In this study, Jennis et al. characterize the first mouse model of an African-specific naturally occurring coding region variant at codon 47 of the p53 tumor suppressor gene (S47). They show that homozygous S47 mice are markedly tumor-prone and that the S47 variant impairs not only p53-mediated cell death but also the ability of p53 to transactivate a subset of genes involved in metabolism and ferroptosis.
Keywords: p53, tumor suppression, metabolism, ferroptosis, polymorphism, Ser46 phosphorylation
Abstract
A nonsynonymous single-nucleotide polymorphism at codon 47 in TP53 exists in African-descent populations (P47S, rs1800371; referred to here as S47). Here we report that, in human cell lines and a mouse model, the S47 variant exhibits a modest decrease in apoptosis in response to most genotoxic stresses compared with wild-type p53 but exhibits a significant defect in cell death induced by cisplatin. We show that, compared with wild-type p53, S47 has nearly indistinguishable transcriptional function but shows impaired ability to transactivate a subset of p53 target genes, including two involved in metabolism: Gls2 (glutaminase 2) and Sco2. We also show that human and mouse cells expressing the S47 variant are markedly resistant to cell death by agents that induce ferroptosis (iron-mediated nonapoptotic cell death). We show that mice expressing S47 in homozygous or heterozygous form are susceptible to spontaneous cancers of diverse histological types. Our data suggest that the S47 variant may contribute to increased cancer risk in individuals of African descent, and our findings highlight the need to assess the contribution of this variant to cancer risk in these populations. These data also confirm the potential relevance of metabolism and ferroptosis to tumor suppression by p53.
The p53 tumor suppressor (TP53) holds the distinction of being the most frequently mutated gene in human cancer. Germline mutations in the TP53 gene exist in individuals with Li-Fraumeni syndrome, who suffer from multiple tumors before the third decade of life (Malkin et al. 1992). An additional 50% of sporadic human tumors contain somatic mutations in the TP53 gene (Hollstein et al. 1991). The majority of these is missense mutations that occur in the DNA-binding domain, which inhibit the ability of this protein to bind to p53 response elements in the promoters of p53 target genes and subsequently transactivate gene expression (Vogelstein et al. 2000). In addition to these changes, there are several functionally significant polymorphisms in the TP53 gene as well as in genes encoding proteins that regulate the p53 pathway (such as MDM2 and MDM4); these variants can decrease p53 pathway activation and contribute to increased cancer risk (Pietsch et al. 2006; Grochola et al. 2010).
p53 is believed to possess at least three subsets of target genes through which it suppresses tumor development: It induces senescence through target proteins like CDKN1A (p21), induces apoptosis through target proteins like PUMA and NOXA (which encode “BH3-only” proteins), and controls metabolism/redox state through target proteins like SCO2, TIGAR, GLS2 (glutaminase 2), and SESTRIN 1/2 (for review, see Budanov 2014). There are compelling data indicating that, depending on cell type and the activated oncogene that is the driver, p53 uses one or more of these pathways for tumor suppression. In addition, more recently, the process of ferroptosis has been implicated in tumor suppression by p53; ferroptosis is an iron-mediated nonapoptotic cell death pathway that requires high reactive oxygen species and likely involves a lipid peroxide intermediate (Dixon et al. 2012, 2015; Dixon and Stockwell 2014; Yang et al. 2014). The regulation of ferroptosis by p53 is mediated in part by p53-dependent negative regulation of the cystine transporter SLC7A11 (Jiang et al. 2015). In addition, the p53 target gene GLS2 and the process of glutaminolysis have recently been implicated in ferroptosis (Gao et al. 2015).
p53 is heavily post-translationally modified, and, in some cases, these modifications have a considerable impact on the ability of p53 to perform its transcriptional, apoptotic, and tumor suppressor functions (Kruse and Gu 2008; Vousden and Prives 2009). Most pertinent to this study, Ser46 phosphorylation is required for efficient p53-mediated cell death in several cell line systems and in mice (Bulavin et al. 1999; Oda et al. 2000; D'Orazi et al. 2002; Feng et al. 2006). Over two decades ago, a naturally occurring polymorphism in TP53 was reported in Africans and African Americans (rs1800371, G/A). This polymorphism converts the proline residue proximal to Ser46 in human p53 to a serine (Felley-Bosco et al. 1993); we noticed that this eliminates the proline required for phosphorylation of Ser46 by the proline-directed kinases that phosphorylate this residue, which include p38MAPK, HIPK2, and DYRK (Felley-Bosco et al. 1993; Bulavin et al. 1999; Hofmann et al. 2002; Taira et al. 2007). We showed that the Ser47 variant (referred to here as S47) is markedly impaired for phosphorylation on Ser46 and also has significantly impaired ability to induce cell death in multiple human cell lines engineered to contain inducible forms of human wild-type p53 and S47 (Li et al. 2005). However, the impact of this variant on the p53 signaling pathway and cancer risk in an intact animal has never been assessed.
The S47 polymorphism appears to be restricted to African-descent populations, with a frequency of ∼1.5% in African Americans and between 6% and 8% in certain African populations; in contrast, this variant has not been detected in Caucasian Americans (see http://evs.gs.washington.edu/EVS). Given the very low allele frequency for S47, population analyses designed to assess the impact of this polymorphism on cancer risk in African-descent populations have lacked sufficient power. We reasoned that the S47 allele may explain part of the increased cancer risk disparities in African Americans relative to Caucasians and other ethnic groups. Therefore, in this study, we sought to determine the impact of the S47 variant on p53 function and cancer risk in a mouse model so as to better inform the rationale that human studies should be done. We found that the S47 variant is modestly or not at all impaired for most p53 functions; however, it is impaired for the ability to transactivate a subset of p53 target genes to induce cell death following cisplatin or ferroptosis inducers. Notably, the S47 variant is also impaired for tumor suppression, and S47 heterozygous and homozygous mice are markedly predisposed to hepatocellular carcinoma and other cancers.
Results
Generation of a mouse model for wild-type p53 and the S47 variant
In order to address the influence of the S47 polymorphism on p53 function within an organism, we generated a knock-in mouse for the S47 allele. For this, we chose the humanized p53 knock-in (Hupki) targeting allele, which replaces mouse exons 4–9 with the corresponding human exons (codons 32–332). Hupki p53 has been shown by several groups to be fully tumor-suppressive and transcriptionally active and to accurately recapitulate the activity of human p53 in mice (Luo et al. 2001; Reinbold et al. 2008; Frank et al. 2011). We previously modeled the codon 72 variants of p53 using the Hupki platform and derived considerable information from these Hupki mice that subsequently held true for human p53 codon 72 variants (Frank et al. 2011). Additionally, a knock-in mouse for p53 substituting Ser46 with alanine (S46A) was created using the Hupki platform, and cells from these mice recapitulate the apoptotic defect evident in inducible human cell line models (Feng et al. 2006).
