Abstract
Viruses use a strategy of high mutational rates to adapt to environmental and therapeutic pressures, circumventing the deleterious effects of random single-point mutations by coevolved compensatory mutations, which restore protein fold, function or interactions damaged by initial ones. This mechanism has been identified as contributing to drug resistance in the HIV-1 Gag polyprotein and especially its capsid proteolytic product, which forms the viral capsid core and plays multifaceted roles in the viral life cycle. Here, we determined the X-ray crystal structure of C-terminal domain of the feline immunodeficiency virus (FIV) capsid and through interspecies analysis elucidate the structural basis of co-evolutionarily and spatially correlated substitutions in capsid sequences, which when otherwise uncoupled and individually substituted into HIV-1 capsid impair virion assembly and infectivity. The ability to circumvent the deleterious effects of single amino acid substitutions by cooperative secondary substitutions allows mutational flexibility that may afford viruses an important survival advantage. The potential of such interspecies structural analysis for preempting viral resistance by identifying such alternative but functionally equivalent patterns is discussed.
In the evolutionary host-virus arms race, interactions are based on an endless cycle of adaptations in which the virus necessarily evolves to manipulate and survive the hostile host environment and the host adapts to disrupt this manipulation. The human immunodeficiency virus (HIV) manages to escape eradication by drugs and immune responses mainly through a strategy of high turnover and extremely high mutational rate. Random mutations, however, rarely correlate directly to survival adaptations and more often have deleterious effects. Mutations can directly impair enzymatic activity or perturb interfaces central to intrinsic folding or extrinsic interactions essential for crafting protein assemblies of virus and viral-host complexes. Auxiliary mutations, which induce compensatory structural and functional changes, can stabilize a mutant protein facilitating its persistence in an evolved virus strain (reviewed in1). Essentially, coevolved second-site compensatory mutations repair the protein fold and/or enzymatic activity, or fix protein interfaces vital for interactions with other proteins2,3. The latter strategy may be achievable via complementary mutations in the binding partner3 or through interactions with alternative partners, which are functionally equivalent and redundantly available, resulting in rerouting-resistance4.
Positions of amino acid pairs evolving in a correlated manner have long been proposed to prescribe protein structure and function, and such correlation rules have been found sufficient to describe protein fold and guide the design of artificial sequences that consequently fold into native structures2. Co-evolution guided structure prediction has neatly been demonstrated in a recent study accurately modeling novel protein structures using distance restraints between amino acid pairs as predicted from co-evolutionary patterns5, a method that can further be enhanced upon combining the strong correlations of amino acid pairs with their weak correlations at the genetic codon-level6.
In addition to providing insight into the basic relationship between amino acid sequence and protein fold, identifying and characterizing correlated substitutions in naturally diverse and coevolved resistant viruses is imperative to drug and vaccine development3. One of the promising therapeutic targets in HIV-1 biology is the Gag polyprotein and especially its capsid (CA) proteolytic product, which forms the viral capsid core and plays multifaceted fundamental roles in the viral life cycle7,8,9,10. Comprehensive analyses of HIV-1 Gag sequences, examining patterns of variability for such correlation rules, have indeed identified numerous coevolved pairs of linked mutations contributing to drug resistance7,8,11,12,13,14 and have been attributed either to natural virus polymorphism7,11 or drug-challenges15,16,17. Although one study demonstrated that HIV-1 CA is an extremely genetically fragile protein, with 70% of the 135 tested single point mutations (covering 44% of CA sequence) yielding replication defective viruses18, another showed that mutating the most conserved residues in HIV-1 CA does not strongly associate with the highest fitness costs19. These apparently contradictory results can be consolidated by considering conserved functional coupling of mutations since focusing on the specific identities of conserved residues or individual mutations, rather than coevolved patterns, could suggest misleading fragility.
To extend our understanding of the functional and spatial correlations of coevolved substitutions, we investigated the primary and tertiary structures of CA from a non-primate lentivirus, the feline immunodeficiency virus (FIV). FIV is the lentivirus that most closely parallels HIV-1 in replication biology and pathogenesis, and is the only non-primate model in which the lentivirus induces an AIDS-like syndrome in its natural host. This makes FIV an attractive small animal model for anti-AIDS vaccine and drug research20,21. While proteins from the feline lentivirus share an abundance of structural and functional commonalities with HIV-1 (reviewed in22), their amino acid sequences highly diverge. Therefore, pinpointing interspecies subtleties can highlight crucially conserved patterns for drug and vaccine targeting, and more importantly, can uncover conceivably latent escape patterns accessible to challenged HIV-1. Here we show the structural basis of co-evolutionarily and spatially correlated substitutions in non-primate CA sequences, which when otherwise uncoupled and individually substituted into HIV-1 CA impair virion assembly and infectivity.
