosaic gain (10%) of chromosome 13, mosaic gain (21%) of chromosome 21, and mosaic gain (21%) of chromosome X (Figure 3, A–G). Although the IGH gene region on chromosome 14 underwent complex structural rearrangements, array studies did not show any imbalance involving this region, thus indicating that, despite the complex nature, the IGH rearrangements are balanced.
Figure 3.
SNP microarray showing the complex abnormalities observed. (A) Mosaic gain of chromosome 2 (blue arrow). (B) Mosaic gain of chromosome 5q11.2q35.3 (blue arrow). (C) Mosaic gain of chromosome 7p22p12.3 (blue arrow) and mosaic loss of chromosome 7p12.3q36.3 (red arrow). (D) Multiple gains and losses of chromosome 12. Mosaic LOH for 12p13.33p12.3 (red line and arrow). Mosaic LOH for 12q21.1q24.33 (red line and arrow). Four copies of 12p12.3 (green line and arrow). Three to four copies of mosaic gain 12p12.3p12.1 (purple line and arrow). Two to three copies of 12p12.1q14.3 (blue line and arrow) and four to five copies of mosaic gain 12q14.3q21.1 (pink line and arrow). (E) Mosaic gain on chromosome 13 (blue arrow). (F) Mosaic gain of chromosome 21. (G) Mosaic gain of X chromosome.
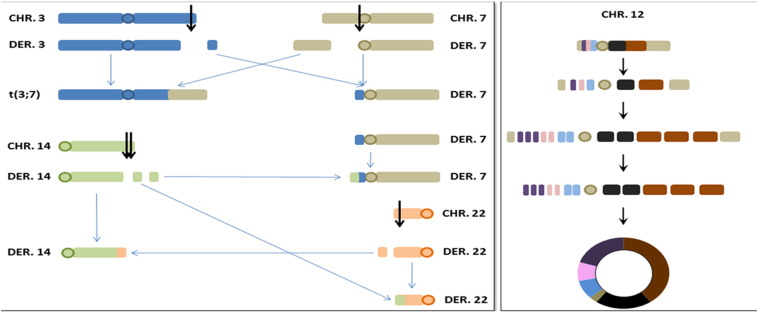
Based on these results, we propose that simultaneous breaks on chromosomes 3, 7, 14, and 22 with concurrent exchange and reassembly of the broken segments lead to a complex chromosomal rearrangement resulting in several derivative chromosomes: a der(3) consisting of 3pter-q29 and 7pter-p11.2, a der(7) consisting of 7p11.2-qter, 3q29-qter and 14q32-qter, a der(14) consisting of 14pter-q32 and 22q12-qter, and a der(22) consisting of 22pter-q12 and 14q32-qter (Figure 4). Of the one to four ring chromosomes observed on karyotypic analysis, based on the array data, the most commonly observed ring chromosome was derived from chromosome 12 (present in more than 10 cells), whereas 2 smaller ring chromosomes (Figure 3C) were interpreted as originating from 7p (present in less than 10 cells) and the remaining 1 ring chromosome (Figure 1) derived from chromosome 13 (present in less than 3 cells). The array and FISH results suggested that simultaneous shattering of chromosome 12 with subsequent duplication and triplication of interstitial segments and loss of telomeric regions lead to the ring formation (Figure 4).
Figure 4.
Schematic representation of complex rearrangements (A) involving chromosomes 3, 7, 14, and 22. (B) Multiple breaks and reassembly leading to formation of r(12).