We previously showed that the S47 variant appears to occur exclusively in cis with the Pro72 variant of p53 (Li et al. 2005). Therefore, we generated S47 embryonic stem cell lines using the P72 Hupki targeting construct. Embryonic stem cell lines with successfully targeted alleles were confirmed by Southern analysis (Supplemental Fig. S1A). Males with germline transmission of the targeted allele were crossed to EIIA-Cre females, and Cre-mediated excision of the neomycin resistance cassette was monitored by Southern analysis (Supplemental Fig. S1B). RNA was isolated from mouse embryonic fibroblasts (MEFs) containing either the wild-type Hupki or S47 Hupki alleles and used to sequence the full-length p53 cDNA; the only difference observed was at codon 47, which encoded proline (CCC) in wild-type p53 and serine (TCC) in the S47 allele (data not shown). To ensure genetic homogeneity, wild-type and S47 Hupki mice were backcrossed to C57Bl/6 mice for over 10 generations and then crossed to each other; for the majority of studies, we analyzed sibling littermate mice derived from heterozygote S47/wild-type crosses.
We generated several independent batches of MEFs from wild-type and S47 mice. We exposed these MEFs to etoposide, which led to significant phosphorylation of p53 on Ser15 (an activating marker) in both wild-type and S47 MEFs, along with clear induction of apoptosis. The induction of apoptosis was modestly reduced in S47 cells, as assessed by Western blots for cleaved lamin A and cleaved caspase-3 (Fig. 1A,B). Similar results were obtained following doxorubicin treatment (Supplemental Fig. S2A). Although we noted occasional decreases in the level of S47 protein induced by DNA damage compared with wild-type p53 (for example, see Supplemental Fig. S2A), these differences were not always seen, and we could find no evidence for a difference in the half-life between wild-type and S47 protein in cells treated with DNA-damaging agents (Supplemental Fig. S2B).
Figure 1.
Reduced DNA damage-mediated apoptosis in cells from the S47 mouse. (A) MEFs were generated from Hupki embryos containing either wild-type (WT) p53 or the S47 variant. MEFs were treated with 20 μM etoposide for 24 h, and protein lysates were analyzed by Western blot for the proteins indicated. The data depicted are representative of multiple experiments in multiple independent batches of MEFs. (B) Quantification of the relative intensity of cleaved lamin A blots from three independent experiments (as depicted in A) in primary MEFs from the wild-type and S47 mice untreated or treated with 20 μM etoposide for 24 h. Error bars mark standard deviation. (C) Immunohistochemical analysis of the small intestine of wild-type (Wt) and S47 Hupki mice 4 h following exposure to 5 Gy of γ irradiation (right panels) or untreated (left panels) for total p53. Bars, 100 μm. (D) Quantification of the number of cells staining positively for p53 antisera in equal millimeters of crypts from the small intestines and colons of mice with wild-type p53 or S47. Error bars mark standard error. IR indicates 5 Gy of ionizing radiation. (E) Immunohistochemical analysis of the small intestine of wild-type and S47 Hupki mice 4 h following exposure to 5 Gy of γ irradiation (right panels) or untreated (left panels) for apoptotic cells (cleaved lamin A). Red arrows mark apoptotic cells. Bars, 100 μm. (F) Quantification of apoptosis in the wild-type and S47 small intestines (left) and colons (right) as cells positive for cleaved lamin A following 5 Gy of radiation. The data depicted are averaged from three fields from three independent experiments in which equal millimeters of crypts were analyzed and quantified. Error bars represent standard deviation. (*) P-value <0.05.
We next assessed p53 induction and apoptosis in response to γ radiation in wild-type and S47 mice, focusing on tissues that show p53-dependent radio sensitivity (thymus, small intestine, and colon). Treatment of wild-type and S47 mice with 5 Gy of γ radiation led to an equal number of cells staining positively for p53 in the small intestine, thymus, and colon in wild-type and S47 mice (Fig. 1C,D; Supplemental Fig. S2C,D). In the small intestine and colon, there were reduced numbers of apoptotic cells in S47 mice following γ radiation compared with wild-type (Fig. 1E,F). In contrast, the levels of apoptotic cells in the thymus were nearly identical (Supplemental Fig. S2C). These combined data indicate that there appears to be a modest but consistent reduction in the apoptotic potential of the S47 protein in response to genotoxic stress in some but not all cell types.
Markedly impaired cell death by cisplatin in human and mouse S47 cells
Phosphorylation of Ser46 occurs in response to many genotoxic stresses, but phosphorylation of this residue by p38MAPK is particularly required for p53-mediated cell death following cisplatin treatment (Hernandez Losa et al. 2003; Zhu et al. 2013). Therefore, we next analyzed the response of wild-type and S47 MEFs and mice to cisplatin (Cis-dichlorodiammineplatinum [CDDP]). We noted significantly reduced cell death by cisplatin in S47 MEFs compared with wild type (Fig. 2A). Cisplatin-mediated cell death at this dose and time point was clearly p53-dependent, as noted by the absence of cleaved caspase-3 in MEFs from the p53 knockout mice (Fig. 2B). These results were corroborated using two independent assays to assess cell death and viability: Annexin V staining for apoptosis (Fig. 2C) and Alamar blue assays to assess cell viability; the latter cell viability assays indicated a 15-fold difference in cisplatin IC50 between wild-type and S47 MEFs (Fig. 2D). To confirm these results in human cells, we analyzed human lymphoblastoid cell lines (LCLs) that we obtained from the Coriell Institute, which had been genotyped as part of the 1000 Genomes Project (http://www.1000genomes.org) and identified and confirmed by us to be homozygous S47 or wild type (see the Materials and Methods). Treatment of homozygous wild-type and S47 LCLs with cisplatin for 24 h resulted in roughly equivalent p53 induction but greatly impaired apoptosis in S47 cells, as assessed by cleaved lamin A and cleaved caspase-3 protein (Fig. 2E). This impaired apoptosis in S47 LCLs was also evident following Annexin V staining (Fig. 2F). Even greater differences between wild-type and S47 LCLs were noted following cisplatin treatment using cell viability assays, and we found a 10-fold difference in the IC50 between wild-type and S47 human LCLs for cisplatin (Fig. 2G). Similar differences in IC50 were noted for adriamycin as well (Fig. 2H).
Figure 2.
