Ghirlando and Felsenfeld review recent major advances in understanding the multiple roles of CTCF in gene regulation and genome organization and especially in how such domains are generated and organized.
Keywords: chromatin, insulators, topologically associated domains
Abstract
The role of the zinc finger protein CTCF in organizing the genome within the nucleus is now well established. Widely separated sites on DNA, occupied by both CTCF and the cohesin complex, make physical contacts that create large loop domains. Additional contacts between loci within those domains, often also mediated by CTCF, tend to be favored over contacts between loci in different domains. A large number of studies during the past 2 years have addressed the questions: How are these loops generated? What are the effects of disrupting them? Are there rules governing large-scale genome organization? It now appears that the strongest and evolutionarily most conserved of these CTCF interactions have specific rules for the orientation of the paired CTCF sites, implying the existence of a nonequilibrium mechanism of generation. Recent experiments that invert, delete, or inactivate one of a mating CTCF pair result in major changes in patterns of organization and gene expression in the surrounding regions. What remain to be determined are the detailed molecular mechanisms for re-establishing loop domains and maintaining them after replication and mitosis. As recently published data show, some mechanisms may involve interactions with noncoding RNAs as well as protein cofactors. Many CTCF sites are also involved in other functions such as modulation of RNA splicing and specific regulation of gene expression, and the relationship between these activities and loop formation is another unanswered question that should keep investigators occupied for some time.
With the advent of chromosome conformation capture (3C) and related methods to measure intranuclear contacts (Dekker and Misteli 2015), it has become clear that, within the nucleus, the genome is engaged in an intimate conversation with itself. Relatively short-range interactions between enhancers and promoters help regulate expression of individual genes or gene families (Tolhuis et al. 2002). Longer-range interactions may organize the genome into topologically distinct regions. In vertebrates, many of these interactions are mediated by contacts involving CTCF. The protein CTCF was first cloned and characterized as a vertebrate transcription factor (Lobanenkov et al. 1990; Klenova et al. 1993). Subsequently, binding sites for CTCF, found at either end of the chicken β-globin locus (Chung et al. 1997; Bell et al. 1999) and later at the imprinted Igf2/H19 locus (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000) in mice and humans, were shown to serve as insulating boundary elements: They blocked interactions between enhancer and promoter when placed between them but not otherwise.
Insulating elements were already well known from work in Drosophila (Udvardy et al. 1985; Geyer and Corces 1992; Kellum and Schedl 1992). Even in these early studies, it was evident that the properties of insulator elements might arise from an ability to form closed loops in which pairs of elements widely separated in the genome come together at the base of the loop (Udvardy et al. 1985; Geyer and Corces 1992; Muravyova et al. 2001). In this manner, interactions between regulatory elements residing in different loops would be inhibited, whereas interactions within a given loop would be favored. This model has been elaborated on theoretically (Doyle et al. 2014) and confirmed in many laboratories; the role of CTCF in organization of such domains has now been explored extensively (Ong and Corces 2014; Vietri Rudan and Hadjur 2015). The past year has seen major advances in understanding the multiple roles of CTCF in gene regulation and genome organization and especially in how such domains are generated and organized.
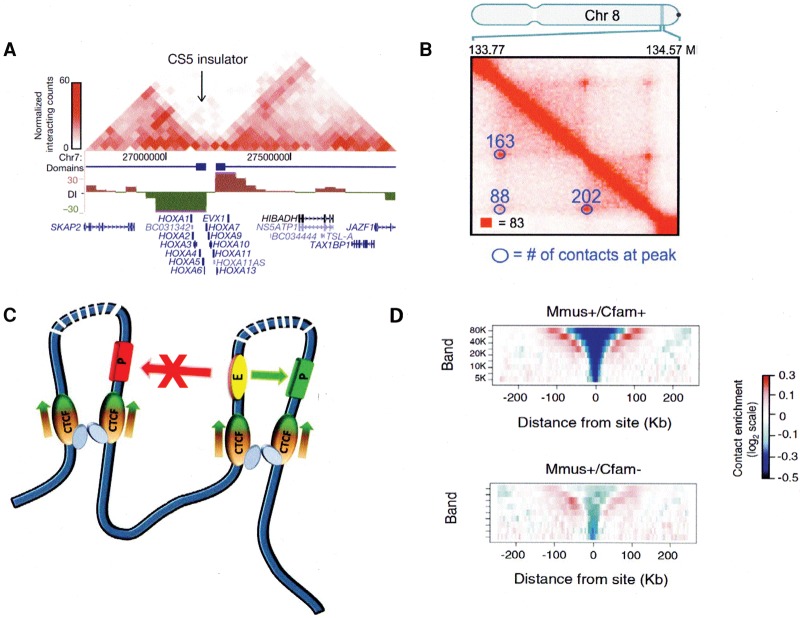
These results, which are the main focus of this review, reflect the increasing resolution of data obtained with high-throughput 3C (Hi-C) methods, allowing Dixon et al. (2012) to show that the genome could be subdivided into ∼2000 “topologically associated domains” (TADs), with contacts strong within each TAD but quite weak between different TADs (Fig. 1A). They found that CTCF in most cases demarcated the individual TAD boundaries, consistent with an ability to block interactions across its binding sites. In other experiments, depletion of CTCF not only reduced intradomain contacts but increased interdomain interactions (Zuin et al. 2014). Higher (4 kb) resolution was achieved by Phillips-Cremins and Corces (2013) using 3C carbon copy (5C) methods to analyze a selected part of the genome in mouse embryonic stem (ES) cells. This allowed them to resolve sub-TADs within the TADs and show that >80% of the interactions that they observed involved some combination of sites for CTCF, SMC1 (a cohesin complex component), and the Mediator complex component Med12. It has been known for some time that a large proportion of bound CTCF is associated with cohesin (Parelho et al. 2008; Rubio et al. 2008; Wendt et al. 2008) and that Mediator recruits cohesin independently of CTCF (Kagey et al. 2010). Loops involving either CTCF+SMC1 or CTCF alone tended to be the longest (on the order of 1 Mb), and comparison of these ES cell data with those from neural progenitor cells showed that such loops were also enriched among those conserved between the two cell types. This led to the proposal that these constitutive CTCF sites mark domains critical for chromosome architecture, while other loops would be associated with more local and specific regulatory tasks. More recent Hi-C studies (Rao et al. 2014) at a remarkable map resolution (see the investigators’ definition) of 1 kb allow an even more detailed description of genome organization. Rao et al. (2014) were able to detect a much larger number of smaller “contact domains” with a distinct preference for interaction within the domain and exclusion of neighbors. Using stringent criteria, they identified Hi-C “cross-peaks” (Fig. 1B) reflecting strong contact between distant sites, generating loops. In 38% of cases, the loop ends correspond to the boundaries of a contact domain, and these regions are referred to as “loop domains.” CTCF and cohesin subunits were found to occupy 86% of contact peak loci, and, in 54% of cases, a CTCF-binding motif, with CTCF and cohesin subunits localized there, was identified. Most important is the observation that almost all of the loops in this subset are anchored at a pair of convergent sites binding CTCF as well as SMC3, RAD21, and, presumably, the rest of the cohesin complex (Fig. 1C). It is difficult to compare the number of loop domains identified by this procedure with earlier or later results because quite stringent signal to noise criteria were applied.