Results and Discussion
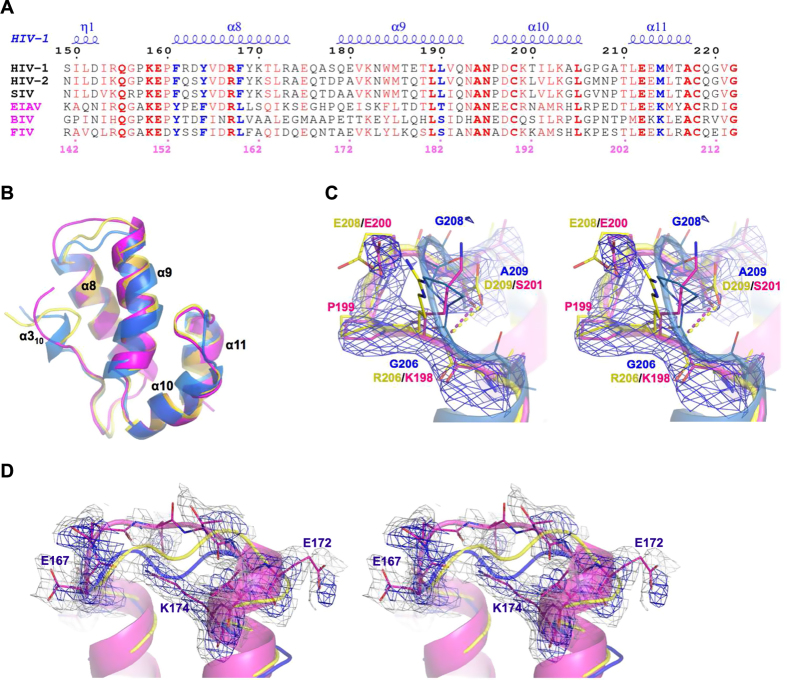
Capsid (CA) proteins from different retroviruses have remarkably low sequence identity23,24. So divergent are the sequences that even amino acids demonstrated as critical in a particular retrovirus are non-conserved in another. For example, in HIV-1 CA the Y164F or G208E mutations attenuate virion production and infectivity18. Despite this essentiality for the function of HIV-1 CA, these amino acids are not individually conserved and CA from feline immunodeficiency virus (FIV) inherently harbors F and E at these positions respectively (Fig. 1A). Nevertheless, the α-helical fold of the CA protein and capsid-core assemblies are highly conserved across the various retroviruses including HIV-1, equine infectious anemia virus (EIAV), human T-cell leukemia virus type I (HTLV) and Rous sarcoma virus (RSV) (reviewed in25). There appears to be a strong selective pressure that necessitates maintenance of a specific functional structure even during the mutation and evolution of CA encoding sequences10,23,26. Indeed, a more holistic sequence comparison demonstrates the significance of coevolved substitutions and resolves the conundrum of the strict conservation of structure and function despite considerable variability in amino acid sequences. A simple sequence analysis immediately highlights that the Y164 mentioned above forms part of a conserved amino acid pair F/Y in primate viruses (HIV-1 F161/Y164) that is switched to Y/F in non-primate viruses (Fig. 1A). This suggests that the deleterious action of the single Y164F mutation in HIV-1 CA might be rescued by the acquisition of a second-site mutation, F161Y, which would restore the conserved amino acid set. To probe for such structurally essential and potentially correlated pairs in the lentiviral CA, we determined the structure of the C-terminal domain (CTD) of FIV CA (CACTD), which naturally evolved to cope with substitutions individually shown deleterious to HIV-1 CA, and compared it to other non-primate and primate lentiviral CA structures available in the Protein Data Bank. Importantly, the CTD segment of CA, unlike the N-terminal domain (NTD), has been reported as biologically equivalent and functionally transferable between primate and non-primate lentiviruses23, making the interspecies comparison between FIV and HIV-1 physiologically relevant and especially convincing.
Figure 1. Preservation of FIV CACTD overall structure.
(A) Sequence alignment of CACTD. Top numbering and secondary structure are shown for HIV-1 (4XFX) and bottom numbering for FIV. Identical residues are shown in bold red font and conserved equivalents in red font. Bold blue font indicates correlated sites that are different between primate (black titles) and non-primate (pink titles) lentiviruses. (B) Superposition of CACTD of FIV (magenta), HIV-1 (blue, 4XFY) and EIAV (yellow, 1EIA). FO-FC map (blue mesh, 2.5 σ) calculated after omitting corresponding fragments of hook-like residues 197–202 (C) or loop (residues 166–174) connecting α-8/α-9 (gray mesh represents 2FO-FC map at 1.0 σ) (D). Panels (C) and (D) are in walleye stereo view.