Marked impairment of cisplatin-mediated apoptosis in S47 cells and mice. (A) Primary MEFs from the wild-type (Wt) or S47 Hupki mouse were treated with 10 μM cisplatin (CDDP) for the time points indicated, and protein lysates were analyzed by Western blot analysis for the proteins indicated. The data depicted are representative of three independent experiments in a minimum of three independent batches of MEFs. (B) Primary MEFs from the wild-type and S47 mice as well as the p53 knockout mouse (p53−/−) were treated with 10 μM CDDP for 24 h, and protein lysates were analyzed by Western blot analysis for the proteins indicated. (C) Flow cytometric analysis of Annexin V-positive cells from primary wild-type and S47 MEFs treated with 10 μM CDDP for 24 h. The totals represent an average of three independent experiments normalized to untreated controls. Error bars represent standard deviations. (D) IC50 analysis for cisplatin (CDDP) in primary wild-type (WT) and S47 MEFs treated with the indicated concentrations of cisplatin for 72 h and analyzed for viability by the Alamar blue assay. The depicted data represent an average of four independent experiments on independent batches of MEFs. Error bars represent standard deviation. (E) Human LCLs homozygous for wild-type p53 and the S47 variant were treated with 10 μM CDDP for 24 h, and protein lysates were analyzed by Western blot for the proteins indicated. (F) Flow cytometric analysis of Annexin V-positive cells from wild-type and S47 human LCLs treated with 10 μM CDDP for 24 h. The totals represent an average of three independent experiments normalized to untreated controls. Error bars represent standard deviations. (G) IC50 analysis for cisplatin (CDDP) in wild-type and S47 LCLs treated with the indicated concentrations of cisplatin for 48 h and analyzed for viability by Alamar blue staining. The depicted data represent an average of three independent experiments. Error bars represent standard deviation. (H) IC50 analysis for adriamycin in wild-type and S47 LCLs treated with the indicated concentrations of adriamycin for 48 h and analyzed for viability by Alamar blue staining. The depicted data represent an average of three independent experiments. Error bars represent standard deviation. (I) Cisplatin-mediated apoptosis, as assessed by cells positive for cleaved lamin A, in the kidneys of wild-type or S47 mice following injection with 20 mg/kg CDDP and analyzed after 48 h. Data are representative of n = 3 per mice group. Bar, 100 μm. (J) Clonogenic survival of shARF immortalized wild-type and S47 MEFs treated with the indicated concentrations of cisplatin (CDDP), plated at equal cell numbers 48 h later, and stained with crystal violet after 7 d. (K) Quantification of clonogenic survival of immortalized wild-type and S47 MEFs following cisplatin treatment. All values were normalized to the untreated control averaged from three independent experiments. Error bars represent standard deviation. (*) P-value < 0.05.
We next extended our findings by assessing cisplatin-mediated cell death in wild-type and S47 mice. Here we focused on the kidney, which is a key site of p53-mediated cell death in response to cisplatin (Sprowl et al. 2014). We noted a marked decrease in apoptotic cells in the kidneys of S47 mice relative to wild-type following cisplatin treatment; also evident were decreased regions of cell clearing (evidence of cell death) in the S47 kidney (Fig. 2I). We next examined the long-term effect of cisplatin on the viability of wild-type and S47 MEFs. To do this, we first immortalized several lines of wild-type and S47 MEFs by stably infecting them with a short hairpin for p19ARF; this short hairpin (shARF) is specific for ARF and not p16INK4a and immortalizes MEFs, thereby effectively eliminating the selection for p53 mutation that occurs during long-term culture (Humbey et al. 2008). Treatment of multiple independent clones of wild-type-shARF and S47-shARF MEFs with increasing doses of cisplatin for 24 h, followed by plating of equal numbers of cells, revealed a dramatically enhanced ability of S47-shARF MEFs to survive following cisplatin (Fig. 2J,K). Similar findings were made following treatment with carboplatin (Supplemental Fig. S3A,C). In contrast, the clonogenic survival of wild-type and S47 cells following etoposide was not markedly different (Supplemental Fig. S3B,D).
We next extended these studies to analyze cells that were heterozygous for the S47 variant S47/wild type. Surprisingly, heterozygous S47/wild-type MEFs more closely resembled homozygous S47 MEFs and were markedly impaired for cell death by cisplatin (Fig. 3A,B). Likewise, the IC50 for cisplatin in S47/wild-type MEFs was markedly increased compared with wild-type MEFs and was more similar to the IC50 in S47 homozygous MEFs (Fig. 3C). Similar decreases in cisplatin-mediated cell death were also evident in human heterozygous S47/wild-type LCLs (Fig. 3D). It is of note that these heterozygous S47/wild-type LCLs were from the child of the homozygous wild-type and S47 LCL donors, so genetic heterogeneity is reduced (see the Materials and Methods).
Figure 3.

Marked impairment of cisplatin-mediated apoptosis in S47/wild-type heterozygous cells. (A) Primary MEFs from the wild-type (Wt) or S47 Hupki mouse or S47/wild-type heterozygous mice were treated with 10 μM cisplatin (CDDP) for 24 h, and protein lysates were analyzed by Western blot analysis for the proteins indicated. The data depicted are representative of three independent experiments in a minimum of three independent batches of MEFs. (B) Quantification of the relative intensity of cleaved caspase-3 and cleaved lamin A blots from three independent experiments (as depicted in A) in primary MEFs from the wild-type (WT), S47, and S47/wild-type heterozygous mice untreated or treated with 10 μM cisplatin (CDDP) for 24 h. The level of signal in wild-type cells treated with CDDP was set to 100. Error bars mark standard deviation. (C) IC50 analysis for cisplatin (CDDP) in primary wild-type, S47, and S47/wild-type MEFs treated with the indicated concentrations of cisplatin for 72 h and analyzed for viability by the Alamar blue assay. The depicted data represent an average of three independent experiments. Error bars represent standard deviation. (D) Quantification of the relative intensity of cleaved lamin A (black box) and cleaved caspase-3 (gray box) signal from three independent experiments in LCLs that are wild type, S47, or heterozygous (S47/wild type). Cells were untreated or treated with 10 μM cisplatin (CDDP) for 24 h. The level of cleaved lamin A in untreated S47 cells was set to 1, and the fold change is depicted relative to that. The averaged results from three independent experiments with standard deviation are depicted.
