SUMMARY
We recently reported that homozygous Presenilin-1 (Psen1) knockin (KI) mice carrying the familial Alzheimer’s disease (FAD) mutation L435F or C410Y recapitulate the phenotypes of Psen1−/− mice. Production and steady-state levels of Aβ40 and Aβ42 are undetectable in KI/KI brains and reduced in KI/+ brains, though the Aβ42/Aβ40 ratio is slightly increased in KI/+ brains. Moreover, the FAD mutation impairs synaptic function, learning and memory, and age-dependent neuronal survival in the adult brain. Here we extend our analysis of the effects of the L435F and C410Y mutations to the generation of Aβ43. Similar to Aβ40 and Aβ42, production of Aβ43 is undetectable in KI/KI brains and reduced in KI/+ brains. These results support our previous conclusions that the L435F and C410Y mutations cause loss of Presenilin function and γ-secretase activity, including impaired Aβ production in the brain. This Matters Arising Response paper addresses the Veugelen et al. Matters Arising paper.
INTRODUCTION
We recently reported the generation and multidisciplinary analysis of two Presenilin-1 (Psen1) knockin (KI) mice, in which the familial Alzheimer’s disease (FAD) mutation L435F or C410Y was introduced into the endogenous mouse Psen1 locus. We found that homozygous L435F and C410Y KI/KI mice recapitulated the phenotypes of Psen1−/− mice, including perinatal lethality, shortened body axis and kinked tail, and impaired neurogenesis and Notch signaling (Handler et al., 2000; Shen et al., 1997; Song et al., 1999; Xia et al., 2015). While Psen1 mRNA expression was normal in the embryonic brain of L435F and C410Y KI/KI mice, only very low levels of Presenilin-1 (PS1) N-terminal fragment (NTF) and C-terminal fragment (CTF) were detected, and PS1 accumulated predominantly as holoprotein.
Using a well-established in vitro γ-secretase assay (Saito et al., 2011; Takahashi et al., 2003; Watanabe et al., 2012), we found that γ-secretase activity was abolished in L435F and C410Y KI/KI brains, as measured by elimination of the production of the Notch intracellular domain (NICD), Aβ40 and Aβ42 (Xia et al., 2015). In addition, the CTFs of the amyloid precursor protein (APP) and N-cadherin accumulated to levels comparable to those observed in Psen1−/− brains (Xia et al., 2015). De novo production of Aβ40 and Aβ42 was reduced (~50%) in the embryonic brain or the adult cerebral cortex of L435F and C410Y KI/+ mice, and the Aβ42/Aβ40 ratio was slightly increased (~15%) due to the greater reduction of the steady-state levels of Aβ40 relative to Aβ42, which exacerbated Aβ deposition when L435F KI/+ mice were crossed with APP transgenic mice overexpressing human mutant APP (Xia et al., 2015). Furthermore, relative to the wild-type Psen1 allele, the L435F mutation impaired hippocampal synaptic plasticity and memory and caused age-dependent neurodegeneration in the cerebral cortex (Xia et al., 2015). We therefore concluded that the FAD L435F and C410Y mutations impair γ-secretase activity in vivo, and suggested that PSEN mutations cause FAD-related phenotypes through a loss-of-function mechanism (Xia et al., 2015).
Here, in response to the challenges raised by Veugelen et al., we extend our work to examine the effects of the L435F and C410Y mutations on Aβ43 production. We found that Aβ43 production is reduced in the cerebral cortex of L435F KI/+ mice, and is undetectable in C410Y KI/KI embryonic brains. These results reinforce our previous conclusion that the L435F and C410Y mutations cause reduced production of Aβ peptides in heterozygosity and undetectable Aβ production in homozygosity.