The highly preserved FIV CACTD fold reveals only minor structural subtleties
Unsurprisingly, FIV CA encoded an absolutely conserved CTD fold comprising the characteristic four α-helical bundle (α-8 to α-11), which perfectly superimposes to CA of EIAV (0.55 Å RMSD) and HIV-1 (1.14 Å RMSD in average of several structures) (Fig. 1B).
Along with maintaining the highly conserved fold, the structure of FIV CACTD also retains the delicately tuned plasticity of CA α-helices and connecting-loops, which drive and support the dynamic structural changes that CA undergoes during the viral lifecycle. In the FIV CACTD structure, the flexible 310-helix adopts a rather extended conformation similar to a previous NMR observation in tubular assemblies of HIV-1 CA27 (Fig. 1B). Since this 310-helix is contained within the inter-domain linker connecting CTD and NTD domains, its dynamic nature has been anticipated to allow the flexible reorganization of the two domains during CA assembly27. The loop connecting α-10 and α-11 forms a distinct hook-like structure in FIV CACTD, a feature that is not seen in the HIV-1 CA structures but is superimposable to that of the EIAV CA structure (Fig. 1B,C). This hook-like configuration appears necessary to accommodate the larger Glu, Lys and Arg side chains in non-primate lentiviruses as compared to the conserved Gly in primate viruses (discussed below). Another elastic connector is the flexible loop connecting α-8 and α-9, which was poorly resolved in the FIV CACTD structure and could not be modeled exactly according to structures from HIV-1 or EIAV as these would clash with the well-structured K174 of FIV CACTD (Fig. 1D). This loop has indeed been shown to display a wide range of conformations in reported CA structures, depending on the arrangement of the dimeric interfaces, and is missing in some crystal structures28 and in the solid-state NMR analysis of tubular CA27. Flexibility of this loop correlates with the crucial pliability of α-9, which in the FIV CACTD structure was best superimposed to α-9 of HIV-1 CA in the CAI-inhibitor bound (PDB: 2BUO29) and the dehydrated crystal (PDB: 4XFY30) states (Fig. 1B). A disulfide bridge, conceivably important for CA flexibility and formed between two conserved lentiviral Cys residues (190 and 210 in FIV), was reduced in a similar manner to structures of HIV-1 CA bound to CAI29 and domain-swapped24.
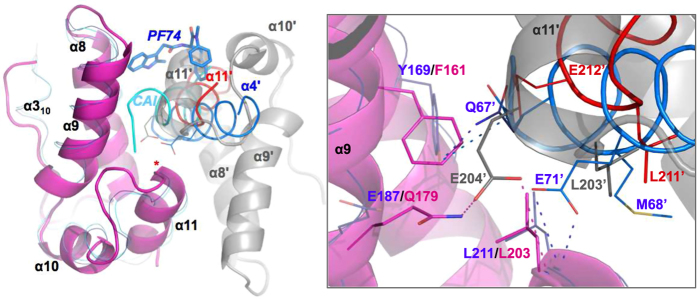
A novel dimeric interface in the FIV CACTD structure mimics the binding of the CAI inhibitor helical-peptide
Flexibility of CA is most appreciated in playing a pivotal role during conformational transitions, which are triggered upon proteolytic maturation of the viral Gag-polypeptide31,32 and result in the trimeric CTD–CTD interface unique for mature capsid formation and stability30,32. Distinct CA interacting interfaces have been implicated at different stages of Gag maturation including NTD α-4 packing against a hydrophobic pocket, which is formed by α-8 and α-9 in the CTD30,33. This hydrophobic pocket was previously shown to bind small molecules30 and CAI peptide29 inhibitors, and it has been proposed to interact with host cofactors10. Mutational analysis of pocket-residues suggested that CTD–CTD dimerization is very sensitive to the detailed contacts within this conserved groove and underscored an allosteric role in regulating CA reorganization33.
The asymmetric unit of the FIV CACTD crystal contains two CTD molecules packed through a novel dimeric interface that docks the α-11′ symmetry mate (denoted with the prime symbol) into this conserved pocket resembling the binding of the CAI helical-peptide and PF74 small molecule (Fig. 2). α-11′ docking at this groove also parallels the packing of α-11′ from symmetry related molecules in the crystal packing of two HIV-1 CA mutants, Y169A and L211S (PDB: 3DS2 and 3DPH33), and comparison with these structures demonstrates the spatial superposition of two conserved residues, Glu (HIV-1 E212′ and FIV E204′) and Leu (HIV-1 L211′ and FIV L203′) (Fig. 2 and inset, red helix). Indeed, a E212A mutation in HIV-1, which would disturb such α-11 docking reduces infectivity by 3-fold25, implicating a potential biologic relevance of such docking, perhaps transient, during capsid maturation or assembly.