Figure 1.
CTCF roles in domain organization within the nucleus. (A) TADs in the human HOXA locus, with a CTCF insulator site between them. (Adapted by permission from Macmillan Publishers Ltd. from Dixon et al. 2012.) (B) High-resolution Hi-C analysis of a small region of human chromosome 8 in GM12878 cells. Contact peaks are circled. (Adapted from Rao et al. 2014 with permission from Elsevier.) (C) Loop domains bordered by CTCF sites typically associated with cohesin. Interactions between enhancers and promoters within the same loop are favored; those between loops are blocked. At loops bordered by the strongest and most conserved CTCF sites, CTCF is oriented as shown, with the N terminus of each protein facing into the loop (see also Fig. 5, below). (D) Contact insulation analysis showing reduced frequency of contacts across CTCF boundary sites conserved between mice and dogs, compared with nonconserved sites. (Adapted from Vietri Rudan et al. 2015 with permission from Elsevier.)
Convergence is critical
Other results during the past year revealed important properties of chromatin domain structures. By comparing syntenic regions in four vertebrates, Vietri Rudan et al. (2015) identified conserved CTCF-binding sites, which they showed are also the ones with the highest affinity for CTCF. They found that such sites tend to mark the borders of conserved large-scale Hi-C domains, in contrast to species-specific CTCF sites, which are located within the larger domains. An analysis of the patterns of interaction across CTCF-binding sites at loop termini shows a striking correlation between conservation and strong insulation. Furthermore, these strong conserved sites have a preferred convergent orientation with respect to one another (Fig. 1D).
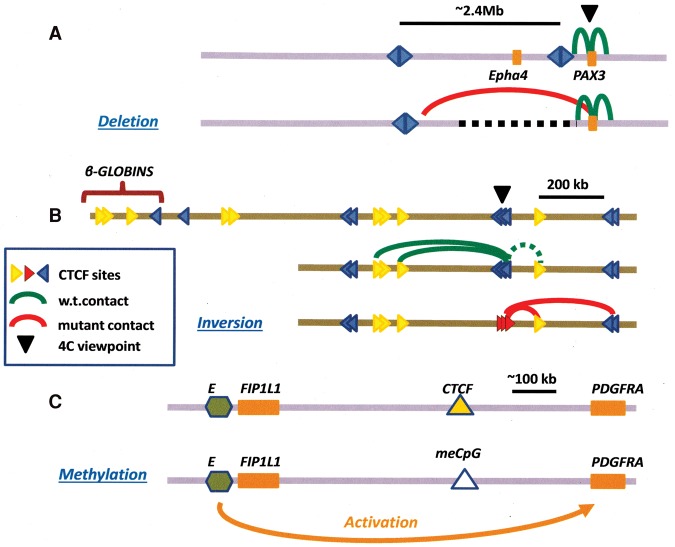
These discoveries of a predominant convergent orientation, now confirmed in other laboratories by complementary techniques (de Wit et al. 2015), have raised many questions and inspired several groups to examine the consequences of deleting or altering the orientation of one of a CTCF-binding site pair. It had been shown (Nora et al. 2012) that deletion of a TAD boundary in the neighborhood of the Xist locus on the X chromosome could result in ectopic long-range contacts and overall misregulation of expression. Recent analysis of the Six homeodomain locus of zebrafish (Gomez-Marin et al. 2015) revealed the presence of oriented CTCF sites (shown in that study as divergent between adjacent TADs and therefore convergent within TADs) at TAD boundaries; deletion of one of these boundaries in BACs leads to inappropriate interdomain enhancer–promoter interactions. Similarly, CRISPR/Cas-mediated deletion of a CTCF site within the Hox clusters in mouse ES cells disrupts a topological boundary, resulting in activation of previously silent Hox genes. (Narendra et al. 2015). The importance of maintaining these boundaries is made clear in experiments deleting a CTCF-associated TAD boundary near the limb enhancers normally associated with the mouse Epha4 gene (Fig. 2A). This results in altered patterns of gene expression, leading to limb malformation. DNA rearrangements that similarly disrupt this boundary are shown to be associated with pathogenic limb formation in humans (Lupianez et al. 2015). The importance of maintaining domain integrity is also implied in the conservation of CTCF-mediated loop domains between naive and primed ESCs (Ji et al. 2016).
Figure 2.
Effects of altering CTCF-binding sites on domain structure and gene expression. (A) Effect on 4C contacts of deleting DNA containing an insulator boundary near the mouse PAX3 gene, showing novel interactions with regions further upstream (Lupianez et al. 2015). Disruption of a TAD boundary had been shown earlier to cause ectopic chromosomal contacts and long-range transcriptional misregulation in the mouse Xist locus (Nora et al. 2012; see also Dowen et al. 2014). (B) Effect of inverting CTCF-binding sites on the pattern of 4C contacts near the mouse β-globin locus. The dotted green interaction line calls attention to the nonconvergent orientation of the CTCF sites marked by the blue triangles and the yellow one immediately downstream. After inversion, contacts between the red (inverted) sites and the yellow site actually strengthened despite the fact that the sites are not facing toward each other on the loop (Guo et al. 2015; see also de Wit et al. 2015) (C) Effect of methylation of a CTCF site on boundary activity. In certain human gliomas, the product of the mutated isocitrate dehydrogenase (IDH) gene interferes with DNA demethylation at a critical CTCF-binding site, resulting in loss of CTCF binding and insulation and inappropriate activation of the PDGFRA gene, a glioma oncogene, by a distal enhancer (green hexagon) (Flavahan et al. 2016).