RESULTS
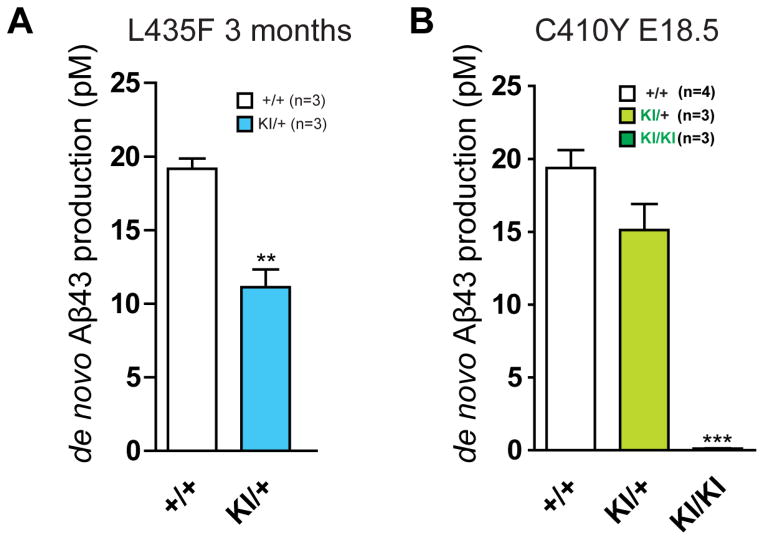
Veugelen et al. described in their Matters Arising paper that retroviral overexpression of PS1 bearing the R278I, L435F or C410Y mutation in immortalized Psen-deficient mouse embryonic fibroblasts (MEFs) resulted in 9- to 75-fold increases of secreted Aβ43, compared to their control cells. We previously reported that de novo production and steady-state levels of Aβ40 and Aβ42 were undetectable in the brain of L435F and C410Y KI/KI embryos at day 18.5 (Figure 3B–C, S2B–C in Xia et al., 2015). To determine whether L435F and C410Y mutations exert similar or distinct effects on Aβ43 production, we measured Aβ43 production using the same antibody as Veugelen et al., which is currently the only available antibody for Aβ43 detection. We selectively evaluated Aβ43 production in the embryonic brain of C410Y KI mice and the adult cerebral cortex of L435F KI mice. Using the same in vitro γ-secretase assay as in our recent study (Xia et al., 2015), we found that de novo Aβ43 production is reduced in the cerebral cortex of L435F KI/+ mice at 3 months of age, and is undetectable in the C410Y KI/KI embryonic brain at day 18.5 (Figure 1). These results are in agreement with our earlier findings showing reduced production of Aβ40 and Aβ42 in the L435F and C410Y KI/+ brains, and undetectable production of Aβ40 and Aβ42 in the L435F and C410Y KI/KI brains (Xia et al., 2015).
Figure 1. Aβ43 production is reduced in L435F KI/+ brains and undetectable in C410Y KI/KI brains.
(A) ELISA measurements of Aβ43 following in vitro γ-secretase assay in L435F KI mice at 3 months of age. De novo production of Aβ43 in cortical homogenates is reduced in L435F KI/+ mice relative to littermate Psen1+/+ controls (**p<0.005). (B) ELISA measurements of Aβ43 following in vitro γ-secretase assay in C410Y KI mice at embryonic day 18.5 (E18.5). Relative to littermate Psen1+/+ controls, de novo production of Aβ43 in brain homogenates is reduced in C410Y KI/+ mice and undetectable in C410Y KI/KI mice (***p<0.001). All data are presented as mean ± SEM.
DISCUSSION
In the Veugelen et al. Matters Arising paper, the authors used retroviral overexpression to examine the effects of the PSEN1 mutations R278I, C410Y and L435F on Aβ production in immortalized MEFs. They found that secreted Aβ43 levels were dramatically increased in Psen-deficient MEFs overexpressing R278I, C410Y or L435F mutant PS1, relative to wild-type PS1 (Figure 1B, 1C in Veugelen et al.). Therefore, they challenged our earlier findings showing that de novo production and steady-state levels of Aβ40 and Aβ42 were undetectable in L435F and C410Y KI/KI brains (Xia et al., 2015). Here we evaluated Aβ43 production in L435F and C410Y KI mice, and found similar loss of Aβ43 production in the brain of these KI mice (Figure 1).
How do we explain the discrepancy in our findings?