Decreased transactivation of GLS2, PMAIP1 (NOXA), and SCO2 in S47 cells
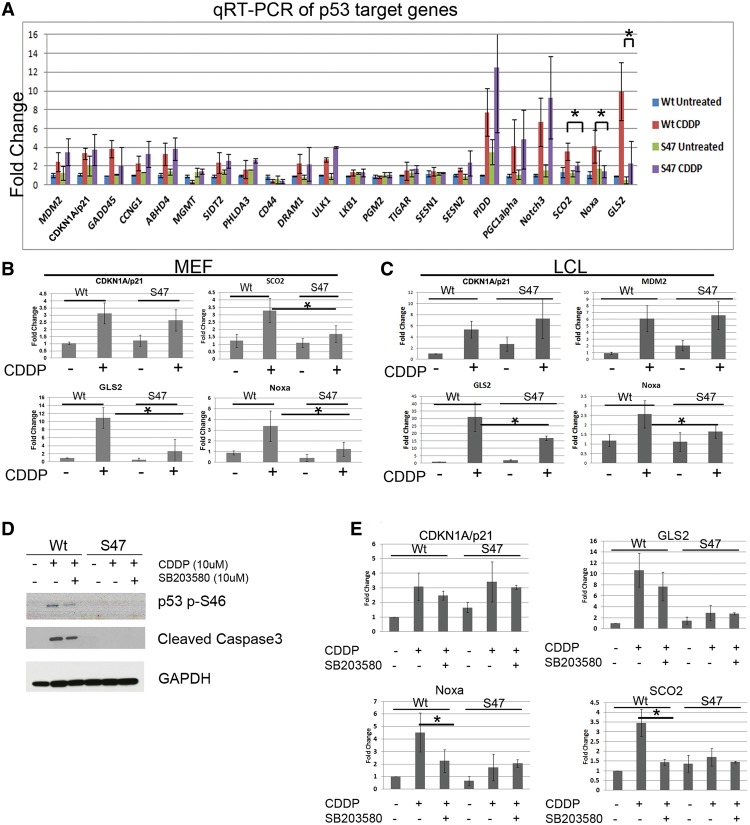
In order to compare the transcriptional potential of wild-type and S47 forms of p53, we performed quantitative RT–PCR (qRT–PCR) of over two dozen known p53 target genes in wild-type and S47 MEFs following treatment with cisplatin. This analysis revealed that the majority of these p53 target genes is induced similarly in wild-type and S47 MEFs following cisplatin; this includes targets responsible for growth arrest (Cdkn1a/p21, Gadd45, and Ccng1), autophagy (Dram1 and Ulk1), and metabolism (Tigar, Sesn1, Sesn2, and Ppargc1a) (Fig. 4A). This also included p53 target genes that are regulated by transactivation domain II of p53 (amino acids 41–83), where the S47 polymorphism resides: Abhd4, Mgmt, Sidt2, and Phlda3 (Brady et al. 2011). Of the over two dozen p53 target genes tested, only three showed significantly impaired transactivation by the S47 variant: These were Gls2, Noxa (Pmaip1), and Sco2 (cytochrome c oxidase assembly protein) (Fig. 4B). The differences in Noxa prompted us to also analyze Bbc3 (Puma), but we saw no difference in Bbc3 induction in S47 and wild-type cells (Supplemental Fig. S4A). Importantly, S47 LCLs treated with cisplatin also showed significantly reduced levels of GLS2 and NOXA mRNA, while the levels of CDKN1A and MDM2 mRNA were comparable in both genotypes (Fig. 4C); SCO2 expression was not detectable in this cell type. To assess the impact of GLS2 and NOXA on the impaired apoptosis in S47 cells, we silenced these genes in wild-type MEFs using either of two different short hairpins. This led to consistent abrogation of cisplatin-mediated cell death in these wild-type MEFs compared with a short hairpin control (Supplemental Fig. S4B,C). These data indicate that the impaired transactivation of Gls2 and Noxa explains at least in part the defect in cisplatin-mediated cell death in S47 cells.
Figure 4.
The S47 variant is impaired for transactivation of a subset of p53 target genes, including Gls2, Noxa (Pmaip1), and Sco2. (A) qRT–PCR analysis of p53 target genes in primary wild-type (Wt) and S47 MEFs treated with 10 μM cisplatin (CDDP) for 24 h. All values were normalized to a control gene (cyclophilin A). Data are averaged from three independent biological replicates. Error bars indicate standard deviation. (*) P < 0.05. (B) qRT–PCR analysis of the p53 target genes indicated in independent batches of primary MEFs from wild-type and S47 mice treated with 10 μM CDDP for 24 h. All values were normalized to a control gene (cyclophilin A). Data are averaged from three independent biological replicates. Error bars indicate standard deviation. (*) P < 0.05. (C) qRT–PCR analysis of the p53 target genes indicated in human LCLs that are homozygous for wild-type p53 or S47 treated with 10 μM CDDP for 24 h. All values were normalized to a control gene (cyclophilin A). Data are averaged from three independent biological replicates. Error bars indicate standard deviation. (*) P ≤0.05. SCO2 was not expressed in LCL cells, so these data are not depicted. (D) Western analysis for the proteins indicated in wild-type and S47 MEFs pretreated with 10 μM p38MAPK inhibitor SB203580 for 2 h followed by 10 μM cisplatin (CDDP) for 24 h. GAPDH served as the loading control. (E) qRT–PCR analysis of the cells in D for the p53 target genes indicated, normalized to control (cyclophilin A). The depicted data represent the average of three independent experiments. Error bars represent standard deviation. (*) P <0.05.
The identification of a subset of p53 target genes exhibiting impaired transactivation in S47 MEFs and LCLs provided us with an opportunity to address a fundamental issue regarding this variant; that is, whether the S47 variant is impaired for apoptosis and transactivation because of lack of phosphorylation on Ser46 or due to a separate, possibly structural, issue. To shed light on this question, we analyzed the transactivation of these three target genes (Sco2, Noxa, and Gls2) by wild-type p53 following cisplatin treatment in the presence and absence of the p38MAPK inhibitor SB203580, which inhibits Ser46 phosphorylation. We were able to confirm the findings of others that SB203580 inhibits Ser46 phosphorylation of wild-type p53 as well as cell death (Fig. 4D). Notably, qRT–PCR of RNA from these MEFs revealed that treatment with SB203580 had no effect on the transactivation of Cdkn1a but led to a significantly decreased ability of p53 to transactivate Sco2 and Noxa (Fig. 4E). Gls2 was decreased as well, but this did not reach statistical significance (Fig. 4E). These data were recapitulated when we silenced p38MAPK using siRNA (Supplemental Fig. S4D). The combined data support the premise that the failure of phosphorylation on Ser46 explains at least part of the decreased ability of the S47 variant to transactivate this subset of p53 target genes.
Decreased binding of S47 to the p53-binding sites in GLS2 and SCO2
We next sought to delineate the mechanism for impaired transactivation of Gls2, Sco2, and Noxa by S47. We first analyzed the ability of LCL lysates from cells containing wild-type and S47 proteins to bind to oligonucleotides containing the p53-binding site in the human CDKN1A promoter using a microsphere protein–DNA-binding assay for p53 (Noureddine et al. 2009). In this in vitro binding assay, we saw a consistently reduced ability of S47 lysates to bind to oligonucleotides containing this p53-binding site compared with wild-type lysates, suggesting that an altered DNA-binding ability might underlie part of the transcriptional defect (Supplemental Fig. S5A–C). These data prompted us to perform chromatin immunoprecipitation (ChIP) in primary MEFs containing wild-type p53 and S47 treated with cisplatin. For Gls2, we identified two consensus p53-binding sites in the murine gene—one in the promoter that overlapped the transcriptional start site and another that was nearly 75 kb upstream of the promoter; both of these sites had significant p53 ChIP-seq (ChIP combined with deep sequencing) peaks in published data sets (Li et al. 2012a, 2013; Kenzelmann Broz et al. 2013). In primary S47 and wild-type MEFs treated with cisplatin, we noted a significantly decreased ability of S47 to chromatin-immunoprecipitate the upstream Gls2 site as well as the previously identified Sco2-binding site; there was also a decreased ability to bind to the Noxa site, but this was not statistically significant (Fig. 5A). In contrast, there was no significant difference in the ability of S47 and wild-type p53 to chromatin-immunoprecipitate the major p53-binding site in the murine Cdkn1a promoter. The latter data suggest that our microsphere-binding assay might be assessing an in vitro binding defect of S47 that was not necessarily reflective of the endogenous murine Cdkn1a site (Fig. 5A). Overall, the impaired ability of S47 to bind to the Gls2 and Sco2 p53 consensus sites was consistent in independent batches of MEFs and was statistically significant (P < 0.05). Furthermore, the impaired binding of S47 to the p53-binding sites in GLS2 and SCO2, but not CDKN1A, was also evident by ChIP in H1299 cell lines containing tetracycline-inducible wild-type and S47 forms of p53 (Fig. 5B). NOXA (PMAIP1) was not expressed in H1299 cells, so we did not perform ChIP for this gene in these cells.