Experiments in the mouse and human protocadherin loci and the human β-globin locus directly address the significance of the orientation of paired CTCF loop sites by reversing the direction of one site (Fig. 2B; Guo et al. 2015). In each case, reversal of orientation results in a new pattern of 4C (circularized 3C) contacts that reflects the disappearance of one loop and formation of a new one that conforms to the CTCF site orientation rules. Similarly, inversion of CTCF sites leads to disruption of looping even though CTCF binding is maintained (de Wit et al. 2015), and the results of an extensive study (Sanborn et al. 2015) of the effect of methodical deletion of individual CTCF sites in a group defining two loop domains are entirely consistent with the requirement that loops be bounded by convergently oriented CTCF sites.
Mechanisms for generating convergence
It is clear that large-scale genome organization is determined by this special set of oriented CTCF sites, but only a subset of CTCF sites is involved in these structures. Many of the shorter-range, CTCF-mediated interactions do not conform to this rule; Guo et al. (2015) and Tang et al. (2015) showed that more weakly formed loop interactions do not all involve convergent sites (see below; Fig. 5, below) but nonetheless display some preferential relative orientation. However, it has been apparent since the discovery of the constraint on major loop CTCF site orientation that equilibrium models might not be sufficient to explain such observations. If convergent orientation at contact sites were energetically preferred, it would overcome any of the relatively small costs of bending the large chromatin region within the loop and there would be no requirement that sites be oriented on the DNA sequence (but see the comment in legend of Fig. 5, below; Arib et al. 2015)
Figure 5.
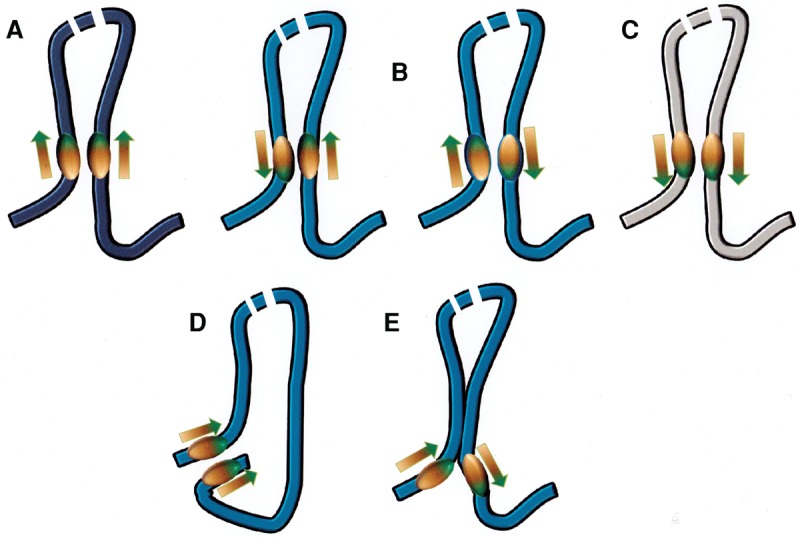
Orientation of CTCF-binding sites at the base of loops (Rao et al. 2014; Guo et al. 2015; Vietri Rudan et al. 2015). (A) The observed preferred convergent orientation for strong, conserved sites. (B) Two equivalent tandem orientations that occur with reduced frequency (see the text). (C) The divergent orientation, which appears to be disfavored. (D) “Coil” arrangement proposed (Tang et al. 2015) to explain how tandemly oriented sites might interact, assuming that where CTCF/cohesins contact each other, they have to align in parallel. (E) Although TADs are large and the energetic costs of deforming chromatin within a domain should be small, there could be cases in which other strong interactions near the base of a loop constrain conformations and make interactions between CTCF sites either less or more energetically favored.
It is apparent that these domains are formed by a nonequilibrium process, and some recent studies indicate what form it might take. In a review of Guo and colleagues (Rao et al. 2014; Guo et al. 2015), Nichols and Corces (2015) suggested that the ability of CTCF to bend DNA at one end of its binding site (Arnold et al. 1996; MacPherson and Sadowski 2010) would create an incipient loop, which could then enlarge until a mating CTCF was encountered. In earlier work, Alipour and Marko (2012) had proposed an extrusion model to explain how condensin-dependent loop domains could be formed on mitotic chromosomes. Sanborn et al. (2015) extended this idea to propose that, in the case of CTCF-mediated loop formation, the loop is stabilized by a pair of cohesin molecules that first form a “handcuff,” generating a small loop. The paired cohesins enlarge the loop as they move away, either carrying along CTCF molecules with them until oriented binding sites are reached or stopping when “properly” oriented bound CTCFs are encountered (Fig. 3). This is an attractive model because its geometry gives rise to exactly the kind of (largely) nonoverlapping pattern of loop domains observed in vivo as well as the intraloop folding patterns deduced from the Hi-C data (Rao et al. 2014; Fudenberg et al. 2015; Sanborn et al. 2015; Dekker and Mirny 2016). Theory and experiment do not necessarily agree in detail, possibly a reflection of the ways in which evolution has elaborated on simple polymer physics. The experimental results do contain some apparent examples of overlapping loops, but this could reflect the presence of different loops in different individual cells rather than their simultaneous presence in a single cell. We do not have enough information at this point to prefer a model in which CTCF is delivered by an advancing processive cohesin complex as opposed to one in which CTCF is already bound to its DNA sites and traps cohesin when it arrives. This raises the separate question of CTCF site occupancy during the cell cycle: Does CTCF remain bound during mitosis? Chromatin immunoprecipitation (ChIP) studies show that some well-characterized CTCF sites do remain occupied (Burke et al. 2005), while others apparently do not (Wendt et al. 2008). Immunofluorescence studies also disagree: Wendt et al. (2008) did not detect CTCF binding in mitotic chromosomes, but Burke et al. (2005) did, perhaps reflecting differences in fixation and staining methods. However, further experiments (Burke et al. 2005) using GFP-tagged CTCF fragments as probes showed that, on mitotic chromosomes, CTCF apparently binds largely to sites that engage the C-terminal zinc fingers. The fact that such sites comprise only 15%–25% of all CTCF sites (see below) suggests that CTCF may not be present at a large proportion of its normal sites during mitosis.