Discussion
DLBCL is the most common malignant lymphoma subtype in adults, accounting for roughly 40% of all cases [21]. DLBCL is characterized by remarkable heterogeneity, affecting several aspects of the disease. This heterogeneity can at least partially be explained by recently identified molecularly defined subtypes [22]. DLBCL can be classified as de novo or as secondary, transformed DLBCL emanating from an indolent lymphoma, most commonly FL. The distinction between transformed and de novo DLBCL is based on the morphological demonstration of low–malignancy grade lymphoma such as FL either before or simultaneous to the DLBCL. In our case, the presence of background nodular follicular dendritic meshwork (CD21 +) suggested that the DLBCL may have arisen from FL. Several secondary genetic abnormalities have been reported to be associated with this histological transformation of FL, including gene rearrangements involving MYC, BCL6, and BCL2 and mutations of TP53 tumor suppressor gene [23]. In addition to these specific genetic changes, transformation has been associated with cytogenetic alterations such as loss or gain of whole chromosomes, deletions, and amplifications [23]. One of such frequent gains is chromosome 18q including the BCL2 gene locus which was also seen in 7% of cells in our case. Other changes acquired exclusively upon transformation included the gain of a segment on chromosome 12 from pter to q12. Trisomy 12 is frequently seen in many B-cell malignancies, with the common region of amplification delineated to 12q12q15 [24]. Copy number gains at 12q12q14 were reported exclusively in DLBCL transformed from FL [25]. In accordance with published studies showing that the 12q amplicon is highly complex and exhibits discontinuous regions of amplification in high-grade lymphomas [26], our case showed multiple and complex rearrangements involving the chromosome 12 with mosaic loss and gain of three to five copies of chromosome 12, especially overlapping the 12q12q14 region. Karyotypic analysis also showed that transformed DLBCLs are cytogenetically more unstable, thereby resulting in multiple numerical and structural alterations. The complex structural rearrangement involving chromosomes 3, 7, 14, and 22 (Figure 4) is an indication of such underlying instability in the genome of these transformed DLBCLs. In addition, several studies have shown overrepresentation of X chromosome in FL and DLBCL [25], and gain of an X chromosome was also seen in our case.
Gene amplification and loss have an important role in the activation of oncogenes and inactivation of tumor suppressor genes. The mechanism underlying the formation of the amplified DNA segment (amplicon) is poorly understood. Microsatellite repeat analysis studies have shown that these amplicons have complex structures with amplified loci interspersed with nonamplified loci [26]. The structure of these amplicons owes its complexity not only to amplifications but also to losses of DNA segments and enables simultaneous activation of cellular oncogenes and inactivation of tumor suppressor genes. It is a common assumption that chromosomal gains and losses exert “dosage effects” on the expression of at least some of the genes within these regions; however, confirmation of this hypothesis especially in the process of FL transformation to DLBCL had been limited. The recently identified phenomenon of “chromoanagenesis” might explain the complex nature of these amplicons because it is known to cause catastrophic shattering of chromosomes or chromosome segments giving rise to chromosomal gains and losses in a single event. The hallmark of chromothripsis is said to be that the catastrophic event involves just a few regions in the genome giving rise to an extremely high local rearrangement density. We propose that the complex chromosome 12 rearrangement in our patient with approximately six breakpoints resulting in multiple gains and losses ultimately leading to the ring formation (Figure 4) is the result of chromothripsis rather than chromoanasynthesis because chromoanasynthesis is reported to occur more frequently in distal parts of chromosome arms and is more common in germline than somatic cells [9], [27]. A second hallmark of chromothripsis is random shattering of chromosomes and reassembly leading to highly mosaic chromosomes from small and large chromosome pieces derived from diverse locations and stitched together in all possible combinations [28]. In our case, the complex rearrangement involving chromosomes 3, 7, 14, and 22 indicates this random shattering and reassembly. Because chromoanasynthesis is reported to be mostly observed within a single chromosome [9], [29], this complex rearrangement in our case most probably is the result of chromothripsis and not chromoanasynthesis. The third hallmark of chromothripsis is said to be presence of two copy number states with the lower number copy state often displaying loss of heterozygosity [28]. In our case, the mosaic loss of heterozygosity on chromosome 12 short and long arms observed with microarray studies is in accordance with this chromothripsis signature. The ability to detect chromothripsis in cancer is limited by the methodology used. For example, the incidence of chromothripsis is reported to be low using SNP-based microarray studies, whereas studies using whole genome sequencing method showed a much higher frequency of chromothripsis in cancer [28].
One of the significant limitations of our study is the lack of sequencing data to confirm chromothripsis or chromoanasynthesis in our patient. Lack of resources does not permit us to undertake sequencing studies to confirm our hypothesis. Nevertheless, our results and the rapid progression of disease provide reasonable evidence of chromoanagenesis in our patient.
In summary, we report a patient with DLBCL possibly arising from FL, and the complex chromosome abnormalities observed in our patient suggest that chromoanagenesis might have led to the complex structural rearrangements. Although several studies have documented multiple structural and numerical chromosome abnormalities, especially in those cases that have transformed from FL to DLBCL, the mechanism resulting in such complex rearrangements has not been postulated. We propose that chromoanagenesis might provide a possible explanation for the chromosome instability and the associated poor prognosis in those patients with transformation from FL to DLBCL.