Figure 5.

Impaired DNA-binding ability of the S47 variant. (A) ChIP of primary wild-type (Wt) and S47 MEFs treated with 10 μM CDDP for 24 h analyzed using antisera to p53 (CM5) or IgG. The percentage binding normalized to input from qPCR analysis is shown. The data depicted are averaged from three independent experiments normalized to input. Error bars represent standard deviation. (*) P-value < 0.05. (B) ChIP analysis of p53 binding to the consensus elements from the genes indicated in human H1299 (p53-null) cells containing doxycycline-inducible wild-type or S47 forms of p53 in the absence and presence of 100 ng/mL doxycycline plus 10 μM cisplatin for 24 h. ChIP was performed using antisera to p53 (fl393G) or normal rabbit IgG. The data depicted are averaged from three independent experiments normalized to input. Error bars represent standard deviation. (*) P-value < 0.05. The IgG results are depicted for the CDKN1A/p21 p53-binding site but were comparable for all other sites analyzed. NOXA was not expressed in this cell line, so this gene was not analyzed.
Increased spontaneous cancer in S47 mice
At the beginning of this study, we set aside a cohort of 20 S47 and wild-type mice in order to analyze life expectancy and fecundity; we were surprised to find a significant percentage of our S47 mice developing spontaneous cancer. In all, 16 out of 20 (80%) of the homozygous S47 mice that were analyzed developed cancer between 12 and 18 mo of age. These cancers were of diverse histological types but predominantly included hepatocellular carcinoma, B-cell lymphoma, and histiocytic sarcoma, along with other tumor types (Fig. 6A–C). The presence of such a high fraction of hepatocellular carcinoma was interesting in light of the fact that high levels of GLS2 protein are normally expressed in this cell type and the data indicating that GLS2 is a tumor suppressor gene in human hepatocellular carcinoma (Liu et al. 2014). We also noted the presence of metastatic lesions in a small fraction of these mice (Fig. 6B,C). In a small cohort of S47/wild-type heterozygote mice, we also noted increased cancer incidence, again with a predisposition for hepatocellular carcinoma (Fig. 6C). Log rank analysis revealed a statistically significant difference in survival between wild-type and S47 mice (P < 0.0001) (Fig. 6D).
Figure 6.
Spontaneous tumorigenesis in S47 but not wild-type mice. (A) Hematoxylin and eosin staining of tumors from the S47 mouse. (Left) Normal liver and liver hepatocellular carcinoma (dotted outline). (Right) Infiltrating B-cell lymphoma in the kidney (arrows). Bar, 100 μm. (B) Hematoxylin and eosin staining of tumors from the S47 mouse. (Top left) Hepatocellular carcinoma. (Top right) Hepatocellular carcinoma metastasized to the lung (dotted outline). (Middle left) Pancreatic ductal adenocarcinoma (asterisks). (Middle right) Pancreatic ductal adenocarcinoma metastasized to a lymph node (asterisk). (Bottom left) Intestinal adenoma. (Bottom right) Stomach adenoma. Bar, 100 μm. (C) Cancer incidence in wild-type and S47 mice and in heterozygous wild-type/S47 mice. The data are representative of a total of 20 S47 and wild-type mice and 12 wild-type/S47 mice; for S47, 16 of these developed cancer. (BCL) B-cell lymphoma; (HCC) hepatocellular carcinoma; (HS) histiocytic sarcoma; (CC) colorectal carcinoma; (PA) pancreatic ductal adenocarcinoma. (D) Kaplan-Meier analysis of survival between wild-type (WT) and S47 mice. n = 20 each. The asterisk represents a single wild-type mouse that died from a noncancerous cause.
Decreased ferroptosis in S47 cells
Although we noted a significant defect in cisplatin-mediated apoptosis in S47 cells, some studies have questioned the relevance of the apoptotic pathway of p53 to tumor suppression (Li et al. 2012b). Moreover, we noted disparities in the amount of cell death induced by cisplatin when we performed assays for apoptosis (Annexin V) versus other cell viability assays (Alamar blue and Trypan blue), suggesting that another, nonapoptotic cell death pathway might be induced by cisplatin. Recent studies have implicated ferroptosis as important for p53-mediated tumor suppression (Jiang et al. 2015). Moreover, recently, the p53 target gene GLS2 was shown to be essential for ferroptosis (Gao et al. 2015). Therefore, we next sought to determine whether S47 cells are impaired for ferroptosis. We first treated S47, wild-type, and S47/wild-type MEFs with erastin, a compound that induces ferroptosis (Dixon et al. 2012). Incubation with 4 µM erastin for 8 h led to profound cell death in wild-type MEFs, consistent with data in the literature (Jiang et al. 2015); in contrast, there was no effect on either S47 or heterozygote S47/wild-type MEFs (Fig. 7A). Cell viability assays to assess the IC50 for erastin indicated that S47 MEFs were 27-fold resistant to this compound, and heterozygote S47/wild-type MEFs were fourfold resistant compared with wild-type MEFs (Fig. 7B). Interestingly, we found that the level of GLS2 was markedly up-regulated by erastin treatment in wild-type MEFs but not in S47 cells and that silencing Gls2 in wild-type MEFs phenocopied the cell death defect in S47 cells treated with erastin (Fig. 7C).
Figure 7.
Impaired ferroptosis in S47 cells. (A) Representative phase-contrast images of primary MEFs containing wild-type p53 (WT), heterozygous S47/wild type, or S47 treated with 4 μM ferroptosis inducer erastin or vehicle (DMSO) for 8 h (magnification 10×). Data represent the average of three independent studies. Bar, 20 μm. (B) Cell viability (Alamar blue) analysis of wild-type, S47/wild-type, or S47 primary MEFs treated with erastin for 72 h. The data represent the average of four independent experiments. Error bars represent standard error of the mean. (C) Western blot analysis for GLS2 in wild-type MEFs, wild-type MEFs infected with a lentiviral short hairpin for Gls2 (shGls2), and S47 MEFs untreated or treated with 4 µM erastin for 24 h. GAPDH served as the loading control. In the bottom panel, the percent viability using the Trypan blue exclusion assay is shown. Error bars represent standard deviation. (D) qRT–PCR analysis of slc7a11 normalized to cyclophilin A. The data are averaged from three independent biological replicates. Error bars represent standard deviation. (E) qRT–PCR analysis of Ptgs2 normalized to cyclophilin A. The data are averaged from three independent biological replicates. Error bars represent standard deviation. (F) Immunoblot analysis for GPX4 in wild-type and S47 MEFs following treatment with 10 µM CDDP for 24 h. (G) Cell viability analysis of wild-type and S47 human LCLs treated with RSL3 for 48 h. The data represent the average of three independent experiments. Error bars represent standard deviation. (H) Trypan blue exclusion analysis of the percent viability in wild-type MEFs or wild-type LCLs exposed to 10 µM CDDP, CDDP plus 2 µM ferrostatin-1 (Fer-1), or CDDP plus 20 µM zVAD-fmk. The data represent the average of three independent experiments. Error bars represent standard deviation. (I) Proposed model depicting the relative abilities of wild-type p53 and S47 to induce senescence, apoptosis, and ferroptosis and suppress spontaneous tumor initiation.