This apparent discrepancy could be explained by key differences in the experimental systems and approaches used in our two studies. First, Veugelen et al. used retrovirus to overexpress wild-type or mutant PS1 at very high levels, whereas wild-type or mutant PS1 is expressed normally under the control of its endogenous regulatory elements in our KI mice. Second, Veugelen et al. used immortalized MEF cell clones stably transduced with retroviral vectors expressing wild-type or mutant PS1. With this approach, the expression of wild-type and mutant PS1 can vary widely among different MEF clones, making it difficult to compare quantitatively the relative effects of the mutations. Unfortunately, Veugelen et al. did not quantify or compare the expression level of wild-type or mutant PS1 in their stable cell lines, nor did they include loading control in their Western analysis. Surprisingly, they also detected PS1 CTFs in Psen-deficient (dKO) MEFs reconstituted with D257A PS1, which is known to abolish γ-secretase activity (Wolfe et al., 1999). In contrast to the varied expression levels of wild-type and mutant PS1 in their stable cell lines, wild-type and mutant PS1 are expressed physiologically at identical levels and with identical spatiotemporal patterns in our KI mice; thus, the effects of wild-type or mutant PS1 on γ-secretase activity and Aβ production can be directly compared. Third, Veugelen et al. used adenovirus to overexpress human Swedish mutant APP, which results in overexpression of APP and excessive production of APP β-CTF and Aβ (Citron et al., 1994; Felsenstein et al., 1994). In our KI mice, APP is expressed under its normal physiological conditions, a situation that is identical to PSEN1 FAD patients.
Lastly, there are likely cell type-specific differences in the regulation of Aβ production between brains and MEFs. Veugelen et al. measured Aβ production and secretion from their MEFs, whereas we measured production and steady-state levels of Aβ in the brain. These discrepancies highlight the importance of investigating FAD pathophysiology in its proper physiological context; we would argue that analysis of the effects of PSEN mutations in the mammalian brain is more relevant to FAD pathophysiology than analysis in virally-transduced MEF cell clones.
In addition to the differences in our experimental systems, a number of observations reported by Veugelen et al. do not make sense. For example, they detected exceptionally high levels of de novo Aβ43 production relative to Aβ40 and Aβ42 in MEFs overexpressing wild-type (WT) PS1 (Figure 1D in Veugelen et al.). Interestingly, they also noticed this unexpected result and commented that “Under these conditions we measured high Aβ43 production in wild type cell extracts, something which is not observed when analyzing intact cells.” However, they didn’t address why they obtained such strikingly different results when measuring secretion (Figure 1C in Veugelen et al.) and production (Figure 1D in Veugelen et al.) of Aβ40, Aβ42 and Aβ43 from the same WT clone. Furthermore, they detected similar levels of Aβ40 and Aβ42 production in their WT MEFs (Figure 1D in Veugelen et al.), even though it is well established that wild-type PS1 produces Aβ40 at much higher levels than Aβ42 (Kretner et al., 2011; Watanabe et al., 2012; Xia et al., 2015).
Since the same MEF clones overexpressing either wild-type or mutant PS1 were used to measure levels of secreted and de novo production of Aβ, one would expect the results from these complementary approaches to correlate with each other. However, there was little correlation between secreted and de novo production of Aβ40, Aβ42 and Aβ43 in MEFs overexpressing either wild-type or mutant PS1 (Figure 1C, 1D in Veugelen et al.). For example, modest levels of secreted Aβ42 were detected in MEFs expressing R278I or C410Y PS1, whereas there was no detectable de novo Aβ42 production in the same MEF clones.
Curiously, Veugelen et al. took their lack of detection of de novo production of Aβ40, Aβ42 and Aβ43 in their mutant cells as evidence to “conclude that the experimental conditions (detergent solubilization) used by Shen and Kelleher compromised the carboxypeptidase processivity of the wild type and mutant enzymes resulting in high Aβ43 production from the wild type enzyme (at the expense of Aβ40) and low Aβ43 generation from mutants.” This statement is conceptually convoluted. It is difficult to fathom that the absence of de novo production of Aβ43 in the KI/KI but not wild-type brain is due to detergent solubilization rather than genotype. Consistent with the expected distribution of Aβ species, we detected the highest level of de novo production of Aβ40, followed by Aβ42 and then Aβ43, in the adult cerebral cortex of wild-type mice (Figure 3D in Xia et al., Figure 1 of this paper).
