Abstract
Helicobacter pylori infection is the strongest risk factor for development of gastric cancer. Host cellular stress responses, including inflammatory and immune responses, have been reported highly linked to H. pylori-induced carcinogenesis. However, whether mitochondrial regulation and metabolic reprogramming, which are potently associated with various cancers, play a role in H. pylori-induced gastric carcinogenesis is largely unknown. Here we revealed that Lon protease (Lonp1), which is a key inductive of mitochondrial unfolded protein response (UPRmt) and is required to maintain the mitochondrial quality, was greatly induced in H. pylori infected gastric epithelial cells. Importantly, we uncovered that knockdown of Lonp1 expression significantly diminished the metabolic switch to glycolysis and gastric cell proliferation associated with low multiplicity of H. pylori infection. In addition, Lonp1 overexpression in gastric epithelial cells also promoted glycolytic switch and cell overgrowth, suggesting H. pylori effect is Lonp1 dependent. We further demonstrated that H. pylori induced Lonp1 expression and cell overgrowth, at least partially, via HIF-1α regulation. Collectively, our results concluded the relevance of Lonp1 for cell proliferation and identified Lonp1 as a key regulator of metabolic reprogramming in H. pylori-induced gastric carcinogenesis.
Introduction
Helicobacter pylori infection is a major cause of chronic gastritis and is the strongest risk factor of gastric cancer [1]. H. pylori has also been defined as a class I carcinogen by the international Agency for Research on Cancer (IARC) [2], [3]. It is well known that H. pylori influences gastric cell proliferation, apoptosis, and cancer cell transformation via production of multiple virulence factors, including CagA, peptidoglycans, VacA, adhesins and outer membrane proteins (OMPs) [4], [5]. However, growing evidence indicated that host chronic gastric stresses also play important roles in H. pylori-induced gastric adenocarcinoma [6]. For example, a few regulators of immune response, inflammation and hypoxia underlying carcinogenesis, like NF-κB and HIF-1α, are activated by H. pylori infection [7], [8]. Beside that, however, the molecular mechanism(s) by which H. pylori-induced stress responses in epithelial cells contribute to carcinogenesis is still not fully characterized.
Lonp1 is a conserved serine peptidase and an important responsive marker for UPRmt, which mounts protective effects to compensate stress-associated mitochondrial injury by inducing the expression of mitochondrial enzymes, chaperons, as well as proteases, to maintain mitochondrial proteostasis and function [9]. Lonp1 plays an essential role in clearance of misfolded or damaged proteins and maintains mitochondrial function and cell viability under diverse physiological conditions, including oxidative stress, endoplasmic reticulum (ER) stress, and hypoxia [10], [11]. In addition, Lonp1 also supports mitochondrial DNA (mtDNA) stability and mitochondrial electron transport chain integrity [12], [13]. Importantly, recent studies indicated that Lonp1 up-regulation in melanoma cells results in robust changes in mitochondrial complexes, leading to impaired mitochondrial respiration and metabolic switch to glycolysis, which are the major features of tumorigenesis [14]. As a consequence, the proliferation rate and viability of melanoma cells, as well as cervical cancer cells, were significantly increased regarding Lonp1 induction [14], [15].
In this study, we performed integrative analysis of published microarray data of H. pylori infection and uncovered the Lonp1, as well as other UPRmt genes, were significantly induced in gastric epithelial cells. We further demonstrated that Lonp1 plays an important role in metabolic switch toward glycolysis and gastric epithelial cell proliferation in response to low multiplicity of H. pylori infection.
Results
Differentially Expressed Genes in H. pylori-Infected Mouse Gastric Epithelial Cells
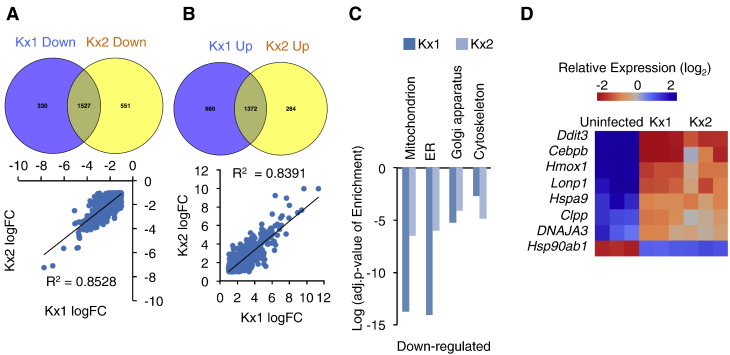
To assess the influence of H. pylori infection on the transcriptional profile of gastric epithelial cells, we analyzed microarray data that was downloaded from NCBI-GEO website (GSE10262). In that study, Gordon and his colleagues treated mouse gastric stem-like cells with chronic atrophic gastritis (ChAG)- and cancer-associated H. pylori strains to examine the transcriptional alterations during the transition from ChAG to gastric adenocarcinoma [16]. In order to obtain a broader range of differentially expressed genes for statistic modeling, we set the cut-off as fold change (FC) > 1.5 and false discovery rate (FDR) < 0.05, which was a little different from the criteria (FC > 2 and FDR < 0.05) in Gordon’s work [16]. Eventually, 2507 (1574 up- and 933 down-regulated) genes and 1483 (956 up- and 527 down-regulated) genes were identified in both Kx1 (ChAG-associated) and Kx2 (cancer-associated) H. pylori infection models, respectively (Figure 1, A–B). We further found that 73% of up-regulated genes and 82% of down-regulated genes in Kx2 model were consistently differentially expressed in Kx1 model (Figure 1, A–B).
Figure 1.
Differentially expressed genes in gastric cells in H. pylori infected mouse gastric epithelial cells. (A-B) Significantly down- and up-regulated genes (fold change > 1.5 and FDR < 0.05) were identified in Kx1 and Kx2 H. pylori infection models. Venn diagram analysis (up) and Pearon’s correlation analysis (down) of significant down- (A) and up-regulated (B) genes. “LogFC” means the value of log2 fold-change for each gene in Kx1 or Kx2 infection model. (C) Significantly enriched (EASE score < 0.05) GO terms regarding cellular compartment were identified in overlapping down-regulated genes. (D) Heatmap showing the significantly deregulated UPRmt genes in both Kx1 and Kx2 H. pylori infection models.
Interestingly, the gene ontology (GO) enrichment analysis of overlapping genes that were differentially expressed in both Kx1 and Kx2 models revealed that GO terms related to phosphorylation regulation, cell growth, stress response, and RNA processing were significantly enriched in up-regulated genes, while GO terms related to transport regulation, cell death, macromolecular complex organization, lipid metabolism and oxidation regulation were significantly enriched in down-regulated genes (Figure S1, A–B). In addition to biological process, we also found that cellular compartment GO terms related to nuclear lumen, endosome, lysosome, proteasome complex, and V-type ATPase complex were significantly enriched in up-regulated genes (Figure S2C). On the other hand, cellular compartment GO terms related to mitochondrion, ER, Golgi apparatus, and cytoskeleton were significantly enriched in down-regulated genes (Figure 1C). The down-regulated mitochondrial genes included those ones encoding components of electron transport chain (ETC), tricarboxylic acid cycle (TCA), as well as beta-oxidation (Figure S2).
We also performed GO enrichment analysis of unique genes that were only differentially expressed in Kx2 models. However, those very few significantly enriched GO terms were all included in Kx1-Kx2-overlapping gene enrichment analysis (data not shown), indicating that cancer-associated H. pylori (Kx2) infection did not cause additional changes of biological processes or cellular compartments as compared with ChAG-associated H. pylori (Kx1) infection. Thus, we speculated that H. pylori-induced carcinogenesis is due to the cumulative effects of chronic stress response in gastric cells.