p53 is required for erastin-mediated ferroptosis, in part due to the ability of this protein to bind and negatively regulate the promoter for the cystine transporter SLC7A11 (Jiang et al. 2015). Therefore, we next analyzed the ability of S47 to negatively regulate this gene and found that the S47 variant is impaired for the ability to negatively regulate SLC7A11 (P = 0.006) (Fig. 7D). Consistent with this, ChIP indicated that the S47 protein had reduced ability to chromatin-immunoprecipitate the p53-binding site identified in the Slc7A11 promoter but not the Cdkn1a promoter (Supplemental Fig. S6A,B). Cisplatin treatment also led to differences in two other markers for ferroptosis between wild-type and S47 cells: Ptgs2 transactivation and GPX4 degradation, both of which were impaired in S47 cells treated with cisplatin (Fig. 7E,F).
We next sought to assess the sensitivity of wild-type and S47 LCLs to ferroptosis. In this case, we found that these human LCLs were considerably resistant to cell death by erastin; however, they were sensitive to cell death by another ferroptosis-inducing agent, RSL3 (Yang et al. 2014). We found that S47 LCLs were markedly resistant to ferroptosis induced by RSL3 (>19-fold) compared with wild-type LCLs (Fig. 7G). Finally, we sought to tie together some of our findings with ferroptosis and cisplatin by testing the possibility that some of the cell death induced by cisplatin might be due to ferroptosis; it is of note that both of these cell death pathways require reactive oxygen species (Zhu et al. 2013; Dixon and Stockwell 2014). Toward this end, we assessed the ability of zVAD-fmk, which inhibits apoptosis, and the compound Fer-1, which inhibits ferroptosis, to inhibit cisplatin-mediated cell death. We found that Fer-1 could partially but significantly rescue a percentage of cisplatin-mediated cell death and that zVAD-fmk was likewise only partially able to rescue cell death by cisplatin (Fig. 7H). Our combined findings suggest that S47 cells are resistant to both cisplatin-mediated cell death and ferroptosis and that cisplatin appears to induce some level of ferroptosis.
Discussion
In this study, we show that the S47 polymorphism in TP53 impairs the ability of p53 protein to induce cell death following cisplatin treatment and to transcribe a subset of target genes due in part to the decreased ability of S47 to bind to the response elements in these promoters. We show that this variant confers increased cancer risk in a mouse model and that it shows a defect in ferroptosis induction. In 2012, the Gu group (Li et al. 2012b) generated a knock-in mouse in which three p53 acetylation sites in the p53 DNA-binding domain were mutated to arginine; this knock-in version of murine p53 failed to transactivate the majority of p53 target genes or induce growth arrest or programmed cell death. Surprisingly, the “3KR” mouse failed to develop cancer, suggesting that tumor suppression by p53 might occur independently of growth arrest or apoptosis (Li et al. 2012b). More recently, this group identified the ability to down-regulate SLC7A11 and induce ferroptosis as one mechanism by which the 3KR mutant may function to suppress tumor development (Jiang et al. 2015). Our data on the S47 variant offer corroboration of the premise that the ferroptosis pathway may be influential in p53-mediated tumor suppression (Fig. 7I). However, we recently completed microarray analysis of the p53 transcriptional program in cisplatin-treated wild-type and S47 LCLs and identified only a dozen genes with defective transactivation by S47. The majority of these genes plays roles in metabolism, making it formally possible that the role of p53 in metabolism may underlie the impaired tumor suppression by the S47 variant.
The importance of Ser46 phosphorylation was identified as an important post-translational modification for p53-mediated cell death many years ago. Despite this fact, it has largely been elusive how this modification affects p53 function. This site is a major binding site for the peptidyl-prolyl isomerase Pin1, which mediates cis–trans isomerization of Pro47 of p53; notably, this binding and isomerization event has been shown recently to be critical for p53-dependent BAX activation and apoptosis (Follis et al. 2015). Specifically, in tamoxifen-inducible versions of the S47 form of p53, the investigators reported that interaction of S47 with Pin1 was reduced compared with wild-type p53 (Follis et al. 2015). We analyzed our wild-type and S47 LCLs for a Pin1–p53 interaction in the absence and presence of cisplatin but did not see an impaired interaction of S47 with Pin1; in fact, the interaction between Pin1 and S47 was increased in both S47 and S47/wild-type cells compared with cells containing wild-type p53 following cisplatin treatment (M Jennis, unpubl.). We cannot explain the differences in our findings from those noted above except that these differences may exist between endogenous and exogenous (inducible) p53 or be subject to genotoxic stress or cell type specificity. Additionally, whereas our data indicate that the S47 variant has enhanced interaction with Pin1, the possibility remains that Pin1 binds to the S47 protein but cannot catalyze cis–trans isomerization and instead interferes with p53 function.
The cancer phenotype of S47 mice is unusual. p53 knockout mice typically develop T-cell lymphoma and sarcoma and almost never develop epithelial cancers (Donehower et al. 1992). Mouse models for Li-Fraumeni syndrome, which contain DNA-binding domain mutants of p53, frequently develop lymphoma and sarcoma but also develop cancers of epithelial origin (Lang et al. 2004). However, hepatocellular carcinoma does not occur in either of these mice. Because of the known role of GLS2 in ferroptosis and the fact that the GLS2 gene is a tumor suppressor in human hepatocellular carcinoma (Liu et al. 2014), we speculate that the impaired ability of S47 to transactivate GLS2 may explain some of the tumor-prone phenotype of the S47 mice. Somewhat surprisingly, we found that S47/wild-type heterozygous cells more closely mirror the cisplatin and ferroptosis defect in homozygous S47 cells; we also found that heterozygous S47/wild-type mice are susceptible to cancer. Presently, there are >50 million African Americans in the United States, and we estimate that up to 1 million of these individuals may be heterozygous for the S47 variant. Furthermore, up to 20,000 individuals may be homozygous for S47. Our data in mice suggest that both of these groups of individuals may be at markedly increased cancer risk. It will be important to perform case control studies on these individuals in order to assess their cancer risk. The studies presented here are powerful rationale for performing genotyping on large African American sample sets for the impact of this variant on cancer risk.