In an effort to overcome the issues associated with overexpression systems, Veugelen et al. used immortalized R278I KI MEFs, in which R278I PS1 is expressed under its endogenous regulatory elements, but human APP was introduced into these cells by adenovirus. Thus, APP as substrate of γ-secretase was presumably overexpressed at varying levels in WT, WT/KI and KI/KI MEF clones, which could affect Aβ production independent of the Psen1 genotype, but no data comparing APP expression levels in these MEF clones were provided. Surprisingly, they reported very high levels of secreted Aβ43 (~46 pM) but little secreted Aβ42 (~1 pM) in KI/KI cells (Figure 1E in Veugelen et al.). Such divergence in the levels of secreted Aβ42 and Aβ43 in R278I KI/KI MEFs is in direct contrast with their findings of similar levels of secreted Aβ42 and Aβ43 in Psen−/− MEFs reconstituted with R278I PS1 (Figure 1C in Veugelen et al.). No explanation or data were provided as to why such divergent results were obtained from these two MEF cell lines expressing only R278I mutant PS1.
In contrast to the statement “our data is in complete agreement with our previous study showing high Aβ43 levels in PsenR278I knock-in mice (Saito et al., 2011)”, the high level of secreted Aβ43 detected in WT/KI MEFs by Veugelen et al. is inconsistent with the undetectable level of secreted Aβ43 previously reported in MEFs of the same KI genotype (Saito et al., 2011). Moreover, Saito et al. reported a strong R278I KI allele dosage-dependent reduction in de novo Aβ43 production, as shown by ~50% reduction in +/KI MEFs and ~90% reduction in KI/KI MEFs (Figure S10c in Saito et al., 2011).
Veugelen et al. also raised objections unrelated to Aβ43 production, including the classical genetic approach we employed to amplify the phenotypic effects of loss of function mutations, namely by reducing the overall gene dosage to minimize compensatory effects. Specifically, we crossed Psen1 L435F KI mice to the Psen2−/− background to enable more direct and accurate analysis of the phenotypic effects of the L435F mutation on PS1 function (Xia et al., 2015). Indeed, we previously found that loss of PS1 expression leads to up-regulation of PS2 expression (Watanabe et al., 2014), which would mask the effects of PS1 loss-of-function mutations. Curiously, Veugelen et al. don’t object to the converse approach of overexpressing PSEN1 and APP. They employed retroviral and adenoviral transduction to overexpress various PS1 mutants and Swedish mutant APP at high levels in their MEFs, despite the well-established potential of non-physiological overexpression systems to yield artifactual results. It should also be noted that overexpression of mutant human APP and massive accumulation of human Aβ42 (e.g. >6,400-fold) in transgenic mice has not been sufficient to cause neurodegeneration (Irizarry et al., 1997a; Irizarry et al., 1997b; Mucke et al., 2000; Saura et al., 2005). By contrast, our reported KI mouse model demonstrated that a clinical PSEN1 mutation triggers age-dependent cerebral cortical neurodegeneration (Xia et al., 2015).
Veugelen et al. also questioned whether mice are suitable models for investigation of AD, because the “mouse brain is not very sensitive to Aβ toxicity for reasons that remain unclear.” By contrast, our results show that the mouse brain is sensitive to loss of Presenilin or γ-secretase function (Saura et al., 2004; Tabuchi et al., 2009; Wines-Samuelson et al., 2010). While mice don’t mimic all end-stage neuropathological features of the human disease, animal models have proven to be useful tools for the investigation of the pathogenic mechanisms. For example, APP transgenic mice provide excellent models of amyloidosis and offer insight into the consequences of Aβ accumulation and deposition (Ashe and Zahs, 2010), whereas Presenilin and Nicastrin mouse models highlight the importance of γ-secretase in learning and memory, synaptic function and neuronal survival (Lee et al., 2014; Saura et al., 2004; Tabuchi et al., 2009; Wines-Samuelson et al., 2010; Yu et al., 2001; Zhang et al., 2009; Zhang et al., 2010), and offer insight into the pathogenic mechanisms of PSEN1 mutations (Xia et al., 2015). The lack of significant neurodegeneration in mouse brains with massive Aβ accumulation may be interpreted as reflecting the lower potency of Aβ accumulation, relative to loss of Presenilin activity, in causing neurodegeneration. Notably, a recent study of >1,300 autosomal dominant AD patients revealed that PSEN1 mutations cause FAD with an earlier mean age of onset than APP mutations, suggesting that PS1 dysfunction is more proximate to disease pathogenesis (Ryman et al., 2014).