H. pylori Infection Up-Regulates UPRmt Genes in Mouse Gastric Epithelial Cells
Impaired expression of genes associated with mitochondrial activity or organelle architecture has been shown to trigger mitochondrial-to-nuclear signaling and increase UPRmt gene expression, including mitochondrial enzymes, chaperons, proteases, as well as transcription factors, to maintain protein quality and mitochondrial function [9], [17]. To assess whether down-regulated functional mitochondrial genes (Figure S2) induces UPRmt signaling, we examined the expression levels of UPRmt genes [18]. Surprisingly, most UPRmt genes were significantly were induced by H. pylori infection in mouse gastric cells. Except Hsp90ab1 (encodes HSP90) that was significantly decreased, Ddit3 (encodes transcriptional factor CHOP), Cebpb (encodes transcriptional factor C/EBP), Hmox1 (encodes heat shock protein HSP32), Lonp1, Hspa9 (encodes mitochondrial HSP70), Clpp (encodes subunit of protease Clpp), and DNAJA3 (encodes mitochondrial HSP40) were significantly elevated by H. pylori infection in both Kx1 and Kx2 models (Figure 1D). Thus, our results strongly indicated that UPRmt signaling was induced in mouse gastric epithelial cells upon H. pylori infection.
H. pylori Infection Significantly Induces Lonp1 Expression in Human Gastric Epithelial Cells
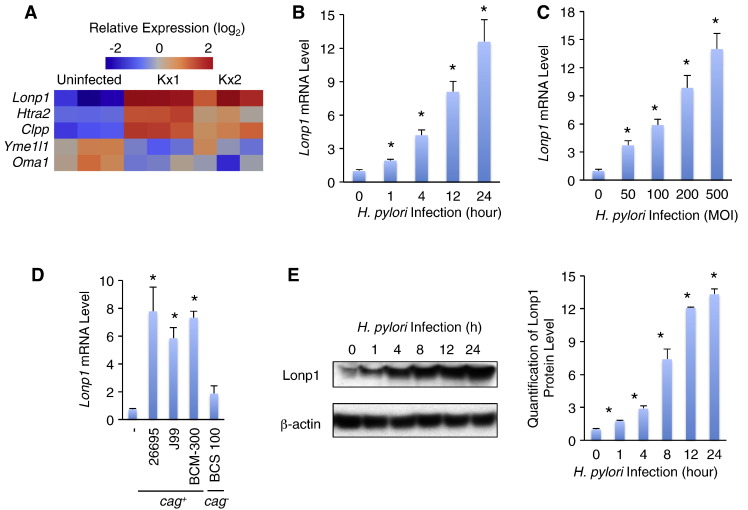
Mitochondrial proteases are important UPRmt indicators and are essential for mitochondrial maintenance and cell survival in response to exogenous stresses [18]. We thus measured the expression of mitochondrial protease genes in the context of H. pylori infection. Yme1l1 (encodes YME1L1) and Oma1 (encodes OMA1) were not significantly affected in both Kx1 and Kx2 infection models. Htra2 (encodes HTRA2 peptidase) was only significantly induced in Kx1 but not Kx2 model (Figure 2A). Only Lonp1 and Clpp were significantly increased in both infection models. Noted that, Lonp1 was with the greatest induction, 9.9 and 7.6 folds in Kx1 and Kx2 infection models, respectively (Figure 2A). Thus, we speculated that, as an important regulator of mitochondrial protection and cell proliferation [14], [15], Lonp1 induction might contribute to H. pylori-induced gastric carcinogenesis.
Figure 2.
Lonp1 expression is induced by H. pylori infection in human gastric epithelial cells. (A) Heatmap showing the expression pattern of mitochondrial proteases in Kx1 and Kx2 H. pylori infection models. (B) Lonp1 mRNA levels in MKN28 gastric cells were indicated by qPCR in response to H. pylori infection (MOI = 50, NCTC 11637) at different time points. (C) Lonp1 mRNA levels in MKN28 gastric cells were induced by different MOIs of H. pylori NCTC 11637 for 4 hours. (D) Lonp1 mRNA levels in MKN28 gastric cells were induced by different H. pylori strains (MOI = 50, 26695, J99, BCM-300, and BCS 100) for 12 hours. (E) Lonp1 proteins that were indicated by western blot (left) and quantified (right) in MKN28 gastric cells were elevated by H. pylori infection (MOI = 50, NCTC 11637) at different time points. Data represent the mean ± SEM from three separate experiments. * P < .05.
Since UPRmt regulation and H. pylori-associated gastric responses are similar across species [18], [19], we next confirmed whether Lonp1 is induced by H. pylori infection in human gastric epithelial cells in a similar manner. Confluent MKN28 cells were infected by H. pylori strain NCTC 11637 (MOI = 50) for 0, 1, 4, 12 and 24 hours. We found that Lonp1 mRNA level was increased by H. pylori infection by 1.9, 4.2, 8.1 and 12.6 folds compared to uninfected cells (Figure 2B). MKN28 cells were then infected with different MOIs of NCTC 11637 H. pylori for 12 hours and Lonp1 mRNA level was increased in a dose-dependent manner (Figure 2C). Importantly, in addition to Lonp1, other UPRmt genes, including transcriptional factors, enzymes, and chaperons, were also induced by H. pylori infection (MOI = 50, 12 hours) (Figure S3). We further treated MKN28 cells with different strains of H. pylori (MOI = 50, 12 hours), 26695, J99, BCM-300, as well as BCS 100. Consistently, similar to NCTC 11637, cag+H. pylori strains, 26695, J99, and BCM-300, potently induced Lonp1 mRNA expression (Figure 2D). However, cag− H. pylori strain BCS 100 failed to increase Lonp1 mRNA level (Figure 2D), suggesting that cag-encoding virulence factors, including CagA, are essential for Lonp1 induction. We next assessed Lonp1 protein level change and found that H. pylori infection significantly elevated Lopn1 protein levels, indicated by western blot, in MKN28 cells in a time-dependent manner (Figure 2E). Taken together, our results indicated that H. pylori infection increases Lonp1 expression in human gastric epithelial cells at both mRNA and protein levels.
Lonp1 Regulates Mitochondrial Function in Response to H. pylori Infection
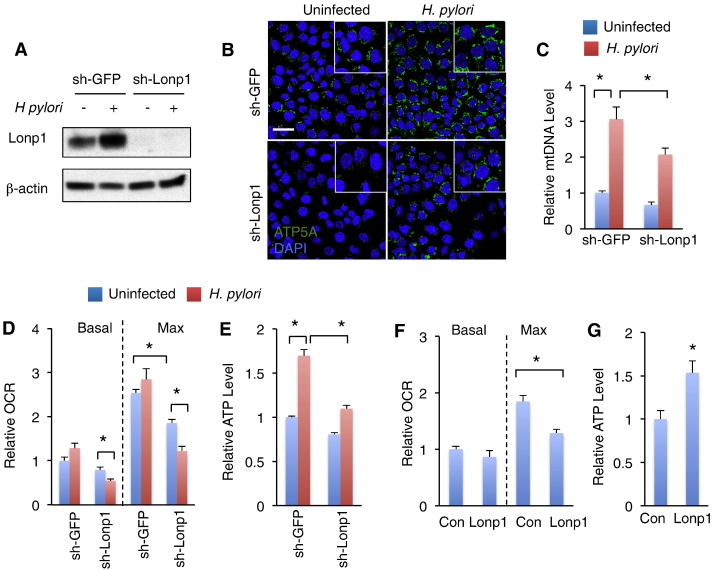
Previous studies have reported that H. pylori infection impairs host cell mitochondrial function via virulence factor production, including CagA and VacA [20]. We also accessed the mitochondrial regulation in response to H. pylori infection by measuring mitochondrial mass. Strikingly, low multiplicity of H. pylori infection resulted in a great increase in mitochondrial mass, including mitochondrial density and mitochondrial DNA level (Figure 3, B–C). Lonp1 has been reported to restore mitochondrial function under diverse stresses [14], [21]. To examine whether Lonp1 is required for mitochondrial mass increase in response to H. pylori infection, we knocked down Lonp1 expression in MKN28 cells using lentivrius-induced RNAi (sh-Lonp1). Compared to sh-GFP, Lonp1 expression was dramatically decreased at both mRNA and protein levels in sh-Lonp1 cells (Figures 3A and S5). Interestingly, Lonp1 deficiency significantly, but partially, diminished H. pylori-associated increase of mitochondrial intensity and mitochondrial DNA level compared to control (sh-GFP) (Figure 3, B–C).