Figure 2. FIV CACTD crystal packing.
FIV CACTD dimer (magenta and gray) compared to HIV-1 CA bound to CAI helical-peptide (cyan ribbon, 2BUO), symmetry NTD α-4 (blue ribbon, 4XFZ) or PF74 inhibitor (blue sticks, 4XFZ). Symmetry HIV E71′ and FIV E204′ are shown in blue and gray lines, respectively. α-11′ from a symmetry mate of HIV Y169A mutant is shown (red ribbon, 3DS2). Red asterisk indicate position of HIV L211 (FIV L203). Inset: close up of the core around the red asterisk. Residues of HIV and FIV are shown in lines (symmetry mates denoted with a prime-symbol). Dashed magenta (FIV) and blue (HIV) lines denote 2.6 – 4.5 Å distances. Two conformations for Q67′, native (blue) and modeled (dark blue), are shown. Residue labels are colored in accordance to structure (HIV: blue, FIV: magenta, EIAV: yellow).
Walling the groove are FIV F161 (HIV-1 Y169) and L203 (HIV-1 L211) that appear essential for packing the α-11 from the adjacent monomer (Fig. 2, inset). These residues are indeed conserved in most retroviruses and while the overall intrinsic structures of HIV-1 Y169A and L211A/S CACTD mutants were not altered, cone-shaped cores and virus infectivity were completely lost, suggesting that these positions are critical for CA reorganization and assembly of mature-like particles33. Further, HIV-1 Y169 and L211 were also implicated in the docking of α-4 of NTD to this CTD groove (Fig. 2, blue helix). This packing of α-4 places HIV-1 vital and conserved E71′ (and Q67′)25 in a close spatial resemblance to the conserved E204′ of FIV α-11, mimicking comparable contacts with the backbone amide of HIV-1 L211 (FIV L203) from adjacent CA monomer (Fig. 2, inset). Therefore, the novel crystal packing of FIV CACTD molecules via the binding groove of CTD from one monomer and α-11′ of the adjacent one, while resembling reported interactions of HIV-1 CA subunits, suggests that the subtle allosteric groove of FIV CACTD is also preserved despite the different amino acid composition.
FIV CACTD coevolved-substitutions preserve the functional fold despite sequence divergence
The high sequence homology (~40% identity) between FIV and HIV-1 CACTD sufficiently encoded for a highly preserved structure. Still, 70% of the 135 tested single point mutations in HIV-1 CA yielded defective viruses18, indicating that the divergent sequences comprise coupled substitutions that compensate for the sequence alterations in delicately tuning and preserving the functional fold of CA. To uncover such compensatory correlation rules, we analyzed the divergent sets of structurally correlated amino acids at the inter- and intra-molecular interfaces facilitating such delicate structural preservation. The amino acid sets examined here were those that can directly modulate intrinsic stability, folding and self-assembly of the CA protein. Amino acid sets that can alter CA patterns in exploiting different cellular cofactors and alternative pathways were beyond the scope of this study.
At the core of HIV-1 CACTD is a conserved Y164. Mutation to Phe, naturally present at this position in other retroviruses24, causes a 36% reduction in HIV-1 single-cycle infectivity and a 69% decrease in spreading fitness18, suggesting a functional role of the Y164 hydroxyl oxygen. Indeed, structural analysis of the available HIV-1 CA structures reveals a conserved interaction (2.5 Å) between this oxygen and the backbone amide of residue 190, an interaction that appears to pin down the C-terminal end of α-9 while leaving its N-terminal end extremely flexible (Fig. 3A). In order to maintain this apparently essential structural feature in the presence of Phe rather than Tyr, non-primate lentiviruses have apparently coevolved the residue at 161 (F161 in HIV-1) as Tyr so preserving (in a flipped direction) the F161/Y164 pattern of HIV-1 as Y153/F156 (FIV CA) (Fig. 1A). This configuration both preserves the pinning interaction between Tyr and the C-terminal end of α-9 (Fig. 3A) and maintains the hydrophobic core features. Therefore, whilst the F/Y pair is imperative for structural integrity, the relative position of either residue is apparently interchangeable.