Given the prevalence of AD and the lack of effective therapies, open-minded debates about the mechanisms of AD pathogenesis are needed and should lead to more productive therapeutic development. Unfortunately, the arguments presented by Veugelen et al. are undermined by numerous technical and conceptual limitations and inconsistencies, which raise significant uncertainties regarding the validity and relevance of their results. It is also regrettable that they present a distorted view of the conclusions advanced in our recent study. In particular, Veugelen et al. claim that we proposed that “Aβ is an epiphenomenon in FAD (Xia et al., 2015)”, even though the paper contains no such statement or conclusion. On the contrary, we performed extensive analysis focused on Aβ production and accumulation, and reported that heterozygosity for the FAD PSEN1 mutation increased the Aβ42/40 ratio and exacerbated Aβ deposition via a loss-of-function mechanism mediated by a greater reduction in steady-state levels of Aβ40 than Aβ42 (Xia et al., 2015). Consistent with this interpretation, the PSEN1 L435F mutation promotes Aβ deposition in the FAD brain in the form of cotton wool plaques, which display strong Aβ42 immunoreactivity but scant Aβ40 immunoreactivity (Heilig et al., 2010). Based on evidence that Aβ40 is protective against Aβ deposition (Kim et al., 2007; Wang et al., 2006), we suggest that preferential loss of Aβ40 can explain Aβ deposition in the brains of mouse models and human FAD patients without any need to invoke excessive production of longer Aβ species. While Aβ43 deposition may occur in the FAD brain via a similar loss-of-function mechanism, the assertion that Aβ43 is “highly amyloidogenic” and “likely pathogenic” warrants critical examination. Aβ43 differs from Aβ42 by an additional C-terminal threonine, a polar residue which should in principle reduce its hydrophobicity relative to Aβ42; indeed, recent biophysical analysis has shown that Aβ43 is less aggregation-prone and seeds amyloid formation less efficiently than Aβ42 (Chemuru et al., 2016).
The fundamental difference between our views and those of Veugelen et al. is how PSEN mutations cause neurodegeneration and dementia in FAD. Veugelen et al. believe that “The hypothesis that β amyloid is triggering the neurodegeneration in FAD patients remains the most parsimonious and consistent explanation for all experimental data.” While we acknowledge a significant role of Aβ in FAD, our genetic findings point to a more important causal role for loss of essential functions of Presenilin in the pathogenesis of neurodegeneration and dementia in FAD. First, Presenilin is essential for learning and memory, synaptic function and age-dependent neuronal survival (Saura et al., 2004; Wines-Samuelson et al., 2010; Yu et al., 2001; Zhang et al., 2009; Zhang et al., 2010). Second, partial loss of Presenilin function is also sufficient to cause age-dependent neurodegeneration, though at a later age of onset and with lesser severity than observed with complete loss of Presenilin function (Watanabe et al., 2014). Third, FAD mutations in PSEN1 cause loss of γ-secretase activity, though the severity of impairment varies among mutations (Heilig et al., 2013; Heilig et al., 2010; Saito et al., 2011; Xia et al., 2015). Lastly, our recent study showed that clinical PSEN1 mutations promote the development of key FAD-related phenotypes, including synaptic and cognitive impairment, amyloid deposition, and cerebral cortical neurodegeneration with accompanying inflammatory changes, through a loss-of-function mechanism (Xia et al., 2015). Thus, the loss-of-function mechanism that we have demonstrated for FAD-linked PSEN1 mutations provides the most coherent explanation for all existing, reproducible human and mouse data. Based on these insights, we have proposed that restoration of normal Presenilin function may offer the most direct approach to devise effective therapies that can combat dementia and neurodegeneration in FAD patients (Shen and Kelleher, 2007; Xia et al., 2015).
EXPERIMENTAL PROCEDURES
Mice
Generation and characterization of Psen1 L435F and C410Y KI mice have been described previously (Xia et al., 2015). Mice were maintained on the C57BL6/J-129 hybrid genetic background and littermate controls were used for all analysis. Timed mating between heterozygous Psen1 KI/+ mice were set up to obtain KI/KI embryos. The tails from adult mice or embryos were removed for genotyping. The experimental analyses were performed in a genotype-blind manner, though homozygous KI/KI mice were grossly abnormal and were easily distinguishable from KI/+ and +/+ mice. All procedures relating to animal care and treatment conformed to institutional and NIH guidelines.