Figure 3.
Induction of Lonp1 maintains mitochondrial function in response to low multiplicity of H. pylori infection. (A) Western blot indicating Lonp1 protein levels in MKN28 cells in response to low multiplicity of H. pylori infection (MOI = 50, NCTC 11637) and lentivrius-induced RNAi. (B-C) Mitochondrial mass, which was indicated by immunostaining of ATP5A (B), and mtDNA level (C) in MKN28 cells under low multiplicity of H. pylori infection for 24 hours (MOI = 50, NCTC 11637). (D-E) Basal and maximal mitochondrial oxygen rates (D) and cellular ATP levels (E) in control (sh-GFP) and Lonp1-deficient (sh-Lonp1) MKN28 cells in response to 24 hours of low multiplicity of H. pylori infection were measured. (F-G) Basal and maximal mitochondrial oxygen rates (F) and cellular ATP levels (G) in control (Con) and Lonp1-overexpressing (Lonp1) MKN28 cells. Data represent the mean ± SEM from three separate experiments. * P < .05.
Respiration rate and ATP production are two major features of mitochondrial activity. We next studied whether Lonp1 regulates mitochondrial respiration in response to H. pylori infection. However, even though high multiplicity of H. pylori infection has been shown to impair mitochondrial respiration, including oxygen consumption and electron transport [22], low multiplicity of H. pylori infection failed to suppress basal or maximal oxygen consumption rate in control MKN28 cells (sh-GFP) (Figure 3D). In Lonp1-deficient cells oxygen consumption was slight decreased without H. pylori infection (Figure 3D). Surprisingly, both basal and maximal oxygen consumption rate in Lonp1-deficient cells were further significantly decreased by 24 hours of low multiplicity of H. pylori infection (Figure 3D), indicating that Lonp1 induction is essential for mitochondrial respiration maintenance in response to H. pylori infection. We next clarified Lonp1 roles in mitochondrial respiration regulation by overexpressing Lonp1 in MKN28 cells and found that, however, maximal oxygen consumption rate in MKN28 cells was also significantly suppressed by Lonp1 overexpression (Figure 3F).
We also measured ATP production in gastric cells regarding H. pylori infection. Slightly different from mitochondrial respiration, ATP production in MKN28 cells was elevated by 24 hours of low multiplicity of H. pylori infection (MOI = 25, 50, 100), but decreased by high multiplicity of H. pylori infection (MOI = 500) (Figure S4A). Interestingly, Lonp1 deficiency mildly decreased ATP levels without H. pylori infection, however, significantly abolished ATP production induced by low multiplicity of H. pylori infection in MKN28 cells (Figure 3E). In addition, Lonp1 overexpression elevated ATP production by ~ 1.5-fold without H. pylori infection (Figure 3F). Collectively, our results indicated that Lonp1 plays a critical role in homeostatic regulation of mitochondrial activity in response to H. pylori infection.
Lonp1 Contributes to H. pylori-Induced Metabolic Switch to Glycolysis
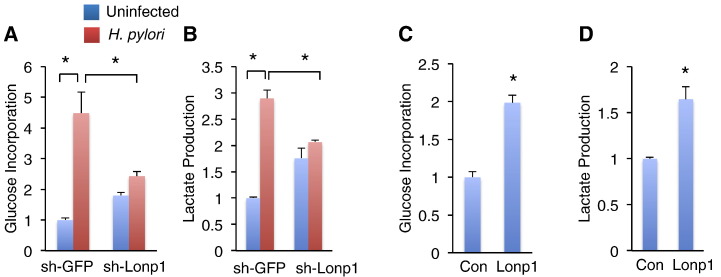
A metabolic switch towards glycolysis over mitochondrial respiration was observed in most solid tumor cells [23]. Our gene expression analysis in muscle gastric cells also indicated that major components involved in glycolysis were significantly increased in H. pylori infection models (Figure S2, right). We therefore hypothesized that H. pylori infection promotes metabolic glycolytic switch to influence gastric carcinogenesis. To address this hypothesis, we measured both glucose incorporation and lactate production, which are indicative of glycolysis [24], in MKN28 gastric epithelial cells. Low multiplicity of H. pylori infection for 24 hours dramatically promoted glucose incorporation and lactate production in sh-GFP MKN28 cells (Figure 4, A–B). Endogenous Lonp1 deficiency in MKN28 cells somehow slightly elevated glucose incorporation and lactate production without H. pylori infection (Figure 4, A–B). Strikingly, both glucose incorporation and lactate production associated with H. pylori infection were potently diminished in Lonp1-deficient cells (Figure 4, A–B), suggesting Lonp1 is essential for H. pylori-induced glycolytic shift. In contrast, Lonp1 overexpression in MKN28 cells significantly promoted both glucose incorporation and lactate production without H. pylori infection (Figure 4, C–D). Taken together, our results indicated that Lonp1 is required to enhance glycolytic switch in response to H. pylori infection.
Figure 4.
Low multiplicity of H. pylori infection results in glycolytic switch via Lonp1 induction. (A-B) Glucose incorporation (A) and lactate production (B) in control (sh-GFP) and Lonp1-deficient (sh-Lonp1) MKN28 cells in response to low multiplicity of H. pylori infection (MOI = 50, NCTC 11637) for 24 hours were indicated. (C-D) Glucose incorporation (C) and lactate production (D) in control (Con) and Lonp1-overexpressing (Lonp1) MKN28 cells were measured. Data represent the mean ± SEM from three separate experiments. * P < .05.
Lonp1 is Required for H. pylori-Induced Gastric Cell Proliferation
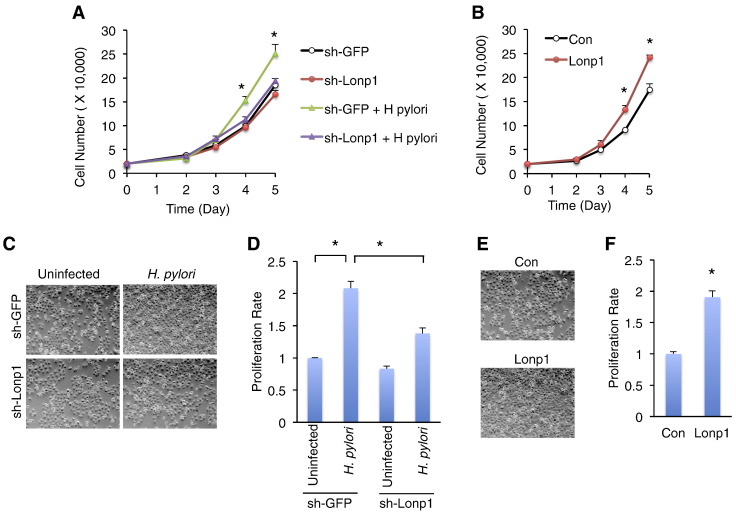
Metabolic switch toward glycolysis is highly associated with tumorigenesis/cell overproliferation [23]. We next examined whether Lonp1 contributes to gastric cell overgrowth in response to H. pylori infection. Previous studies indicated that low MOI (< 100) of H. pylori promotes, whereas high MOI (> 400) suppresses, cell proliferation [25], [26], [27]. To obtain an optimal gastric cell proliferative condition for H. pylori infection, we tested different MOIs of H. pylori using a 3-dimensional model system that provides interactions between cells, growth factors, and an extracellular matrix. Consistent with previous reports, we found that MOI = 50 of H. pylori, which potently promotes glycolysis, manifested a maximal proliferative effect on MKN28 gastric epithelial cells (Figure S4B). In order to examine whether Lonp1 is important for H. pylori-induced gastric cell proliferation, we measured proliferation rate in Lonp1-deficient cells in the context of low multiplicity of H. pylori infection (MOI = 50). Strikingly, Lonp1 deficiency significantly diminished the increase in both cell growth and proliferation rate associated with H. pylori infection (Figure 5A, C–D). On the other hand, we tested whether elevated Lonp1 expression is sufficient to promote gastric epithelial cell proliferation. Consistent with the previous studies in cervical cancer cells and melanoma cells [14], [15], overexpression of Lonp1 significantly increased cell growth and proliferation rate of gastric epithelial cells without H. pylori infection (Figure 5B, E–F). Thus, our results demonstrated that low multiplicity of H. pylori infection results in gastric cell proliferation via Lonp1 function.