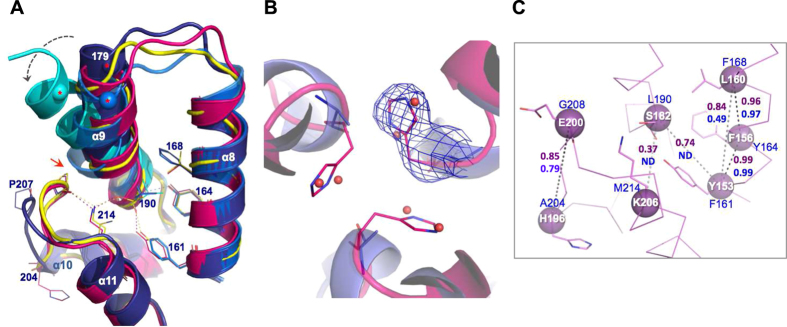
Figure 3. Structural basis of correlated substitutions.
(A) Superposition of CACTD of FIV (magenta), EIAV (yellow, 1EIA), HIV-1 native (dark blue, 4XFX), dehydrated (blue, 4XFY) and domain-swapped (cyan, 2ONT) forms. HIV-1 residue numbering is shown. Dynamic N-terminus of α-9 (spheres with red asterisks: Cα of E179) is highlighted with a dashed gray arrow. Red arrow stresses steric hindrance between HIV P207 and α-9 bulge. Interactions are shown in dashed lines colored yellow (EIAV), blue (HIV) or magenta (FIV). Residue 190 denotes the static C-terminus base of α-9. (B) Trimeric CA interface of FIV (magenta) modeled per native HIV-1 structure (dark blue, 4XFX). HIV A204 (blue lines) and water molecules (red spheres) and FIV H196 (magenta lines) are shown. FO-FC map (blue mesh, 2.5 σ) calculated after omitting H196 of FIV CA. (C) Correlated residues of FIV CA (magenta ribbon) are presented in magenta spheres (Cα) and side chains (lines). Residue numbering of FIV (white font) and HIV-1 (blue font) are shown. Correlated pairs are coupled with dashed lines of black (high score) or gray (weaker score) colors and the Gremlin’s probability score5 is shown in magenta (FIV) and blue (HIV-1) font. ND: not detected.
Indeed, comparison of primate and non-primate CACTD amino acid sequences highlights a sequence pattern, which extends beyond this F/Y or Y/F pair, to include F/Y/F/L/M in primate viruses and Y/F/L/S(T)/K in FIV and other non-primate viruses (Fig. 1A). Switching the L190/M214 hydrophobic packing pair of primate viruses into the S(T)/K polar pair in non-primate viruses creates a potential hydrogen bond in the latter structure (Fig. 3A). Creation of such a polar interaction apparently requires the presence of both S(T) and K since a single substitution to L190S or M214L in HIV-1 CA resulted in either non-viable viruses or viruses with diminished virus viability and infectivity18. Therefore, the FIV inherent S182 (T190 in EIAV), at this HIV-1 190 position (Fig. 1A), may have been tolerated in the non-primate viruses by the co-acquisition of the compensatory Lys partner (K206 in FIV, and K214 in EIAV, instead of HIV-1 M214) (Fig. 3A). The stabilizing effect of the HIV-1 L/M packing or FIV S/K interaction appears crucial in the accurate positioning and pinning of the α-9 C-terminal base in a similar manner to the F/Y or Y/F pair discussed above.
FIV K206 also interacts (2.7 Å) with the carbonyl oxygen of the conserved FIV P199 (HIV-1 P207) stabilizing the above-described hook-like structure of the loop connecting α-10 and α-11. In HIV-1, this loop, which has been shown to mediate the trimeric interface30 and significantly change conformation27 during CA assembly, harbors a crucial G208 residue and a G208E mutation resulted in diminished HIV-1 infectivity and spreading fitness18. FIV CA inherently possesses E200 (E208 in EIAV) at this position, however the detrimental effect of HIV-1 G208E appears to have been compensated by the coevolution of a polar side-chain in FIV, S201 (D209 in EIAV), which substitutes HIV-1 A209 and interacts (2.9 Å) with the backbone amide of residue 198 in FIV (206 of HIV-1/EIAV), which forms the hook-like structure accommodating and “pulling-back” the larger E200 residue (Fig. 1C). Indeed, a polar side-chain substitution with A209T in HIV-1 retained wild type infectivity (108%) and spreading fitness while substitution with a nonpolar side-chain, A209V, retained only 72% infectivity18.
The smaller non-primate L160 (Y/F/L/S(T)/K/H) substituting the primate F168 (F/Y/F/L/M/A) correlates better with the hook-like structure since F168 in HIV-1 causes a bulge in the middle of α-9 that would conceivably clash with conserved P207 in a non-primate hook-like structure(Fig. 3A, red arrow).