Figure 3.
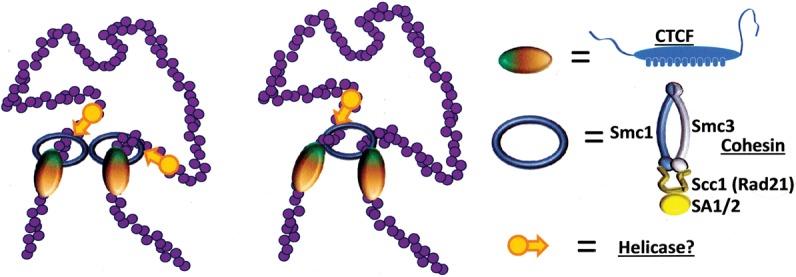
Proposed mechanisms (Sanborn et al. 2015) for generating loop domains terminated by convergently oriented CTCF sites (see Fig. 5, below). Cohesin bound to chromatin extrudes a loop and continues until it reaches a properly oriented CTCF site on each arm of the loop. It then stops searching; CTCF either comigrates with cohesin or is prebound, but cohesin is deposited only when CTCFs are properly oriented. Two possible configurations of cohesin are shown, corresponding to proposed models of cohesin interaction with chromatin (Nasmyth 2011). This process would require an energy source, suggested here to be an as yet unspecified helicase, shown as orange arrows.
The discovery during the past few years of conserved and quite selective CTCF-mediated interaction patterns had immediately raised the questions: How are some CTCF interactions selected in preference to others? Are loop domain structures maintained during replication and cell division or instead regenerated de novo? The mechanism proposed by Sanborn et al. (2015) certainly supports the latter model. New questions then arise: How much of the large-scale structure is disrupted during cell division? When is it disrupted, and at what stage of the cell cycle is it regenerated? The cohesin handcuff model is attractive, but what would be the energy source required to propel cohesin along the loop it is in the process of enlarging?
Evidence from Hi-C and 5C measurements of cells arrested during mitosis shows that little if any of the large-scale loop domain structure survives, implying that higher-order chromatin structures have to form de novo in early G1 (Naumova et al. 2013). If a cohesin “handcuff” (or some variant of it) is responsible for this pattern regeneration, ATP-dependent helicases such as RUVBL1/RUVBL2, known to be required for decondensation of mitotic chromosomes (Magalska et al. 2014), might be recruited to drive loop extrusion (Fig. 3). This still leaves unsettled the question of how such structures are maintained during DNA replication. It has been shown, for example, that cohesin remains bound at transcription factor cluster sites through replication and in the absence of CTCF (Yan et al. 2013). It is also unclear what structures are responsible for maintaining contacts, once formed, between sites at the base of the loop domains. Presumably, CTCF has to be present. The evidence is strong that cohesin is also required. A number of studies (Seitan et al. 2013; Sofueva et al. 2013; Zuin et al. 2014) in which cohesin components were depleted have explored in detail the role of cohesin in the maintenance of higher-order structure.
The nature of CTCF-binding sites
The next problem is to understand what makes the loops such stable structures. If we assume that both CTCF and cohesin are required, the determining factor might be the stability of CTCF binding to DNA (since cohesin is recruited to DNA by CTCF). There is considerable information about DNA sequence motifs that bind CTCF and the dissociation constants associated with those motifs. The DNA-binding protein CTCF is restricted to bilaterians and is highly conserved across most of the animal evolutionary tree (Heger et al. 2012; Vietri Rudan et al. 2015), and the presence of multiple zinc fingers suggests that it can engage DNA in multiple ways (Filippova et al. 1996; Nakahashi et al. 2013). The earliest CTCF-binding site to be identified as part of an insulating boundary element is located upstream of the chicken β-globin locus (Bell et al. 1999). CTCF binds there in vitro with subnanomolar affinity, and Renda et al. (2007) have shown that four of the central zinc fingers, 4–8, are required for this high-affinity interaction. Subsequent studies have demonstrated that this belongs to a set of nonpalindromic CTCF-binding sites with a sequence consensus referred to as M1 (Holohan et al. 2007; Kim et al. 2007; Xie et al. 2007; Schmidt et al. 2012), which is proposed to engage zinc fingers 4–7 in vivo (Nakahashi et al. 2013). This 20-base-pair (bp) core motif is present in most of the known CTCF-binding sites identified by ChIP (ChIP-seq [ChIP combined with deep sequencing] and ChIP-exo [ChIP exonuclease]) (Fig. 4), and the nonspecific engagement of zinc fingers other than 4–7 by the flanking DNA sequence is thought to further stabilize CTCF binding.
Figure 4.
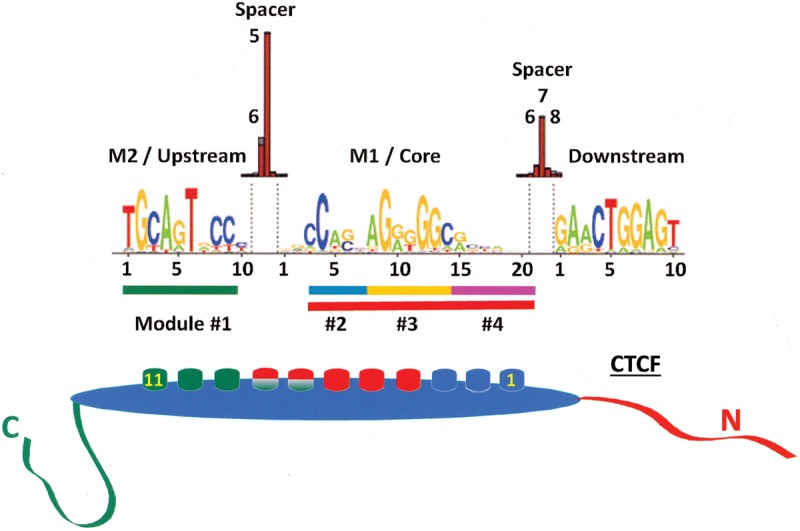
CTCF-binding motifs showing the M1/core that specifically engages fingers 4–8 and the M2/upstream sequence that engages fingers 7–11, with overlapping binding of the middle fingers to M1 and M2 (Nakahashi et al. 2013). Fingers not engaged in sequence-specific contacts may nonetheless contribute to overall binding stability through nonspecific interactions. Note that the sequence as shown binds CTCF with the N terminus facing downstream (Renda et al. 2007; Nakahashi et al. 2013). The DNA-binding modules described by Rhee and Pugh (2011) based on a ChIP-exo study are highlighted as colored bars at the bottom of the motif.