References
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 3.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 5.Rabbits TH. Commonality but diversity in cancer gene fusions. Cell. 2009;137:391–395. doi: 10.1016/j.cell.2009.04.034. [DOI] [PubMed] [Google Scholar]
- 6.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, McDonald TY, Ghandi M. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA. Chromosome ctastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcutt PA. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell CR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA. Massive genomic rearrangement acquired in a single catastrophic even during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–670. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 13.Jones MJ, Jallepalli PV. Chromothripsis: chromosomes in crisis. Dev Cell. 2012;23:908–917. doi: 10.1016/j.devcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyerson M, Pellman D. Cancer genomes evolve by pulverizing single chromosomes. Cell. 2011;144:9–10. doi: 10.1016/j.cell.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 16.Tubio JM, Estivill X. Cancer: when catastrophe strikes a cell. Nature. 2011;470:476–477. doi: 10.1038/470476a. [DOI] [PubMed] [Google Scholar]
- 17.Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatalica Z, Velagaleti G, Kuivaniemi H, Tromp G, Palazzo J, Graves KM, Guigneaux M, Wood T, Sinha M, Luxon B. Gene expression profile of an adenomyoepithelioma of the breast with a reciprocal translocation involving chromosomes 8 and 16. Cancer Genet Cytogenet. 2005;156:14–22. doi: 10.1016/j.cancergencyto.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 19.Pearson P. The use of new staining techniques for human chromosome identification. J Med Genet. 1972;9:264–275. doi: 10.1136/jmg.9.3.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webb BD, Scharf RJ, Spear EA, Edelmann LJ, Stroustrup A. Evaluation of the Affymetrix Cytoscan® Dx assay for developmental delay. Expert Rev Mol Diagn. 2015;15:185–192. doi: 10.1586/14737159.2015.975213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardim JW. IARC Press; Lyon, France: 2008. WHO Classification of Tumours and Haematopoietic and Lymphoid Tissues. [Google Scholar]
- 22.Lenz G. Insights into the molecular pathogenesis of activated B-cell-like diffuse large B-cell lymphoma and its therapeutic implications. Cancers. 2015;7:811–822. doi: 10.3390/cancers7020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez-Climent JA, Alizadeh AA, Segraves R, Blesa D, Rubio-Moscardo F, Albertson DG, Garcia-Conde J, Dyer MJS, Ley R, Pinkel D. Transformation of follicular lymphoma to diffuse large cell lymphoma is associated with heterogeneous set of DNA copy number and gene expression alterations. Blood. 2003;101:3109–3117. doi: 10.1182/blood-2002-07-2119. [DOI] [PubMed] [Google Scholar]
- 24.Rao PH, Houldsworth J, Dyomina K, Parsa NZ, Cigudosa JC, Louie DC, Popplewell L, Offit K, Jhanwar SC, Chaganti RS. Chromosomal and gene amplification in diffuse large B-cell lymphoma. Blood. 1998;92:234–240. [PubMed] [Google Scholar]
- 25.Hough RE, Goepel JR, Alcock HE, Hancock BW, Lorigan PC, Hammond DW. Copy number gain at 12q12-14 may be important in the transformation from follicular lymphoma to diffuse large B cell lymphoma. Br J Cancer. 2011;84:499–503. doi: 10.1054/bjoc.2000.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf M, Aaltonen LA, Szymanska J, Tarkkanen M, Blomqvist C, Berner JM, Myklebost O, Knuutila S. Complexity of 12q13-22 amplicon in liposarcoma: microsatellite repeat analysis. Genes Chromosomes Cancer. 1997;18:66–70. doi: 10.1002/(sici)1098-2264(199701)18:1<66::aid-gcc8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 27.Malhotra A, Lindberg M, Faust GG, Leibowitz ML, Clark RA, Layer RM, Quinlan AR, Hall IM. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res. 2013;5:762–776. doi: 10.1101/gr.143677.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kloosterman WP, Koster J, Molenaar JJ. Prevalence and clinical implications of chromothripsis in cancer genomes. Curr Opin Oncol. 2014;26:64–72. doi: 10.1097/CCO.0000000000000038. [DOI] [PubMed] [Google Scholar]
- 29.Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, van Roosmalen MJ, van Lieshout S, Nijman IJ, Roessingh W. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]