Similarly, the overall structural conservation at the delicate trimeric 3-fold interface also conceals the significant sequence variations and corresponding adaptations. The HIV-1 A204 is substituted with a bulkier H196 in FIV CA (Fig. 1A). While hydrophobic substitutions of A204 to V/L retained HIV-1 infectivity and capsid stability, A204D replacement resulted in the production of non-infectious virions with unstable and abnormal cores30,34. Interestingly, two strategically structured water molecules, positioned near A204 at the trimeric interface of native CA (Fig. 3B), were lost upon crystal dehydration resulting in tighter packing of CA subunits30. The hydration layer, especially at the two- and threefold interfaces, has been proposed to strategically stabilize these variable interfaces and complement the flexible surface of CA30. Superposing the FIV structure to the trimeric interface of HIV-1 native CA structure (PDB: 4XFX) reveals the accurate placement of the FIV H196 imidazole ring onto the two waters from each HIV-1 CA monomer (Fig. 3B). This supports the strategic location and role of these two waters in CA flexibility and highlights a strategic role for H196 in FIV CA flexibility.
Analyzing coevolved residues using the Gremlin pseudolikelihood method, which previously predicted protein structures based on coevolved distance restraints5, revealed a correlation probability of ~1.0 for the adjacent pairs within HIV-1 F/Y/F and FIV Y/F/L supporting our structural-based correlation analysis (Fig. 3C). Likewise, a correlation between the FIV H196 (or HIV A204), at the trimeric interface (Fig. 3B), and the covariant FIV E200 residue (G208 in HIV) of the hook-like loop, was successfully detected (Fig. 3C). However, a correlation between the S/K pair, which was revealed in our FIV CACTD structural analysis, was poorly predicted with 0.37 probability in FIV and missed for HIV-1 (Fig. 3C), emphasizing the power of structural analysis in revealing spatially coupled correlations.
Here we provide a simple demonstration of the power of interspecies structural analysis in elucidating functional coupling of co-evolutionarily and spatially correlated pairs of substitutions, which when otherwise uncoupled and individually assessed can mistakenly implicate fragility and propose misleading hot-spots for therapeutic targeting. Structural comparison of the FIV and HIV-1 CA reveals the mechanistic basis of functional coupling of coevolved sets that spatially cooperate to preserve a viable structure, protecting the virus from assumed genetic fragility. The ability to circumvent deleterious effects of single amino acid substitutions by cooperative secondary substitutions allows mutational flexibility that may afford viruses an important survival advantage. This mechanism provides the virus with flexibility to exploit alternative but functionally equivalent patterns when the default ones are impaired by mutations, especially during the acquisition of resistance to antiviral therapeutics and cellular restrictions. The accessibility of such hidden escape mechanisms to particular viruses can be uncovered by identifying distinct but functionally equivalent interactions naturally coevolved in relative viruses. Intrinsic differences in the different selective pressures of cell culture and whole organisms underscore the need for an adequate animal model to evaluate potential risks from newly emerging resistant virus strains, and in this regard, the ability to investigate FIV coevolution in its natural host offers a unique opportunity.
Methods
Protein Expression and Purification
Capsid (CA) C-terminal domain (CTD, residues 143–211) of Feline immunodeficiency virus (FIV) Petaluma strain was subcloned into pET28b (Novagen) with a thrombin-cleavable His-tag at the N-terminus.
ArcticExpress cells (Agilent Technologies Genomics) containing the CACTD construct were grown to an O.D600 of 0.6 (LB media, 37 °C), induced with 0.5 mM IPTG and grown for a further ~20 hours at 16 °C. Harvested cells were resuspended in lysis buffer (50 mM potassium phosphate buffer pH7.5, 5 mM Imidazole, 5 mM β-mercaptoethanol and 500 mM NaCl), disrupted and the His-tagged proteins from the supernatant were captured using HisPur cobalt resin (Pierce) according to the manufacturer instructions. Protein eluted in 150 mM Immidazole was dialyzed to 20 mM HEPES pH 7.4, 1 mM DTT, and 150 mM NaCl. Thrombin (2 units/mg, Novagen) was used to cleave the His-tag (4 °C, overnight), and was subsequently removed using benzamidine sepharose resin (GE Healthcare Life Sciences). FIV CACTD protein was further purified using Superdex-200 size exclusion column (GE Healthcare Life Sciences) in dialysis buffer.
Protein Crystallization, Data Collection and Structure Determination
Purified FIV CACTD (~7 mg/ml) was mixed with an equal volume of crystal screen well buffer and crystals were grown (20 °C) using the sitting-drop vapor-diffusion method. Diffraction quality crystals were readily obtained in the B7 condition of Index (Hampton Research) containing 56 mM Sodium phosphate monobasic monohydrate and 1.344 M potassium phosphate dibasic (1.4 M buffer at pH 8.2). Prior to data collection the crystal was flash frozen in the B7 condition of Index (Hampton Research) supplemented with 20% (v/v) glycerol.