Figure 5.
Lonp1 is required for low multiplicity of H. pylori-induced gastric cell proliferation. (A-B) Cell growth curves of control (sh-GFP) and Lonp1-deficient (sh-Lonp1) MKN28 cells in response to low multiplicity of H. pylori infection (MOI = 50, NCTC 11637) (A) and Lonp1-overexpressing MKN28 cells (B). (C-D) Images (C) and proliferation rates indicated by MTT assays (D) of control (sh-GFP) and Lonp1-deficient (sh-Lonp1) MKN28 cells in response to low multiplicity of H. pylori infection at day 5 (MOI = 50, NCTC 11637). (E-F) Images (E) and proliferation rates indicated by MTT assays (F) of control (Con) and Lonp1-overexpressing (Lonp1) MKN28 cells. Each group was compared with sh-GFP (A) or Con (B) at each time point in growth curve experiment. Data represent the mean ± SEM from three separate experiments. * P < .05.
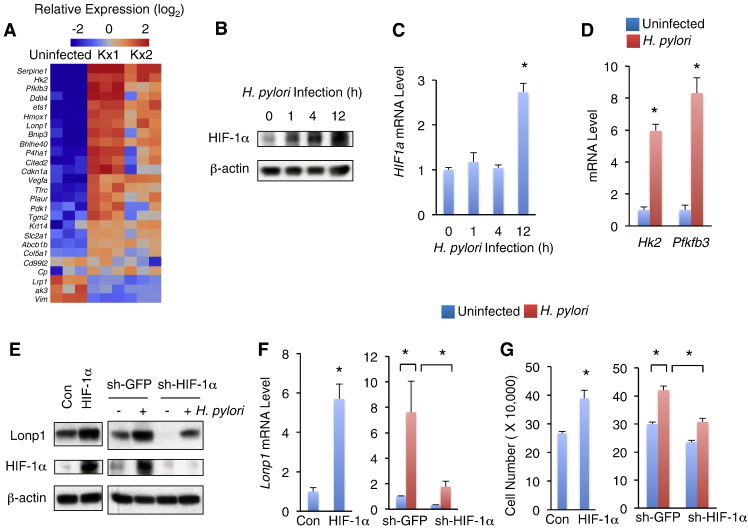
HIF-1α Regulates Lonp1 Expression in Response to H. pylori Infection
We next assessed the molecular mechanism(s) by which Lonp1 expression is induced by H. pylori infection. HIF-1α has been shown to directly target Lonp1 promoter and regulate Lonp1 expression in HeLa cells under hypoxia conditions [28]. The activity and protein level of HIF-1α are increased in gastric epithelial cells upon H. pylori infection [7]. In addition, mRNA profiling from H. pylori infected mouse gastric cells also indicated that well-known HIF-1α target genes [29] were up-regulated in both Kx1 and Kx2 models (Figure 6A). Thus, we speculated that HIF-1α might regulate Lonp1 expression upon H. pylori infection. To address this hypothesis, we first examined the HIF-1α expression in response to H. pylori infection and found that low multiplicity of H. pylori infection elevated HIF-1α protein level at 1 hour and in a time-dependent manner (Figure 6B). However, in contract to protein level, low multiplicity of H. pylori infection failed to elevated HIF1a mRNA level at early time points (1 hour and 4 hours) (Figure 6C), suggesting that H. pylori infection elevates HIF-1α expression, at least, via post-transcriptional regulation. Consistent to HIF-1α protein level, two HIF-1α target genes, Hk2 and Pfkfb3, were dramatically up-regulated in MKN28 gastric cells in response to H. pylori infection (Figure 6D).
Figure 6.
HIF-1α contributes to low MOI of H. pylori-induced Lonp1 induction and gastric cell proliferation. (A) Heatmap showing differential expression of known HIF-1α targets in Kx1 and Kx2 H. pylori infection models. (B-C) HIF-1α protein levels indicated by western blot (B) and HIF1a mRNA levels indicated by qPCR (C) in MKN28 gastric cells in response to low multiplicity of H. pylori infection at different time points (MOI = 50, NCTC 11637). (D) Hk2 and Pfkfb3, two HIF-1α targets, in MKN28 cells were transcriptionally elevated by low multiplicity of H. pylori infection (MOI = 50, NCTC 11637) for 12 hours. (E-F) Lonp1 protein levels (E) and mRNA levels (F) in HIF-1α-overexpressing MKN 28 cells (left) or HIF-1α-knockdown MKN28 cells in response to low multiplicity of H. pylori infection for 12 hours (MOI = 50, NCTC 11637) (right). (G) Cell growth of control and HIF-1α-overexpressing MKN28 gastric cells (left) or HIF-1α-knockdown cells in response to low multiplicity of H. pylori infection (right). Data represent the mean ± SEM from three separate experiments. * P < .05.
Then we clarified the HIF-1α role in Lonp1 transcriptional regulation in gastric epithelial cells by overexpressing HIF-1α. Interestingly, lenti-viral-induced HIF-1α overexpression significantly induced Lonp1 expression in MKN28 cells at both mRNA and protein levels (Figure 6, E–F, left). To confirm whether HIF-1α directly regulates Lonp1 transcription in gastric cells, we performed luciferase report assay by transfecting luciferase reporter plasmids that contained Lonp1 promoter regions into HIF-1α overexpressing or control MKN28 gastric cells. We found that HIF-1α dramatically enhanced Lonp1 promoter activity (Figure S6), suggesting HIF-1α directly regulates Lonp1 transcription. Similar to Lonp1 proliferative roles, HIF-1α overexpression also promoted MKN28 gastric epithelial cell growth (Figure 6G, left).
We therefore assessed whether HIF-1α is essential for Lonp1 expression and gastric epithelial cell proliferation in response to H. pylori infection by knocking down HIF-1α expression (Figure 6E). Strikingly, H. pylori-induced Lonp1 expression at both mRNA and protein levels were dramatically, but not completely, diminished in HIF-1α knockdown cells (Figure 6, E–F). In addition, HIF-1α knockdown also significantly alleviated H. pylori-induced gastric cell proliferation (Figure 6G, right). Taken together, our results demonstrated that HIF-1α is required for Lonp1 expression and cell proliferation in response to H. pylori infection.
Discussion
Differential gene expression has been shown to link to various biological processes, including aging, stem cell proliferation, metabolism and tumorigenesis, across species [30], [31], [32], [33]. Using bioinformatics method to unify the representation of gene-related biological processes and molecular regulation will help us systemically understand molecular modulation under diverse physiological conditions. The mechanism how H. pylori infection results in gastric carcinogenesis is still not fully understood. Thus, in this study, to get a comprehensive understanding of H. pylori-induced molecular change regarding gastric carcinogenesis, we analyzed transcriptional prolife in a H. pylori-infection gastric cancer model. Even though the dataset has been published and Gordon and his colleagues have hypothesized that some unique genes, which were induced only in cancer-associated (Kx2) H. pylori-infected mouse gastric cells, might contribute to gastric carcinogenesis [16], we surprisingly found that most of deregulated genes in Kx2 infected cells were also included in deregulated genes in ChAG-associated (Kx1) infected cells (Figure 1, A–B). Our GO enrichment analysis further indicated that no additional GO terms were significantly enriched in Kx2-assoicated gene expression compared to Kx1, suggesting cancer-associated H. pylori infection failed to cause additional changes in biological process or cellular compartment as compared with ChAG-associated H. pylori. Thus, we hypothesized that H. pylori-induced gastric carcinogenesis relies on the cumulative effects of chronic stress response in gastric cells. Then we focused on the overlapping deregulated genes in both Kx1 and Kx2 models and uncovered that genes associated to UPRmt, glycolysis, as well as HIF-1α targets, were significantly increased in mouse gastric epithelial cells. The similar regulation was further confirmed in human gastric cells in response to H. pylori infection and then shed the light on our molecular mechanism investigation.