A second 10-bp CTCF motif (Fig. 4), referred to as M2, found upstream of M1 has been identified (Rhee and Pugh 2011; Schmidt et al. 2012; Nakahashi et al. 2013), and this alone engages zinc fingers 9–11 with nanomolar affinity (Xiao et al. 2015). Genome-wide studies indicate that motif M2 is found in conjunction with M1 in 15%–25% of the CTCF sites that possess M1, and it is expected that CTCF will bind to these sites with extremely high affinity, although this may depend on the spacer between these sites. The unusually high affinities (which typically reflect slow off rates and diffusion limited binding rates) are responsible for the long residence time on chromatin, which is ∼11 min, approximately an order of magnitude longer than observed for most transcription factors (Nakahashi et al. 2013). It must be kept in mind that the method used to make these measurements, fluorescence recovery after photobleaching (FRAP) of GFP-tagged CTCF, might not account for “nonexchangeable” CTCF that binds with the highest of affinities. Interestingly, Nakahashi et al. (2013) have also identified a 10-bp motif that, when found downstream from M1, results in destabilization of CTCF binding, possibly through the disengagement of zinc fingers 1–2 (Fig. 4).
DNA methylation and CTCF binding
It has been known for a long time from studies of imprinting at the Igf2/H19 locus that cytosine methylation at a CpG within the CTCF motif greatly lowers binding affinity (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000) and that a particular site within M1 is critical (Renda et al. 2007). Only some CTCF motifs contain a CpG at the right place, where its presence can be used for regulation of CTCF binding. A very recent report (Flavahan et al. 2016) provided a striking example of both the importance of CTCF-mediated domain formation for cell function and the potential role of DNA methylation in regulating domain architecture. The investigators showed that a mutation in the isocitrate dehydrogenase (IDH) gene, associated with certain classes of human gliomas, exerts its effect by inhibiting pathways that normally lead to demethylation of CpGs. The resulting increase in methylation of susceptible CTCF sites and loss of CTCF binding disrupts a TAD boundary. The loss of insulation in turn allows a previously blocked constitutive enhancer ∼900 kb away to interact with and activate PDGFRA, a known glioma oncogene (Fig. 2C). As the investigators point out, similar mechanisms may be at work at other CTCF boundary elements in these cells. Furthermore, it seems likely that other kinds of malignant cells with aberrant methylation pathways will also suffer disruptions in domain organization. There is good reason to think that, in many kinds of cells, the subset of CTCF sites marked by a CpG that interacts with zinc finger 7 (Renda et al. 2007) will be sensitive to local or global methylation changes, with consequences that could be varied and dramatic. However, not all such sites will necessarily be methylated in vivo because CTCF binding could protect against methylation (Stadler et al. 2011). More generally, mutations in individual CTCF sites (Tang et al. 2015) can lead to loss of binding and disruption of loop formation, with important consequences for disease susceptibility. CTCF-binding sites are major hot spots for mutations in the cancer genome (Katainen et al. 2015), and oncogenes can be activated by mutations that disrupt CTCF binding at the boundaries of loop domains (Hnisz et al. 2016).
CTCF does not work alone
CTCF exhibits a range of affinities for DNA, depending on the particular sequence within the canonical binding motifs or within noncanonical motifs not yet fully characterized (Plasschaert et al. 2014). It has already been noted that evolutionarily conserved CTCF sites demarcating domain structures are usually those with high affinity (Guo et al. 2015; Vietri Rudan et al. 2015). The implicit identification of weaker affinity sites and the possible lack of a CTCF-binding motif at sites occupied by both CTCF and cohesin (Rao et al. 2014) suggests that other cofactors may be required for at least some CTCF functions. Neighboring binding sites for other regulatory factors may augment or modulate CTCF function (for review, see Weth and Renkawitz 2011). It has also been known for many years (Wallace and Felsenfeld 2007) that a variety of other proteins is recruited to particular binding sites by CTCF and may play important and diverse roles in its activities. For example, Smad proteins are associated with CTCF at the Igf2/H19 imprinted control region (Bergstrom et al. 2010) and at many sites in Drosophila (Van Bortle et al. 2014). The general transcription factor II-I (TFII-I) helps stabilize CTCF binding at certain promoter-proximal regions (Pena-Hernandez et al. 2015). The DEAD-box helicase p68 is associated in HeLa cells with 7% of CTCF sites (Yao et al. 2010). At the Igf2/H19 locus, p68 helps, in association with the long noncoding RNA SRA, to stabilize cohesin binding and create an effective insulator. At many genomic sites in ES cells, DNA-bound CTCF/cohesin can recruit the core promoter factor TAF3 and mediate its contact with promoters through TAF3-dependent loop formation (Liu et al. 2011). In addition, CTCF undergoes modifications such as poly(ADP) ribosylation (Guastafierro et al. 2013), phosphorylation (Klenova et al. 2001), and sumoylation (MacPherson et al. 2009) that are important for its activity. CTCF also interacts with the enzyme poly-ADP-ribose (PARP1) itself to help establish interchromosomal contacts during the circadian cycle between active loci enriched in circadian genes and repressed lamina-associated domains (LADs) (Zhao et al. 2015).
Recent reports also made it clear that many RNAs bind to CTCF to modulate its regulatory functions. Of note are studies (Saldana-Meyer et al. 2014; Kung et al. 2015) showing that CTCF interacts with many endogenous RNAs. Saldana-Meyer et al. (2014) reported that at least 17,000 genomic RNAs interact with CTCF. They identified an RNA-binding domain within the CTCF C terminus, which, together with CTCF zinc fingers 10 and 11, interacts with Wrap53 RNA, the p53 antisense transcript; the CTCF–RNA interaction appears to be important for regulation of p53 expression. Kung et al. (2015) similarly reported that a wide variety of genomic RNAs interacts with CTCF, with binding strengths that appear in some cases to exceed those seen for interactions with CTCF-binding motifs on DNA. Complexes of CTCF with Tsix and Xite RNAs target CTCF to the X inactivation center, providing a pathway for specific deposition of CTCF at selected sites. In an earlier study of mouse X chromosome inactivation (Sun et al. 2013), this laboratory had shown conversely that Jpx RNA, expressed from a site neighboring Xist, is able to interact with and remove bound CTCF from DNA, resulting in up-regulation of Xist expression. In these mechanisms, which appear to play an important role in X chromosome pairing and X inactivation, DNA and RNA compete for binding to CTCF. It is still not clear under what circumstances this competition for CTCF between particular RNA and DNA sites is won by one or the other.