Diffraction data was collected on a Rigaku FR-X rotating anode generator coupled to VariMax-Cu/Cr DW confocal optics and an R-AXIS HTC image plate detector, and processed using HKL300035. The structure was solved by molecular replacement using Phaser36 and 3DTJ33 as a search model. The resulting model was fitted to the electron densities using COOT37 and refined, in CCP4 suite38, using REFMAC539. Table 1 summarizes data collection and refinement statistics. Figures presenting structures, as well as structural superposition, were prepared using PyMOL Molecular Graphics System (Schrödinger, LLC).
Table 1. Data collection and refinement statistics.
| FIV CA CTD | |
|---|---|
| Data collection | |
| Space group | P 61 2 2 |
| Mol/ASU Cell dimensions | 2 |
| a, b, c (Å) | 64.19, 64.19, 178.56 |
| α,β,γ (°) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 40.63–2.29 (2.37–2.29)* |
| Rsym or Rmerge | 0.108 (0.529) |
| I/σI | 17.91 (2.92) |
| Completeness (%) | 99.71 (96.96) |
| Redundancy | 16.33 (6.44) |
| Refinement | |
| Resolution (Å) | 2.29 |
| No. unique reflections | 10456 (988) |
| Rwork / Rfree (5% test set) | 0.235 (0.241)/0.287 (0.353) |
| No. atoms | |
| Protein | 1141 |
| Water | 98 |
| B-factors | |
| Protein | 42.4 |
| Water | 41.0 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.015 |
| Bond angles (°) | 1.52 |
| Ramachandran (%) | |
| Favored | 97 |
| Outliers | 1.4 |
| PDB code | 5DCK |
*Number in parentheses is for highest resolution shell.
Additional Information
Accession numbers: Atomic coordinates and structure factors were deposited in the Protein Data Bank with the accession number 5DCK.
How to cite this article: Khwaja, A. et al. Structure of FIV capsid C-terminal domain demonstrates lentiviral evasion of genetic fragility by coevolved substitutions. Sci. Rep. 6, 24957; doi: 10.1038/srep24957 (2016).
Acknowledgments
This research benefited from use of the Technion Center of Structural Biology facility of the. Lorry I. Lokey Center for Life Sciences and Engineering and the Russell Berrie Nanotechnology. Institute. We thank Ms. Jasmin Ackermann for assistance in preliminary cloning and crystallization experiments.
Footnotes
Author Contributions A.K., M.G., A.M. and A.A. designed and conducted the experiments, analyzed the data and prepared the manuscript.
References
- Marx A. & Alian A. The Road Less Traveled: HIV’s Use of Alternative Routes through Cellular Pathways. J Virol 89, 5204–5212, doi: 10.1128/JVI.03684-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolich M. et al. Evolutionary information for specifying a protein fold. Nature 437, 512–518, doi: 10.1038/nature03991 (2005). [DOI] [PubMed] [Google Scholar]
- Kowarsch A., Fuchs A., Frishman D. & Pagel P. Correlated mutations: a hallmark of phenotypic amino acid substitutions. Plos Comput Biol 6, doi: 10.1371/journal.pcbi.1000923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A. & Alian A. Rerouting Resistance: Escaping Restriction Using Alternative Cellular Pathways. Trends Microbiol 23, 595–597, doi: 10.1016/j.tim.2015.08.005 (2015). [DOI] [PubMed] [Google Scholar]
- Ovchinnikov S. et al. Large-scale determination of previously unsolved protein structures using evolutionary information. Elife 4, doi: 10.7554/eLife.09248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob E., Unger R. & Horovitz A. Codon-level information improves predictions of inter-residue contacts in proteins by correlated mutation analysis. Elife 4, doi: 10.7554/eLife.08932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. et al. Functional conservation of HIV-1 Gag: implications for rational drug design. Retrovirology 10, 126, doi: 10.1186/1742-4690-10-126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn W. F. et al. Deep sequencing of protease inhibitor resistant HIV patient isolates reveals patterns of correlated mutations in Gag and protease. Plos Comput Biol 11, e1004249, doi: 10.1371/journal.pcbi.1004249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sage V., Mouland A. J. & Valiente-Echeverria F. Roles of HIV-1 capsid in viral replication and immune evasion. Virus Res 193, 116–129, doi: 10.1016/j.virusres.2014.07.010 (2014). [DOI] [PubMed] [Google Scholar]
- Campbell E. M. & Hope T. J. HIV-1 capsid: the multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13, 471–483, doi: 10.1038/nrmicro3503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P. A. et al. Correlation of naturally occurring HIV-1 resistance to DEB025 with capsid amino acid polymorphisms. Viruses 5, 981–997, doi: 10.3390/v5030981 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahirel V. et al. Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc Natl Acad Sci USA 108, 11530–11535, doi: 10.1073/pnas.1105315108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina O. V. et al. Computational identification of novel amino-acid interactions in HIV Gag via correlated evolution. Plos One 7, e42468, doi: 10.1371/journal.pone.0042468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wang Y., Gao Y., Li G. & Huang J. Integrated analysis of residue coevolution and protein structures capture key protein sectors in HIV-1 proteins. Plos One 10, e0117506, doi: 10.1371/journal.pone.0117506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. et al. Compensatory substitutions in the HIV-1 capsid reduce the fitness cost associated with resistance to a capsid-targeting small-molecule inhibitor. J Virol 89, 208–219, doi: 10.1128/JVI.01411-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Price A. J., Halambage U. D., James L. C. & Aiken C. HIV-1 Resistance to the Capsid-Targeting Inhibitor PF74 Results in Altered Dependence on Host Factors Required for Virus Nuclear Entry. J Virol 89, 9068–9079, doi: 10.1128/JVI.00340-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R. et al. Second-site suppressors of HIV-1 capsid mutations: restoration of intracellular activities without correction of intrinsic capsid stability defects. Retrovirology 9, 30, doi: 10.1186/1742-4690-9-30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn S. J. et al. Extreme genetic fragility of the HIV-1 capsid. Plos Pathog 9, e1003461, doi: 10.1371/journal.ppat.1003461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland M. et al. HIV-1 conserved-element vaccines: relationship between sequence conservation and replicative capacity. J Virol 87, 5461–5467, doi: 10.1128/JVI.03033-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienzle D. FIV in cats–a useful model of HIV in people? Vet Immunol Immunopathol 159, 171–179, doi: 10.1016/j.vetimm.2014.02.014 (2014). [DOI] [PubMed] [Google Scholar]
- Poeschla E. M. Primate and feline lentiviruses in current intrinsic immunity research: the cat is back. Vet Immunol Immunopathol 143, 215–220, doi: 10.1016/j.vetimm.2011.06.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galilee M. & Alian A. Identification of Phe187 as a crucial dimerization determinant facilitates crystallization of a monomeric retroviral integrase core domain. Structure 22, 1512–1519, doi: 10.1016/j.str.2014.08.001 (2014). [DOI] [PubMed] [Google Scholar]
- Esteva M. J., Affranchino J. L. & Gonzalez S. A. Lentiviral Gag assembly analyzed through the functional characterization of chimeric simian immunodeficiency viruses expressing different domains of the feline immunodeficiency virus capsid protein. Plos One 9, e114299, doi: 10.1371/journal.pone.0114299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D. et al. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc Natl Acad Sci USA 104, 4353–4358, doi: 10.1073/pnas.0609477104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler U. K., Stray K. M., Garrus J. E. & Sundquist W. I. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77, 5439–5450, doi: 10.1128/JVI.77.9.5439-5450.2003 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obal G. et al. Conformational plasticity of a native retroviral capsid revealed by x-ray crystallography. Science 349, 95–98, doi: 10.1126/science.aaa5182 (2015). [DOI] [PubMed] [Google Scholar]
- Bayro M. J., Chen B., Yau W. M. & Tycko R. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. J Mol Biol 426, 1109–1127, doi: 10.1016/j.jmb.2013.12.021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O., Ganser-Pornillos B. K., Banumathi S., Hua Y. & Yeager M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J Mol Biol 401, 985–995, doi: 10.1016/j.jmb.2010.06.042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternois F., Sticht J., Duquerroy S., Krausslich H. G. & Rey F. A. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol 12, 678–682, doi: 10.1038/nsmb967 (2005). [DOI] [PubMed] [Google Scholar]
- Gres A. T. et al. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 349, 99–103, doi: 10.1126/science.aaa5936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E. O. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13, 484–496, doi: 10.1038/nrmicro3490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. et al. Protease cleavage leads to formation of mature trimer interface in HIV-1 capsid. Plos Pathog 8, e1002886, doi: 10.1371/journal.ppat.1002886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartonova V. et al. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem 283, 32024–32033, doi: 10.1074/jbc.M804230200 (2008). [DOI] [PubMed] [Google Scholar]
- Zhao G. et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646, doi: 10.1038/nature12162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W., Cymborowski M., Otwinowski Z. & Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica. Section D, Biological crystallography 62, 859–866, doi: 10.1107/S0907444906019949 (2006). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674, doi: 10.1107/S0021889807021206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67, 235–242, doi: 10.1107/S0907444910045749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography 67, 355–367, doi: 10.1107/S0907444911001314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]