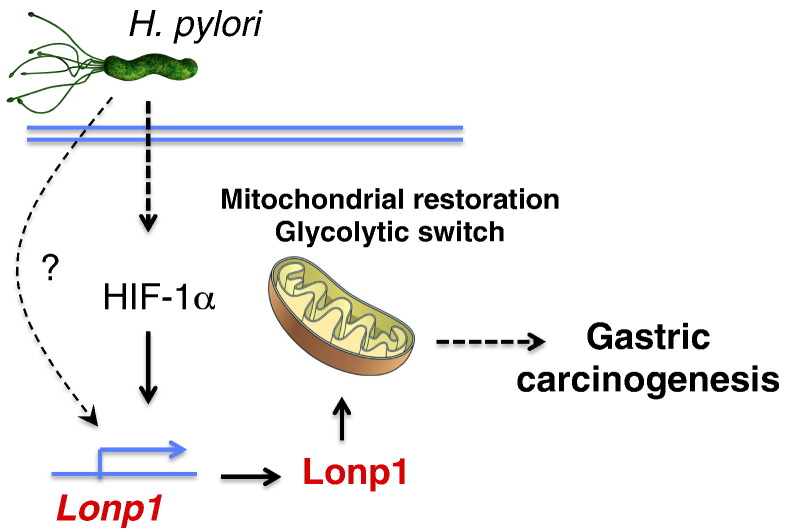
H. pylori infection has been shown to cause mitochondrial dysfunction or perturbation in gastric epithelial cells in a dose-dependent manner [34], [35]. Although severe mitochondrial dysfunction is detrimental, the salutary effects of mild mitochondrial perturbation have been reported in multiple organisms [17], [36], [37], [38]. Consistent with this notion, we observed that low MOI of H. pylori promotes, but high MOI of H. pylori suppresses, gastric cell proliferation, suggesting the homeostatic interaction of mitochondrial injury and compensatory protection effects. Growing evidence suggested that UPRmt signaling networks, which induce expression of chaperons and proteases to maintain mitochondrial quality and function, could be a contributing factor. For example, mitochondrial heat shock proteins, HSP60/70/90, and protease, Clpp & Lonp1, have been implicated in cancer cell protection from stresses [9], [14], [39], [40], [41], [42]. The compensatory UPRmt signaling might provide novel insights into the molecular mechanisms of gastric cell overproliferation associated with low multiplicity of H. pylori infection. In this study, we first characterized that mitochondrial protease Lonp1 is induced by H. pylori infection in gastric epithelial cells in a dose- and time-dependent manner. Furthermore, consistent with previous reports of Lonp1 role in mitochondrial maintenance and glycolysis switch [14], [15], we demonstrated that Lonp1 is essential for mitochondrial restoration, metabolic shift to glycolysis and gastric cell overproliferation associated with low multiplicity of H. pylori infection (Figure 7). Our findings manifested the novel molecular mechanisms by which H. pylori infection promotes gastric carcinogenesis and offered potential therapeutic opportunities for H. pylori-induced gastric cancer by targeting Lonp1.
Figure 7.
Schematic of molecular mechanism of Lonp1 induction and gastric carcinogenesis in response to H. pylori infection. Low multiplicity of H. pylori infection increases Lonp1 expression in gastric cells via HIF-1α regulation and other unknown mechanisms. Subsequently, excess Lonp1 results in mitochondrial restoration and metabolic switched toward glycolysis and contribute to gastric cell overproliferation and gastric carcinogenesis.
UPRmt signaling as well as Lonp1 function have been shown to contribute to delicate modulation of mitochondrial activity and hemostatic transition between oxidative and glycolytic metabolism [14], [17]. In this study, we have found that both overexpression and knockdown of Lonp1 promote glycolytic switch, including glucose incorporation and lactate production, in gastric epithelial cells (Figure 4). The similar regulation has been coincidently observed in López-Otín’s work [14]. Their results demonstrated that Lonp1 overexpression triggers metabolic reprogramming and promotes cellular glycolytic switch via gene expression regulation. Meanwhile, mitochondrial dysfunction caused by genetic or pharmaceutical manipulation results in a compensatory increase in glycolysis to maintain energy homeostasis [43], [44], [45]. We’ve exhibited that Lonp1 knockdown decreased mitochondrial mass and oxygen consumption in gastric cells, thus we speculated that the glycolytic increase in Lonp1 knockdown cells could be an obligate consequence of the mitochondrial dysfunction.
HIF-1α is an important hypoxia responsive factor and is found to enhance Lonp1 expression in HeLa cells [28]. H. pylori infection also stabilizes HIF-1α and increases its activity in gastric cells [7]. In this study, we confirmed that HIF-1α enhances Lonp1 expression in response to H. pylori infection, as HIF-1α overexpression potently increased Lonp1 expression, whereas HIF-1α knockdown significantly diminished Lonp1 induction associated with H. pylori infection. However, we observed that knockdown of HIF-1α failed to completely abolish Lonp1 induction in response to H. pylori infection, suggesting other unknown regulators are also involved in Lonp1 induction. UPRmt transcriptional factor CHOP/C/EBPβ is not likely to regulate Lonp1 expression, as its cognate binding elements are absent in Lonp1 promoter region [46]. Interestingly, Lonp1 expression is robustly induced by PERK signaling in response to ER stress [47]. Since transmission of cell stress from ER to mitochondria is largely unknown in H. pylori-induced gastric carcinogenesis, it is worth studying how PERK is involved in Lonp1 induction in response to H. pylori infection in our future study.
In addition to Lonp1, other UPRmt responsive genes were also significantly induced in H. pylori infected gastric epithelial cells (Figures 1D and S3), indicating these genes and their associated biological processes might be involved in H. pylori-induced gastric carcinogenesis. Didt3 and Cebpb encode CHOP and C/EBPβ, respectively, to form hetero-dimers and activate transcription of UPRmt responsive genes [46]. Hmox1 encodes a heme oxygenase and exhibits anti-oxidation effects [48]. Hspa9 and DNAJA3 have been shown to encode HSP70 and HSP40, respectively, and Clpp encodes a mitochondrial protease to maintain mitochondrial proteostasis and activity [9]. Thus, in the future we will characterize the roles of each of these UPRmt responsive genes in H. pylori-induced gastric cell proliferation and uncover more potential mechanism(s) for gastric carcinogenesis.
Material and Methods
Bioinformatics Analysis of Microarray Data
Microarray data was obtained from NCBI-GEO (http://www.ncbi.nlm.nih.gov/geo/) (GSE10262). Expression levels of probes were calculated using RMA normalization and mapped in CDF version Affymetrix Mouse 430_2. Genes with fold change > 1.5, false discovery rate (FDR) < 0.05 were considered as significant. All comparisons were made between infected and uninfected. The significantly up- and down-regulated genes were uploaded separately to DAVID Bioinformatics Resource (https://david.ncifcrf.gov). The mouse genome 430 2.0 was used as a background for the GO analysis. The GO terms with EASE score < 0.05 were selected for interpretation [49].