Preparation of CHAPSO-solubilized microsomal fractions
Mice were anesthetized by intraperitoneal injection of sodium pentobarbital and then transcardially perfused with 20 ml cold PBS (pH 7.4). The adult cortices or embryonic brains were dissected for purification of microsomal fractions or immediately frozen in liquid nitrogen. Embryonic brains or adult cortices were homogenized in homogenization buffer [20 mM PIPES, pH 7.0, 140 mM KCl, 0.25 M sucrose, 5 mM EGTA, EDTA-free complete protease inhibitor cocktail (Roche)] using a glass/Teflon tissue grinder. The homogenates were centrifuged at 800 g for 10 min to remove nuclei and cell debris. The postnuclear supernatants were recentrifuged at 100,000 g for 1 h, and the resulting pellets were washed with 0.1 M sodium carbonate, pH 11.3, and then centrifuged again. The membrane pellets were solubilized with freshly prepared 1% 3-[(3-cholamidopropyl) dimethylammonio]-2-hydroxy-1- propanesulfonate (CHAPSO) buffer (50 mM PIPES, pH 7.0, 0.25 M sucrose, 1 mM EGTA) for 1 h on ice and then centrifuged at 100,000g for 1 h, and the final supernatants were saved as crude γ-secretase fractions and stored at −80°C.
In vitro γ-secretase assay
γ-secretase-mediated de novo Aβ43 generation was measured using a method described previously (Takahashi et al., 2003; Watanabe et al., 2012; Xia et al., 2015). Briefly, 1% CHAPSO-solubilized microsomal fractions (7.5 μg per reaction) from E18.5 brains or adult cortices at 3 months of age were mixed with the assay buffer [final concentration: 150 μg/ml γ-secretase fraction, 0.3% CHAPSO, 10 mM HEPES (pH 7.3), 150 mM NaCl, 5 mM EDTA, complete protease inhibitor cocktail (Roche), 5 mM 1,10-phenanthroline, 10 μg/ml phosphoramidon, and 0.1% (w/v) phosphatidylcholine], and incubated with 2 μM recombinant C100-FmH (substrate for de novo Aβ generation) at 37°C for 14 hours. Recombinant C100-FmH (APP C100 recombinant protein tagged with Flag-Myc-Histidine) was produced in DH5α, which was transformed with the pTrcHis2A-C100-FmH plasmid, and induced with 0.1 mM IPTG for 3 hours at 37°C, and C100-FmH was purified with Ni2+-chelated HiTrap Chelating HP column (GE Healthcare).
Enzyme-linked Immunosorbent Assay (ELISA)
Aβ43 generated in the in vitro assay was quantified using the human amyloid (1-43) (FL) assay kit purchased from IBL-America (Cat#27710). In this kit, the capture antibody [anti-Human Aβ (38-43) Rabbit IgG] is precoated on the plate and the detection antibody [anti-Human Aβ (N) (82E1) mouse IgG] is labeled with HRP. To increase the sensitivity of Aβ43 detection, the protocol from the manufacturer was modified according to Saito et al. (Saito et al., 2011). Briefly, The HRP signal was quantified by incubation with 50 μM Amplex UltraRed reagent (Invitrogen, Cat#A36006) for 30 minutes, followed by detection of the Amplex UltraRed fluorescent signal, using a 530 nm excitation filter and 590 nm emission filter in Synergy HT Multi-Mode Microplate Reader. Each sample was measured in duplicate. The detection range of the assay for Aβ43 is from 0.51 pM to 32.5 pM.
Statistical Analysis
Statistical analysis was performed using two-tailed unpaired Student’s t-test for all comparisons of the ELISA results. A value of p < 0.05 is considered significant. All data are represented as mean ± SEM.
HIGHLIGHTS.
Reduced Aβ43 production in heterozygous Psen1 knockin brains
Undetectable Aβ43 production in homozygous Psen1 knockin brains
Acknowledgments
We would like to thank our lab members for helpful discussions. This work was supported by grants from the National Institutes of Health (R01NS041783 and R01NS042818 to JS, and R01NS075346 to RJK) and an award from the MetLife Foundation (to JS).