Materials and methods
Cell culture, reagents, and plasmids
Human LCL cultures GM18870 (G/G, wild-type p53), GM18871 (A/A, homozygous S47), and GM18872 (A/G, heterozygous child from GM18870 and GM18871 parents) were identified for genotype at rs1800371 using the 1000 Genomes Web site and were obtained from the Coriell Institute. These were grown in RPMI supplemented with 15% fetal bovine serum (FBS) and 1% 100 IU/mL penicillin/100 µg/mL streptomycin (pen/strep). The genotype of these lines was confirmed by DNA sequencing. H1299 p53-null human non-small-cell lung carcinoma cells containing a tetracycline regulatory element (H1299 T-Reg) were provided by Steven McMahon (Thomas Jefferson University). The tetracycline-inducible p53 plasmid (Plenti4/TO/V5-DEST) was subjected to site-directed mutagenesis to generate the S47 variant. H1299 T-Reg wild-type or S47 p53 cells were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM; Cellgro), 1% pen/strep (Cellgro, 30-002-CI), and 10% tetracycline approved FBS (Clontech, 631106). MEFs were obtained from 12.5-d-old Hupki mice and were cultured at 37°C in DMEM with 1% pen/step and 10% FBS (Gemini, 100–106). Etoposide (Sigma, E1383), CDDP (Acros Organics, 193760010), and carboplatin (Sigma, C2538) were used at the indicated concentrations. Doxycycline (BD Biosciences, 631311) was used at a concentration of 100 ng/mL. RSL3 (Aobious, Inc., AO1514), erastin (Sigma Aldrich, 571203-78-6), and Fer-1 (Sigma Aldrich, SML0583) were used at the concentrations indicated.
Generation of S47 Hupki mice
Wild-type Hupki mice were generated previously. S47 mice were generated with the Hupki targeting construct as described (Luo et al. 2001) following site-directed mutagenesis to create serine at amino acid 47. All mice were backcrossed to C57Bl/6 for >10 generations, and, in the majority of cases, sibling littermates were used for analyses. All studies were performed in accordance with federal and institutional guidelines according to Institutional Animal Care and Use Committee protocols. Mice were housed in plastic cages with ad libitum diet and maintained with a 12-h dark/12-h light cycle at 22°C. For irradiation experiments, mice were exposed to a cesium-137 γ irradiation source (The Wistar Institute), and tissues were harvested 4 h later. Mice were injected intraperitoneally with cisplatin at 20 mg/kg.
Western blotting and immunohistochemistry
Primary antibodies used for Western blotting included p53 (ab6) (Calbiochem, OP43), p53 Ser-46-P (Abcam, ab122898), p53 Ser-15-P (Cell Signaling, 9284), MDM2 (ab1 and ab2) (Calbiochem, OP46T and OP115), p21 (ab6) (Calbiochem, OP79), cleaved lamin A (Cell Signaling, 2035), cleaved caspase-3 (Cell Signaling, 9061), GAPDH (14C10) (Cell Signaling, 2118), GPX4 (Abcam, 125066), and GLS2 (Abcam, ab113509). Secondary antibodies conjugated to horseradish peroxidase were used at a dilution of 1:10,000 (Jackson Immunochemicals). ECL was from Amersham (RPN2232). Densitometry quantification of protein signals was performed using ImageJ software (Taira et al. 2007). Immunohistochemistry of formalin-fixed tissues and Western blotting were performed as described (Frank et al. 2011).
qRT–PCR
Cells were lysed using QIAshredder columns (Qiagen). Total RNA was isolated using RNeasy minikits (Qiagen) and on-column DNase digestions (Qiagen) following the manufacturer's protocol. Equal amounts of RNA from these samples were then used to create cDNA using a high-capacity reverse transcription kit (Applied Biosciences, 4368814). qPCR was performed using Brilliant III UltraFast SYBR Green qPCR mix kits (Agilent Technologies) on a Stratagene Mx3005P device (Agilent Technologies). Data analysis of fold changes in gene transcription levels was done using the MxPro program (Stratagene). RNA expression levels were normalized to the housekeeping gene cyclophilin A.
Gene silencing of Noxa, GLS2, and p38MAPK
Cell lines with shRNA knockdown of Noxa and GLS2 were generated by infection with the lentiviral vector pLKO.1-puro carrying a shRNA sequence against mouse Noxa (shRNA1 [CGTGTGTTATTGTAAGCATTA; TRCN0000009706] and shRNA2 [GCACCGGACATAACTGTGGTT; TRCN0000009709]) or GLS2 (shRNA1 [GAACCTGCTATTTGCTGCATA; TRCN0000177991] and shRNA2 [CTCCCTCAATGAGGAAGGAAT; TRCN0000198612]) (Open Biosystems). VSVG-pseudotyped lentivirus was generated by cotransfection of 293-FT cells with shRNA constructs and packaging vectors according to the manufacturer's protocols (Invitrogen, K4960-00). Lentivirus was added to cells with 6 μg/mL polybrene for maximum viral transduction. Stable cells were selected using 1 μg/mL puromycin, and gene knockdown was confirmed by qRT–PCR. p38MAPK (Mapk14) was silenced by transfection with siControl or SMARTpool Accell Mapk14 siRNA (A-040125-13).
Cell viability assays, Annexin V staining, clonogenic survival
For cell viability assays, cells were plated at a density of 2000 cells per well on a 96-well plate and left to grow overnight in complete growth medium at 37°C. Cells were then treated with the indicated drugs at the indicated concentrations and left to incubate for 72 h. Cells were then incubated with Alamar blue (Life Technologies, DAL1025) for 2 h at 37°C, and viability was read out according to the manufacturer's protocol on a SynergyHT plate reader (BioTek), or the number of cells staining with Trypan Blue was counted using a hemacytometer. Data analysis (including logarithmic transformation, graphical analysis, and IC50 calculation) was conducted using Prism software (GraphPad). Annexin V staining to detect apoptotic cells and cell viability assay were performed using Guava Nexin reagent (Millipore, 4500-0450) on the Guava EasyCyte HT system according to the manufacturer's protocols. For clonogenic survival assays, 5000 viable cells were plated on 10-cm plates in complete growth medium and allowed to grow for 1 wk. The medium was then removed, cells were stained using Crystal Violet (Sigma, C3886), and colonies were captured by camera and counted.
DNA-binding assays
For ChIP analysis, primary MEFs or H1299 T-Reg wild-type or S47 cells were grown and treated on 150-mm plates to 60%–80% confluency before being harvested. Assays were carried out as described (Frank et al. 2011), and qPCR was performed and analyzed as described above. Primers for ChIP-qPCR were designed based on established literature (the human cells used were p21, GLS2, and SCO2; the mouse cells used were p21, SCO2, and Noxa) or by using a p53 response element-predictive algorithm (Su et al. 2015) for previously unknown p53 response elements in murine GLS2. For the microsphere assay for protein–DNA binding, allele-specific p53 binding was evaluated using the MAPD assay, as previously described (Noureddine et al. 2009; Bandele et al. 2011). All binding reactions were performed in triplicate.