Cell Culture, Bacteria Culture and Reagents
MKN28 human gastric epithelial cell line (JCRB Cell Bank, JCRB0253) was a kind gift from Cyril Benes. MKN28 cells were cultured in growth medium, RPMI-1640 medium containing 10% FBS (Invitrogen) and 100 U/mL penicillin and 100 μg/mL streptomycin, at 37°C with 100% humidity and 5% CO2. H. pylori strains NCTC 11637 (ATCC 43504), 26695 (ATCC 700392), J99 (ATCC 700824), BCM-300 (ATCC BAA-1606) and BCS 100 (ATCC BAA-945) were obtained from ATCC and were grown in Brucella broth with 5% FBS at 37°C. For H. pylori infection assays, bacteria cells were harvested by centrifugation at 8000 rpm for 5 minutes and added to gastric cells at multiplicity of infection (MOI) of bacteria to cell from 25:1 to 500:1. Lonp1 polyclonal antibody (HPA002192), β-actin monoclonal antibody (A5441), and pLKO.1-puromysin shRNA lentiviral vectors for Lonp1 (TRCN0000291803) and HIF-1α (TRCN0000003810) were purchased from Sigma. ATP5A monoclonal antibody used for tracking mitochondria (ab14748) was obtained from Abcam. HIF-1α antibody was purchased from Novus Biologicals (NB100-105). Lentiviral overexpression vector pLJM1-EGFP (19319) and pcDNA3 vector containing full-length HIF-1α (18949) were obtained from Addgene. pCMV6-XL4 vector containing full-length Lonp1 (SC117111) was purchased from Origene.
Lentivirus-Induced RNAi and Overexpression
HIF-1α and Lonp1 were subcloned from commercial vectors containing full-length cDNAs into lentiviral pLJM1-EGFP vector. pLKO.1-puromysin vectors containing shRNAs and pLJM1-EGFP-puromysin overexpression vectors were used for knocking down and overexpressing target genes (Lonp1 and HIF-1α), respectively. Lentivrius packaging was performed as previously described [50]. For lentivrius transduction, MKN28 cells at 50% confluence were incubated with packed viruses in growth medium containing 8 μg/mL Polybrene (Sigma) overnight. Infected cells were cultured for 48 hours and then selected with 3 μg/mL puromycin for 72 hours to generate stable cells. Knockdown efficiency and overexpression was further confirmed with western blot.
Real-Time Quantitative PCR
Cells were washed three times with PBS and immediately homogenized in Trizol (Invitrogen) and processed through alcohol precipitation according to the manufacturer’s instructions. RNAs were dissolved in nucleas-free water (Ambion) and were quantified using a NanoDrop spectrophotometer. cDNA was synthesized from 1 μg of total RNA using iScript cDNA Synthesis Kit (BioRad). qPCR was performed in 20 μL reaction that contains 10 μL iQ™ SYBR Green Supermix (2X), 250 nM forward or reverse primer and 10 ng cDNA. The following program was run in CFX96 Real-time PCR system (Bio-Rad):
| Polymerase activation and DNA denaturation at 95°C | Amplification |
Melt curve analysis 55-95°C 0.5°C increment |
||
|---|---|---|---|---|
| Denaturation at 95 °C | Annealing/extension + plate read at 60°C | Cycles | ||
| 3 min | 15 s | 30 s | 40 | 3 s/step |
Relative mRNA level (target gene mRNA/Gapdh mRNA) was calculated by a comparative Ct method, using the following equation: target gene mRNA/Gapdh mRNA = 2− ΔCt, where ΔCt=Cttarget gene − CtGapdh. The following primers for each gene with 90% to 110% efficiencies were used:
Lonp1-forward: CCTGACTGCAGAGATCGTGA
Lonp1-reverse: CCCATGTCGCTCAGGTAGAT
Ddit3-forward: CAGAACCAGCAGAGGTCACA
Ddit3-reverse: AGCTGTGCCACTTTCCTTTC
Cebpb-forward: CTCGCAGGTCAAGAGCAAG
Cebpb-reverse: GACAGCTGCTCCACCTTCTT
Hmox1-forward: CTCAAACCTCCAAAAGCC
Hmox1-reverse: TCAAAAACCACCCCAACCC
Hspa9-forward: GGACTATCGCTCCATGCCAA
Hspa9-reverse: CTTTACTTGGGGCTCTGCCA
Clpp-forward: CTCATTCCCATCGTGGTGGA
Clpp-reverse: GATAACAAGGCTGGCAACGC
DNAJA3-forward: GGTGTCAGCCTTACAGGAAGAT
DNAJA3-reverse: ACCCCCTTTGCAGCTTGATT
Htra2-forward: AGAGTTTCTGCATCGTGGGG
Htra2-reverse: AGACCAGCCCTGGGACTC
Hsp90ab1-forward: ATGGAAGAGAGCAAGGCAAA
Hsp90ab1-reverse: GCAGCAAGGTGAAGACACAA
Hif1a-forward: CTGACCCTGCACTCAATCAA
Hif1a-reverse: TCCATCGGAAGGACTAGGTG
Hk2-forward: ACCCACATTCGAGTTGGAAG
Hk2-reverse: CAGTGGGTTGTCATGAGTGG
Pfkfb3-forward: GACAAATGCGACAGGGACTT
Pfkfb3-reverse: TAGTACACGATGCGGCTCTG
Gapdh-forward: CAGCCTCAAGATCATCAGCA
Gapdh-reverse: TGTGGTCATGAGTCCTTCCA
Luciferase Assay
LONP1 promoter sequences were amplified from genomic DNA of MKN28 cells by PCR using the following primers with MluI and BglII restriction sites, digested, and ligated into pGL3-Basic luciferase report vector (Promega), which encodes firefly luciferase. HIF-1α stable or control cells were transfected with firefly luciferase reporter plasmid (200 ng) and control reporter plasmid pAct-Renilla (40 ng), which encodes Renilla luciferase, using Effectene (Qiagen). The ratio of firefly to Renilla luciferase activity was determined using the Dual-Luciferase Assay System (Promega).
− 435/− 415-Forward: GCTCTTACGCGTGGGAGTCGCTGCACAATCCGA
− 338/− 318-Forward: GCTCTTACGCGTCTCCGCCTGAATCTTGAGCAT
+ 1/+ 22-Forward: ATGCTCTTACGCGTAGCTCCCTGAAGCGGCTGTTTC
+ 71/+ 92-Reverse: TTACTTAGATCTTCATTTCCGCTCGCCGCGAAAC
Western Blot
Cells were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% Na-deoxycholate) containing protease inhibitor and phosphatase inhibitor cocktails (ThermoFisher). 40 μg total proteins were separated by SDS-PAGE gel electrophoresis and transferred to PVDF membranes. Total protein levels were assessed by western blot using primary antibodies and HRP-conjugated secondary (1:3000) antibodies.
Immunostaining
Cells were planted and infected with or without H. pylori in glass chamber slides and were washed three times with PBS. Then cells were fiexed with 4% paraformaldehyde in PBS for 30 minutes and washed with PBS containing 0.2% Triton-X (PBST). After being blocked with PBST containing 1% BSA, cells were immunostained with monoclonal anti-ATP5A antibody (1:500) overnight at 4°C. Slides were washed and incubated with AlexaFluor 488-conjugated secondary antibody (1:500, Invitrogen) and DAPI (1:5000) for 1 hour at room temperature and mounted for confocal imaging.
mtDNA Analysis
Similar to qPCR, mtDNA was also qualified using iQ™ SYBR Green Supermix with CFX96 Real-time PCR system (Bio-Rad). Total DNA was used as a template and relative mtDNA copy numbers were assessed after mitochondrial gene Nd1 normalization by the single-copy nuclear gene Pk. Relative mtDNA copy number (mtDNA amount/nDNA amount) was calculated by a comparative Ct method, using the following equation: mtDNA/nDNA=2− ΔCt, where ΔCt=Ctmitochondrial− Ctnuclear. The primers with 90% to 110% efficiencies are listed like below.