Footnotes
The authors declare no competing financial interest.
AUTHOR CONTRIBUTIONS
D.X., R.J.K. and J.S. designed experiments and wrote the paper, and D.X. performed the experiments.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashe KH, Zahs KR. Probing the biology of Alzheimer’s disease in mice. Neuron. 2010;66:631–645. doi: 10.1016/j.neuron.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemuru S, Kodali R, Wetzel R. C-Terminal Threonine Reduces Abeta Amyloidogenicity Compared with Abeta. J Mol Biol. 2016;428:274–291. doi: 10.1016/j.jmb.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron M, Vigo-Pelfrey C, Teplow DB, Miller C, Schenk D, Johnston J, Winblad B, Venizelos N, Lannfelt L, Selkoe DJ. Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proc Natl Acad Sci U S A. 1994;91:11993–11997. doi: 10.1073/pnas.91.25.11993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein KM, Hunihan LW, Roberts SB. Altered cleavage and secretion of a recombinant beta-APP bearing the Swedish familial Alzheimer’s disease mutation. Nat Genet. 1994;6:251–255. doi: 10.1038/ng0394-251. [DOI] [PubMed] [Google Scholar]
- Handler M, Yang X, Shen J. Presenilin-1 regulates neuronal differentiation during neurogenesis. Development. 2000;127:2593–2606. doi: 10.1242/dev.127.12.2593. [DOI] [PubMed] [Google Scholar]
- Heilig EA, Gutti U, Tai T, Shen J, Kelleher RJ., 3rd Trans-dominant negative effects of pathogenic PSEN1 mutations on gamma-secretase activity and Abeta production. J Neurosci. 2013;33:11606–11617. doi: 10.1523/JNEUROSCI.0954-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig EA, Xia W, Shen J, Kelleher RJ., 3rd A presenilin-1 mutation identified in familial Alzheimer disease with cotton wool plaques causes a nearly complete loss of gamma-secretase activity. J Biol Chem. 2010;285:22350–22359. doi: 10.1074/jbc.M110.116962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry MC, McNamara M, Fedorchak K, Hsiao K, Hyman BT. APPSw transgenic mice develop age-related A beta deposits and neuropil abnormalities, but no neuronal loss in CA1. J Neuropathol Exp Neurol. 1997a;56:965–973. doi: 10.1097/00005072-199709000-00002. [DOI] [PubMed] [Google Scholar]
- Irizarry MC, Soriano F, McNamara M, Page KJ, Schenk D, Games D, Hyman BT. Abeta deposition is associated with neuropil changes, but not with overt neuronal loss in the human amyloid precursor protein V717F (PDAPP) transgenic mouse. J Neurosci. 1997b;17:7053–7059. doi: 10.1523/JNEUROSCI.17-18-07053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Onstead L, Randle S, Price R, Smithson L, Zwizinski C, Dickson DW, Golde T, McGowan E. Abeta40 inhibits amyloid deposition in vivo. J Neurosci. 2007;27:627–633. doi: 10.1523/JNEUROSCI.4849-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretner B, Fukumori A, Gutsmiedl A, Page RM, Luebbers T, Galley G, Baumann K, Haass C, Steiner H. Attenuated Abeta42 responses to low potency gamma-secretase modulators can be overcome for many pathogenic presenilin mutants by second-generation compounds. J Biol Chem. 2011;286:15240–15251. doi: 10.1074/jbc.M110.213587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sharma M, Sudhof TC, Shen J. Synaptic function of nicastrin in hippocampal neurons. Proc Natl Acad Sci U S A. 2014;111:8973–8978. doi: 10.1073/pnas.1408554111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L, Masliah E, Yu GQ, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L. High-level neuronal expression of abeta 1-42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci. 2000;20:4050–4058. doi: 10.1523/JNEUROSCI.20-11-04050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman DC, Acosta-Baena N, Aisen PS, Bird T, Danek A, Fox NC, Goate A, Frommelt P, Ghetti B, Langbaum JB, et al. Symptom onset in autosomal dominant Alzheimer disease: a systematic review and meta-analysis. Neurology. 2014;83:253–260. doi: 10.1212/WNL.0000000000000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Suemoto T, Brouwers N, Sleegers K, Funamoto S, Mihira N, Matsuba Y, Yamada K, Nilsson P, Takano J, et al. Potent amyloidogenicity and pathogenicity of Abeta43. Nat Neurosci. 2011;14:1023–1032. doi: 10.1038/nn.2858. [DOI] [PubMed] [Google Scholar]