Supplementary Material
Acknowledgments
We acknowledge the Cell Culture Facility and Transgenic Facility at The Fox Chase Cancer Center as well as the members of the Microscopy Facility (Fred Keeney), the Genomics Facility (Tran Nguyen and Celia Chang), and the Laboratory Animal Facility at The Wistar Institute for assistance with these studies. Support for Core Facilities used in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute. This research was funded by R01 CA102184 (M.E.M.) and PO1 CA114046 (to D.L.G). This work was supported in part by funding from the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health Z01-ES-100475 (to D.A.B.). M.J., C.-P.K., S.B., A.B.-K., J.I.-J.L., D.A.B., Q.L., M.H., D.L.G., and M.E.M. conceived and designed the study. K.Q.C., X.L., D.S.G., and M.H. provided reagents and expertise. M.J., C.-P.K., S.B., A.B.-K., J.I.-J.L., S.K., J.P.S., M.R.C., D.K.P., X.W., D.A.B., Q.L., M.H., D.L.G., and M.E.M. acquired and analyzed the data. M.J., D.L.G., and M.E.M. drafted the manuscript, and all authors provided critical revisions.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.275891.115.
References
- Bandele OJ, Wang X, Campbell MR, Pittman GS, Bell DA. 2011. Human single-nucleotide polymorphisms alter p53 sequence-specific binding at gene regulatory elements. Nucleic Acids Res 39: 178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV. 2014. The role of tumor suppressor p53 in the antioxidant defense and metabolism. Subcell Biochem 85: 337–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ Jr. 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J 18: 6845–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Stockwell BR. 2014. The role of iron and reactive oxygen species in cell death. Nat Chem Biol 10: 9–17. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. 2012. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Winter GE, Musavi LS, Lee ED, Snijder B, Rebsamen M, Superti-Furga G, Stockwell BR. 2015. Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem Biol 10: 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. [DOI] [PubMed] [Google Scholar]
- D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4: 11–19. [DOI] [PubMed] [Google Scholar]
- Felley-Bosco E, Weston A, Cawley HM, Bennett WP, Harris CC. 1993. Functional studies of a germ-line polymorphism at codon 47 within the p53 gene. Am J Hum Genet 53: 752–759. [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hollstein M, Xu Y. 2006. Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle 5: 2812–2819. [DOI] [PubMed] [Google Scholar]
- Follis AV, Llambi F, Merritt P, Chipuk JE, Green DR, Kriwacki RW. 2015. Pin1-induced proline isomerization in cytosolic p53 mediates BAX activation and apoptosis. Mol Cell 59: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AK, Leu JI, Zhou Y, Devarajan K, Nedelko T, Klein-Szanto A, Hollstein M, Murphy ME. 2011. The codon 72 polymorphism of p53 regulates interaction with NF-κB and transactivation of genes involved in immunity and inflammation. Mol Cell Biol 31: 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. 2015. Glutaminolysis and transferrin regulate ferroptosis. Mol Cell 59: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochola LF, Zeron-Medina J, Meriaux S, Bond GL. 2010. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2: a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez Losa J, Parada Cobo C, Guinea Viniegra J, Sanchez-Arevalo Lobo VJ, Ramon y, Cajal S, Sanchez-Prieto R. 2003. Role of the p38 MAPK pathway in cisplatin-based therapy. Oncogene 22: 3998–4006. [DOI] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol 4: 1–10. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. 1991. p53 mutations in human cancers. Science 253: 49–53. [DOI] [PubMed] [Google Scholar]
- Humbey O, Pimkina J, Zilfou JT, Jarnik M, Dominguez-Brauer C, Burgess DJ, Eischen CM, Murphy ME. 2008. The ARF tumor suppressor can promote the progression of some tumors. Cancer Res 68: 9608–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. 2015. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzelmann Broz D, Spano Mello S, Bieging KT, Jiang D, Dusek RL, Brady CA, Sidow A, Attardi LD. 2013. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev 27: 1016–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. 2008. SnapShot: p53 posttranslational modifications. Cell 133: 930–930 e931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119: 861–872. [DOI] [PubMed] [Google Scholar]
- Li X, Dumont P, Della Pietra A, Shetler C, Murphy ME. 2005. The codon 47 polymorphism in p53 is functionally significant. J Biol Chem 280: 24245–24251. [DOI] [PubMed] [Google Scholar]
- Li M, He Y, Dubois W, Wu X, Shi J, Huang J. 2012a. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell 46: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. 2012b. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu J, McLaughlin N, Bachvarov D, Saifudeen Z, El-Dahr SS. 2013. Genome-wide analysis of the p53 gene regulatory network in the developing mouse kidney. Physiol Genomics 45: 948–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhang C, Lin M, Zhu W, Liang Y, Hong X, Zhao Y, Young KH, Hu W, Feng Z. 2014. Glutaminase 2 negatively regulates the PI3K/AKT signaling and shows tumor suppression activity in human hepatocellular carcinoma. Oncotarget 5: 2635–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Yang Q, Tong WM, Hergenhahn M, Wang ZQ, Hollstein M. 2001. Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 20: 320–328. [DOI] [PubMed] [Google Scholar]
- Malkin D, Jolly KW, Barbier N, Look AT, Friend SH, Gebhardt MC, Andersen TI, Borresen AL, Li FP, Garber J, et al. 1992. Germline mutations of the p53 tumor-suppressor gene in children and young adults with second malignant neoplasms. N Engl J Med 326: 1309–1315. [DOI] [PubMed] [Google Scholar]
- Noureddine MA, Menendez D, Campbell MR, Bandele OJ, Horvath MM, Wang X, Pittman GS, Chorley BN, Resnick MA, Bell DA. 2009. Probing the functional impact of sequence variation on p53–DNA interactions using a novel microsphere assay for protein-DNA binding with human cell extracts. PLoS Genet 5: e1000462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862. [DOI] [PubMed] [Google Scholar]
- Pietsch EC, Humbey O, Murphy ME. 2006. Polymorphisms in the p53 pathway. Oncogene 25: 1602–1611. [DOI] [PubMed] [Google Scholar]
- Reinbold M, Luo JL, Nedelko T, Jerchow B, Murphy ME, Whibley C, Wei Q, Hollstein M. 2008. Common tumour p53 mutations in immortalized cells from Hupki mice heterozygous at codon 72. Oncogene 27: 2788–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprowl JA, Lancaster CS, Pabla N, Hermann E, Kosloske AM, Gibson AA, Li L, Zeeh D, Schlatter E, Janke LJ, et al. 2014. Cisplatin-induced renal injury is independently mediated by OCT2 and p53. Clin Cancer Res 20: 4026–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Wang X, Campbell MR, Song L, Safi A, Crawford GE, Bell DA. 2015. Interactions of chromatin context, binding site sequence content, and sequence evolution in stress-induced p53 occupancy and transactivation. PLoS Genet 11: e1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. 2007. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 25: 725–738. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408: 307–310. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. 2009. Blinded by the light: the growing complexity of p53. Cell 137: 413–431. [DOI] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. 2014. Regulation of ferroptotic cancer cell death by GPX4. Cell 156: 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Regunath K, Jacq X, Prives C. 2013. Cisplatin causes cell death via TAB1 regulation of p53/MDM2/MDMX circuitry. Genes Dev 27: 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.