Nd1-forward: CCCTAAAACCCGCCACATCT
Nd1-reverse: GAGCGATGGTGAGAGCTAAGGT
Pk-forward: AGCCCAAATGGCCTTGAA
Pk-reverse: AGAGACAGAATGCCAGTGAGC
Lactate, Glucose and ATP Measurements
Cells seeded in triplicated in 24-well plates were infected with or without H. pylori for 24 hours. After that, the cells were washed and lysed with passive lysis buffer (Promega). The lysates were centrifuged at 12,000 rpm at 4°C for 5min, and supernatants were collected. The protein concentration of the supernatant was measured with Bradford kit (Sigma). The ATP level of supernatant was determined using the ATP determination kit (Invitrogen) according to the instructions in a SpectraMax plate reader (Molecular Devices) and normalized to protein levels. The media were collected and the content of lactate was determined using lactate assay kit (Sigma) and normalized to cell protein levels. As for glucose incorporation, the cells were infected with or without H. pylori for 24 hours in growth medium containing 1 μCi/mL 2-deoxy-D-[3H] glucose (PerkinElmer). Then, the cells were washed and lysed in 0.4 mL 1% Na-dodecyl sulfate. Intracellular [3H] glucose content was determined in 4 mL scintillant using a Beckman scintillation counter and normalized to cell protein level.
Mitochondrial Oxygen Consumption Analysis
After H. pylori infection for 24 hours, mitochondrial oxygen consumption in intact cells was measured using a Seahorse Bioscience extracellular flux analyzer according to manufacturer’s instructions. Briefly, after 15 minutes equilibration, three measurements of 3 minutes were performed, separated by 3 minutes of mixing. Maximal membrane potential was assessed by adding 1 μM oligomycin, and uncoupled mitochondrial respiration was induced with 1 μM CCCP. Both 1 μM rotenone and antimycin were used to stop the mitochondrial oxygen consumption.
Cell Proliferation Assay
Tissue culture plates were coated with Matrigel™ Matrix (BD) containing growth factors at 37°C for 1 hour. Then, 20,000 cells were planted per well on Matrigel™ matrix and 12 hours later were infected with or without H. pylori. After trypsinization, cells were re-suspended in an equal volume of growth medium containing 0.05% trypan blue. The number of live cells in triplicated wells that excluded trypan blue was counted for 5 days to generate cellular growth curve. MTT assay was also used for measuring cell proliferation rate. Briefly, after 5-day cell growth with or without H. pylori infection, cells were incubated with 1 mg/mL MTT (Roche) in growth medium for 2 hours at 37°C and then with extraction buffer (20% SDS, 50% dimethylformamide) overnight at 37°C. Absorbance was measured at 590 nm in SpectraMax plate reader and the proliferation rate was normalized to control cells.
Statistical Analysis
All experimental results are indicated as mean ± error bars represent SEM. Unpaired Student’s t test and one-way ANOVA followed by post-hoc test were performed to assess the differences. P < .05 was considered statistically significant and shown with asterisks.
Author Contributions
MH. P. and K. W. conceived and design the experiments. B. L., MG. W., NY. H. and X. H. performed most of the biochemical and cell proliferation experiments. GQ. J. and XP. Q. performed immunostaining and confocal imaging. XF. Z., Y. L. and K. L. performed cloning and H. pylori culture. W. S. performed bioinformatics analysis of microarray data. MH. P. and K. W. discussed the results and wrote the manuscript.
The following are the supplementary data related to this article.
GO analysis of significantly deregulated genes in Kx1 and Kx2 models. Significantly enriched (EASE score < 0.05) GO terms were identified in both models (A, B, biological process; C, cellular compartment).
Mitochondrial and glycolytic gene expression in Kx1 and Kx2 models. Heatmap showing the genes that encode components of ETC., TCA cycle, beta-oxidation, as well as glycolysis, were differentially deregulated in H. pylori infection model.
UPRmt genes were induced by H. pylori infection in human gastric cells. mRNA levels indicated by qPCR of UPRmt genes in MKN28 gastric cells in response to H. pylori infection (MOI = 50, NCTC 11637) for 12 hours. Data represent the mean ± SEM from three separate experiments. * P < .05.
Low multiplicity of H. pylori infection induces ATP production and cell proliferation. (A) Cellular ATP levels in MKN28 gastric cells under different MOIs of H. pylori for 24 hours (NCTC 11637). (B) 20,000 MKN28 cells were planted in pre-coated plates and then infected by different MOIs of H. pylori for 5 days (NCTC 11637). Proliferation rate was indicated by MTT assay. Data represent the mean ± SEM from three separate experiments. * P < .05.
Knockdown of Lonp1 expression in human gastric cells. MKN28 gastric cells were infected with lenti-viruses (sh-GFP and sh-Lonp1) for 48 hours and selected with puromycin for another 72 hours. Lonp1 mRNA level (A) and protein level (B) were analyzed with qPCR and western blot, respectively. Data represent the mean ± SEM from three separate experiments. * P < .05.
HIF-1α directly regulates Lonp1 transcription. (A) Predicted HIF-1α binding sites in Lonp1 promoter region. (B) pAct-Renilla and firefly luciferase reporter pGL2 plasmids with indicated Lonp1 promoter sequences containing predicted HIF-1α binding sites were transfected into control (Con) and HIF-1α overexpression (HIF-1α) MKN28 cells. Transfected cells were cultured for 48 hours and lysed to measure firefly: Renilla luciferase activity.
Acknowledgement
We thank Dr. Cyril Benes for providing gastric cancer cells. This work was supported by the Key Projects in Sichuan Provincial Scientific Technology (2011SZ0332,2013JY0104) and the Sichuan Provincial health Department Grant (110167,100481).
Contributor Information
Kang Wang, Email: pangmh2000@163.com.
Minghui Pang, Email: pangshattuck@126.com.