This is different from the situation reported at the p53/Wrap53 locus (Saldana-Meyer et al. 2014), where the investigators proposed that CTCF can bind simultaneously to DNA through its more N-terminal zinc fingers and to RNA through its C terminus. They also showed that addition of RNA can result in formation of CTCF multimers in solution, suggesting one way in which CTCF loop interactions might be stabilized in vivo. Other interactions that could help stabilize loops were implied in the model proposed by Sanborn et al. (2015) for cohesin binding at loop boundaries. If cohesin forms a handcuff involving a pair of cohesin molecules or simply a single closed circle surrounding both arms of the loop, this could create a tether for the ends of the loop (Fig. 3). That depends, of course, on the stability of the cohesin ring structure during interphase. We know that, during mitosis, a single cohesin molecule forms a quite stable ring around sister chromatids, the opening of which requires a specific set of chemical reactions (Nasmyth 2011). Less is known about the stability of cohesin binding to chromatin in interphase cells. In mice, the protein Wapl is required for release of cohesin from chromatin during all stages of the cell cycle (Tedeschi et al. 2013). Exchange rates for bound cohesin, measured in rat kidney cells by inverse FRAP, not surprisingly vary with cell cycle stage (Gerlich et al. 2006). In G2, 30% of cohesin is bound to chromatin with a residence time of ∼6 h, probably representing those complexes involved in tethering of sister chromatids. In contrast, during G1, 44% of cohesin complexes are bound with a residence time of 24 min, and longer times were not reported. Interestingly, these are of the same order of magnitude as times reported for CTCF exchange. Taken at face value, the results would suggest that cohesin handcuff structures, although relatively stable, could not alone provide long-term stability of loop structures during interphase. It is important to remember, however, that perhaps the majority of cohesin complexes is attached to chromatin through Mediator, rather than CTCF, and the observed values may reflect this population. The stability of loop domains ultimately may depend on a mixture of the stability of CTCF binding to DNA, the strength of its interaction with cohesin, the topological constraints conferred by the closed cohesin ring, and the stability of that ring. One important step will be to obtain experimental evidence that cohesin actually forms rings around chromatin during interphase. It is also possible that the structures at the base of the loop are labile so that contacts are broken and reformed but that the loop ends are held near each other by different, shorter-range interactions within the loop that function as a kind of molecular Velcro.
Convergence is not universal
Given the variability in binding strength of CTCF motifs and the effect of local environment and bound cofactors, it is difficult to envisage a single mechanism for CTCF action. Different definitions of loop domains or TADs necessarily give rise to varying estimates in the number of CTCFs known to be involved in such structures. Using the most stringent definition of contact peaks (marking strong contacts between distant sites) in GM12878 cells, Rao et al. (2014) associated 54% of a total of 12,903 contact peaks with the presence of a CTCF motif. If this is taken strictly, it indicates that a considerable number of contact peaks are not associated with CTCF, and, given that there are (according to Encode ChIP-seq data) >40,000 sites occupied by CTCF in these cells, it would also mean that many CTCF-binding sites are not involved in contact peaks. A different method of identifying paired sites (Guo et al. 2015) uses published ChIA-PET (chromatin interaction analysis with paired-end tag sequencing) data (Handoko et al. 2011) to count only those CTCF sites in K562 cells that are actually occupied by CTCF. This yields an estimate of a total of ∼25,000 ChIA-PET interactions, of which ∼78% are associated with bound CTCF at both members of the pair. Of these, ∼76% involve convergently oriented sites, and most of the rest are tandem (i.e., motifs facing in the same direction along the DNA).
A new ChIA-PET analysis (Tang et al. 2015) in GM12878 cells gave quite similar results: 64% of sites are convergent, and 33% are tandem. Interestingly, in both cases, only 2% of sites are paired in the divergent orientation, which provides another constraint on possible mechanisms (Fig. 5). Tang et al. (2015) suggested that the tandem sites interact to form a “coil” rather than the “hairpin” generated by the interaction of convergent sites (Fig. 5), preserving the parallel spatial orientation of the two CTCF motifs, which could well be required if cohesins bound to the two CTCFs had to interact. Consistent with earlier results, they reported that the convergent sites are associated with TADs, whereas the interactions involving tandem sites are weaker and associated with loops formed within TADs. The latter are likely to be more transient contacts. Are these contacts generated also by a loop extrusion mechanism? If it is assumed (Sanborn et al. 2015) that a processive mechanism deposits CTCF preferentially (but not exclusively) when it encounters a “properly” oriented binding domain, that could result in something like the observed frequencies of convergent, tandem, and divergent paired loop sites. The deposition mechanism could sometimes (tandem orientation) deliver one of the two CTCFs to a site facing in the wrong direction but would be even less likely to do it if both sites were divergent. However, this does not explain in itself why those contacts, once established, should differ in strength.
Still another method of evaluating data is to calculate TAD strength, defined by the ratio of intra- versus inter-TAD interaction frequencies, which, in principle, allows for inclusion of the entire range of interaction strengths (Van Bortle et al. 2014). This may be particularly useful for categorizing weaker contacts; for example, in Drosophila, where, unlike the situation in vertebrates, there are, in addition to CTCF, a number of other proteins associated with architectural activity, and site occupancy by these factors is correlated with TAD border strength. Because of the multiplicity of factors involved, it is more difficult in Drosophila to isolate the contribution of CTCF to domain organization. Drosophila CTCF has N-terminal and C-terminal domains quite different from those in the vertebrate protein, although it shares with vertebrates the same DNA-binding motifs and strong zinc finger homologies. As a result, it recruits, to a considerable extent, a different set of cofactors (Van Bortle et al. 2014). Nonetheless, there is a clear TAD organization in Drosophila (Sexton et al. 2012; Van Bortle et al. 2014; Eagan et al. 2015; Li et al. 2015) in which CTCF plays a major role. Furthermore, the bands seen in Drosophila polytene chromosomes correspond to TADs (Eagan et al. 2015), and the same TAD organization is present in diploid cells.