- Saura CA, Chen G, Malkani S, Choi SY, Takahashi RH, Zhang D, Gouras GK, Kirkwood A, Morris RG, Shen J. Conditional inactivation of presenilin 1 prevents amyloid accumulation and temporarily rescues contextual and spatial working memory impairments in amyloid precursor protein transgenic mice. J Neurosci. 2005;25:6755–6764. doi: 10.1523/JNEUROSCI.1247-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ, 3rd, Kandel ER, Duff K, et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ., 3rd The presenilin hypothesis of Alzheimer’s disease: evidence for a loss-of-function pathogenic mechanism. Proc Natl Acad Sci U S A. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Nadeau P, Yuan M, Yang X, Shen J, Yankner BA. Proteolytic release and nuclear translocation of Notch-1 are induced by presenilin-1 and impaired by pathogenic presenilin-1 mutations. Proc Natl Acad Sci U S A. 1999;96:6959–6963. doi: 10.1073/pnas.96.12.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Chen G, Sudhof TC, Shen J. Conditional forebrain inactivation of nicastrin causes progressive memory impairment and age-related neurodegeneration. J Neurosci. 2009;29:7290–7301. doi: 10.1523/JNEUROSCI.1320-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Hayashi I, Tominari Y, Rikimaru K, Morohashi Y, Kan T, Natsugari H, Fukuyama T, Tomita T, Iwatsubo T. Sulindac sulfide is a noncompetitive gamma-secretase inhibitor that preferentially reduces Abeta 42 generation. J Biol Chem. 2003;278:18664–18670. doi: 10.1074/jbc.M301619200. [DOI] [PubMed] [Google Scholar]
- Wang R, Wang B, He W, Zheng H. Wild-type presenilin 1 protects against Alzheimer disease mutation-induced amyloid pathology. J Biol Chem. 2006;281:15330–15336. doi: 10.1074/jbc.M512574200. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Iqbal M, Zheng J, Wines-Samuelson M, Shen J. Partial loss of presenilin impairs age-dependent neuronal survival in the cerebral cortex. J Neurosci. 2014;34:15912–15922. doi: 10.1523/JNEUROSCI.3261-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Xia D, Kanekiyo T, Kelleher RJ, 3rd, Shen J. Familial frontotemporal dementia-associated presenilin-1 c.548G>T mutation causes decreased mRNA expression and reduced presenilin function in knock-in mice. J Neurosci. 2012;32:5085–5096. doi: 10.1523/JNEUROSCI.0317-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wines-Samuelson M, Schulte EC, Smith MJ, Aoki C, Liu X, Kelleher RJ, 3rd, Shen J. Characterization of age-dependent and progressive cortical neuronal degeneration in presenilin conditional mutant mice. PLoS One. 2010;5:e10195. doi: 10.1371/journal.pone.0010195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- Xia D, Watanabe H, Wu B, Lee SH, Li Y, Tsvetkov E, Bolshakov VY, Shen J, Kelleher RJ., 3rd Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer’s disease. Neuron. 2015;85:967–981. doi: 10.1016/j.neuron.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Saura CA, Choi SY, Sun LD, Yang X, Handler M, Kawarabayashi T, Younkin L, Fedeles B, Wilson MA, et al. APP processing and synaptic plasticity in presenilin-1 conditional knockout mice. Neuron. 2001;31:713–726. doi: 10.1016/s0896-6273(01)00417-2. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wu B, Beglopoulos V, Wines-Samuelson M, Zhang D, Dragatsis I, Sudhof TC, Shen J. Presenilins are essential for regulating neurotransmitter release. Nature. 2009;460:632–636. doi: 10.1038/nature08177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Zhang C, Ho A, Kirkwood A, Sudhof TC, Shen J. Inactivation of presenilins causes pre-synaptic impairment prior to post-synaptic dysfunction. J Neurochem. 2010;115:1215–1221. doi: 10.1111/j.1471-4159.2010.07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]