References
- 1.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Anwar WA, B.K., Correa P, Forman D, Gentile JM. Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 4.Wroblewski LE, Peek RM, Jr., Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamaoka Y, Graham DY. Helicobacter pylori virulence and cancer pathogenesis. Future Oncol. 2014;10:1487–1500. doi: 10.2217/fon.14.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sepulveda AR. Helicobacter, Inflammation, and Gastric Cancer. Curr Pathobiol Rep. 2013;1:9–18. doi: 10.1007/s40139-013-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharyya A, Chattopadhyay R, Hall EH, Mebrahtu ST, Ernst PB, Crowe SE. Mechanism of hypoxia-inducible factor 1 alpha-mediated Mcl1 regulation in Helicobacter pylori-infected human gastric epithelium. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1177–G1186. doi: 10.1152/ajpgi.00372.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamb A, Chen LF. The many roads traveled by Helicobacter pylori to NFkappaB activation. Gut Microbes. 2010;1:109–113. doi: 10.4161/gmic.1.2.11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochim Biophys Acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bezawork-Geleta A, Brodie EJ, Dougan DA, Truscott KN. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep. 2015;5:17397. doi: 10.1038/srep17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochim Biophys Acta. 2012;1823:56–66. doi: 10.1016/j.bbamcr.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA + Lon protease. Mol Cell. 2013;49:121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Q, Li T, Hou W, Zheng J, Schrum LW, Bonkovsky HL. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J Biol Chem. 2011;286:26424–26430. doi: 10.1074/jbc.M110.215772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quiros PM, Espanol Y, Acin-Perez R, Rodriguez F, Barcena C, Watanabe K, Calvo E, Loureiro M, Fernandez-Garcia MS, Fueyo A. ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 2014;8:542–556. doi: 10.1016/j.celrep.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Nie X, Li M, Lu B, Zhang Y, Lan L, Chen L, Lu J. Down-regulating overexpressed human Lon in cervical cancer suppresses cell proliferation and bioenergetics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannakis M, Chen SL, Karam SM, Engstrand L, Gordon JI. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc Natl Acad Sci U S A. 2008;105:4358–4363. doi: 10.1073/pnas.0800668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 19.Peek RM. Helicobacter pylori infection and disease: from humans to animal models. Dis Model Mech. 2008;1:50–55. doi: 10.1242/dmm.000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvino Fernandez M, Parra Cid T. H. pylori and mitochondrial changes in epithelial cells. The role of oxidative stress. Rev Esp Enferm Dig. 2010;102:41–50. doi: 10.4321/s1130-01082010000100006. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B, Shen XL, Liang R, Li Y, Huang K, Zhao C, Luo Y, Xu W. Protective role of the mitochondrial Lon protease 1 in ochratoxin A-induced cytotoxicity in HEK293 cells. J Proteomics. 2014;101:154–168. doi: 10.1016/j.jprot.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Chen M, Andersen LP, Zhai L, Kharazmi A. Characterization of the respiratory chain of Helicobacter pylori. FEMS Immunol Med Microbiol. 1999;24:169–174. doi: 10.1111/j.1574-695X.1999.tb01278.x. [DOI] [PubMed] [Google Scholar]
- 23.Jang M, Kim SS, Lee J. Cancer cell metabolism: implications for therapeutic targets. Exp Mol Med. 2013;45 doi: 10.1038/emm.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bussiere FI, Chaturvedi R, Asim M, Hoek KL, Cheng Y, Gainor J, Scholz A, Khan WN, Wilson KT. Low multiplicity of infection of Helicobacter pylori suppresses apoptosis of B lymphocytes. Cancer Res. 2006;66:6834–6842. doi: 10.1158/0008-5472.CAN-05-4197. [DOI] [PubMed] [Google Scholar]
- 26.Li N, Han L, Chen J, Lin X, Chen H, She F. Proliferative and apoptotic effects of gastric epithelial cells induced by coccoid Helicobacter pylori. J Basic Microbiol. 2013;53:147–155. doi: 10.1002/jobm.201100370. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Sun Y, Liu S, Yu H, Li W, Zeng J, Chen C, Jia J. Upregulation of progranulin by Helicobacter pylori in human gastric epithelial cells via p38MAPK and MEK1/2 signaling pathway: role in epithelial cell proliferation and migration. FEMS Immunol Med Microbiol. 2011;63:82–92. doi: 10.1111/j.1574-695X.2011.00833.x. [DOI] [PubMed] [Google Scholar]
- 28.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 29.Liu W, Shen SM, Zhao XY, Chen GQ. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol. 2012;3:165–178. [PMC free article] [PubMed] [Google Scholar]
- 30.Amcheslavsky A, Song W, Li Q, Nie Y, Bragatto I, Ferrandon D, Perrimon N, Ip YT. Enteroendocrine cells support intestinal stem-cell-mediated homeostasis in Drosophila. Cell Rep. 2014;9:32–39. doi: 10.1016/j.celrep.2014.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song W, Ren D, Li W, Jiang L, Cho KW, Huang P, Fan C, Song Y, Liu Y, Rui L. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 2010;11:427–437. doi: 10.1016/j.cmet.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 34.Huang XW, Luo RH, Zhao Q, Shen ZZ, Huang LL, An XY, Zhao LJ, Wang J, Huang YZ. Helicobacter pylori induces mitochondrial DNA mutation and reactive oxygen species level in AGS cells. Int J Med Sci. 2011;8:56–67. doi: 10.7150/ijms.8.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machado AM, Desler C, Boggild S, Strickertsson JA, Friis-Hansen L, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection affects mitochondrial function and DNA repair, thus, mediating genetic instability in gastric cells. Mech Ageing Dev. 2013;134:460–466. doi: 10.1016/j.mad.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, Perrimon N. Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev Cell. 2015;33:36–46. doi: 10.1016/j.devcel.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, Mattson R, Hurren R, Babovic S, Maclean N. Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell. 2015;27:864–876. doi: 10.1016/j.ccell.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 41.Sherman MY, Gabai VL. Hsp70 in cancer: back to the future. Oncogene. 2015;34:4153–4161. doi: 10.1038/onc.2014.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 43.Hu Y, Lu W, Chen G, Wang P, Chen Z, Zhou Y, Ogasawara M, Trachootham D, Feng L, Pelicano H. K-ras(G12V) transformation leads to mitochondrial dysfunction and a metabolic switch from oxidative phosphorylation to glycolysis. Cell Res. 2012;22:399–412. doi: 10.1038/cr.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelicano H, Lu W, Zhou Y, Zhang W, Chen Z, Hu Y, Huang P. Mitochondrial dysfunction and reactive oxygen species imbalance promote breast cancer cell motility through a CXCL14-mediated mechanism. Cancer Res. 2009;69:2375–2383. doi: 10.1158/0008-5472.CAN-08-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rafikov R, Sun X, Rafikova O, Louise Meadows M, Desai AA, Khalpey Z, Yuan JX, Fineman JR, Black SM. Complex I dysfunction underlies the glycolytic switch in pulmonary hypertensive smooth muscle cells. Redox Biol. 2015;6:278–286. doi: 10.1016/j.redox.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hori O, Ichinoda F, Tamatani T, Yamaguchi A, Sato N, Ozawa K, Kitao Y, Miyazaki M, Harding HP, Ron D. Transmission of cell stress from endoplasmic reticulum to mitochondria: enhanced expression of Lon protease. J Cell Biol. 2002;157:1151–1160. doi: 10.1083/jcb.200108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida T, Maulik N, Ho YS, Alam J, Das DK. H(mox-1) constitutes an adaptive response to effect antioxidant cardioprotection: A study with transgenic mice heterozygous for targeted disruption of the Heme oxygenase-1 gene. Circulation. 2001;103:1695–1701. doi: 10.1161/01.cir.103.12.1695. [DOI] [PubMed] [Google Scholar]
- 49.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagy TA, Wroblewski LE, Wang D, Piazuelo MB, Delgado A, Romero-Gallo J, Noto J, Israel DA, Ogden SR, Correa P. beta-Catenin and p120 mediate PPARdelta-dependent proliferation induced by Helicobacter pylori in human and rodent epithelia. Gastroenterology. 2011;141:553–564. doi: 10.1053/j.gastro.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GO analysis of significantly deregulated genes in Kx1 and Kx2 models. Significantly enriched (EASE score < 0.05) GO terms were identified in both models (A, B, biological process; C, cellular compartment).
Mitochondrial and glycolytic gene expression in Kx1 and Kx2 models. Heatmap showing the genes that encode components of ETC., TCA cycle, beta-oxidation, as well as glycolysis, were differentially deregulated in H. pylori infection model.
UPRmt genes were induced by H. pylori infection in human gastric cells. mRNA levels indicated by qPCR of UPRmt genes in MKN28 gastric cells in response to H. pylori infection (MOI = 50, NCTC 11637) for 12 hours. Data represent the mean ± SEM from three separate experiments. * P < .05.
Low multiplicity of H. pylori infection induces ATP production and cell proliferation. (A) Cellular ATP levels in MKN28 gastric cells under different MOIs of H. pylori for 24 hours (NCTC 11637). (B) 20,000 MKN28 cells were planted in pre-coated plates and then infected by different MOIs of H. pylori for 5 days (NCTC 11637). Proliferation rate was indicated by MTT assay. Data represent the mean ± SEM from three separate experiments. * P < .05.
Knockdown of Lonp1 expression in human gastric cells. MKN28 gastric cells were infected with lenti-viruses (sh-GFP and sh-Lonp1) for 48 hours and selected with puromycin for another 72 hours. Lonp1 mRNA level (A) and protein level (B) were analyzed with qPCR and western blot, respectively. Data represent the mean ± SEM from three separate experiments. * P < .05.
HIF-1α directly regulates Lonp1 transcription. (A) Predicted HIF-1α binding sites in Lonp1 promoter region. (B) pAct-Renilla and firefly luciferase reporter pGL2 plasmids with indicated Lonp1 promoter sequences containing predicted HIF-1α binding sites were transfected into control (Con) and HIF-1α overexpression (HIF-1α) MKN28 cells. Transfected cells were cultured for 48 hours and lysed to measure firefly: Renilla luciferase activity.