Other roles for CTCF
Many of the more local domains help regulate interactions between enhancers and promoters and employ weaker and less conserved binding sites. Some loops associated with regulation of gene expression (for example, those associated with TAF3) (Liu et al. 2011) involve CTCF at only one end of the loop. Recent results implicate other CTCF sites in various mechanisms associated with RNA splicing. The protocadherin locus takes advantage of CTCF interactions to bring together multiple combinations of variable and constant exons, with a resulting great diversity in the RNA and protein products (Guo et al. 2012). CTCF, in some cases, also plays a less exotic role in the RNA splicing mechanism (Shukla et al. 2011; Paredes et al. 2013; Agirre et al. 2015) by slowing the progression of transcribing RNA polymerase II (Pol II), which, as is known for some other bound factors, can result in a different choice of exons in the spliced product. The CTCF site associated with this function is correlated with the presence of HP1α and AGO1 near regulated exons (Agirre et al. 2015). It is not clear whether such sites are involved in loop formation. There is evidence that this need not be a part of the mechanism: Slowing of elongation can also be observed in vitro with templates carrying only a single CTCF-binding site (Shukla et al. 2011). CTCF is also found upstream of the transcription start site in unidirectionally transcribed genes, where it acts together with cohesin as a barrier to antisense transcription (Bornelov et al. 2015). This ability to impede Pol II is presumably connected to CTCF's slow exchange time: The polymerase has to wait for the CTCF to leave before it can advance. Interestingly, similar mechanisms were among the early alternative proposals for how insulators might work.
CTCF can perform other architectural functions, such as bringing together widely separated DNA sequences during V(D)J (Medvedovic et al. 2013; Ebert et al. 2015; Gerasimova et al. 2015; Lin et al. 2015; Narendra et al. 2015) and class switch (Birshtein 2012) recombination. One class of CTCF sites that does not fit neatly into this picture has been found in the α-satellite repeats of pericentromeric regions. Unusually, these sites engage only the C-terminal zinc fingers of CTCF (Burke et al. 2005; Xiao et al. 2015), and CTCF in turn recruits the centromeric protein CENP-E (Xiao et al. 2015). It remains to be determined at this site and no doubt at other sites in the genome whether CTCF has still further ways in which to affect genome organization. While the studies discussed here (largely published during the last 2 years, and many published within the past few months) provide us with astonishing amounts of information about large-scale genome organization, we still have a lot to learn about the processes that create that organization and the details of the local molecular interactions that hold it all together.
Acknowledgments
We thank members of our laboratory for their suggestions and encouragement. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.277863.116.
References
- Agirre E, Bellora N, Allo M, Pages A, Bertucci P, Kornblihtt AR, Eyras E. 2015. A chromatin code for alternative splicing involving a putative association between CTCF and HP1α proteins. BMC Biol 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour E, Marko JF. 2012. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res 40: 11202–11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arib G, Cleard F, Maeda RK, Karch F. 2015. Following the intracellular localization of the iab-8ncRNA of the bithorax complex using the MS2-MCP-GFP system. Mech Dev 138(Pt 2): 133–140. [DOI] [PubMed] [Google Scholar]
- Arnold R, Burcin M, Kaiser B, Muller M, Renkawitz R. 1996. DNA bending by the silencer protein NeP1 is modulated by TR and RXR. Nucleic Acids Res 24: 2640–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396. [DOI] [PubMed] [Google Scholar]
- Bergstrom R, Savary K, Moren A, Guibert S, Heldin CH, Ohlsson R, Moustakas A. 2010. Transforming growth factor β promotes complexes between Smad proteins and the CCCTC-binding factor on the H19 imprinting control region chromatin. J Biol Chem 285: 19727–19737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birshtein BK. 2012. The role of CTCF binding sites in the 3′ immunoglobulin heavy chain regulatory region. Front Genet 3: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelov S, Komorowski J, Wadelius C. 2015. Different distribution of histone modifications in genes with unidirectional and bidirectional transcription and a role of CTCF and cohesin in directing transcription. BMC Genomics 16: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke LJ, Zhang R, Bartkuhn M, Tiwari VK, Tavoosidana G, Kurukuti S, Weth C, Leers J, Galjart N, Ohlsson R, et al. 2005. CTCF binding and higher order chromatin structure of the H19 locus are maintained in mitotic chromatin. EMBO J 24: 3291–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Bell AC, Felsenfeld G. 1997. Characterization of the chicken β-globin insulator. Proc Natl Acad Sci 94: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Misteli T. 2015. Long-range chromatin interactions. Cold Spring Harb Perspect Biol 7: a019356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164: 1110– 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Vos ES, Holwerda SJ, Valdes-Quezada C, Verstegen MJ, Teunissen H, Splinter E, Wijchers PJ, Krijger PH, de Laat W. 2015. CTCF binding polarity determines chromatin looping. Mol Cell 60: 676–684. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. 2014. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159: 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle B, Fudenberg G, Imakaev M, Mirny LA. 2014. Chromatin loops as allosteric modulators of enhancer–promoter interactions. PLoS Comput Biol 10: e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagan KP, Hartl TA, Kornberg RD. 2015. Stable chromosome condensation revealed by chromosome conformation capture. Cell 163: 934–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, Hill L, Busslinger M. 2015. Spatial regulation of V-(D)J recombination at antigen receptor loci. Adv Immunol 128: 93–121. [DOI] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16: 2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2015. Formation of chromosomal domains by loop extrusion. bioRxiv 10.1101/024620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T, Guo C, Ghosh A, Qiu X, Montefiori L, Verma-Gaur J, Choi NM, Feeney AJ, Sen R. 2015. A structural hierarchy mediated by multiple nuclear factors establishes IgH locus conformation. Genes Dev 29: 1683–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. 2006. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol 16: 1571–1578. [DOI] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Gomez-Marin C, Tena JJ, Acemel RD, Lopez-Mayorga M, Naranjo S, de la Calle-Mustienes E, Maeso I, Beccari L, Aneas I, Vielmas E, et al. 2015. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci 112: 7542–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastafierro T, Catizone A, Calabrese R, Zampieri M, Martella O, Bacalini MG, Reale A, Di Girolamo M, Miccheli M, Farrar D, et al. 2013. ADP-ribose polymer depletion leads to nuclear Ctcf re-localization and chromatin rearrangement. Biochem J 449: 623–630. [DOI] [PubMed] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. 2012. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci 109: 21081–21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489. [DOI] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. 2012. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci 109: 17507–17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. 2016. Activation of proto-oncogenes by disruption chromosome neighborhoods. Science 351: 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet 3: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell PE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 2016. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C, Pant V, Loukinov D, Pugacheva E, Qi CF, Wolffe A, Ohlsson R, Lobanenkov VV. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr Biol 10: 853–856. [DOI] [PubMed] [Google Scholar]
- Katainen R, Dave K, Pitkanen E, Palin K, Kivioja T, Valimaki N, Gylfe AE, Ristolainen H, Hanninen UA, Cajuso T, et al. 2015. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet 47: 818–821. [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol 12: 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova EM, Nicolas RH, Paterson HF, Carne AF, Heath CM, Goodwin GH, Neiman PE, Lobanenkov VV. 1993. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol 13: 7612–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova EM, Chernukhin IV, El-Kady A, Lee RE, Pugacheva EM, Loukinov DI, Goodwin GH, Delgado D, Filippova GN, Leon J, et al. 2001. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol Cell Biol 21: 2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung JT, Kesner B, An JY, Ahn JY, Cifuentes-Rojas C, Colognori D, Jeon Y, Szanto A, del Rosario BC, Pinter SF, et al. 2015. Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell 57: 361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong CT, Cubenas-Potts C, Hu M, Lei EP, Bosco G, et al. 2015. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell 58: 216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SG, Guo C, Su A, Zhang Y, Alt FW. 2015. CTCF-binding elements 1 and 2 in the Igh intergenic control region cooperatively regulate V(D)J recombination. Proc Natl Acad Sci 112: 1815–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R. 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. 1990. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene 5: 1743–1753. [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson MJ, Sadowski PD. 2010. The CTCF insulator protein forms an unusual DNA structure. BMC Mol Biol 11: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson MJ, Beatty LG, Zhou W, Du M, Sadowski PD. 2009. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol 29: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalska A, Schellhaus AK, Moreno-Andres D, Zanini F, Schooley A, Sachdev R, Schwarz H, Madlung J, Antonin W. 2014. RuvB-like ATPases function in chromatin decondensation at the end of mitosis. Dev Cell 31: 305–318. [DOI] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Tamir IM, Schwickert TA, Novatchkova M, Sun Q, Huis In ‘t Veld PJ, Guo C, Yoon HS, et al. 2013. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity 39: 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291: 495–498. [DOI] [PubMed] [Google Scholar]
- Nakahashi H, Kwon KR, Resch W, Vian L, Dose M, Stavreva D, Hakim O, Pruett N, Nelson S, Yamane A, et al. 2013. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep 3: 1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra V, Rocha PP, An D, Raviram R, Skok JA, Mazzoni EO, Reinberg D. 2015. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347: 1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2011. Cohesin: a catenase with separate entry and exit gates? Nat Cell Biol 13: 1170–1177. [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. 2013. Organization of the mitotic chromosome. Science 342: 948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MH, Corces VG. 2015. A CTCF code for 3D genome architecture. Cell 162: 703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes SH, Melgar MF, Sethupathy P. 2013. Promoter-proximal CCCTC-factor binding is associated with an increase in the transcriptional pausing index. Bioinformatics 29: 1485–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. 2008. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433. [DOI] [PubMed] [Google Scholar]
- Pena-Hernandez R, Marques M, Hilmi K, Zhao T, Saad A, Alaoui-Jamali MA, del Rincon SV, Ashworth T, Roy AL, Emerson BM, et al. 2015. Genome-wide targeting of the epigenetic regulatory protein CTCF to gene promoters by the transcription factor TFII-I. Proc Natl Acad Sci 112: E677–E686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Corces VG. 2013. Chromatin insulators: linking genome organization to cellular function. Mol Cell 50: 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert RN, Vigneau S, Tempera I, Gupta R, Maksimoska J, Everett L, Davuluri R, Mamorstein R, Lieberman PM, Schultz D, et al. 2014. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res 42: 774–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda M, Baglivo I, Burgess-Beusse B, Esposito S, Fattorusso R, Felsenfeld G, Pedone PV. 2007. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J Biol Chem 282: 33336–33345. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. 2008. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci 105: 8309–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldana-Meyer R, Gonzalez-Buendia E, Guerrero G, Narendra V, Bonasio R, Recillas-Targa F, Reinberg D. 2014. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev 28: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148: 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. 2013. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res 23: 2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. 2013. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J 32: 3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. [DOI] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. 2013. Jpx RNA activates Xist by evicting CTCF. Cell 153: 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. 2015. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163: 1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A, Wutz G, Huet S, Jaritz M, Wuensche A, Schirghuber E, Davidson IF, Tang W, Cisneros DA, Bhaskara V, et al. 2013. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature 501: 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P. 1985. The 87A7 chromomere: identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185: 341–358. [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Nichols MH, Li L, Ong CT, Takenaka N, Qin ZS, Corces VG. 2014. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol 15: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M, Hadjur S. 2015. Genetic tailors: CTCF and cohesin shape the genome during evolution. Trends Genet 31: 651–660. [DOI] [PubMed] [Google Scholar]
- Vietri Rudan M, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, Hadjur S. 2015. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep 10: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. 2007. We gather together: insulators and genome organization. Curr Opin Genet Dev 17: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801. [DOI] [PubMed] [Google Scholar]
- Weth O, Renkawitz R. 2011. CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol 89: 459–468. [DOI] [PubMed] [Google Scholar]
- Xiao T, Wongtrakoongate P, Trainor C, Felsenfeld G. 2015. CTCF recruits centromeric protein CENP-E to the pericentromeric/centromeric regions of chromosomes through unusual CTCF-binding sites. Cell Rep 12: 1704–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci 104: 7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. 2013. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154: 801–813. [DOI] [PubMed] [Google Scholar]
- Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. 2010. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 24: 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sifakis EG, Sumida N, Millan-Arino L, Scholz BA, Svensson JP, Chen X, Ronnegren AL, Mallet de Lima CD, Varnoosfaderani FS, et al. 2015. PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol Cell 59: 984–997. [DOI] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci 111: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]